Breast carcinoma is a global health problem that
affects women's quality of life. The World Health Organization
estimates that there are >2.26 million new cases and >684,000
deaths worldwide each year (1).
The incidence of breast carcinoma varies widely across populations
and regions and is influenced by factors such as age, ethnicity and
socioeconomic status (2). Despite
advances in diagnostic imaging and biomarker research, limitations,
such as the low sensitivity of dense breast tissue and variations
in screening accessibility, hinder its early detection (3). Furthermore, treatment resistance and
metastatic recurrence pose major challenges, emphasizing the need
for new diagnostic and therapeutic strategies. In this context,
exosomal microRNAs (miRNAs) have emerged as critical regulators of
breast carcinoma progression, with potential applications in early
diagnosis, prognosis and targeted therapy (4,5).
Previous studies have shown that exosomes are small
extracellular vesicles secreted by cells that play a key role in
intercellular communication by transporting biologically active
molecules including proteins, lipids and nucleic acids (6,7).
Among them, miRNAs, short non-coding RNA involved in
post-transcriptional gene regulation, have been identified as key
mediators of tumor growth, metastasis and therapy resistance
(8). There is growing evidence
that exosomal miRNAs secreted by breast cancer cells act as
oncogenes that promote malignant tumor behavior by inhibiting tumor
suppressor genes or activating pro-tumor signaling pathways. For
example, miR-221/222 can mediate endocrine therapy resistance
(9) and high expression of
miR-10b is closely associated with distant metastasis in breast
cancer (10). The secretion and
uptake of these exosomal miRNAs are highly specific and can reshape
the tumor microenvironment and drive disease progression.
Furthermore, their presence in biological fluids and
disease-specific expression patterns make exosomal miRNAs promising
non-invasive biomarkers for breast carcinoma (11).
Building on previous findings, recent studies have
explored the mechanistic roles of exosomal miRNAs in modulating the
tumor microenvironment (TME), facilitating drug resistance and
promoting calcification in breast carcinoma. Moreover, the
integration of nanotechnology and exosome-based drug delivery
systems, such as synthetic miRNA mimics and inhibitors, has opened
new avenues for miRNA-targeted therapies. These emerging insights
bridge the gap between basic research and clinical applications,
bringing exosomal miRNAs closer to becoming practical oncology
tools. The aim of the present review was to provide a comprehensive
overview of the role of exosomal miRNAs in breast carcinoma,
summarize past studies, highlight recent advances and assess their
potential in precision medicine. By exploring their diagnostic,
prognostic and therapeutic applications, this study contributes to
the development of novel strategies to improve breast carcinoma
management and patient prognosis.
A variety of risk factors associated with breast
carcinoma development have been identified. Germline pathogenic
variants in the BRCA1 and BRCA2 genes confer a
markedly elevated risk of breast carcinoma development (12). Endogenous hormonal factors,
particularly elevated estrogen and progesterone levels or prolonged
exposure to these hormones through early menarche (<12 years),
late menopause (>55 years), nulliparity, or advanced maternal
age at first childbirth (>30 years), have been mechanistically
linked to oncogenesis (13). In
addition, lifestyle factors, including physical inactivity, obesity
(body mass index ≥30 kg/m2), smoking and excessive
alcohol consumption (>three drinks/day), have been linked to an
increased risk of breast carcinoma development (14). Furthermore, environmental factors,
including ionizing radiation and exposure to certain chemicals, are
associated with the development of breast carcinoma (15).
The primary diagnostic modalities for the early
detection of breast carcinoma include histopathological analysis,
radiographic imaging and molecular biomarker assays. Conventional
mammography demonstrates limited sensitivity (58-72%) in dense
breast tissue (Breast Imaging Reporting and Data System density
category C/D) (16), leading to
false-negative result interpretations and diagnostic delays.
Radiographically dense parenchyma, characterized by >50%
fibroglandular tissue composition, not only obscures tumor
visualization but also demonstrates an independent correlation with
2-6-fold increased breast carcinoma risk through stromal-epithelial
interactions (17). While
population-based screening programs have reduced mortality rates by
15-40%, they incur substantial healthcare expenditures and induce
screening-related anxiety (18).
Moreover, equitable implementation remains challenging in
low-resource settings due to infrastructure limitations and trained
personnel shortages, compounded by heterogeneous international
guidelines. Therefore, current early diagnostic technologies for
breast cancer are limited by imaging sensitivity and the stability
of molecular markers. Future diagnostic strategies should be a
combination of imaging and liquid biopsy models, in which exosomal
miRNAs are expected to be a key bridge between the two.
Exosomes are a class of small membranous vesicles
secreted by cells, usually 30-150 nm in diameter and belong to a
subgroup of extracellular vesicles. They are found in various body
fluids, including blood, saliva and urine and are secreted by a
wide range of cell types (19).
Proteomic profiling has revealed the complexity of the exosomal
cargo, including thousands of proteins, various miRNAs and lipids.
The protein composition of exosomes varies depending on their
cellular origin and includes cytoplasmic enzymes, fusion proteins,
membrane transporter proteins, molecular chaperones and structural
proteins (20). The main function
of exosomes is to mediate intercellular communication and influence
the function of recipient cells. In breast carcinoma, exosomes have
been shown to be involved in tumor progression, metastasis and
immune evasion.
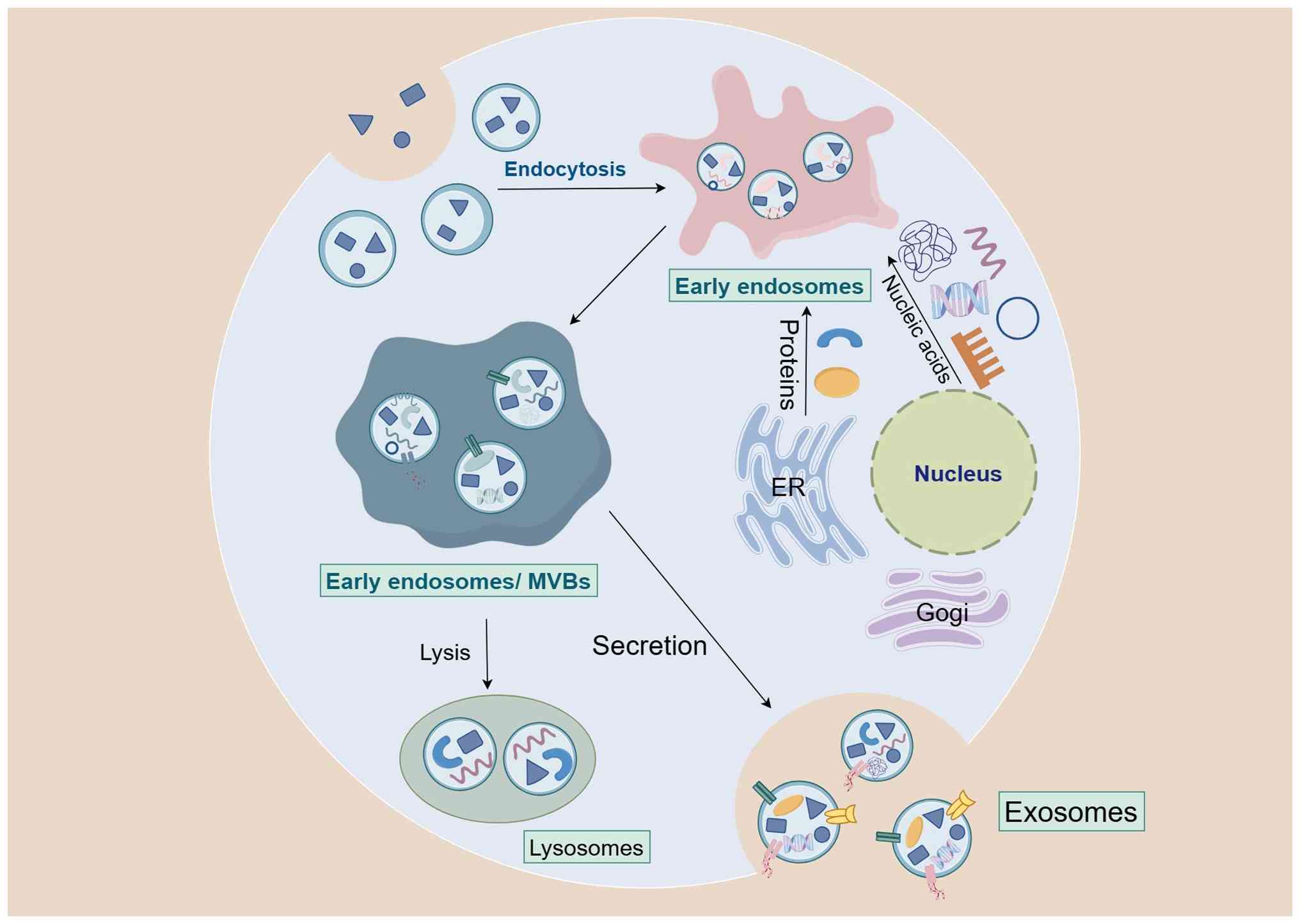
Studies have revealed several key mechanisms
underlying exosome formation, including endosomal sorting complexes
required for transport (ESCRT)-dependent and ESCRT-independent
pathways, the role of quadruple transmembrane proteins and the
formation of atypical exosomes (21,22). Fig.
1 illustrates the process of exosome formation.
Exosome formation primarily relies on the ESCRT
mechanism. This process begins with t early endosomes, which are
formed through the endocytosis of the plasma membrane. As the
endosomes mature into multivesicular bodies (MVBs), their membrane
folds inwards to form intraluminal vesicles (ILVs). These ILVs
become exosomes when MVBs fuse with the plasma membrane and are
released into the extracellular space (23).
Exosomes are rich in cholesterol,
phosphatidylcholine, phosphatidylserine and other lipids that are
not only involved in exosome biosynthesis, but also affect their
uptake and function (24).
Intracellular cholesterol levels regulate exosome release through
the PI3K/AKT signaling pathway: elevated intracellular cholesterol
inhibits this pathway, resulting in decreased exosome release,
whereas decreased intracellular cholesterol activates this pathway
and increases exosome release (25). Additionally, neutral
sphingomyelinase 2 promotes ILV and exosome formation by converting
sphingomyelin into sphingosine. Moreover, the autophagy-related
protein LC3 promotes sphingosine-mediated ILV formation by
recruiting the nSMase activator onto endosomal membranes (26).
Tetraspanins, such as CD9, CD63 and CD81 not only
play crucial roles in endosomal membrane invagination and ILV
formation but also influence exosome stabilization and facilitate
cell-cell communication (27). In
terms of cargo sorting, tetraspanins can recognize and bind to
specific proteins and nucleic acids and regulate the cargo loading
of exosomes (28).
Studies have revealed some mechanisms underlying
atypical exosome formation. For instance, certain cell types can
generate exosomes via the Golgi pathway or calcium-dependent
secretory autophagy (29). In
addition, some viruses can hijack the exosome formation mechanism
of host cells for their propagation (30).
Exosomal miRNAs are a class of small non-coding RNAs
that are usually 18-22 nucleotides in length and are encapsulated
in exosomes. These miRNAs are involved in the post-transcriptional
regulation of gene expression by binding to complementary sequences
on target miRNAs, leading to miRNA degradation or translational
repression (31,32). Exosomal miRNAs are selectively
packaged into exosomes, which allows them to be protected from
degradation by RNases and transported over long distances through
body fluids, such as blood, urine and milk (19). This intercellular communication
mechanism is critical for various physiological and pathological
processes including cancer progression, immune regulation and
tissue homeostasis (33). This
property makes exosomal miRNAs potential biomarkers for disease
diagnosis and prognosis as well as possible targets for the
treatment of cancers, including breast carcinoma.
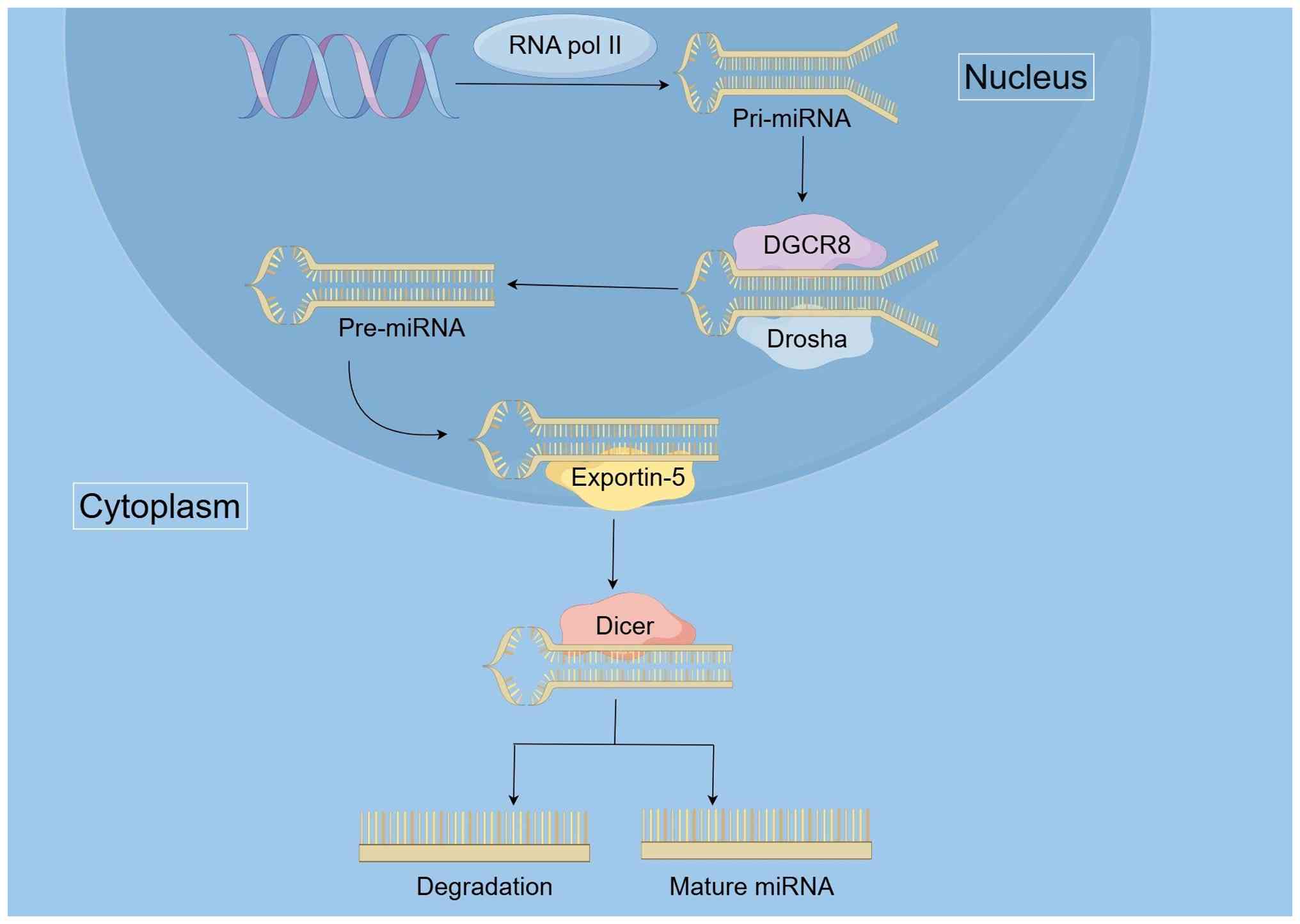
Exosomal miRNA biosynthesis is a multi-step process
that involves the transcription, processing and selective packaging
of miRNAs into exosomes. Studies have shown that precursor miRNAs
(pre-miRNAs) associated with processing complexes such as Dicer,
Argonaute 2 and TAR RNA-binding protein can be detected in exosomes
derived from breast carcinoma cells. These pre-miRNAs are further
processed into mature miRNAs within exosomes, thereby establishing
a new mechanism for the integration of miRNAs into exosomes
(34). Typically, miRNA
biosynthesis begins in the nucleus, where miRNA genes are
transcribed by RNA polymerase II to produce primary miRNAs
(pri-miRNAs). These pri-miRNAs are then processed by the
Drosha-DGCR-8 complex into pre-miRNAs, which is a critical step in
the miRNA maturation process (35). The pre-miRNAs are transported from
the nucleus to the cytoplasm via Exportin-5, a transport protein
that facilitates the movement of pre-miRNAs across the nuclear
envelope. In the cytoplasm, Dicer enzymes further process the
pre-miRNAs into mature miRNA duplexes (36). This step is essential for the
functionalization of miRNAs so that they can be integrated into
exosomes. Fig. 2 depicts miRNA
biosynthesis.
Several mechanisms have been proposed to explain how
specific miRNAs are categorized into exosomes. One of the key
mechanisms involves the recognition of exosomal miRNA sequences or
motifs by RNA-binding proteins such as hnRNPA2B1, Y-box-binding
protein 1 and synaptotagmin-binding cytoplasmic RNA-interacting
protein (SYNCRIP). SYNCRIP is involved in the exosomal partitioning
of miRNAs by recognizing specific sequences such as the hEXO motif,
which enhances miRNA loading into exosomes. This process is crucial
for cell-cell communication and is regulated by the NURR domain of
SYNCRIP, which binds to miRNA targets with high affinity (37,38). HnRNPA2B1 binds to specific
sequence motifs in miRNAs, thereby controlling their loading into
exosomes. Its binding activity is influenced by SUMOylation and
alterations in either the motif or hnRNPA2B1 expression levels can
modulate the efficiency of miRNA sorting. Notably, hnRNPA2B1 is
essential for the selective sorting of miRNAs, such as miR-486a-5p,
which has cardioprotective effects (39,40). YBX 1 also facilitates the sorting
of miRNAs (such as miR-223) into exosomes by binding to specific
sequence motifs. It shuttles between different cellular
compartments, suggesting its dynamic role in miRNA sorting and
exosome formation (41). In
addition, the ESCRT machinery and associated proteins such as ALIX
and TSG101 play key roles in exosome biosynthesis and miRNA
integration. The ESCRT machinery and related proteins, such as ALIX
and TSG101, promote the budding of ILVs into MVBs. miRNAs are
selectively sorted into ILVs during this process, which is followed
by the formation of MVBs. When MVBs fuse to the cell membrane, ILVs
are released as an exosome (42,43).
Studies have also highlighted the role of
post-transcriptional modifications, such as uridylation and
adenylation, in the sorting of miRNAs into exosomes. For example,
the addition of uridine residues to the 3'terminus of miRNAs
enhances their incorporation into the exosomes (44). Furthermore, the lipid composition
of the exosomal membrane, particularly in the presence of ceramide,
has been implicated in the sorting process (45). Once packaged into exosomes, miRNAs
are protected from degradation by RNases and can be transported to
recipient cells via circulation or other body fluids. After uptake
by recipient cells, exosomal miRNAs regulate gene expression and
influence various cellular processes, including proliferation,
apoptosis and metastasis, thus playing an important role in breast
carcinoma progression and therapy resistance (46).
A variety of miRNAs have been identified as key
factors in the development and progression of breast carcinoma.
These oncogenic miRNAs exert their oncogenic effects through a
variety of mechanisms, including the repression of tumor suppressor
genes, activation of pro-tumor signaling pathways and promotion of
epithelial-mesenchymal transition (47). High expression of oncogenic miRNAs
is associated with poor prognosis and advanced disease stage,
highlighting their potential as therapeutic targets and diagnostic
biomarkers for breast carcinoma. Table I lists miRNAs that contribute to
breast carcinoma development and progression (9,10,48-60).
Calcification, particularly microcalcification, is a
common radiological feature observed in mammographic imaging of
breast carcinoma. Calcifications are generally categorized into two
types: Macrocalcifications and microcalcifications.
Macrocalcifications are larger, coarser calcium deposits that are
typically benign and often related to aging, inflammation, or past
injuries. Radiologically, microcalcifications appear as fine,
granular specks, while macrocalcifications present as larger, often
round or irregular, dense opacities. Microcalcifications are
predominantly composed of hydroxyapatite, consisting of small
calcium phosphate deposits within the breast tissue and are often
associated with malignant lesions (94). Their chemical composition,
particularly the carbonate content and protein-matrix ratio,
correlates with the grading of pathological breast carcinoma and
may serve as a diagnostic and prognostic indicator (95). These calcifications are thought to
originate from cellular debris, necrosis, or active mineralization
processes mediated by tumor cells and the TME.
Exosomal miRNAs have been increasingly recognized as
important regulators of tumor progression and the TME. Although
there have been few direct studies on the relationship between
exosomal miRNAs and breast carcinoma calcification in existing
literature, the following potential mechanisms can be hypothesized
based on their known biological functions and associated signaling
pathways.
Downregulation of miR-30b has been observed in
recurrent breast carcinoma, whereas overexpression of its target
gene, CCNE2, may influence calcium channel activity through
the PI3K/AKT signaling pathway (99). Disruption of calcium homeostasis
via this mechanism may contribute to the formation of
microcalcifications. Additionally, miR-16 (103) exhibits antiangiogenic properties
by inhibiting the expression of vascular endothelial growth factor.
Impaired angiogenesis may result in localized hypoxia, which
activates hypoxia-inducible factors that subsequently upregulate
the expression of calcium-binding proteins.
Recent dual strategy advances in miRNA-based
therapeutics have emphasized the use of synthetic miRNA mimics to
restore tumor suppressor miRNAs. For example, the use of miRNAs
from the let-7 family (104) and
anti-miRNA oligonucleotides to inhibit oncogenic miRNAs such as
miR-21 (105) and miR-155
(106). A phase I clinical trial
demonstrated that MRX34, a liposomal miRNA-34a mimic, exhibited
antitumor activity in metastatic breast carcinoma by targeting the
PD-L1 and MET pathways, although challenges in delivery efficiency
remain (107,108). Novel nanoparticle-based delivery
systems such as exosome-like vesicles can be effective in improving
the stability and tumor targeting of miRNA analogs in TNBC models
(109). By contrast, anti-miRNA
inhibitors such as anti-miR-21 and anti-miR-155 oligonucleotides
inhibit oncogenic miRNAs. Preclinical studies have shown that
anti-miR-155 acts synergistically with immune checkpoint inhibitors
to reprogram the TME, reduce immunosuppressive myeloid cell
infiltration and enhance T cell responses in metastatic models
(110).
The synergistic effects of miRNA therapeutics with
conventional therapies have gained increasing attention. Hyaluronic
acid-based nanocarriers can target CD44-overexpressing TNBC cells
and efficiently co-deliver doxorubicin (DOX) and miR-542-3p. This
system provides an effective strategy for reversing TNBC resistance
by enhancing intracellular drug accumulation, upregulating the
expression of the p53 tumor suppressor protein, directly
downregulating the apoptosis inhibitory protein survivin,
overcoming chemotherapy resistance through multiple pathways and
markedly inducing synergistic apoptosis in vitro (111). Functional exosome-mediated
co-delivery system of DOX and hm-miR159 provides a promising and
innovative combination therapy to address the challenge of TNBC
chemoresistance (112).
Furthermore, emerging strategies that combine miRNA inhibitors with
immune checkpoint inhibitors can reprogram the TME in metastatic
breast carcinoma, thus offering a potentially powerful avenue for
enhancing immunotherapeutic efficacy (113).
Recent progress in the targeted delivery of
therapeutic miRNAs has greatly advanced their therapeutic
potential. In terms of carrier design, lipid nanoparticles and
exosome-mimetic vesicles have emerged as prominent platforms for
enhancing the stability, bioavailability and tumor-targeting
capability of therapeutic miRNAs. For instance, Ohno et al
(114) engineered exosomes
derived from HEK293 cells by displaying the GE11 peptide via
a Lamp2b fusion protein on their surface, constructing engineered
exosomes that specifically recognize EGFR. These exosomes
efficiently delivered miRNAs to EGFR-positive breast tumors in
vivo and markedly suppressed tumor growth. Similarly,
RGD-modified exosomes, which target αvβ3 integrin, have been used
to co-deliver miR-34a to TNBC. This approach not only directly
induces tumor cell apoptosis but also remodels the
immunosuppressive microenvironment by downregulating PD-L1, thereby
enhancing CD8+ T cell infiltration (115,116). Although several miRNA
formulations have entered early phase clinical trials, challenges
remain regarding delivery efficiency, immune tolerance and
large-scale production. Continued optimization of targeted delivery
systems is therefore crucial for the successful clinical
translation of miRNA-based therapeutics.
Exosomes carry various biomarkers associated with
breast carcinoma, such as carcinoembryonic antigen and carbohydrate
antigen 125, which have potent therapeutic diagnostic properties
(117). A systematic review
conducted up to March 2023 analyzed 46 articles and found that
exosomal miRNAs were markedly associated with the
clinicopathological features of breast carcinoma (118). The present review emphasized
that exosomal miRNAs such as miR-16, miR-21 and miR-155, have been
analyzed in at least three studies and are suitable for selection
as potential biomarkers. These miRNAs could provide information
regarding early diagnosis, disease progression, recurrence,
treatment response and metastasis. Analyzing exosomal miRNAs in
blood samples is a simple and noninvasive method that is important
for the early detection of breast carcinoma (118). A study published in 2024
explored the advancement of serum exosomal miR-21 as a molecular
diagnostic marker for breast carcinoma and its efficacy was
assessed for the early detection and clinical diagnosis of breast
carcinoma (119). However, its
application in early diagnosis presents several challenges. The
primary bottleneck is that the identification technology itself is
complex, time-consuming and expensive. In addition, its diagnostic
specificity must be improved to accurately differentiate breast
cancer from other malignancies. Finally, whether serum or plasma
should be preferred for cancer detection using exosomal miRNAs
needs to be studied further.
Exosomal miRNAs have been recognized as important
biomarkers for the prognostic assessment of breast carcinoma
because of their high stability and specificity. For example,
miRNA-373 is a specific biomarker for TNBC and its expression level
correlates with the degree of malignancy and prognosis of breast
carcinoma (120). Similarly, the
expression levels of miR-1246 and miR-155 are markedly elevated in
drug-resistant breast carcinoma cells compared to drug-sensitive
cells and this differential expression has been associated with
poor progression-free survival and overall survival in patients
(121). Additionally, dynamic
monitoring of exosomal miRNAs (such as miR-141, miR-182 and
miR-183) during neoadjuvant therapy can predict pathologic complete
response, thereby enabling more informed and personalized treatment
decisions (122).
Breast carcinoma remains an enormous global health
challenge that requires the continuous exploration of novel
diagnostic and therapeutic strategies. Exosomal miRNAs have emerged
as pivotal regulators of breast carcinoma pathogenesis, providing
valuable insights into tumor progression, metastasis and therapy
resistance. The present study reviewed the role of exosomal miRNAs
in breast carcinoma, highlighting their dual functions as oncogenic
drivers and tumor suppressors, as well as their utility as
diagnostic biomarkers and therapeutic potential. In the present
review, the dual oncogenic and inhibitory roles of exosomal miRNAs
were described, focusing on their multilevel regulatory mechanisms
in TME, drug tolerance and pathological calcification. Based on the
analysis of relevant literature, the present review proposed a
conceptual model of exosomal miRNA calcification axis, which
suggests that exosomal miRNAs may play a bridging role in breast
cancer calcification and progression by regulating cell metabolism,
immune inflammatory response and osteogenic signaling pathways.
However, there are several challenges and opportunities in
translating these findings into clinical practice. For example,
there is no perfect methodology for the extraction and purification
of exosomal RNAs. The differences in miRNA expression levels
between studies may be related to the heterogeneity of sample
sources, analytical methodologies and patient populations. Further
studies are needed to fully elucidate the role of exosomal miRNAs
in breast carcinoma calcification. The combination of advanced
imaging techniques, such as high-resolution mammography and
molecular imaging, as well as exosomal miRNA analysis, may provide
new insights into the mechanisms of calcification and its clinical
significance. Additionally, the development of exosomal miRNA-based
therapeutics may provide new avenues for the prevention and
treatment of breast carcinoma calcification. Overall, the present
review provided a systematic framework for the study of exosomal
miRNAs in breast cancer and demonstrates their potential value in
pathological calcification, precise diagnosis and treatment. In the
future, with technological progress and multidisciplinary research,
exosomal miRNAs are expected to become an important breakthrough in
understanding the complex biology of breast cancer and achieving
personalized treatment.
Not applicable.
ZMC was responsible for research conceptualization,
original draft preparation and reviewing the entire manuscript. PH
conducted supplementary analysis, responded to reviewers' comments
and updated and integrated relevant literature during the revision.
DYY performed calibration of the full text, organized the logical
flow and improved the language. SL contributed to the conception
and design of the study, participated in developing the manuscript
framework, critically revised the manuscript for important
intellectual content and provided final approval of the version to
be published. SQC guided and oversaw the entire research process,
supervised the completion of the present review and provided final
approval of the manuscript. Data authentication is not applicable.
All authors read and approved the final manuscript.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
During the preparation of this work, DeepSeek was
used to provide suggestions for the logical organization and
language polishing of some paragraphs and ChatGPT was utilized to
assist in optimizing sentence expressions and enhancing text
fluency. Using these tools, the content was reviewed sentence by
sentence. The accuracy of the professional concepts was verified
and the expressions generated by the AI were adjusted to conform to
the research and the writing style of the authors. The originality
and scientific nature of the content were ensured and full
responsibility for the content of the publication was assumed by
the authors.
Not applicable.
No funding was received.
|
1
|
Arnold M, Morgan E, Rumgay H, Mafra A,
Singh D, Laversanne M, Vignat J, Gralow JR, Cardoso F, Siesling S
and Soerjomataram I: Current and future burden of breast cancer:
Global statistics for 2020 and 2040. Breast. 66:15–23. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Giaquinto AN, Sung H, Miller KD, Kramer
JL, Newman LA, Minihan A, Jemal A and Siegel RL: Breast cancer
statistics, 2022. CA Cancer J Clin. 72:524–541. 2022.PubMed/NCBI
|
|
3
|
Tornai M, Hugg J, Patt B, et al: Abstract
PO5-18-01: Cancer detection rate meta-analysis comparison of
contemporary dense-breast supplemental screening modalities. Cancer
Res. 2024, https://aacrjournals.org/cancerres/article/84/9_Supplement/PO5-18-01/744391/Abstract-PO5-18-01-Cancer-Detection-Rate-Meta.
View Article : Google Scholar
|
|
4
|
Li J, He D, Bi Y and Liu S: The emerging
roles of exosomal miRNAs in breast cancer progression and potential
clinical applications. Breast Cancer (Dove Med Press). 15:825–840.
2023.PubMed/NCBI
|
|
5
|
Thakur P, Dahiya H, Kaushal A, Gupta VK,
Saini AK and Saini RV: Exosomal miRNAs as Next-generation therapy
vehicles in breast cancer. Curr Gene Ther. 23:330–342. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Di Bella MA: Overview and update on
extracellular vesicles: Considerations on exosomes and their
application in modern medicine. Biology. 11:8042022. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Simeone P, Bologna G, Lanuti P,
Pierdomenico L, Guagnano MT, Pieragostino D, Del Boccio P, Vergara
D, Marchisio M, Miscia S and Mariani-Costantini R: Extracellular
vesicles as signaling mediators and disease biomarkers across
biological barriers. Int J Mol Sci. 21:25142020. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
He B, Zhao Z, Cai Q, Zhang Y, Zhang P, Shi
S, Xie H, Peng X, Yin W, Tao Y and Wang X: miRNA-based biomarkers,
therapies, and resistance in cancer. Int J Biol Sci. 16:2628–2647.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Di Martino MT, Arbitrio M, Caracciolo D,
Cordua A, Cuomo O, Grillone K, Riillo C, Caridà G, Scionti F,
Labanca C, et al: miR-221/222 as biomarkers and targets for
therapeutic intervention on cancer and other diseases: A systematic
review. Mol Ther Nucleic Acids. 27:1191–1224. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kim J, Siverly AN, Chen D, Wang M, Yuan Y,
Wang Y, Lee H, Zhang J, Muller WJ, Liang H, et al: Ablation of
miR-10b suppresses oncogene-induced mammary tumorigenesis and
metastasis and reactivates tumor-suppressive pathways. Cancer Res.
76:6424–6435. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ashekyan O, Abdallah S, Shoukari AA,
Chamandi G, Choubassy H, Itani ARS, Alwan N and Nasr R: Spotlight
on exosomal Non-coding RNAs in breast cancer: An in silico analysis
to identify potential lncRNA/circRNA-miRNA-target axis. Int J Mole
Sci. 23:83512022. View Article : Google Scholar
|
|
12
|
Lee A, Moon BI and Kim TH: BRCA1/BRCA2
pathogenic variant breast cancer: Treatment and prevention
strategies. Ann Lab Med. 40:114–121. 2020. View Article : Google Scholar :
|
|
13
|
Admoun C and Mayrovitz HN: The etiology of
breast cancer. Breast Cancer. Mayrovitz HN: Exon Publications;
Brisbane: 2022, View Article : Google Scholar
|
|
14
|
Obeagu EI and Obeagu GU: Breast cancer: A
review of risk factors and diagnosis. Medicine (Baltimore).
103:e369052024. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bazyka DA, Lytvynenko OO and Litvinenko
OO: Influence of ionizing radiation on the development of breast
cancer. Probl Radiac Med Radiobiol. 28:22–48. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Petrović D, Šćepanović B, Spirovski M,
Nikin Z and Prvulović Bunović N: Comparative diagnostic efficacy of
four breast imaging modalities in dense breasts: A Single-center
retrospective study. Biomedicines. 13:17502025. View Article : Google Scholar
|
|
17
|
Wanders JO, Holland K, Veldhuis WB, Mann
RM, Pijnappel RM, Peeters PH, van Gils CH and Karssemeijer N:
Volumetric breast density affects performance of digital screening
mammography. Breast Cancer Res Treat. 162:95–103. 2017. View Article : Google Scholar :
|
|
18
|
Brill JV: Screening for cancer: The
economic, medical, and psychosocial issues. Am J Manag Care.
26(Suppl): S300–S306. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kalluri R and LeBleu VS: The biology,
function, and biomedical applications of exosomes. Science.
367:eaau69772020. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zemanek T, Danisovic L and Nicodemou A:
Exosomes, their sources, and possible uses in cancer therapy in the
era of personalized medicine. J Cancer Res Clin Oncol. 151:162024.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Tabatabai TS, Alizadeh M, Rezakhani L,
Tabatabai TS, Ehterami A, Kloucheh SG, Kebria MM, Vaez A and Salehi
M: Unlocking the potential of EXOs in regenerative medicine: A
comprehensive review. Tissue Cell. 97:1030682025. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Larios J, Mercier V, Roux A and Gruenberg
J: ALIX- and ESCRT-III-dependent sorting of tetraspanins to
exosomes. J Cell Biol. 219:e2019041132020. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Arya SB, Collie SP and Parent CA: The
ins-and-outs of exosome biogenesis, secretion, and internalization.
Trends Cell Biol. 34:90–108. 2024. View Article : Google Scholar
|
|
24
|
Fayyazpour P, Fayyazpour A, Abbasi K,
Vaez-Gharamaleki Y, Zangbar MS, Raeisi M and Mehdizadeh A: The role
of exosomes in cancer biology by shedding light on their lipid
contents. Pathol Res Pract. 250:1548132023. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Abdullah M, Nakamura T, Ferdous T, Gao Y,
Chen Y, Zou K and Michikawa M: Cholesterol regulates exosome
release in cultured astrocytes. Front Immunol. 12:7225812021.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lee YJ, Shin KJ and Chae YC: Regulation of
cargo selection in exosome biogenesis and its biomedical
applications in cancer. Exp Mol Med. 56:877–889. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Toribio V and Yáñez-Mó M: Tetraspanins
interweave EV secretion, endosomal network dynamics and cellular
metabolism. Eur J Cell Biol. 101:1512292022. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Gurung S, Perocheau D, Touramanidou L and
Baruteau J: The exosome journey: From biogenesis to uptake and
intracellular signalling. Cell Commun Signal. 19:472021. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ariotti N, Wu Y, Okano S, Gambin Y,
Follett J, Rae J, Ferguson C, Teasdale RD, Alexandrov K and Meunier
FA: An inverted CAV1 (caveolin 1) topology defines novel
autophagy-dependent exosome secretion from prostate cancer cells.
Autophagy. 17:2200–2216. 2021. View Article : Google Scholar :
|
|
30
|
Chu YD, Chen MC, Yeh CT and Lai MW:
Hijacking host extracellular vesicle machinery by hepatotropic
viruses: Current understandings and future prospects. J Biomed Sci.
31:972024. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Sarkar N and Kumar A: Paradigm shift:
MicroRNAs interact with target gene promoters to cause
transcriptional gene activation or silencing. Exp Cell Res.
444:1143722025. View Article : Google Scholar
|
|
32
|
Li C, Zhou T, Chen J, Li R, Chen H, Luo S,
Chen D, Cai C and Li W: The role of Exosomal miRNAs in cancer. J
Trans Med. 20:62022. View Article : Google Scholar
|
|
33
|
Zhang Y, Liu Y, Liu H and Tang WH:
Exosomes: Biogenesis, biologic function and clinical potential.
Cell Biosci. 9:192019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Tran N: Cancer Exosomes as miRNA
Factories. Trends Cancer. 2:329–331. 2016. View Article : Google Scholar
|
|
35
|
Tang J, He J, Feng C and Tu C: Exosomal
MiRNAs in osteosarcoma: Biogenesis and biological functions. Front
Pharmacol. 13:9020492022. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Sun Z, Shi K, Yang S, Liu J, Zhou Q, Wang
G, Song J, Li Z, Zhang Z and Yuan W: Effect of exosomal miRNA on
cancer biology and clinical applications. Mol Cancer. 17:1472018.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Hóbor F, Dallmann A, Ball N, Cicchini C,
Battistelli C, Ogrodowicz RW, Christodoulou E, Martin SR, Castello
A, Tripodi M, et al: A cryptic RNA-binding domain mediates Syncrip
recognition and exosomal partitioning of miRNA targets. Nat Commun.
9:8312018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Santangelo L, Giurato G, Cicchini C,
Montaldo C, Mancone C, Tarallo R, Battistelli C, Alonzi T, Weisz A
and Tripodi M: The RNA-binding protein SYNCRIP is a component of
the hepatocyte exosomal machinery controlling MicroRNA sorting.
Cell Rep. 173:799–808. 2016. View Article : Google Scholar
|
|
39
|
Jones K, Phan A, Zhang C, Haar L and Lynch
T: Abstract MP219: Hnrnpa2b1-dependent selective sorting of
Mir-486a-5p Into Msc-derived exosomes contributes to
cardioprotection. Circulation Res. 129:2021. View Article : Google Scholar
|
|
40
|
Zhou X, Wang L, Zou W, Chen X, Roizman B
and Zhou G: hnRNPA2B1 associated with recruitment of RNA into
exosomes plays a key role in herpes simplex virus 1 release from
infected cells. J Virology. 94:e00367–20. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Ma L, Singh J and Schekman R: Two
RNA-binding proteins mediate the sorting of miR223 from
mitochondria into exosomes. Elife. 12:e858782023. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Colombo M, Raposo G and Théry C:
Biogenesis, secretion, and intercellular interactions of exosomes
and other extracellular vesicles. Annu Rev Cell Dev Biol.
30:255–289. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Hessvik NP and Llorente A: Current
knowledge on exosome biogenesis and release. Cell Mol Life Sci.
75:193–208. 2018. View Article : Google Scholar
|
|
44
|
Wani S and Kaul D: Cancer cells govern
miR-2909 exosomal recruitment through its 3'- end
post-transcriptional modification. Cell Biochem Funct. 36:106–111.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Horbay R, Hamraghani A, Ermini L, Holcik
S, Beug ST and Yeganeh B: Role of ceramides and lysosomes in
extracellular vesicle biogenesis, cargo sorting and release. Int J
Mol Sci. 23:153172022. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Hoshino A, Kim HS, Bojmar L, Gyan KE,
Cioffi M, Hernandez J, Zambirinis CP, Rodrigues G, Molina H,
Heissel S, et al: Extracellular vesicle and particle biomarkers
define multiple human cancers. Cell. 182:1044–1061.e1018. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
O'Bryan S, Dong S, Mathis M and Alahari S:
The roles of oncogenic miRNAs and their therapeutic importance in
breast cancer. Eur J Cancer. 72:1–11. 2017. View Article : Google Scholar
|
|
48
|
Syed RU, Banu H, Alshammrani A, Alshammari
MD, G SK, Kadimpati KK, Khalifa AAS, Aboshouk NAM, Almarir AM,
Hussain A and Alahmed FK: MicroRNA-21 (miR-21) in breast cancer:
From apoptosis dysregulation to therapeutic opportunities. Pathol
Res Pract. 262:1555722024. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Giordo R, Ahmadi FAM, Husaini NA,
Al-Nuaimi NRAM, Ahmad SMS, Pintus G and Zayed H: microRNA 21 and
long non-coding RNAs interplays underlie cancer pathophysiology: A
narrative review. Noncoding RNA Res. 9:831–852. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Zhang W, Chen CJ and Guo GL: MiR-155
promotes the proliferation and migration of breast cancer cells via
targeting SOCS1 and MMP16. Eur Rev Med Pharmacol Sci. 22:7323–7332.
2018.PubMed/NCBI
|
|
51
|
Grimaldi AM, Nuzzo S, Condorelli G,
Salvatore M and Incoronato M: Prognostic and clinicopathological
significance of MiR-155 in breast cancer: A systematic review. Int
J Mol Sci. 21:58342020. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Liu Y, Zhao Q, Xi T, Zheng L and Li X:
MicroRNA-9 as a paradoxical but critical regulator of cancer
metastasis: Implications in personalized medicine. Genes Dis.
8:759–768. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Li X, Zeng Z, Wang J, Wu Y, Chen W, Zheng
L, Xi T, Wang A and Lu Y: MicroRNA-9 and breast cancer. Biomed
Pharmacother. 122:1096872020. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Ueda S, Takanashi M, Sudo K, Kanekura K
and Kuroda M: miR-27a ameliorates chemoresistance of breast cancer
cells by disruption of reactive oxygen species homeostasis and
impairment of autophagy. Lab Invest. 100:863–873. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Zhang H, Lang TY, Zou DL, Zhou L, Lou M,
Liu JS, Li YZ, Ding DY, Li YC, Zhang N, et al: miR-520b promotes
breast cancer stemness through Hippo/YAP signaling pathway. Onco
Targets Ther. 12:11691–11700. 2019. View Article : Google Scholar
|
|
56
|
Soheilifar MH, Vaseghi H, Seif F, Ariana
M, Ghorbanifar S, Habibi N, Papari Barjasteh F and Pornour M:
Concomitant overexpression of mir-182-5p and mir-182-3p raises the
possibility of IL-17-producing Treg formation in breast cancer by
targeting CD3d, ITK, FOXO1, and NFATs: A meta-analysis and
experimental study. Cancer Sci. 112:589–603. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Murugesan M and Premkumar K: Integrative
miRNA-mRNA functional analysis identifies miR-182 as a potential
prognostic biomarker in breast cancer. Mol Omics. 17:533–543. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Fei, Zhang J, Zhong L, Wang L, Liu Y, Wang
Y, Peng L and Guo B: Upregulated microRNA-301a in breast cancer
promotes tumor metastasis by targeting PTEN and activating
Wnt/β-catenin signaling. Gene. 535:191–197. 2014. View Article : Google Scholar
|
|
59
|
Santana T, de Oliveira Passamai L, de
Miranda FS, Borin TF, Borges GF, Luiz WB and Campos LCG: The role
of miRNAs in the prognosis of Triple-Negative breast cancer: A
systematic review and meta-analysis. Diagnostics (Basel).
13:1272022. View Article : Google Scholar
|
|
60
|
Wang Y, Zeng G and Jiang Y: The emerging
roles of miR-125b in cancers. Cancer Manag Res. 12:1079–1088. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Chu C, Liu X, Bai X, Zhao T, Wang M, Xu R,
Li M, Hu Y, Li W, Yang L, et al: MiR-519d suppresses breast cancer
tumorigenesis and metastasis via targeting MMP3. Int J Biol Sci.
14:228–236. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Deng X, Zhao Y and Wang B:
miR-519d-mediated downregulation of STAT3 suppresses breast cancer
progression. Oncol Rep. 34:2188–2194. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Ma H, Liu T, Xu Y, Wang X, Wang J and Liu
X: MiR-519d and miR-328-3p combinatorially suppress breast cancer
progression. Onco Targets Ther. 13:12987–12997. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Li J, Li Y and Cheng H: Circ-RPPH1
knockdown retards breast cancer progression via miR-328-3p-mediated
suppression of HMGA2. Clin Breast Cancer. 22:e286–e295. 2022.
View Article : Google Scholar
|
|
65
|
Yin M, Zhang Z and Wang Y: Anti-tumor
effects of miR-34a by regulating immune cells in the tumor
microenvironment. Cancer Med. 12:11602–11610. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Li L, Yuan L, Luo J, Gao J, Guo J and Xie
X: MiR-34a inhibits proliferation and migration of breast cancer
through down-regulation of Bcl-2 and SIRT1. Clin Exp Med.
13:109–117. 2013. View Article : Google Scholar
|
|
67
|
Li ZH, Weng X, Xiong QY, Tu JH, Xiao A,
Qiu W, Gong Y, Hu EW, Huang S and Cao YL: miR-34a expression in
human breast cancer is associated with drug resistance. Oncotarget.
8:106270–106282. 2017. View Article : Google Scholar :
|
|
68
|
Fontana A, Barbano R, Dama E, Pasculli B,
Rendina M, Morritti MG, Melocchi V, Castelvetere M, Valori VM,
Ravaioli S, et al: Combined analysis of miR-200 family and its
significance for breast cancer. Sci Rep. 11:29802021. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Song C, Liu LZ, Pei XQ, Liu X, Yang L, Ye
F and Xie X, Chen J, Tang H and Xie X: miR-200c inhibits breast
cancer proliferation by targeting KRAS. Oncotarget. 6:34968–34978.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Hong T, Ding J and Li W: miR-7 reverses
breast cancer resistance to chemotherapy by targeting MRP1 and
BCL2. Onco Targets Ther. 12:11097–11105. 2019. View Article : Google Scholar
|
|
71
|
Kalinowski FC, Brown RAM, Ganda C, Giles
KM, Epis MR, Horsham J and Leedman PJ: microRNA-7: A tumor
suppressor miRNA with therapeutic potential. Int J Biochem Cell
Biol. 54:312–317. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Wang Z, Hu S, Li X, Liu Z, Han D, Wang Y,
Wei L, Zhang G and Wang X: MiR-16-5p suppresses breast cancer
proliferation by targeting ANLN. BMC Cancer. 21:11882021.
View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Ni Q, Qian Y, Yi T, Zhou J, Sang K and Pan
C: miR-16-5p may modulate migration and proliferation through TP53
and LncRNA-NEAT1 in triple-negative breast cancer. Gene Rep.
37:1020382024. View Article : Google Scholar
|
|
74
|
Piergentili R, Marinelli E, Cucinella G,
Lopez A, Napoletano G, Gullo G and Zaami S: miR-125 in breast
cancer etiopathogenesis: An emerging role as a biomarker in
differential diagnosis, regenerative medicine, and the challenges
of personalized medicine. Noncoding RNA. 10:162024.PubMed/NCBI
|
|
75
|
Luo Y, Wang X, Niu W, Wang H, Wen Q, Fan
S, Zhao R, Li Z, Xiong W, Peng S, et al: Elevated microRNA-125b
levels predict a worse prognosis in HER2-positive breast cancer
patients. Oncol Lett. 13:867–874. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Pan S, Zhao X, Shao C, Fu B, Huang Y,
Zhang N, Dou X, Zhang Z, Qiu Y, Wang R, et al: STIM1 promotes
angiogenesis by reducing exosomal miR-145 in breast cancer
MDA-MB-231 cells. Cell Death Dis. 12:382021. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Contreras-Sanzón E, Carlos-Reyes Á,
Sierra-Martínez M, Acosta-Altamirano G, Luna-Rivero C, Núñez-Corona
D, García-Hernández AP, Ibarra-Sierra E, Vidrio-Morgado H,
Alvarez-Sánchez ME, et al: Metastatic breast tumors downregulate
miR-145 regulating the hypoxia-induced vasculogenic mimicry. Transl
Oncol. 33:1016802023. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Baxter DE, Allinson LM, Al Amri WS,
Poulter JA, Pramanik A, Thorne JL, Verghese ET and Hughes TA:
MiR-195 and its target SEMA6D regulate chemoresponse in breast
cancer. Cancers (Basel). 13:59792021. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Wang Y, Zhang X, Zou C, Kung HF, Lin MC,
Dress A, Wardle F, Jiang BH and Lai L: miR-195 inhibits tumor
growth and angiogenesis through modulating IRS1 in breast cancer.
Biomed Pharmacother. 80:95–101. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Chaudhari R, Nasra S, Meghani N and Kumar
A: MiR-206 conjugated gold nanoparticle based targeted therapy in
breast cancer cells. Sci Rep. 12:47132022. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Babaeenezhad E, Abdolvahabi Z, Asgharzadeh
S, Abdollahi M, Shakeri S, Moradi Sarabi M and Yarahmadi S:
Potential function of microRNA miRNA-206 in breast cancer
pathogenesis: Mechanistic aspects and clinical implications. Pathol
Res Pract. 260:1554542024. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Sun D, Li C and Zhang F: MicroRNA-206
suppresses growth and metastasis of breast cancer stem cells via
blocking EVI-1-mediated CALR expression. PLoS One. 17:e02749192022.
View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Tian Y, Chen ZH, Wu P, Zhang D, Ma Y, Liu
XF, Wang X, Ding D, Cao XC and Yu Y: MIR497HG-derived miR-195 and
miR-497 mediate tamoxifen resistance via PI3K/AKT signaling in
breast cancer. Adv Sci (Weinh). 10:e22048192023. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Tao S, Li H, Ma X, Lian B, He J, Gao Y and
Li J: Methylation-mediated silencing of MicroRNA-497 promotes
breast cancer progression through up-regulation of Mucin1. Front
Oncol. 10:5520992020. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Plantamura I, Cataldo A, Cosentino G and
Iorio MV: miR-205 in breast cancer: State of the art. Int J Mol
Sci. 22:272021. View Article : Google Scholar :
|
|
86
|
Shen Y, Xu Y, Huang L, Chi Y and Meng L:
MiR-205 suppressed the malignant behaviors of breast cancer cells
by targeting CLDN11 via modulation of the epithelial-to-mesenchymal
transition. Aging (Albany NY). 13:13073–13086. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Ouyang B, Bi M, Jadhao M, Bick G and Zhang
X: miR-205 regulates tamoxifen resistance by targeting estrogen
receptor coactivator MED1 in human breast cancer. Cancers.
16:39922024. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Hong BS, Ryu HS, Kim N, Kim J, Lee E, Moon
H, Kim KH, Jin MS, Kwon NH, Kim S, et al: Tumor suppressor
miRNA-204-5p regulates growth, metastasis, and immune
microenvironment remodeling in breast cancer. Cancer Res.
79:1520–1534. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Yang F, Bian Z, Xu P, Sun S and Huang Z:
MicroRNA-204-5p: A pivotal tumor suppressor. Cancer Med.
12:3185–3200. 2023. View Article : Google Scholar :
|
|
90
|
Bermúdez M, Martínez-Barajas MG,
Bueno-Urquiza LJ, López-Gutiérrez JA, Villegas-Mercado CE and
López-Camarillo C: Role of MicroRNA-204 in regulating the hallmarks
of breast cancer: An update. Cancers. 16:28142024. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Soung YH, Chung H, Yan C, Fesler A, Kim H,
Oh ES, Ju J and Chung J: Therapeutic potential of chemically
modified miR-489 in triple-negative breast cancers. Cancers.
12:22092020. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Soni M, Patel Y, Markoutsa E, Jie C, Liu
S, Xu P and Chen H: Autophagy, cell viability, and chemoresistance
are regulated by miR-489 in breast cancer. Mol Cancer Res.
16:1348–1360. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Menon A, Abd-Aziz N, Khalid K, Poh C and
Naidu R: miRNA: A promising therapeutic target in cancer. Int J Mol
Sci. 23:115022022. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Nahmias Y, Grobman GY and Vidavsky N:
Inhibiting pathological calcium phosphate mineralization:
Implications for disease progression. ACS Appl Mater Interfaces.
16:18344–18359. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Shin K, Laohajaratsang M, Men S, Figueroa
B, Dintzis S and Fu D: Quantitative chemical imaging of breast
calcifications in association with neoplastic processes.
Theranostics. 10:5865–5878. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Li XJ, Ren ZJ, Tang JH and Yu Q: Exosomal
MicroRNA MiR-1246 promotes cell proliferation, invasion and drug
resistance by targeting CCNG2 in breast cancer. Cell Physiol
Biochem. 44:1741–1748. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Hannafon BN, Trigoso YD, Calloway CL, Zhao
YD, Lum DH, Welm AL, Zhao ZJ, Blick KE, Dooley WC and Ding WQ:
Plasma exosome microRNAs are indicative of breast cancer. Breast
Cancer Res. 18:902016. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Eichelser C, Stückrath I, Müller V,
Milde-Langosch K, Wikman H, Pantel K and Schwarzenbach H: Increased
serum levels of circulating exosomal microRNA-373 in
receptor-negative breast cancer patients. Oncotarget. 5:9650–9663.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Sueta A, Yamamoto Y and Iwase H: The role
of exosomal microRNAs; focus on clinical applications in breast
cancer. Cancer Drug Resist. 2:847–861. 2019.PubMed/NCBI
|
|
100
|
Tang LB, Ma SX, Chen ZH, Huang QY, Wu LY,
Wang Y, Zhao RC and Xiong LX: Exosomal microRNAs: Pleiotropic
impacts on breast cancer metastasis and their clinical
perspectives. Biology (Basel). 10:3072021.PubMed/NCBI
|
|
101
|
Kan JY, Shih SL, Yang SF, Chu PY, Chen FM,
Li CL, Wu YC, Yeh YT, Hou MF and Chiang CP: Exosomal microRNA-92b
is a diagnostic biomarker in breast cancer and targets
survival-related MTSS1L to promote tumorigenesis. Int J Mol Sci.
25:12952024. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Xie QH, Zheng JQ, Ding JY, Wu YF, Liu L,
Yu ZL and Chen G: Exosome-mediated immunosuppression in tumor
microenvironments. Cells. 11:19462022. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Singh T, Kaushik M, Mishra LC, Behl C,
Singh V and Tuli HS: Exosomal miRNAs as novel avenues for breast
cancer treatment. Front Genet. 14:11347792023. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Xu J, Zhao X, Liang X, Guo D, Wang J, Wang
Q and Tang X: Development of miRNA-based PROTACs targeting Lin28
for breast cancer therapy. Sci Adv. 10:eadp03342024. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Gomez IG, MacKenna DA, Johnson BG, Kaimal
V, Roach AM, Ren S, Nakagawa N, Xin C, Newitt R, Pandya S, et al:
Anti-microRNA-21 oligonucleotides prevent Alport nephropathy
progression by stimulating metabolic pathways. J Clin Invest.
125:141–156. 2015. View Article : Google Scholar :
|
|
106
|
Dogra P, Shinglot V, Ruiz-Ramírez J, Cave
J, Butner JD, Schiavone C, Duda DG, Kaseb AO, Chung C, Koay EJ, et
al: Translational modeling-based evidence for enhanced efficacy of
standard-of-care drugs in combination with anti-microRNA-155 in
non-small-cell lung cancer. MedRxiv. Mar 15–2024. View Article : Google Scholar
|
|
107
|
Hong DS, Kang YK, Borad M, Sachdev J,
Ejadi S, Lim HY, Brenner AJ, Park K, Lee JL, Kim TY, et al: Phase 1
study of MRX34, a liposomal miR-34a mimic, in patients with
advanced solid tumours. Br J Cancer. 122:1630–1637. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Abate M, Lombardi A, Luce A, Porru M,
Leonetti C, Bocchetti M, Campani V, De Rosa G, Graziano SF, Nele V,
et al: Fluorescent nanodiamonds as innovative delivery systems for
MiR-34a replacement in breast cancer. Mol Ther Nucleic Acids.
33:127–141. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Yan G, Xiao Q, Zhao J, Chen H, Xu Y, Tan M
and Peng L: Brucea javanica derived exosome-like nanovesicles
deliver miRNAs for cancer therapy. J Control Release. 367:425–440.
2024. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Wang J, Wang Q, Guan Y, Sun Y, Wang X,
Lively K, Wang Y, Luo M, Kim JA, Murphy EA, et al: Breast cancer
cell-derived microRNA-155 suppresses tumor progression via
enhancing immune cell recruitment and antitumor function. J Clin
Invest. 132:e1572482022. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Wang S, Zhang J, Wang Y and Chen M:
Hyaluronic acid-coated PEI-PLGA nanoparticles mediated co-delivery
of doxorubicin and miR-542-3p for triple negative breast cancer
therapy. Nanomedicine. 12:411–420. 2016. View Article : Google Scholar
|
|
112
|
Kousar K, Ahmad T, Abduh MS, Kanwal B,
Shah SS, Naseer F and Anjum S: miRNAs in regulation of tumor
microenvironment, chemotherapy resistance, immunotherapy modulation
and miRNA therapeutics in cancer. Int J Mol Sci. 23:138222022.
View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Zhou H, Jia W, Lu L and Han R: MicroRNAs
with multiple targets of immune checkpoints, as a potential
sensitizer for immune checkpoint inhibitors in breast cancer
treatment. Cancers. 15:8242023. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Ohno S, Takanashi M, Sudo K, Ueda S,
Ishikawa A, Matsuyama N, Fujita K, Mizutani T, Ohgi T, Ochiya T, et
al: Systemically injected exosomes targeted to EGFR deliver
antitumor microRNA to breast cancer cells. Mol Ther. 21:185–191.
2013. View Article : Google Scholar
|
|
115
|
Deng S, Wang M, Wang C, Zeng Y, Qin X, Tan
Y, Liang B and Cao Y: p53 downregulates PD-L1 expression via
miR-34a to inhibit the growth of triple-negative breast cancer
cells: A potential clinical immunotherapeutic target. Mol Biol Rep.
50:577–587. 2023. View Article : Google Scholar
|
|
116
|
Huang X and Xie X, Wang H, Xiao X, Yang L,
Tian Z, Guo X, Zhang L, Tang H and Xie X: PDL1 And LDHA act as
ceRNAs in triple negative breast cancer by regulating miR-34a. J
Exp Clin Cancer Res. 36:1292017. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Khanicheragh P, Abbasi-Malati Z, Saghebasl
S, Hassanpour P, Milani SZ, Rahbarghazi R and Hasani A: Exosomes
and breast cancer angiogenesis; Highlights in intercellular
communication. Cancer Cell Int. 24:4022024. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Linares-Rodríguez M, Blancas I and
Rodríguez-Serrano F: The predictive value of blood-derived exosomal
miRNAs as biomarkers in breast cancer: A systematic review. Clin
Breast Cancer. 25:e48–e55.e15. 2025. View Article : Google Scholar
|
|
119
|
Li H and Tie XJ: Exploring research
progress in studying serum exosomal miRNA-21 as a molecular
diagnostic marker for breast cancer. Clin Transl Oncol.
26:2166–2171. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Piasecka D, Braun M, Kordek R, Sądej R and
Romanska H: MicroRNAs in regulation of triple-negative breast
cancer progression. J Cancer Res Clin Oncol. 144:1401–1411. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Zhang Z, Zhang L, Yu G, Sun Z, Wang T,
Tian X, Duan X and Zhang C: Exosomal miR-1246 and miR-155 as
predictive and prognostic biomarkers for trastuzumab-based therapy
resistance in HER2-positive breast cancer. Cancer Chemother
Pharmacol. 86:761–772. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Todorova VK, Byrum SD, Gies AJ, Haynie C,
Smith H, Reyna NS and Makhoul I: Circulating exosomal microRNAs as
predictive biomarkers of neoadjuvant chemotherapy response in
breast cancer. Curr Oncol. 29:613–630. 2022. View Article : Google Scholar : PubMed/NCBI
|