Introduction
Lung cancer is the most common cause of
cancer-related mortality worldwide. A number of theories and
multiple causes for lung cancer development have been proposed and
it was suggested that multiple genetic modifications and signaling
pathway alterations may be involved in lung oncogenesis (1). Rapid advances in this field have lead
to targeted therapy, including the FDA-approved epidermal growth
factor receptor and vascular endothelial growth factor inhibitors
erlotinib (2) and bevacizumab
(3), which have significantly
improved the outcome in patients with non-small-cell lung cancer.
The identification of additional molecular markers specific to lung
cancer may improve early detection, which is critical for improving
survival.
A major role for epigenetic mechanisms has been
suggested in cancer development (4). Specifically, methylation patterns are
significantly altered in cancer cells. Hypermethylation of certain
CpG islands is observed in the majority of tumor cells and is
associated with gene silencing. This is more prominent for certain
tumor suppressor genes whose loss-of-function promotes
carcinogenesis (5). The DNA
methylation status may also provide a highly sensitive and specific
marker for early cancer diagnosis.
To investigate the pathogenesis of radiation-induced
lung cancer and further provide evidence for clinical application,
we established a model of transformed human bronchial epithelial
cells (BEP2D) induced by α-particles (6) and the methylation status in the
promoter regions of multiple tumor suppressor genes was analyzed by
methylation-specific polymerase chain reaction (PCR).
Materials and methods
Cell culture
The BEP2D cell line is a human papillomavirus
18-immortalized human bronchial epithelial cell line and was kindly
provided by Dr Curtis C. Harris (National Cancer Institute, MD,
USA) (7). The BERP35T1 malignant
transformant cell line was derived from the BEP2D cell line through
α-particle irradiation (6). The
cells were cultured in serum-free LHC-8 medium (Biofluids Inc.,
Rockville, MD, USA) at 37°C under a 95% air/5% CO2
atmosphere.
SssI methyltransferase treatment
Genomic DNA was extracted from normal lung tissue
and treated with SssI methyltransferase (New England Biolabs,
Ipswich, MA, USA). SssI-treated DNA was then used as a substrate
for methylation-specific PCR.
Methylation-specific PCR analysis
We extracted and purified genomic DNA from BEP2D
cells and their malignant transformants, BERP35T1 cells. Following
modification by treatment with sodium bisulfite,
methylation-specific PCR (8) was
performed with 2 μl of bisulfite-treated DNA with methylation- and
unmethylation-specific primers for each tumor suppressor gene. The
amplified products were separated electrophoretically on 3% agarose
gels and visualized using ethidium bromide staining. The primer
sets and size of the amplified products are listed in Table I.
 | Table IPrimers used for methylation-specific
polymerase chain reaction. |
Table I
Primers used for methylation-specific
polymerase chain reaction.
| Gene | Sense primer
(5′-3′) | Antisense primer
(5′-3′) | Annealing temp
(°C) | Size (bp) |
|---|
|
p16INK4a | M:
TTATTAGAGGGTGGGGCGGATCGC | M:
GACCCCGAACCGCGACCGTAA | 65 | 150 |
| U:
TTATTAGAGGGTGGGGTGGATTGT | U:
CAACCCCAAACCACAACCATAA | 65 | 151 |
| MGMT | M:
TTTCGACGTTCGTAGGTTTTCGC | M:
GCACTCTTCCGAAAACGAAACG | 66 | 81 |
| U:
TTTGTGTTTTGATGTTTGTAGGTTTTTGT | U:
AACTCCACACTCTTCCAAAAACAAAACA | 66 | 93 |
| DAPK | M:
GGATAGTCGGATCGAGTTAACGTC | M:
CCCTCCCAAACGCCGA | 64 | 98 |
| U:
GGAGGATAGTTGGATTGAGTTAATGTT | U:
CAAATCCCTCCCAAACACCAA | 64 | 106 |
|
p14ARF | M:
GTGTTAAAGGGCGGCGTAGC | M:
AAAACCCTCACTCGCGACGA | 64 | 122 |
| U:
TTTTTGGTGTTAAAGGGTGGTGTAGT | U:
CACAAAAACCCTCACTCACAACAA | 64 | 132 |
| GSTP1 | M:
TTCGGGGTGTAGCGGTCGTC | M:
GCCCCAATACTAAATCACGACG | 55 | 91 |
| U:
GATGTTTGGGGTGTAGTGGTTGTT | U:
CCACCCCAATACTAAATCACAACA | 55 | 97 |
Results
Verification of methylation detection
system
The analyses of the methylation status of the genes
were performed by methylation-specific PCR. For the purpose of
verification of our methylation detection system, we extracted DNA
from normal lung tissue and then treated it with SssI methylase.
SssI methylase-treated and unmodified DNAs were used as substrates
for the PCR reaction using p14ARF gene
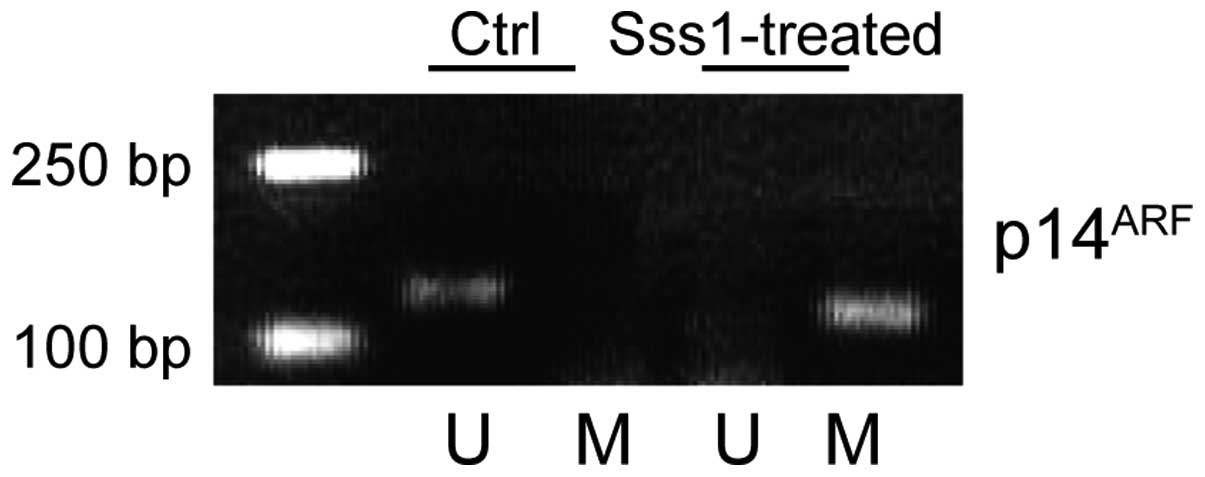
methylation-specific PCR primers. As shown in Fig. 1, a positive PCR product appeared in
the SssI methylase-treated DNA sample, but not in the untreated
control group, reflecting the specificity of our detection system.
The results were further confirmed by direct DNA sequencing.
Methylation-specific PCR of tumor
suppressor genes
In view of the critical role played by tumor
suppressors in cell cycle control and tumor genesis, we
investigated the methylation status of multiple tumor suppressor
genes in BEP2D cells and their malignant transformant BERP35T1
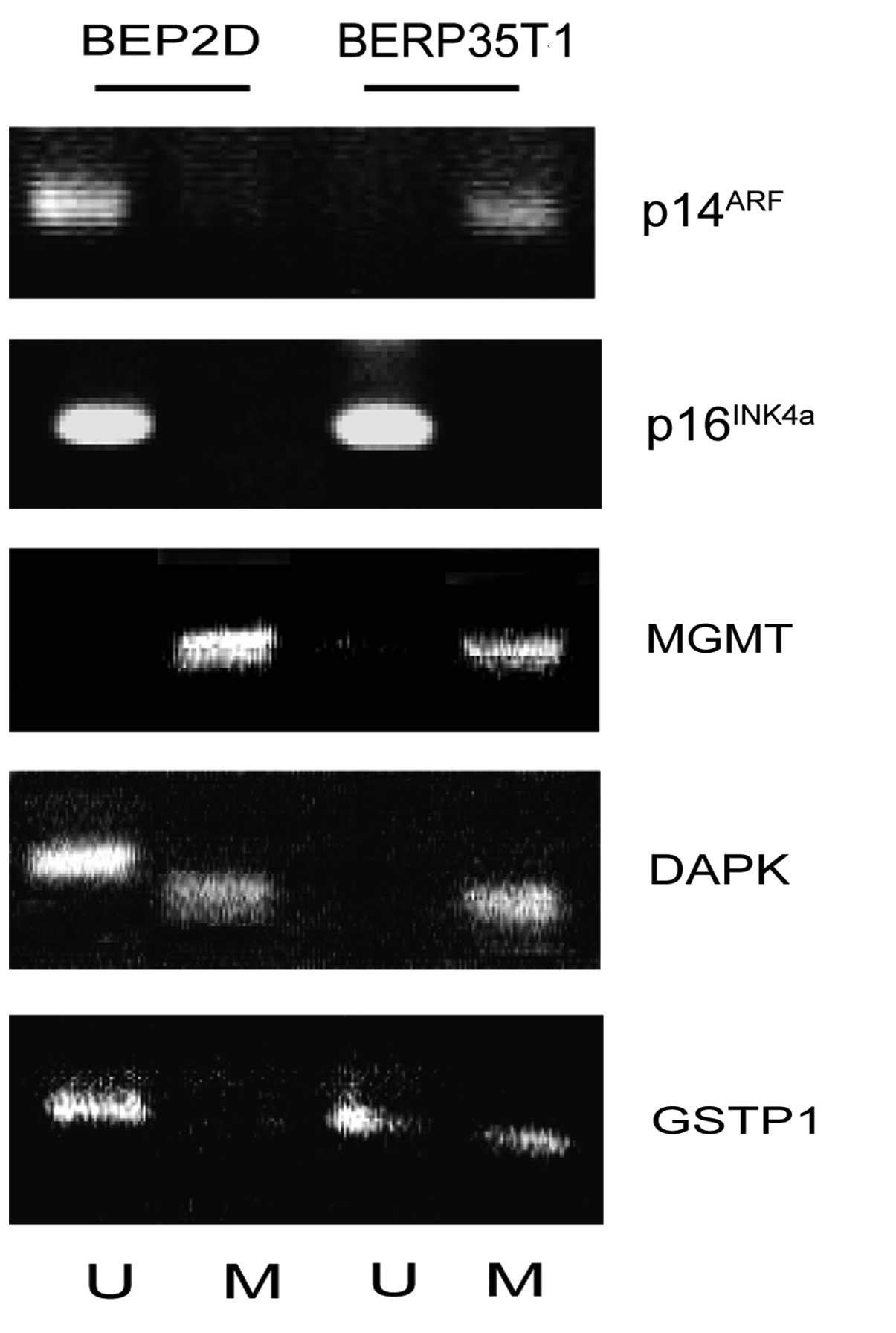
cells induced by α-particles. The CpG island in the 5′ promoter
region of the p14ARF gene was unmethylated in BEP2D
cells, but was modified through methylation in the malignant
transformant BERP35T1 cells. Of note, the p16INK4a gene,
which shares two exons with the p14ARF gene, was not
methylated in the promoter CpG islands.
The O6-methylguanine-DNA
methyltransferase (MGMT) gene is a specific DNA repair enzyme,
which removes the alkyl group from the O6-position of
guanine, preventing its mutagenic and carcinogenic effects
(9,10). The MGMT gene was found to be
methylated in BEP2D cells, as well as in malignant transformant
BERP35T1 cells (Fig. 2).
Death-associated protein kinase (DAPK) is a
multi-domain serine/threonine protein kinase that possesses
apoptotic and tumor-suppressive functions (11). The DAPK gene was partially
methylated in BEP2D cells and completely methylated in BERP35T1
cells (Fig. 2).
Methylation of the glutathione S-transferase P1
(GSTP1) gene promoter region is the most common epigenetic change
in prostate cancer (12). The GSTP1
gene was unmethylated in BEP2D cells and partially methylated in
BERP35T1 cells, suggesting that it may also involved in lung
oncogenesis (Fig. 2).
Discussion
DNA methylation in the promoter region is an
important mechanism of tumor suppressor gene inactivation in human
cancers. An alteration in the methylation status, particularly
hypermethylation, typically occurs at CpG islands in the promoter
region and is associated with tumor suppressor gene inactivation,
ultimately leading to carcinogenesis (13,14).
We used the α-particle-induced malignant
transformant BERP35T1 cell line as a cell culture model for lung
carcinogenesis; this cell line was derived from the BEP2D cell
line, a human papillomavirus 18-immortalized human bronchial
epithelial cell line (7). Ionizing
radiation was selected to establish BERP35T1, as it may induce a
high incidence of large chromosomal deletions or translocations as
a result of strand breakage (15).
When compared to BEP2D cells, malignant transformant BERP35T1 cells
exhibited altered growth kinetics, resistance to serum-induced
terminal differentiation and anchorage-independent growth.
To elucidate the molecular mechanism underlying
radiation-mediated carcinogenesis, we used methylation-specific PCR
to detect aberrant promoter methylation of multiple tumor
suppressor genes, including p14ARF and
p16INK4a (cell cycle regulators), MGMT and GSTP1 (DNA
repair) and DAPK (cell apoptosis) genes in BEP2D cells and
malignant transformant BERP35T1 cells. As shown in Fig. 2, these genes exhibited different
methylation patterns. For example, p16INK4A and
p14ARF are two tumor suppressors encoded by the
cyclin-dependent kinase inhibitor 2A gene locus. Through
alternative splicing, p16INK4A and p14ARF
have distinct promoter regions and first exons, but share the
second and third exons. The p16INK4A and
p14ARF proteins have important functions in cell cycle
regulation and generally act as negative regulators of cell cycle
progression. p16INK4A mainly acts via the retinoblastoma
pathway as an inhibitor of the cyclin-dependent kinases 4 and 6
(16) and p14ARF is
crucial for the p53 pathway through inhibiting mouse double minute
2 homolog (17,18). Our findings demonstrated that the
methylation status of p16INK4A and p14ARF was
regulated in a distinctive pattern, suggesting that
p16INK4A and p14ARF are separately involved
in tumor development.
Our results also demonstrated a distinctive
methylation pattern among other tumor suppressors in
α-particle-induced malignant transformant BERP35T1 cells, when
compared to their wild-type counterparts. For example, the MGMT
gene was methylated in both cell lines; the DAPK gene was only
partially methylated in wild-type cells and completely modified
through methylation in the malignant transformed cell line; as
regards the GSTP1 gene, it was unmethylated in wild-type cells and
partially methylated in BERP35T1 cells.
Our final goal is to apply DNA methylation markers
in early lung cancer diagnosis. Achieving this goal largely depends
on the sensitivity and specificity of these epigenetic markers. Our
present study demonstrated the practicality of this approach, since
the methylation patterns are quite distinctive for the malignant
transformed cell line compared to its wild-type counterpart. Our
ongoing efforts are aimed at screening high-risk groups for lung
cancer using these epigenetic signature and suggest a future
direction for manipulating epigenetic alterations towards lung
cancer therapy.
Acknowledgements
This study was supported by a grant from the
National High Technology Research and Development Program of China
(863 Program) (no. 2001AA221271), a grant from the Major State
Basic Research Development Program of China (973 Program) (no.
G1998051207), a grant from the Military Scientific Research
Foundation for Returned Scholars (no. 98H037) and a grant from the
Military Science Foundation for The Excellent Youth Scholars during
the 9th Five-Year Plan Period (no. 01J006).
References
|
1
|
Herbst RS, Heymach JV and Lippman SM: Lung
cancer. N Engl J Med. 359:1367–1380. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Shepherd FA, Rodrigues Pereira J, Ciuleanu
T, et al: Erlotinib in previously treated non-small-cell lung
cancer. N Engl J Med. 353:123–132. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sandler A, Gray R, Perry MC, et al:
Paclitaxel-carboplatin alone or with bevacizumab for non-small-cell
lung cancer. N Engl J Med. 355:2542–2550. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Boehm JS and Hahn WC: Towards systematic
functional characterization of cancer genomes. Nat Rev Genet.
12:487–498. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Esteller M: CpG island hypermethylation
and tumor suppressor genes: a booming present, a brighter future.
Oncogene. 21:5427–5440. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lou T, Xiang X and Wu D: Transformation of
human bronchial epithelial cells BEP2D induced by 238Pu
α-particles. Chin J Lung Canc. 3:428–431. 2000.(In Chinese).
|
|
7
|
Willey JC, Broussoud A, Sleemi A, et al:
Immortalization of normal human bronchial epithelial cells by human
papillomaviruses 16 or 18. Cancer Res. 51:5370–5377.
1991.PubMed/NCBI
|
|
8
|
Herman JG, Graff JR, Myohanen S, Nelkin BD
and Baylin SB: Methylation-specific PCR: a novel PCR assay for
methylation status of CpG islands. Proc Natl Acad Sci USA.
93:9821–9826. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Esteller M, Hamilton SR, Burger PC, Baylin
SB and Herman JG: Inactivation of the DNA repair gene
O6-methylguanine-DNA methyltransferase by promoter
hypermethylation is a common event in primary human neoplasia.
Cancer Res. 59:793–797. 1999.
|
|
10
|
Wolf P, Hu YC, Doffek K, Sidransky D and
Ahrendt SA: O6-Methylguanine-DNA methyltransferase
promoter hypermethylation shifts the p53 mutational spectrum in
non-small cell lung cancer. Cancer Res. 61:8113–8117. 2001.
|
|
11
|
Bialik S and Kimchi A: The
death-associated protein kinases: structure, function, and beyond.
Annu Rev Biochem. 75:189–210. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Esteller M, Corn PG, Urena JM, Gabrielson
E, Baylin SB and Herman JG: Inactivation of glutathione
S-transferase P1 gene by promoter hypermethylation in human
neoplasia. Cancer Res. 58:4515–4518. 1998.PubMed/NCBI
|
|
13
|
Widschwendter M and Jones PA: The
potential prognostic, predictive, and therapeutic values of DNA
methylation in cancer. Clin Cancer Res. 8:17–21. 2002.PubMed/NCBI
|
|
14
|
Hatziapostolou M and Iliopoulos D:
Epigenetic aberrations during oncogenesis. Cell Mol Life Sci.
68:1681–1702. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hall EJ and Hei TK: Genomic instability
and bystander effects induced by high-LET radiation. Oncogene.
22:7034–7042. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kashiwabara K, Oyama T, Sano T, Fukuda T
and Nakajima T: Correlation between methylation status of the
p16/CDKN2 gene and the expression of p16 and Rb proteins in primary
non-small cell lung cancers. Int J Cancer. 79:215–220. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhang Y, Xiong Y and Yarbrough WG: ARF
promotes MDM2 degradation and stabilizes p53: ARF-INK4a locus
deletion impairs both the Rb and p53 tumor suppression pathways.
Cell. 92:725–734. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sherr CJ: Tumor surveillance via the
ARF-p53 pathway. Genes Dev. 12:2984–2991. 1998. View Article : Google Scholar : PubMed/NCBI
|