Introduction
Parkinson’s disease (PD; OMIM: 168600) is the second
most common neurodegenerative disorder, affecting ~2% of the global
population aged ≥65 years (1,2). The
clinical features of PD consist of resting tremor, muscular
rigidity, bradykinesia and postural instability (3). PD can lead to pain (4), depression (5,6),
visual hallucinations (6), dementia
(7) and other non-motor symptoms
(8–11). PD can cause damage and can even
cause the human body to collapse.
The pathogenesis of PD is known to be associated
with environmental and genetic factors. The environmental
hypothesis of PD was popular in the 20th century (3). A number of environmental factors were
found to be significantly associated with PD, including oxidative
stress (12), smoking (13) and environmental toxins (14). In addition, genetic factors play an
essential role in this complex disease. Twin (15) and family (16) studies have shown a higher PD
susceptibility in twins and the first-degree relatives,
respectively. Genome-wide linkage analysis provided evidence that
the gene-by-gene interactions are important in PD susceptibility
(17). A number of genetic markers
have been identified for the risk of PD (18) and were shown to be potential
therapeutic targets to PD (19,20).
Grb10-interacting GYF protein-2 (GIGYF2) is
located within the PARK11 locus, an established locus of PD
(21). The Grb10 adapter protein
interacts with GIGYF2, which regulates the insulin-like growth
factor-1 (IGF-1), stimulating the growth of the insulin signal
(22,23). The IGFs affect the development of
the nervous system by preventing the apoptosis of neuronal and
brain-derived cells (24–26). A previous study found an association
between serum IGF-1 and the progression of motor symptoms in the
early stage of PD (27). In
addition, IGF-1 was shown to correlate with the clinical variables
and diagnosis of PD (27–29). Therefore, GIGYF2 may be a
candidate factor for the risk of PD through its interaction with
IGF-1.
Previously, several studies have performed an
association study between the GIGYF2 polymorphisms and PD
(30–43). Among them, four studies showed a
positive association of the GIGYF2 polymorphisms with PD
(30–33), whereas the other 10 studies showed a
negative association (34–43). These inconsistent results indicated
that the exact role that GIGYF2 played in the
pathophysiology of PD remains to be elucidated. Meta-analysis is
able to enhance the reliability of the conclusion from individual
studies by combining the data from various studies. To determine
the genetic effect of GIGYF2 on PD, a comprehensive
meta-analysis was performed among various case-control association
studies with available genotypic and allelic frequencies.
Materials and methods
Data collection
Studies were selected from PubMed using the
following key words: ‘Parkinson GIGYF2 association’ and
‘Parkinson GIGYF2 polymorphism’. Eligible studies for the
meta-analysis were required to meet the following criteria: i) An
original case-control study with the assessment of the association
between GIGYF2 and PD; ii) the study provides enough data
for obtaining or calculating the odds ratios (ORs) and 95%
confidence intervals (CIs) with the data of the study; iii)
contains genotype distribution of each polymorphism that meet the
Hardy-Weinberg equilibrium (HWE); and iv) the study is involved
with polymorphisms reported by >3 independent studies. As shown
in the previous studies (44–47),
the following information was extracted or calculated from each
study: Genetic locus, first author’s name, year of publication,
country, numbers of cases and controls, ethnicity, reported
association results, power of each case-control study and the minor
allele frequency (MAF) of controls.
Statistical analysis
HWE was tested by the Arlequin program (48). The power of each study was
calculated by the Power and Sample Size Calculation program.
Statistical heterogeneity across the studies included in the
meta-analysis was assessed by Cochran’s Q statistic and
I2 test (49) to
determine the type of analysis. In the meta-analysis, the
fixed-effect model was used for the studies with minimal to
moderate heterogeneity (I2<50%) and the random-effect
model was used for the studies with significant heterogeneity
(I2≥50%). Funnel plots were also generated to observe
the potential publication bias. The statistical analyses of
meta-analyses were performed in Review Manager 5 (50).
Results
Associations between PD and GIGYF2
polymorphisms
As shown in Fig. 1,
20 studies regarding the association of GIGYF2 with PD were
obtained from PubMed. There were no relevant studies found in the
Chinese database WanFang, WeiPu and China National Knowledge
Infrastructure. In total, four duplicates, two non-case-control and
eight without genotyping information studies were removed and three
studies were added that were obtained from the references.
Therefore, there were nine studies selected regarding seven
GIGYF2 polymorphisms, which were C.1378C>A, C.167G>A,
C.1554G>A, C.2940A>G, C.1370C>A, C.3630A>G and
C.3651G>A (Table I). In
particular, there were six studies with 2,281 cases and 1,815
controls for C.1378C>A, five with 5,519 cases and 6,316 controls
for C.167G>A, four with 1,611 cases and 1,460 controls for
C.1554G>A, four with 1,611 cases and 1,460 controls for
C.2940A>G, three with 3,876 cases and 4,688 controls for
C.1370C>A, three with 1,311 cases and 1,260 controls for
C.3630A>G and three studies with 1,311 cases and 1,260 controls
for the C.3651G>A.
 | Table ICharacteristics of the case-control
studies in the current meta-analyses. |
Table I
Characteristics of the case-control
studies in the current meta-analyses.
| First author | Year | Country | Cases/controls | Ethnicity | Resultsa | Power | MAF | (Refs.) |
|---|
| GIGYF2
C.1378C>A |
| Lautier | 2008 | Italy | 249/227 | European | NS | 0.068 | 0.0198 | (40) |
| Guo | 2009 | USA | 310/120 | European | NS | 0.053 | 0.0167 | (33) |
| Cao | 2010 | China | 510/481 | Asian | NS | 0.313 | 0.1455 | (35) |
| Guella | 2011 | Italy | 552/552 | European | NS | 0.114 | 0.0281 | (54) |
| Wang | 2011 | China | 300/200 | Asian | NS | 0.207 | 0.1975 | (30) |
| Meeus | 2011 | Belgium | 305/360 | European | NS | 0.112 | 0.0680 | (43) |
| GIGYF2
C.167G>A |
| Lautier | 2008 | Italy | 249/227 | European | NS | NA | 0.0000 | (40) |
| Zimprich | 2009 | Austria | 699/1051 | European | NS | 0.056 | 0.0005 | (38) |
| Bonetti | 2009 | Italy | 2928/3410 | European | NS | 0.053 | 0.0001 | (41) |
|
Vilarino-Guell | 2009 | Norway &
USA | 1338/1268 | European | S | NA | 0.0000 | (42) |
| Meeus | 2011 | Belgium | 305/360 | European | NS | 0.059 | 0.0056 | (43) |
| GIGYF2
C.1554G>A |
| Lautier | 2008 | Italy | 249/227 | European | NS | 0.083 | 0.0352 | (40) |
| Cao | 2010 | China | 510/481 | Asians | S | 0.254 | 0.1071 | (35) |
| Wang | 2011 | China | 300/200 | Asian | NS | 0.197 | 0.1825 | (30) |
| Guella | 2011 | Italy | 552/552 | European | NS | 0.147 | 0.0426 | (54) |
| GIGYF2
C.2940A>G |
| Lautier | 2008 | Italy | 249/227 | European | NS | 0.264 | 0.3194 | (40) |
| Cao | 2010 | China | 510/481 | Asian | NS | 0.298 | 0.1351 | (35) |
| Wang | 2011 | China | 300/200 | Asian | S | 0.275 | 0.3550 | (30) |
| Guella | 2011 | Italy | 552/552 | European | NS | 0.505 | 0.2870 | (54) |
| GIGYF2
C.1370C>A |
| Lautier | 2008 | Italy | 249/227 | Europeans | NS | NA | 0.0000 | (40) |
| Zimprich | 2009 | Austria | 699/1051 | European | NS | 0.059 | 0.0014 | (38) |
| Bonetti | 2009 | Italy | 2928/3410 | European | NS | 0.053 | 0.0006 | (41) |
| GIGYF2
C.3630A>G |
| Lautier | 2008 | Italy | 249/227 | European | NS | 0.070 | 0.0220 | (40) |
| Cao | 2010 | China | 510/481 | Asian | S | 0.297 | 0.1341 | (35) |
| Guella | 2011 | Italy | 552/552 | European | NS | 0.056 | 0.0027 | (54) |
| GIGYF2
C.3651G>A |
| Lautier | 2008 | Italy | 249/227 | European | NS | 0.223 | 0.2247 | (40) |
| Cao | 2010 | China | 510/481 | Asian | NS | 0.335 | 0.1611 | (35) |
| Guella | 2011 | Italy | 552/552 | European | NS | 0.463 | 0.2391 | (54) |
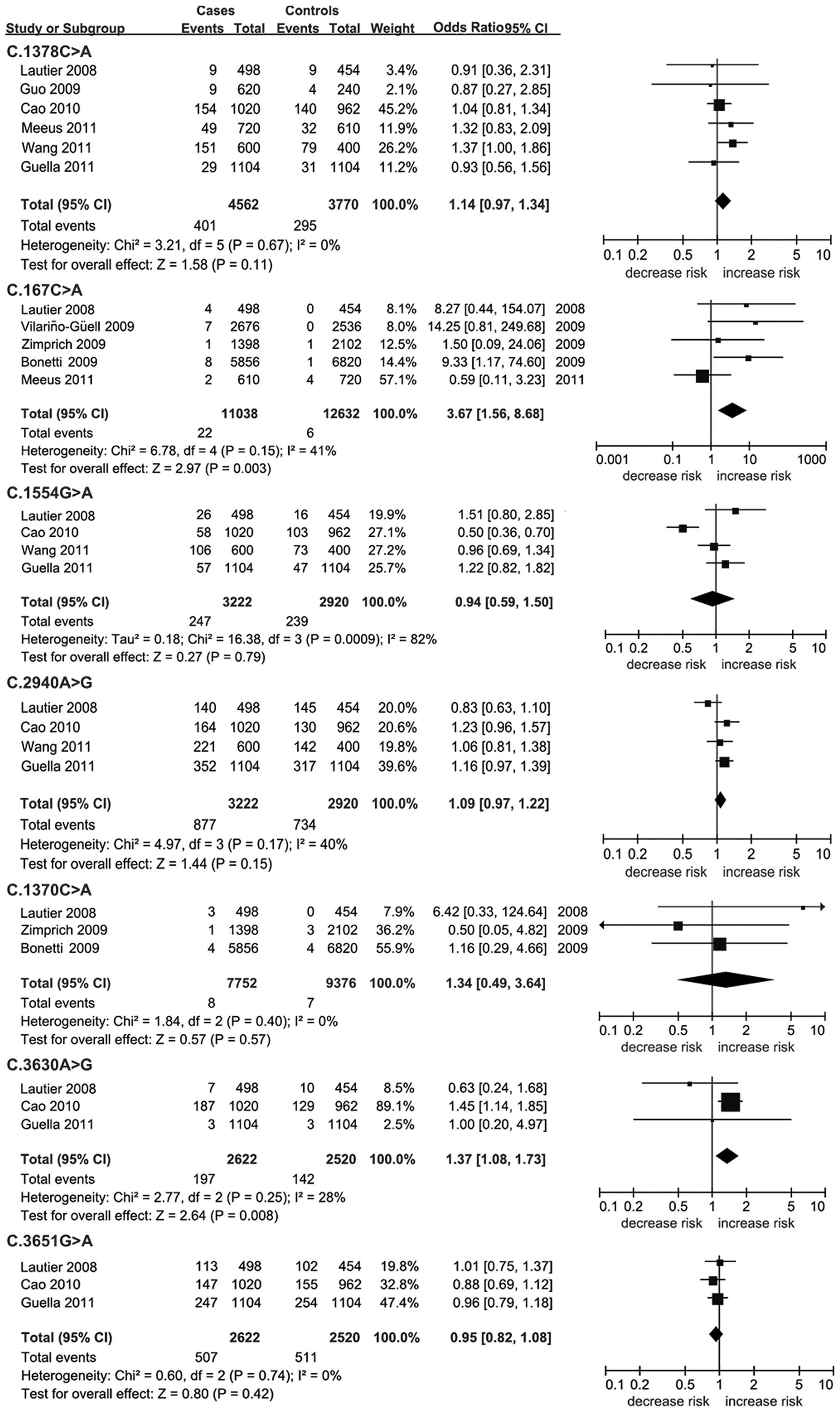
A significant association was found between the
C.3630A>G (P=0.008; OR, 1.37; 95% CI, 1.08–1.73; Table II; Fig.
2) and C.167G>A (P=0.003; OR, 3.67; 95% CI, 1.56–8.68;
Table II; Fig. 2) polymorphisms and PD. There were no
other positive results in the remaining allelic analysis
(P>0.05; Table II; Fig. 2). Low heterogeneity was found for
C.1378C>A (I2=0%), C.167G>A (I2=41%),
C.3651G>A (I2=0%), C.1370C>A (I2=0%),
C.3630A>G (I2=28%) and C.2940A>G
(I2=40%). By contrast, a significant statistical
heterogeneity was observed in the meta-analysis of C.1554G>A
(I2=82%). There was no publication bias for all the
meta-analyses (Fig. 3).
 | Table IIMeta-analyses of the GIGYF2
polymorphisms with Parkinson’s disease. |
Table II
Meta-analyses of the GIGYF2
polymorphisms with Parkinson’s disease.
| Genetic locus | Cases/controls | Genetic model | S | OR (95% CI) | P-value | I2,
% | Power |
|---|
| GIGYF2
C.1378C>A | 2281/1815 | Overall (C vs.
A) | 6 | 1.14
(0.97–1.34) | 0.110 | 0 | 0.634 |
| GIGYF2
C.167C>A | 5519/6316 | Overall (G vs.
A) | 5 | 3.67
(1.56–8.68) | 0.003a | 41 | 0.063 |
| GIGYF2
C.1554G>A | 1611/1460 | Overall (G vs.
A) | 4 | 0.94
(0.59–1.50) | 0.790 | 82 | 0.526 |
| GIGYF2
C.2940A>G | 1611/1460 | Overall (A vs.
G) | 4 | 1.09
(0.97–1.22) | 0.150 | 40 | 0.885 |
| GIGYF2
C.1370C>A | 3876/4688 | Overall (C vs.
A) | 3 | 1.34
(0.49–3.64) | 0.570 | 0 | 0.065 |
| GIGYF2
C.3630A>G | 1311/1260 | Overall (A vs.
G) | 3 | 1.37
(1.08–1.73) | 0.008a | 28 | 0.347 |
| GIGYF2
C.3651G>A | 1311/1260 | Overall (G vs.
A) | 3 | 0.95
(0.82–1.08) | 0.420 | 0 | 0.769 |
As shown in Tables I
and II, the present meta-analyses
showed a much stronger power than for each of the individual
studies. There was sufficient power (Power>0.8) for the
meta-analyses of C.2940A>G (Power=0.885) and C.3651G>A
(Power=0.824). By contrast, relatively lower power values were
found for the meta-analyses of C.1378C>A (Power=0.634),
C.1554G>A (Power=0.547), C.3630A>G (Power=0.357), C.167G>A
(Power=0.063) and C.1370C>A (Power=0.065).
Discussion
GIGYF2 is potentially involved in the
pathogenesis of PD due to the effect of IGF-1 in the insulin
signaling in the central nervous system (51–53).
In the present study, a meta-analyses was performed among 7,246
cases and 7,544 controls to evaluate the association between seven
polymorphisms of GIGYF2 and PD.
An increased risk of PD by 37% was observed for
C.3630A>G. GIGYF2 C.3630A>G is a key polymorphism in
the previous PD studies (35,40,54).
Since the power of C.3630A>G was moderate, further studies
should be conducted to confirm this positive finding. The C.3630G
allele frequency was 13.4% in the Asian population, which was much
higher compared to the European population (Table I), although a low heterogeneity was
found for this polymorphism. In addition, meta-analysis of
C.167C>A from five studies (38,40–43)
was also shown to be significantly associated with the risk of PD.
No significant association was found for the other five
GIGYF2 polymorphisms, which were C.1378C>A, C.1554G>A,
C.2940A>G, C.1370C>A and C.3651G>A. The power was strong
in the association studies of C.2940A>G and C.3651G>A
polymorphisms, but was relatively weak for the remaining three
polymorphisms, which were C.1378C>A, C.1554G>A and
C.1370C>A. A significant difference was found between
ethnicities for the C.1378C>A polymorphism (Fst=0.192), although
there was minimal heterogeneity according to the meta-analysis of
this polymorphism.
Several limitations of the present meta-analysis
should be taken with caution. Firstly, only nine studies were
included in the meta-analysis. The power values of the
meta-analyses were low due to the MAF of certain polymorphisms.
Secondly, PD is a complex and growing disease with a different
physiological status existing in the PD cases. As the pathogenesis
of familial and sporadic PD are not identical (55), family history as an independent risk
factor for PD (56) should be
emphasized in PD association studies. There were at least 1,818
sporadic and 861 familial PD patients involved in the present
meta-analysis. Notably, two studies did not provide the information
(41,42). A previous study strongly supported
that GIGYF2 was a causal factor of PD in the familial study
(40). However, other studies
showed a lack of association in sporadic PD studies (33,35,54).
Subgroup analysis by the PD family history is required to establish
the role of genetic factors in the pathogenesis of PD. Thirdly,
only seven polymorphisms of GIGYF2 were investigated, which
may not fully reflect the function of GIGYF2 in PD. There
are 9571 variants in the GIGYF2 according to the NCBI dbSNP
database (http://www.ncbi.nlm.nih.gov/snp/?term=GIGYF2). Certain
other variants, including C.684T>A, C.1219A>G and
C.3583C>T, have been reported to be significantly associated
with PD (37). In the present
meta-analysis, those variants were not included due to a lack of
relative information.
In conclusion, the present study found that the
GIGYF2 C.3630A>G and C.167G>A polymorphisms were
associated with PD. Future investigations of other ethnic
populations are required to establish the contribution of
GIGYF2 to the risk of PD.
Acknowledgements
The present study was supported by the grants from
the National Natural Science Foundation of China (grant nos.
31100919 and 81371469), Natural Science Foundation of Zhejiang
Province (grant no. LR13H020003), K.C. Wong Magna Fund in Ningbo
University and the Program for Professor of Special Appointment
(Eastern Scholar) at Shanghai Institutions of Higher Learning (to
Miss Mingqing Xu) and the Key Basic Research Foundation of Science
and Technology Commission of Shanghai Municipality (grant no.
13JC1403700) (to Miss Mingqing Xu).
References
|
1
|
Van Den Eeden SK, Tanner CM, Bernstein AL,
et al: Incidence of Parkinson’s disease: variation by age, gender,
and race/ethnicity. Am J Epidemiol. 157:1015–1022. 2003.
|
|
2
|
Elbaz A, Bower JH, Maraganore DM, et al:
Risk tables for parkinsonism and Parkinson’s disease. J Clin
Epidemiol. 55:25–31. 2002.
|
|
3
|
Dauer W and Przedborski S: Parkinson’s
disease: mechanisms and models. Neuron. 39:889–909. 2003.
|
|
4
|
Mylius V, Engau I, Teepker M, et al: Pain
sensitivity and descending inhibition of pain in Parkinson’s
disease. J Neurol Neurosurg Psychiatry. 80:24–28. 2009.
|
|
5
|
Grachev ID: Dopamine transporter imaging
with [123I]FP-CIT (DaTSCAN) in Parkinson’s disease with depressive
symptoms: a biological marker for causal relationships? J Neurol
Neurosurg Psychiatry. 85:130–131. 2014.
|
|
6
|
Blonder LX, Slevin JT, Kryscio RJ, et al:
Dopaminergic modulation of memory and affective processing in
Parkinson depression. Psychiatry Res. 210:146–149. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Aarsland D, Zaccai J and Brayne C: A
systematic review of prevalence studies of dementia in Parkinson’s
disease. Mov Disord. 20:1255–1263. 2005.
|
|
8
|
Gan EC, Lau DP and Cheah KL: Stridor in
Parkinson’s disease: a case of ‘dry drowning’? J Laryngol Otol.
124:668–673. 2010.
|
|
9
|
Klebe S, Golmard JL, Nalls MA, et al;
French Parkinson’s Disease Genetics Study Group; International
Parkinson’s Disease Genomics Consortium (IPDGC). The Val158Met COMT
polymorphism is a modifier of the age at onset in Parkinson’s
disease with a sexual dimorphism. J Neurol Neurosurg Psychiatry.
84:666–673. 2013.PubMed/NCBI
|
|
10
|
Rode J, Bentley A and Parkinson C:
Paraganglial cells of urinary bladder and prostate: potential
diagnostic problem. J Clin Pathol. 43:13–16. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Najafi MR, Chitsaz A, Askarian Z and
Najafi MA: Quality of sleep in patients with Parkinson’s disease.
Int J Prev Med. 4(Suppl 2): S229–S233. 2013.
|
|
12
|
Olanow CW and Tatton WG: Etiology and
pathogenesis of Parkinson’s disease. Annu Rev Neurosci. 22:123–144.
1999.
|
|
13
|
Wirdefeldt K, Adami HO, Cole P,
Trichopoulos D and Mandel J: Epidemiology and etiology of
Parkinson’s disease: a review of the evidence. Eur J Epidemiol.
26(Suppl 1): S1–S58. 2011.
|
|
14
|
Vaglini F, Viaggi C, Piro V, et al:
Acetaldehyde and parkinsonism: role of CYP450 2E1. Front Behav
Neurosci. 7:712013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tanner CM, Ottman R, Goldman SM, et al:
Parkinson disease in twins: an etiologic study. JAMA. 281:341–346.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Alonso ME, Otero E, D’Regules R and
Figueroa HH: Parkinson’s disease: a genetic study. Can J Neurol
Sci. 13:248–251. 1986.
|
|
17
|
Pankratz N, Nichols WC, Uniacke SK, et al:
Genome-wide linkage analysis and evidence of gene-by-gene
interactions in a sample of 362 multiplex Parkinson disease
families. Hum Mol Genet. 12:2599–2608. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Dai D, Wang Y, Wang L, et al:
Polymorphisms of DRD2 and DRD3 genes and Parkinson’s disease: A
meta-analysis. Biomed Rep. 2:275–281. 2014.
|
|
19
|
Corti O, Lesage S and Brice A: What
genetics tells us about the causes and mechanisms of Parkinson’s
disease. Physiol Rev. 91:1161–1218. 2011.PubMed/NCBI
|
|
20
|
Singleton AB, Farrer MJ and Bonifati V:
The genetics of Parkinson’s disease: progress and therapeutic
implications. Mov Disord. 28:14–23. 2013.
|
|
21
|
Cleary SF and Marciano-Cabral F: Soluble
amoebicidal factors mediate cytolysis of Naegleria fowleri
by activated macrophages. Cell Immunol. 101:62–71. 1986. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Morrione A: Grb10 adapter protein as
regulator of insulin-like growth factor receptor signaling. J Cell
Physiol. 197:307–311. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Dufresne AM and Smith RJ: The adapter
protein GRB10 is an endogenous negative regulator of insulin-like
growth factor signaling. Endocrinology. 146:4399–4409. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Russo VC, Gluckman PD, Feldman EL and
Werther GA: The insulin-like growth factor system and its
pleiotropic functions in brain. Endocr Rev. 26:916–943. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Folli F, Ghidella S, Bonfanti L, Kahn CR
and Merighi A: The early intracellular signaling pathway for the
insulin/insulin-like growth factor receptor family in the mammalian
central nervous system. Mol Neurobiol. 13:155–183. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Cardona-Gómez GP, Mendez P, DonCarlos LL,
Azcoitia I and Garcia-Segura LM: Interactions of estrogens and
insulin-like growth factor-I in the brain: implications for
neuroprotection. Brain Res Brain Res Rev. 37:320–334. 2001.
|
|
27
|
Picillo M, Erro R, Santangelo G, et al:
Insulin-like growth factor-1 and progression of motor symptoms in
early, drug-naïve Parkinson’s disease. J Neurol. 260:1724–1730.
2013.PubMed/NCBI
|
|
28
|
Numao A, Suzuki K, Miyamoto M, Miyamoto T
and Hirata K: Clinical correlates of serum insulin-like growth
factor-1 in patients with Parkinson’s disease, multiple system
atrophy and progressive supranuclear palsy. Parkinsonism Relat
Disord. 20:212–216. 2014.
|
|
29
|
Godau J, Knauel K, Weber K, et al: Serum
insulinlike growth factor 1 as possible marker for risk and early
diagnosis of Parkinson disease. Arch Neurol. 68:925–931. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang L, Guo JF, Zhang WW, et al: Follow-up
study of variants of the GIGYF2 gene in Chinese patients with
Parkinson’s disease. J Clin Neurosci. 18:1699–1701. 2011.PubMed/NCBI
|
|
31
|
Dos Santos AV, Pestana CP, Diniz KR, et
al: Mutational analysis of GIGYF2, ATP13A2 and GBA genes in
Brazilian patients with early-onset Parkinson’s disease. Neurosci
Lett. 485:121–124. 2010.PubMed/NCBI
|
|
32
|
Wang L, Guo JF, Zhang WW, et al: Novel
GIGYF2 gene variants in patients with Parkinson’s disease in
Chinese population. Neurosci Lett. 473:131–135. 2010.
|
|
33
|
Guo Y, Jankovic J, Zhu S, et al: GIGYF2
Asn56Ser and Asn457Thr mutations in Parkinson disease patients.
Neurosci Lett. 454:209–211. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Li L, Funayama M, Tomiyama H, et al: No
evidence for pathogenic role of GIGYF2 mutation in Parkinson
disease in Japanese patients. Neurosci Lett. 479:245–248. 2010.
View Article : Google Scholar
|
|
35
|
Cao L, Zhang T, Zheng L, et al: The GIGYF2
variants are not associated with Parkinson’s disease in the
mainland Chinese population. Parkinsonism Relat Disord. 16:294–297.
2010.
|
|
36
|
Tan EK, Lin CH, Tai CH, et al:
Non-synonymous GIGYF2 variants in Parkinson’s disease from two
Asian populations. Hum Genet. 126:425–430. 2009.
|
|
37
|
Nichols WC, Kissell DK, Pankratz N, et al;
Parkinson Study Group-PROGENI Investigators. Variation in GIGYF2 is
not associated with Parkinson disease. Neurology. 72:1886–1892.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zimprich A, Schulte C, Reinthaler E, et
al: PARK11 gene (GIGYF2) variants Asn56Ser and Asn457Thr are not
pathogenic for Parkinson’s disease. Parkinsonism Relat Disord.
15:532–534. 2009.PubMed/NCBI
|
|
39
|
Bras J, Simón-Sánchez J, Federoff M, et
al: Lack of replication of association between GIGYF2 variants and
Parkinson disease. Hum Mol Genet. 18:341–346. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Lautier C, Goldwurm S, Dürr A, et al:
Mutations in the GIGYF2 (TNRC15) gene at the PARK11 locus in
familial Parkinson disease. Am J Hum Genet. 82:822–833. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Bonetti M, Ferraris A, Petracca M,
Bentivoglio AR, Dallapiccola B and Valente EM: GIGYF2 variants are
not associated with Parkinson’s disease in Italy. Mov Disord.
24:1867–1869. 2009.
|
|
42
|
Vilariño-Güell C, Ross OA, Soto AI, et al:
Reported mutations in GIGYF2 are not a common cause of Parkinson’s
disease. Mov Disord. 24:619–620. 2009.
|
|
43
|
Meeus B, Nuytemans K, Crosiers D, et al:
GIGYF2 has no major role in Parkinson genetic etiology in a Belgian
population. Neurobiology Aging. 32:308–312. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Yu X, Huang Y, Li C, Yang H, Lu C and Duan
S: Positive association between lymphotoxin-alpha variation
rs909253 and cancer risk: a meta-analysis based on 36 case-control
studies. Tumour Biol. 35:1973–1983. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Huang Y, Yu X, Wang L, et al: Four genetic
polymorphisms of lymphotoxin-alpha gene and cancer risk: a
systematic review and meta-analysis. PLoS One. 8:e825192013.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Tang L, Wang L, Liao Q, et al: Genetic
associations with diabetes: meta-analyses of 10 candidate
polymorphisms. PLoS One. 8:e703012013. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Xu X, Wang Y, Wang L, et al: Meta-analyses
of 8 polymorphisms associated with the risk of the Alzheimer’s
disease. PLoS One. 8:e731292013.
|
|
48
|
Excoffier L, Laval G and Schneider S:
Arlequin (version 3.0): an integrated software package for
population genetics data analysis. Evol Bioinform Online. 1:47–50.
2007.
|
|
49
|
Coory MD: Comment on: Heterogeneity in
meta-analysis should be expected and appropriately quantified. Int
J Epidemiol. 39:932–933. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Kawalec P, Mikrut A, Wísniewska N and Pilc
A: The effectiveness of tofacitinib, a novel Janus kinase
inhibitor, in the treatment of rheumatoid arthritis: a systematic
review and meta-analysis. Clin Rheumatol. 32:1415–1424. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Giovannone B, Lee E, Laviola L, Giorgino
F, Cleveland KA and Smith RJ: Two novel proteins that are linked to
insulin-like growth factor (IGF-I) receptors by the Grb10 adapter
and modulate IGF-I signaling. J Biol Chem. 278:31564–31573. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Laviola L, Giorgino F, Chow JC, et al: The
adapter protein Grb10 associates preferentially with the insulin
receptor as compared with the IGF-I receptor in mouse fibroblasts.
J Clin Invest. 99:830–837. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Mori K, Giovannone B and Smith RJ:
Distinct Grb10 domain requirements for effects on glucose uptake
and insulin signaling. Mol Cell Endocrinol. 230:39–50. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Guella I, Pistocchi A, Asselta R, et al:
Mutational screening and zebrafish functional analysis of GIGYF2 as
a Parkinson-disease gene. Neurobiol Aging. 32:1994–2005. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Gasser T: Genetics of Parkinson’s disease.
J Neurol. 248:833–840. 2001.
|
|
56
|
Sellbach AN, Boyle RS, Silburn PA and
Mellick GD: Parkinson’s disease and family history. Parkinsonism
Relat Disord. 12:399–409. 2006.
|