Introduction
Retinoblastoma is the most frequent intraocular
malignant tumor type to occur in childhood (1). Retinoblastoma is a heritable cancer
caused by mutations or deletions of the tumor suppressor gene,
retinoblastoma 1 (Rb1) (2). Although
the allelic inactivation of the Rb1 gene serves an important role
in the tumorigenesis of retinoblastoma, other oncogenes or tumor
suppressors may also be important in the development and
progression of the disease (3,4). Further
delineating the molecular events involved in the oncogenesis of
retinoblastoma should provide novel insights into the etiology of
this tumor.
MicroRNAs (miRNAs/miRs) are highly conserved,
endogenous, small non-coding RNAs of ~22 nucleotides in length
(5). miRNAs pair imperfectly with
the 3′-untranslated region (3′UTR) of specific mRNAs to regulate
the transcriptional and post-transcriptional expression of target
genes (6). Mounting evidence has
demonstrated that miRNAs are involved in numerous physiological and
pathological processes (7,8). Of note, the dysregulation of miRNAs
serves a vital role in almost every aspect of tumor biology
(9,10). miRNAs may act as oncogenes or tumor
suppressors by regulating different target genes (11,12).
Martin et al (13)
investigated differentially expressed miRNAs in 12 retinoblastomas
as compared with three normal human retina samples using the Taqman
Low Density Array and identified that miR-129-3p, miR-382, miR-504,
miR-22 and miR-129-5p were significantly downregulated in
retinoblastoma. Furthermore, Huang et al (14) used microarrays to profile three
retinoblastoma samples and one healthy retina sample and the
results indicated that the expression levels of let-7b, let-7c,
miR-24, miR-125b, miR-191, miR-181a and miR-423 were downregulated
in retinoblastoma. Additionally, microarray data provided by Zhao
et al (15) reported that
miR-494, let-7e, miR-513-1, miR-513-2, miR-518c-5p, miR-129-1,
miR-129-2, miR-198, miR-492, miR-498, miR-320, miR-503 and
miR-373-5p were upregulated in retinoblastoma. Therefore, it is
imperative to further elucidate the roles of miRNAs in the
tumorigenesis of retinoblastoma.
miR-101-3p has been reported to be downregulated in
various types of cancer, and functions as a tumor suppressor via
targeting multiple oncogenes. For instance, it has been
demonstrated that miR-101-3p loss in prostate cancer increases the
expression of SUB1, activating genes associated with an aggressive
tumor phenotype (16). Zhang et
al (17) reported that
miR-101-3p suppresses cholangiocarcinoma angiogenesis via targeting
vascular endothelial growth factor. Furthermore, in human colon
cancer cells, the downregulation of miR-101-3p was observed, which
was associated with the upregulation of cyclooxygenase-2 (18). In addition, the expression of
miR-101-3p in non-small cell lung cancer was reported to be
significantly decreased (19).
However, the biological function and underlying mechanisms of
miR-101-3p in retinoblastoma are largely unknown.
The aim of the present study was to determine the
role of miR-101-3p in retinoblastoma, and investigate whether
miR-101-3p targeted enhancer of zeste homolog (EZH2) and histone
deacetylase 9 (HDAC9) to exert its role in retinoblastoma.
Materials and methods
Cell culture
The human retinoblastoma cell lines WERI-Rb-1 and
Y79 were obtained from the American Type Culture Collection
(Manassas, VA, USA). The cell lines were grown in RPMI-1640 medium
supplemented with 10% fetal bovine serum and
penicillin-streptomycin (all Gibco; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA). All cells were maintained at 37°C in a
humidified atmosphere containing 5% CO2.
Plasmids
pEnter (cat. no. P100001), pEnter-EZH2 (cat. no.
CH882009) and pEnter-HDAC9 (cat. no. CH804907) were purchased from
Vigene Biosciences, Inc. (Rockville, MD, USA). Wild-type and mutant
3′UTRs of EZH2 and HDAC9 were synthesized by Genewiz (Suzhou,
Jiangsu, China) and were inserted into a pMIR-REPORT™ luciferase
plasmid (cat. no. AM5795; Thermo Fisher Scientific, Inc.).
Patients and tissues
A total of 12 human retinoblastoma tissue samples (7
males and 5 females; median age, 31 months), and 3 normal retinal
tissues (2 males and 1 female; median age, 56 months) from
individuals who succumbed to conditions other than ophthalmologic
diseases, were obtained from the Second Affiliated Hospital of
Nanchang University (Nanchang, China) between January 2015 and
December 2016. All patients with retinoblastoma were diagnosed and
treated for the first time, and had not received adjuvant
treatments prior to this study in order to avoid treatment-induced
expression changes. All specimens were histopathologically
diagnosed by two pathologists. The current study was approved by
the Ethics Committee of the Second Affiliated Hospital of Nanchang
University, and written informed consent was obtained from the
parents or guardians of patients.
Transfection
MicrON® miR-101-3p agomir (cat. no.
miR40000099-1-10) and an miR-control (cat. no. miR04201-1-10) were
purchased from Guangzhou Ribobio Co., Ltd. (Guangzhou, Guangdong,
China). WERI-Rb-1 and Y79 cells were seeded in in 6-well plates
(5×104 cells per well) and transfected with 50 nM
miR-101-3p agomir or miR-control using Lipofectamine®
3000 (Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. At 48 h post-transfection, the cells were
subjected to subsequent analysis. To determine the roles of target
genes on the effect of miR-101-3p, co-transfection of 50 nM
miR-101-3p agomir+1 µg control plasmid (pEnter) or 50 nM miR-101-3p
agomir+1 µg pEnter-EZH2/pEnter-HDAC9 plasmid in WERI-Rb-1 and Y79
cells was performed using Lipofectamine 3000 (Thermo Fisher
Scientific, Inc.). At 5 days post co-transfection an MTT assay was
performed.
RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was isolated from prepared tissue or
cultured cells using TRIzol (Invitrogen; Thermo Fisher Scientific,
Inc.). For miRNA a specific Bulge-Loop™ miR-101-3p RT primer (cat.
no. ssD809230011) and a U6 RT primer (cat. no. ssD0904071008; both
Guangzhou Ribobio Co., Ltd.) were used for the RT reaction. For
mRNA, an Oligo(dT)15 primer was used to reverse
transcribe the mRNA into cDNA. The RT reaction was performed using
an RT system (cat. no. A3500; Promega Corporation, Madison, WI,
USA). The temperature protocol for RT was as follows: 42°C for 30
min, 70°C for 10 min and hold at 4°C. qPCR was performed using an
SYBR-Green PCR Master mix (Takara Biotechnology, Co., Ltd., Dalian,
China) on an ABI7900HT Fast Real-Time PCR system (Applied
Biosystems; Thermo Fisher Scientific, Inc.). Bulge-Loop™ miRNA qPCR
primer sets for miR-101-3p (cat. no. miRQ0000099-1-1) and RNU6B
(cat. no. MQP-0202) were purchased from Guangzhou Ribobio Co., Ltd.
The primers used for detecting EZH2, HDAC9 and GAPDH were as
follows: EZH2 forward, 5′-GGAACAACGCGAGTCGG-3′ and reverse,
5′-CTGATTTTACACGCTTCCGC-3′; HDAC9 forward,
5′-GAACTCTAAGCCAGATGGGG-3′ and reverse, 5′-GCCCACAGGAACTTCTGACT-3′;
GAPDH forward, 5′-ATGGTGAAGGTCGGTGTGAA-3′ and reverse,
5′-GAGTGGAGTCATACTGGAAC-3′. RNU6B or GAPDH served as internal
controls. The PCR protocol for detection of miR-101-3p and RNU6B
was as follows: 95°C for 10 min, followed by 40 cycles of 95°C for
2 sec, 60°C for 20 sec and 70°C for 10 sec. The PCR protocol for
detection of EZH2, HDAC9 and GAPDH was as follows: 95°C for 5 min,
followed by 40 cycles of 95°C for 10 sec, 60°C for 30 sec, and 72°C
for 15 sec. Cq values were calculated, and fold change was
determined using the 2−ΔΔCq method (20).
Western blot analysis
The cells were lysed with radioimmunoprecipitation
assay lysis buffer [50 mM TrisHCl (pH 7.6), 150 mM NaCl, 1 mM EGTA,
1% Triton NP-40, 1% sodium deoxycholate, 0.1% SDS and 50 mM NaF].
The protein concentration in the extract was measured by the
Bradford assay (Bio-Rad, Richmond, CA, USA). Equal amounts (30 µg)
of protein were separated by 10% SDS-PAGE. The proteins were then
transferred electrophoretically onto polyvinylidene fluoride
membranes. Subsequently, membranes were incubated with primary
antibody at 4°C overnight. The primary antibodies used included
rabbit monoclonal anti-EZH2 (cat. no. ab150433; 1:1,000),
anti-HDAC9 (cat. no. ab109446; 1:1,000) and anti-α-tubulin (cat.
no. ab4074; 1:2,000; all Abcam, Cambridge, UK). Membranes were then
incubated at room temperature for 1 h with goat anti-rabbit
horseradish peroxidase-conjugated secondary antibody (cat. no.
ab205718; 1:2,000; Abcam). An enhanced chemiluminescence detection
system (Amersham; GE Healthcare, Chicago, IL, USA) was used for
developing bands, and the membrane was exposed on X-ray film
(Kodak, Tokyo, Japan). α-tubulin was used as the loading
control.
MTT assay
Cell viability was assessed with an MTT assay. Cells
(103 cells/well) were cultured in 96-well plates. At the
same time each day for 5 days, 20 µl MTT (Sigma-Aldrich; Merck
KGaA, Darmstadt, Germany) was added to each well and incubated for
a further 4 h. Following removal of the culture medium, 200 µl
dimethyl sulfoxide (Sigma-Aldrich; Merck KGaA) was added and the
optical density at 490 nm was measured using an El×808 enzyme
immunoassay analyzer (Bio-Tek Instruments, Inc., Winooski, VT,
USA).
Flow cytometry
Flow cytometry was performed to analyze cell cycle
distribution. Cells were harvested, washed with cold PBS and fixed
in cold 70% ethanol overnight at 4°C, followed by treatment with
0.25 mg/ml RNase A for 30 min at room temperature. After staining
with propidium iodide (PI; 0.05 mg/ml; Sigma-Aldrich; Merck KGaA)
for 15 min at room temperature, the samples were analyzed using a
flow cytometer (Beckman Coulter, Inc., Brea, CA, USA) and was
analyzed using ModFit LT 4.1 (Verity Software House, Topsham, ME,
USA).
Animal experiments
The animal studies were approved by the Ethical
Committee of the Second Affiliated Hospital of Nanchang University.
A total of 10 male BALB/cA nude mice (age, 5 weeks; weight,
15.2–17.8 g) were purchased from Charles River Laboratories, Inc.
(Beijing, China). All animals were maintained at 22°C and 80%
relative humidity with a 12-h day/night cycle and were provided
food and water ad libitum. Y79 cells (106) were
subcutaneously (s.c.) transplanted into the flank of each nude
mouse. Once tumors reached ~40 mm3, nude mice were
assigned to two groups (n=5 per group). Agomir and miR-control (30
µl PBS with 10 nmol/l) were injected into the tumor tissue in each
group by multipoint intratumoral injection every 2 days. The tumor
sizes were monitored, and the tumor volume was calculated using the
following formula: Tumor volume=0.5× long radius × short
radius2. At day 16 following the first miR injection,
mice were sacrificed and the weight of the tumors was measured.
Luciferase reporter assay
Cells were co-transfected with the reporter
constructs (pMIR-REPORT-EZH2-3′UTR-WT, pMIR-REPORT-EZH2-3′UTR-Mut,
pMIR-REPORT-HDAC9-3′UTR-WT or pMIR-REPORT- HDAC9-3′UTR-Mut) and
miR-101-3p agomir or miR-control, using Lipofectamine 3000
according to the manufacturer's protocol. pRL-TK Renilla (Promega
Corporation) was used to normalize the efficiency of the
transfection. At 48 h, cells were harvested and the luciferase
activity was analyzed using the Dual Luciferase Assay kit (Promega
Corporation) according to the manufacturer's protocol. Renilla
luciferase activity was used as an internal control.
Bioinformatics analysis
Targetscan software (version 7.1;
targetscan.org/vert_71/) was used to predict the putative targets
of miR-101-3p.
Statistical analysis
The results were analyzed using SPSS 16.0 (SPSS,
Inc., Chicago, IL, USA). All experiments were performed three
times. Data are presented as the mean ± standard deviation.
Differences between two groups were compared using Student's
t-tests. P<0.05 was considered to indicate a statistically
significant difference.
Results
miR-101-3p is downregulated in
retinoblastoma and suppresses proliferation in retinoblastoma
cells
Previous studies have demonstrated that miR-101-3p
is downregulated in several types of cancer, prompting the
investigation of the expression of miR-101-3p in retinoblastoma in
the present study. RT-qPCR assays were performed for 12
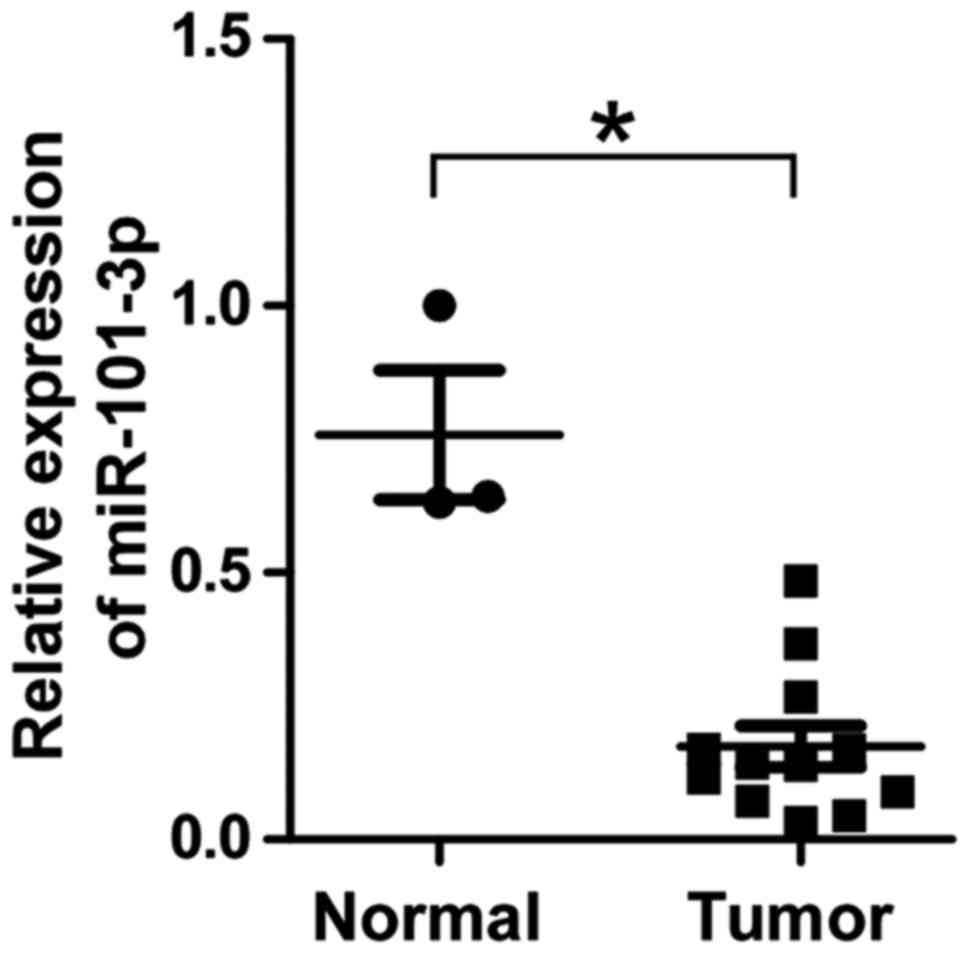
retinoblastoma and 3 normal retina specimens. As demonstrated in
Fig. 1, the expression of miR-101-3p
was significantly downregulated in retinoblastoma samples compared
with in the normal retina samples (P<0.05), suggesting that
miR-101-3p may serve a functional role in retinoblastoma
development. Next, the effects of miR-101-3p on retinoblastoma
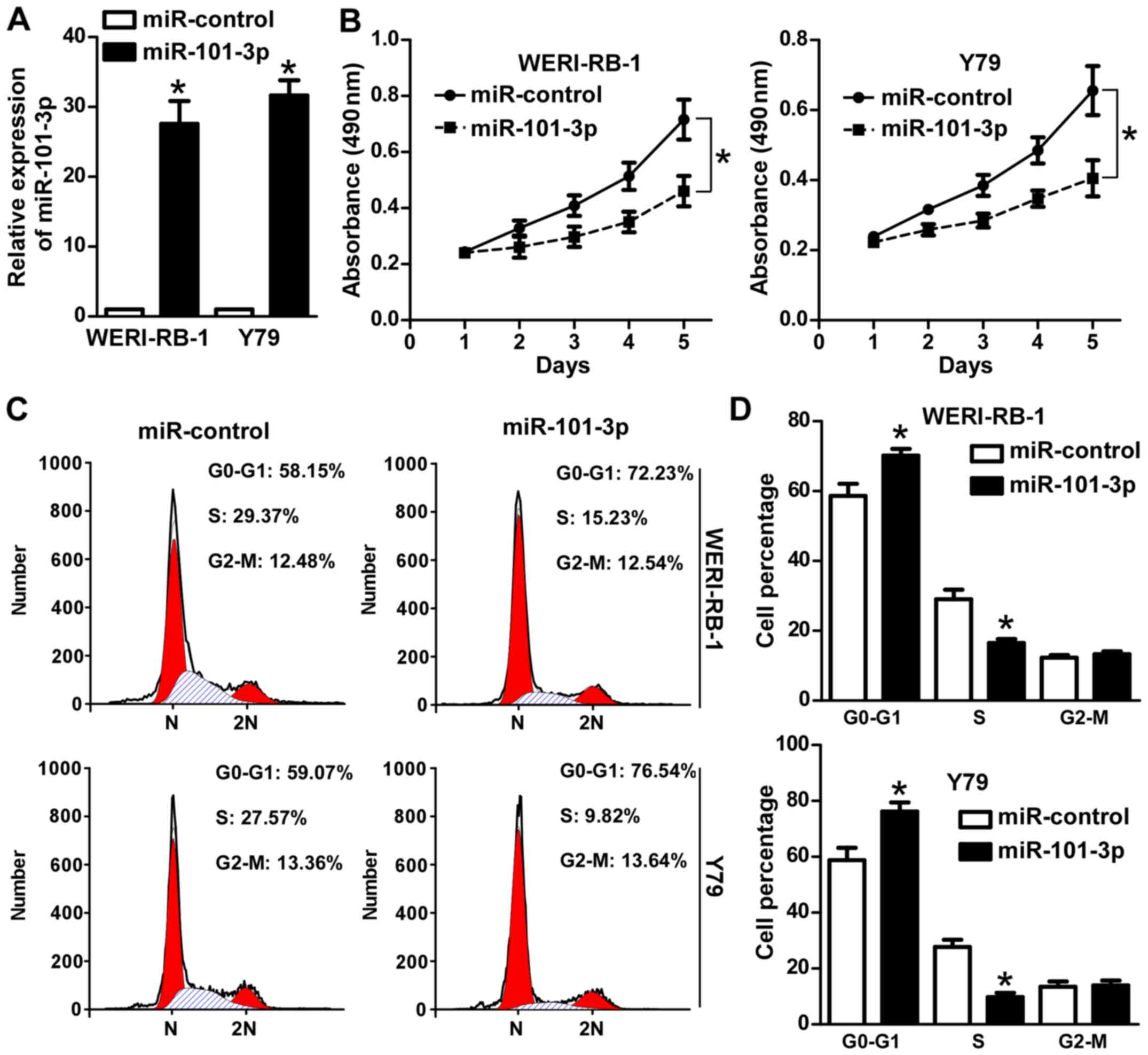
cells were investigated. Transfection with an miR-101-3p agomir
(Fig. 2A) significantly attenuated
the viability of WERI-RB-1 and Y79 cells (P<0.05; Fig. 2B). PI staining and flow cytometry
were subsequently performed to analyze the effect of miR-101-3p on
cell cycle progression. As demonstrated in Fig. 2C and D, miR-101-3p overexpression
significantly increased the proportion of WERI-RB-1 and Y79 cells
in G1 phase (P<0.05). Collectively, these data
suggest that miR-101-3p serves an anti-proliferative role in
retinoblastoma cells.
miR-101-3p inhibits tumor growth in
nude mice
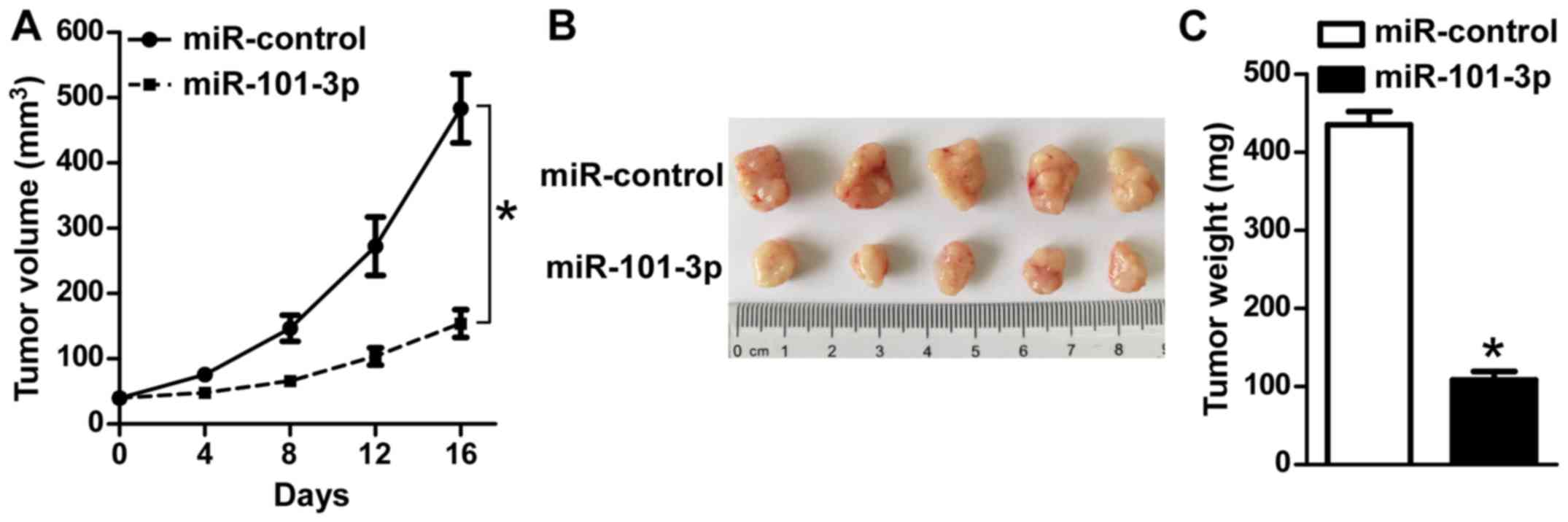
To further explore the function of an miR-101-3p
agomir in in vivo tumor growth, a tumor model was
established by transplanting (s.c.) Y79 cells into the flanks of
nude mice. Once tumors reached ~40 mm3, 30 µl PBS with
10 nmol/l agomir or miR-control was injected into the tumor tissue
every 2 days, and the tumor sizes were monitored. As indicated in
Fig. 3A and B, treatment with
miR-101-3p agomir significantly inhibited tumor growth in nude mice
compared with in the controls (P<0.05). Consistently, the mean
tumor weight in the Y79/miR-101-3p agomir mice was significantly
decreased in comparison with the control (P<0.05; Fig. 3C). These in vivo findings
further indicate that miR-101-3p markedly suppresses tumor growth
in retinoblastoma.
miR-101-3p directly targets EZH2 and
HDAC9 in retinoblastoma cells
Next, the role of miR-101-3p in regulating HDAC9 in
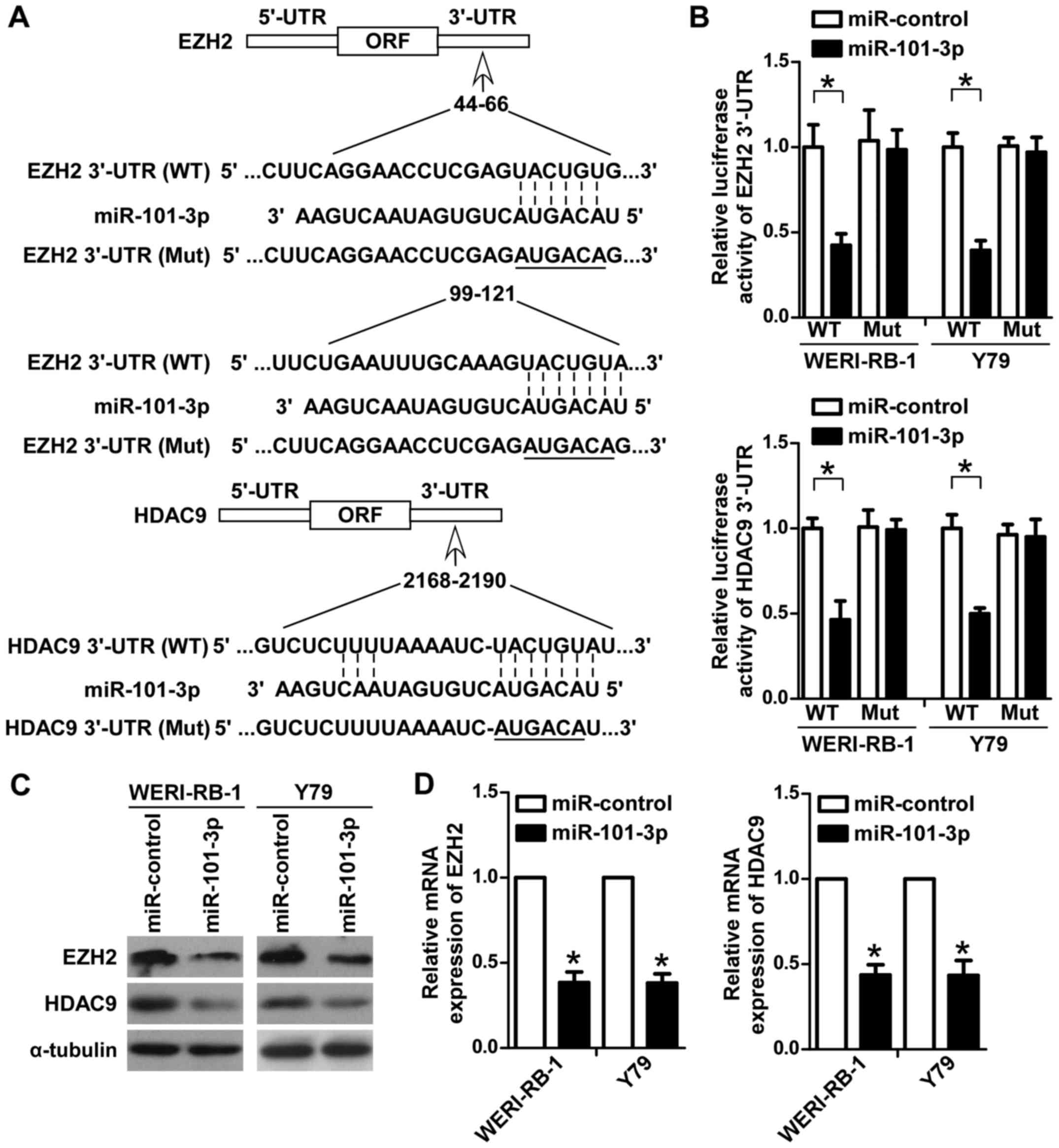
retinoblastoma cells was explored. Initially, bioinformatics
analysis with TargetScan was performed to predict the potential
targets of miR-101-3p. Among these targets, the key roles of EZH2
and HDAC9 in retinoblastoma have been well demonstrated (21–23). As
indicated in Fig. 4A, potential
miR-101-3p binding sites were identified in the 3′UTRs of EZH2 and
HDAC9. To determine whether miR-101-3p directly targets EZH2 and
HDAC9 in retinoblastoma cells, luciferase reporters containing
wild-type or mutant miR-101-3p binding sites in the 3′UTRs of EZH2
and HDAC9 were constructed (Fig.
4A). The reporter vectors were co-transfected with miR-101-3p
or the control miRNA into WERI-RB-1 and Y79 cells. As demonstrated
in Fig. 4B, miR-101-3p significantly
inhibited the luciferase activity of the reporters containing EZH2
and HDAC9 wild-type 3′UTRs, but not of those containing respective
mutant 3′UTRs (P<0.05). To further confirm the regulatory role
of miR-101-3p on EZH2 and HDAC9 in WERI-RB-1 and Y79 cells, western
blotting and RT-qPCR assays were performed. As indicated in
Fig. 4C, the protein levels of EZH2
and HDAC9 were markedly decreased in WERI-RB-1 and Y79 cells
transfected with the miR-101-3p agomir compared with the control
group. Consistently, the mRNA expression levels of EZH2 and HDAC9
were significantly decreased in WERI-RB-1 and Y79 cells transfected
with the miR-101-3p agomir compared with the control group
(P<0.05; Fig. 4D). Collectively,
these data suggested that miR-101-3p directly targeted EZH2 and
HDAC9 in retinoblastoma cells.
Restoration of EZH2 or HDAC9 hinders
the anti-proliferative role of miR-101-3p in WERI-RB-1 and Y79
cells
To determine whether EZH2 and HDAC9 were involved in
the anti-proliferative role of miR-101-3p in retinoblastoma cells,
a rescue experiment was performed by transfecting WERI-RB-1 and Y79
cells with miR-101-3p or the control miRNA along with pEnter,
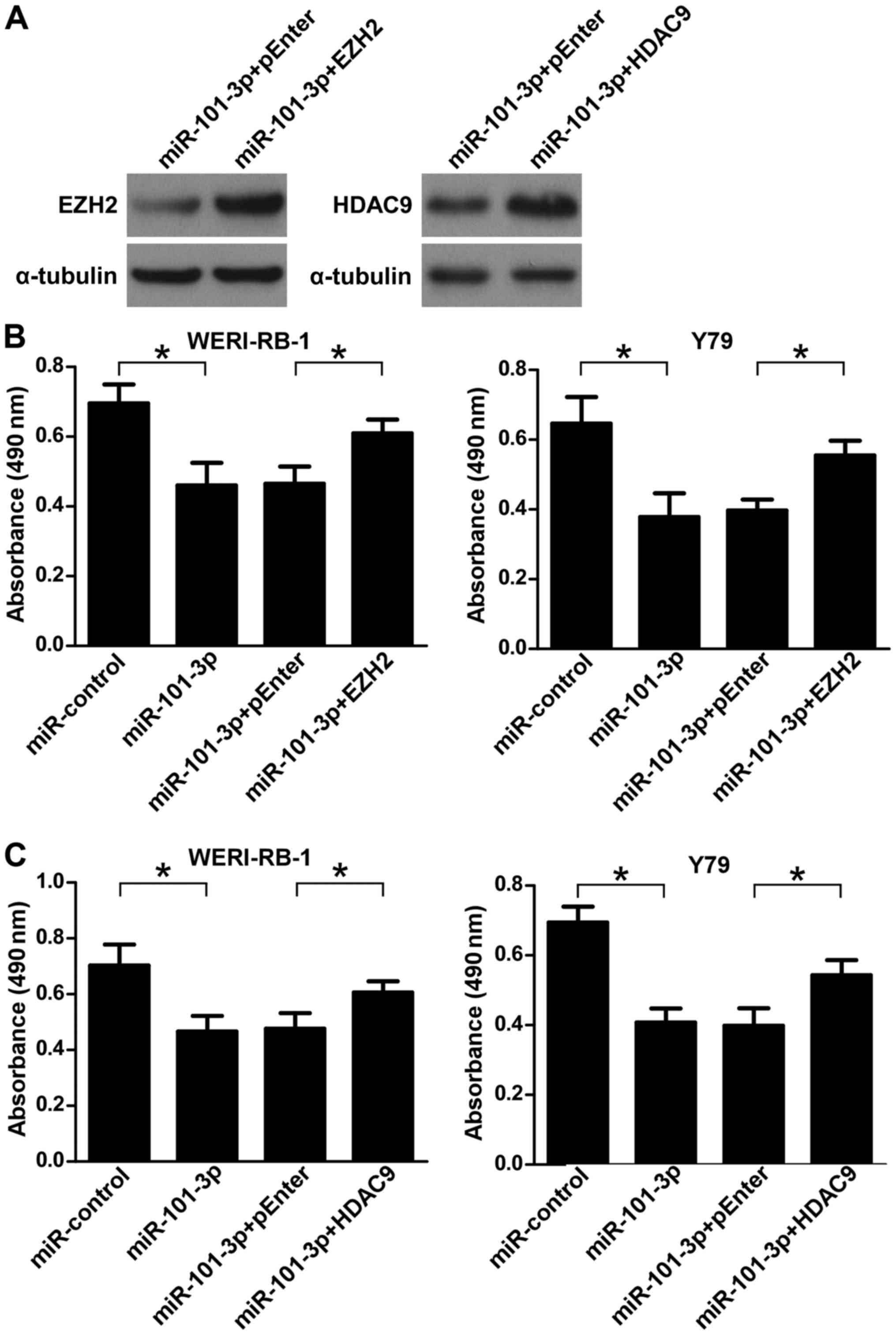
pEnter-EZH2 or pEnter-HDAC9 (Fig.
5A). As illustrated in Fig. 5B and
C, MTT assays demonstrated that the ectopic overexpression of
EZH2 or HDAC9 partially reversed the miR-101-3p-induced suppression
of retinoblastoma cell viability (P<0.05). These data indicated
that downregulation of EZH2 and HDAC9 was involved in mediating the
anti-proliferative role of miR-101-3p in retinoblastoma cells.
Discussion
Accumulating evidence has revealed that the aberrant
expression of miRNAs may be involved in the development and
progression of retinoblastoma (24,25).
Thus, a greater understanding of the specific miRNAs associated
with retinoblastoma tumorigenesis may be crucial for exploring
novel therapeutic strategies for this malignancy. In the present
study, it was identified that the expression level of miR-101-3p
was significantly downregulated in retinoblastoma. The
overexpression of miR-101-3p inhibited retinoblastoma cell
proliferation in vitro and in vivo. Furthermore, data
based on two retinoblastoma cell lines indicated that miR-101-3p
directly targets EZH2 and HDAC9 to modulate retinoblastoma cell
proliferation. These implicate miR-101-3p as a tumor suppressor in
retinoblastoma.
Several lines of evidence have demonstrated that
miR-101-3p is downregulated in various types of cancer, including
hepatocellular carcinoma, non-small cell lung cancer and gastric
cancer (19,26,27). In
line with these studies, the downregulation of miR-101-3p was also
identified in retinoblastoma, suggesting that miR-101-3p may serve
an important role in the development and progression of this
disease. In the present study, it was identified that the ectopic
overexpression of miR-101-3p significantly inhibited the viability
of WERI-RB-1 and Y79 retinoblastoma cells. Accordingly, flow
cytometry analyses demonstrated that miR-101-3p suppressed cell
cycle progression. In addition, in vivo mouse experiments
further confirmed the anti-proliferative role of miR-101-3p on
retinoblastoma cells. These results strongly indicate that
miR-101-3p is involved in the proliferation of retinoblastoma
cells.
It is well documented that miRNAs exert their
functions by regulating multiple target genes. A study by Lin et
al (28) demonstrated that
miR-135b promoted lung cancer metastasis by regulating multiple key
components in the Hippo pathway, including LATS2, β-TrCP and NDR2.
Le et al (29) demonstrated
that a subset of miR-125b target genes affect neuronal
differentiation. In the present study, it was identified that
miR-101-3p inhibited retinoblastoma cell proliferation by directly
targeting EZH2 and HDAC9. EZH2, the catalytic core protein of
polycomb repressor complex 2, is a highly conserved histone
methyltransferase (30). EZH2
suppresses the expression of target genes by catalyzing the
trimethylation of K27 of histone H3 (31). The upregulation of EZH2 has been
identified in various types of malignancy, including colorectal,
breast and prostate cancer (32–34).
Khan et al (21) observed
that EZH2 was overexpressed in retinoblastoma specimens. The
pro-proliferative role of EZH2 in cancer cells is also well
established. Lian et al (35)
demonstrated that EZH2 promotes laryngeal cancer cell proliferation
by regulating Runt-related transcription factor 3 expression.
Additionally, the inhibition of EZH2 by a small-molecule inhibitor
suppresses tumor cell growth via induction of the tumor-suppressor
protein p16INK4A (36).
Previous studies have also reported the expression and functional
roles of HDAC9 in various types of cancer. Upregulated expression
of HDAC9 was identified to promote oral squamous cell carcinoma
growth and cell cycle progression through targeting of the
transcription factors myocyte enhancer factor 2D and nuclear
receptor subfamily 4 group A member 1 (37). Additionally, a study by Zhao et
al (38) demonstrated that HDAC9
epigenetically repressed the transcription of p53 via binding to
its proximal promoter region, thus enhancing the proliferation of
osteosarcoma cells.
In the present study, it was revealed that
miR-101-3p negatively regulated the expression of EZH2 and HDAC9 in
retinoblastoma cells. Furthermore, the results of luciferase
reporter assays further validated that miR-101-3p inhibited the
expression of EZH2 and HDAC9 by directly binding to their 3′UTRs.
The restoration of EZH2 or HDAC9 expression partially reversed the
anti-proliferative effect of miR-101-3p in retinoblastoma cells.
This strongly indicates that the inhibition of cell proliferation
induced by miR-101-3p is mediated by regulating the expression of
EZH2 and HDAC9.
In conclusion, it was demonstrated that miR-101-3p
is downregulated in retinoblastoma. In addition, the overexpression
of miR-101-3p suppresses the proliferation of retinoblastoma cells
via directly targeting EZH2 and HDAC9. These data highlight an
important role for miR-101-3p in the tumorigenesis of
retinoblastoma, and may inform the development of novel therapeutic
methods.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed in this study are
available within the published article.
Authors' contributions
QJ, WH and LC performed the in vitro
experiments and analyzed the data. YY performed the animal
experiments. KS collected the clinical tissue samples. QJ and ZY
designed the study, wrote the manuscript and revised the
manuscript.
Ethics approval and consent to
participate
The current study was approved by the Ethics
Committee of the Second Affiliated Hospital of Nanchang University
and written informed consent was obtained from the parents or
guardians of all patients. The animal studies were approved by the
Ethical Committee of the Second Affiliated Hospital of Nanchang
University.
Patient consent for publication
Written informed consent for the publication of
their data was obtained from the parents or guardians of patients
involved in the present study.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
EZH2
|
enhancer of zeste homolog 2
|
|
HDAC9
|
histone deacetylase 9
|
|
miRNA
|
microRNA
|
|
PI
|
propidium iodide
|
|
qPCR
|
quantitative polymerase chain
reaction
|
|
UTR
|
untranslated region
|
References
|
1
|
Ortiz MV and Dunkel IJ: Retinoblastoma. J
Child Neurol. 31:227–236. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kalsoom S, Wasim M, Afzal S, Shahzad MS,
Ramzan S, Awan AR, Anjum AA and Ramzan K: Alterations in the RB1
gene in Pakistani patients with retinoblastoma using direct
sequencing analysis. Mol Vis. 21:1085–1092. 2015.PubMed/NCBI
|
|
3
|
Busch M, Große-Kreul J, Wirtz JJ, Beier M,
Stephan H, Royer-Pokora B, Metz K and Dünker N: Reduction of the
tumorigenic potential of human retinoblastoma cell lines by TFF1
overexpression involves p53/caspase signaling and miR-18a
regulation. Int J Cancer. 141:549–560. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chu WK, Law KS, Chan SO, Yam JC, Chen LJ,
Zhang H, Cheung HS, Block NL, Schally AV and Pang CP: Antagonists
of growth hormone-releasing hormone receptor induce apoptosis
specifically in retinoblastoma cells. Proc Natl Acad Sci USA.
113:14396–14401. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Salmena L, Poliseno L, Tay Y, Kats L and
Pandolfi PP: A ceRNA hypothesis: The Rosetta Stone of a hidden RNA
language? Cell. 146:353–358. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Mihanfar A, Fattahi A and Nejabati HR:
MicroRNA-mediated drug resistance in ovarian cancer. J Cell
Physiol. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Friedrich M, Pracht K, Mashreghi MF, Jäck
HM, Radbruch A and Seliger B: The role of the miR-148/−152 family
in physiology and disease. Eur J Immunol. 47:2026–2038. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chandra S, Vimal D, Sharma D, Rai V, Gupta
SC and Chowdhuri DK: Role of miRNAs in development and disease:
Lessons learnt from small organisms. Life Sci. 185:8–14. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Fabbri M: MicroRNAs and cancer: Towards a
personalized medicine. Curr Mol Med. 13:751–756. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ceppi P and Peter ME: MicroRNAs regulate
both epithelial-to-mesenchymal transition and cancer stem cells.
Oncogene. 33:269–278. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chen CY, Chang JT, Ho YF and Shyu AB:
MiR-26 down-regulates TNF-α/NF-κB signalling and IL-6 expression by
silencing HMGA1 and MALT1. Nucleic Acids Res. 44:3772–3787. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lou C, Xiao M, Cheng S, Lu X, Jia S, Ren Y
and Li Z: MiR-485-3p and miR-485-5p suppress breast cancer cell
metastasis by inhibiting PGC-1α expression. Cell Death Dis.
7:e21592016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Martin J, Bryar P, Mets M, Weinstein J,
Jones A, Martin A, Vanin EF, Scholtens D, Costa FF, Soares MB and
Laurie NA: Differentially expressed miRNAs in retinoblastoma. Gene.
512:294–299. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Huang JC, Babak T, Corson TW, Chua G, Khan
S, Gallie BL, Hughes TR, Blencowe BJ, Frey BJ and Morris QD: Using
expression profiling data to identify human microRNA targets. Nat
Methods. 4:1045–1049. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhao JJ, Yang J, Lin J, Yao N, Zhu Y,
Zheng J, Xu J, Cheng JQ, Lin JY and Ma X: Identification of miRNAs
associated with tumorigenesis of retinoblastoma by miRNA microarray
analysis. Childs Nerv Syst. 25:13–20. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chakravarthi BV, Goswami MT, Pathi SS,
Robinson AD, Cieślik M, Chandrashekar DS, Agarwal S, Siddiqui J,
Daignault S, Carskadon SL, et al: MicroRNA-101 regulated
transcriptional modulator SUB1 plays a role in prostate cancer.
Oncogene. 35:6330–6340. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhang J, Han C, Zhu H, Song K and Wu T:
miR-101 inhibits cholangiocarcinoma angiogenesis through targeting
vascular endothelial growth factor (VEGF). Am J Pathol.
182:1629–1639. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Strillacci A, Griffoni C, Sansone P,
Paterini P, Piazzi G, Lazzarini G, Spisni E, Pantaleo MA, Biasco G
and Tomasi V: MiR-101 downregulation is involved in
cyclooxygenase-2 overexpression in human colon cancer cells. Exp
Cell Res. 315:1439–1447. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhang X, He X, Liu Y, Zhang H, Chen H, Guo
S and Liang Y: MiR-101-3p inhibits the growth and metastasis of
non-small cell lung cancer through blocking PI3K/AKT signal pathway
by targeting MALAT-1. Biomed Pharmacother. 93:1065–1073. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Khan M, Walters LL, Li Q, Thomas DG,
Miller JM, Zhang Q, Sciallis AP, Liu Y, Dlouhy BJ, Fort PE, et al:
Characterization and pharmacologic targeting of EZH2, a fetal
retinal protein and epigenetic regulator, in human retinoblastoma.
Lab Invest. 95:1278–1290. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ganguly A and Shields CL: Differential
gene expression profile of retinoblastoma compared to normal
retina. Mol Vis. 16:1292–1303. 2010.PubMed/NCBI
|
|
23
|
Zhang Y, Wu D, Xia F, Xian H, Zhu X, Cui H
and Huang Z: Downregulation of HDAC9 inhibits cell proliferation
and tumor formation by inducing cell cycle arrest in
retinoblastoma. Biochem Biophys Res Commun. 473:600–606. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Castro-Magdonel BE, Orjuela M, Camacho J,
García-Chéquer AJ, Cabrera-Muñoz L, Sadowinski-Pine S,
Durán-Figueroa N, Orozco-Romero MJ, Velázquez-Wong AC,
Hernández-Ángeles A, et al: miRNome landscape analysis reveals a 30
miRNA core in retinoblastoma. BMC Cancer. 17:4582017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Montoya V, Fan H, Bryar PJ, Weinstein JL,
Mets MB, Feng G, Martin J, Martin A, Jiang H and Laurie NA: Novel
miRNA-31 and miRNA-200a-mediated regulation of retinoblastoma
proliferation. PLoS One. 10:e01383662015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sheng Y, Ding S, Chen K, Chen J, Wang S,
Zou C, Zhang J, Cao Y, Huang A and Tang H: Functional analysis of
miR-101-3p and Rap1b involved in hepatitis B virus-related
hepatocellular carcinoma pathogenesis. Biochem Cell Biol.
92:152–162. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yan K, Tian J, Shi W, Xia H and Zhu Y:
LncRNA SNHG6 is associated with poor prognosis of gastric cancer
and promotes cell proliferation and EMT through epigenetically
silencing p27 and sponging miR-101-3p. Cell Physiol Biochem.
42:999–1012. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lin CW, Chang YL, Chang YC, Lin JC, Chen
CC, Pan SH, Wu CT, Chen HY, Yang SC, Hong TM and Yang PC:
MicroRNA-135b promotes lung cancer metastasis by regulating
multiple targets in the Hippo pathway and LZTS1. Nat Commun.
4:18772013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Le MT, Xie H, Zhou B, Chia PH, Rizk P, Um
M, Udolph G, Yang H, Lim B and Lodish HF: MicroRNA-125b promotes
neuronal differentiation in human cells by repressing multiple
targets. Mol Cell Biol. 29:5290–5305. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kuzmichev A, Jenuwein T, Tempst P and
Reinberg D: Different EZH2-containing complexes target methylation
of histone H1 or nucleosomal histone H3. Mol Cell. 14:183–193.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Hansen KH, Bracken AP, Pasini D, Dietrich
N, Gehani SS, Monrad A, Rappsilber J, Lerdrup M and Helin K: A
model for transmission of the H3K27me3 epigenetic mark. Nat Cell
Biol. 10:1291–1300. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Fluge Ø, Gravdal K, Carlsen E, Vonen B,
Kjellevold K, Refsum S, Lilleng R, Eide TJ, Halvorsen TB, Tveit KM,
et al: Expression of EZH2 and Ki-67 in colorectal cancer and
associations with treatment response and prognosis. Br J Cancer.
101:1282–1289. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kleer CG, Cao Q, Varambally S, Shen R, Ota
I, Tomlins SA, Ghosh D, Sewalt RG, Otte AP, Hayes DF, et al: EZH2
is a marker of aggressive breast cancer and promotes neoplastic
transformation of breast epithelial cells. Proc Natl Acad Sci USA.
100:11606–11611. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Varambally S, Dhanasekaran SM, Zhou M,
Barrette TR, Kumar-Sinha C, Sanda MG, Ghosh D, Pienta KJ, Sewalt
RG, Otte AP, et al: The polycomb group protein EZH2 is involved in
progression of prostate cancer. Nature. 419:624–629. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Lian R, Ma H, Wu Z, Zhang G, Jiao L, Miao
W, Jin Q, Li R, Chen P, Shi H and Yu W: EZH2 promotes cell
proliferation by regulating the expression of RUNX3 in laryngeal
carcinoma. Mol Cell Biochem. 439:35–43. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Mohammad F, Weissmann S, Leblanc B, Pandey
DP, Højfeldt JW, Comet I, Zheng C, Johansen JV, Rapin N, Porse BT,
et al: EZH2 is a potential therapeutic target for H3K27M-mutant
pediatric gliomas. Nat Med. 23:483–492. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Rastogi B, Raut SK, Panda NK, Rattan V,
Radotra BD and Khullar M: Overexpression of HDAC9 promotes oral
squamous cell carcinoma growth, regulates cell cycle progression,
and inhibits apoptosis. Mol Cell Biochem. 415:183–196. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhao YX, Wang YS, Cai QQ, Wang JQ and Yao
WT: Up-regulation of HDAC9 promotes cell proliferation through
suppressing p53 transcription in osteosarcoma. Int J Clin Exp Med.
8:11818–11823. 2015.PubMed/NCBI
|