Introduction
Coronary artery disease (CAD) is a leading cause of
death worldwide. Atherosclerosis is an important cause for the
development of CAD. In addition, factors including age, sex,
hypertension, dyslipidemia, smoking, and diabetes, can increase the
risk of CAD (1). CAD increases the
occurrence of myocardial infarction, which may lead to serious
complications or even death if the patient has not been treated
timely and effectively. Except for the development of efficient
drugs, the progression of early prediction and diagnosis are also
the direction for CAD prevention. Many studies indicate that
microRNAs (miRNAs) are widely involved in physiological and
pathological processes of CAD (2–8).
miRNAs are a class of small non-coding RNAs first
identified in 1993. miRNAs could either initiate translational
inhibition or degradation of messenger RNAs (mRNAs), and adjust
gene expression at the post-translational stage, by binding to the
3′-untranslated region of target mRNAs. Compared with protein-based
biomarkers, miRNAs have some good characteristics, including
stability in the circulation, higher sensitivity, higher
specificity, tissue- and pathology-specific regulation, to be the
circulating biomarkers. Previosu findings showed that numerous
miRNAs, such as miRNA-1 (2),
miRNA-19 (3), miRNA-92 (4), miRNA-133 (5), miRNA-146a (6), miRNA-155 (7) and miRNA-199 (8) are associated with CAD. However, each
miRNA has numerous target mRNAs, and the effective recognition of
the interaction remains a challenge, limiting the application in
CAD prediction. Therefore, bioinformatic methods have been
developed and used to predict the effective miRNA-mRNA
interaction.
Original TargetScan software was developed by Lewis
et al, and it is the first algorithm to predict miRNA
targets in vertebrates (9). The
first edition of TargetScan obtained miRNA sequences from the Rfam
database. The TargetScan method generated a 3′-UTR dataset for
mammalian and vertebrate genes to confirm miRNA-target
interactions. The TargetScan method examined the data of seed
matches including the expected frequency, the observed count and
the predicted free energy of the seed to assign a Z score to each
3′-UTR. However, the seed matches were not always sufficient to
confer repression. The grade of repression was extremely unstable
in different UTR contexts if repression occurred. In order to avoid
this problem, Friedman et al created a modified algorithm
known as TargetScan probabilities of conserved targeting (PCT) for
quantitatively valuing site preservation (10). The new method incorporates the
phylogenetic tree based on the UTR genomic regions, and assesses
motif conservative by branch-length metric. Additionally,
TargetScan PCT widely improved the computation of the
conservation-specific backgrounds of single seed-match sites. In
2014, Nam et al developed an advanced prediction model based
on TargetScan, termed weighted context+ or wContext+ (11). We termed this new TargetScan method,
as targetscore. This algorithm primarily differed from the other
TargetScan algorithm in three aspects. Firstly, the model of
targetscore was especially designed for miRNA-overexpression
experiments to explore targets of a specific miRNA in a particular
cell condition. Secondly, the model inferred miRNA-targets solely
based on their individual high-dimensional modes of expression
fold-changes and sequence features. Finally, this method operated
on the whole gene expression profile to more closely imitate the
overall likelihood rather than the previous TargetScan method. In
summary, the targetscore algorithm could afford independent and
supplementary information by PCT values and the context scores
which are useful for predicting the target gene of each miRNA.
Thus, the targetscore algorithm was used to predict
the target gene for miRNA-1, miRNA-155 and miRNA-146a, which are
closely related to the pathogenesis of CAD.
Materials and methods
Gene expression profile microarray
data
The gene expression profile dataset no. GSE7638,
stored in the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/) by Pascal and
Meier based on the GPL571 platform, was analyzed by bioinformatics
analysis in our study. The dataset is comprised of 160 samples,
including 110 cases of total RNAs obtained from patients with CAD
and 50 cases of total RNAs obtained from healthy individuals. The
ratio of male to female is moderate in this dataset. We used the
pre-processing software to integrate data after the different
samples were grouped.
Gene expression profile matrix
The expression values of the gene expression profile
were pre-processed to facilitate the calculation of the
differential expression of genes. The values after pre-processing
exclude the missing, the unvaried, the maximum and the minimum
values. We removed the unsuitable values to obtain the relative
expression and form a new expression dataset. The database of gene
pre-treatment was selected as hgu133a. The first step in data
processing was to merge multiple probes of the same gene, and the
average of multiple probes was chosen as the expression of the
gene. The second step was to modify the annotation information
(rename the columns to CAD and control group). Finally, we renamed
the row of the expression matrix to GENESYMBOL after reprocessing
the gene expression values.
After the gene expression was pre-processed, we used
the limma package to calculate the logFC values for all genes. The
limma package has been certified a common choice for the analysis
of data from the experiments on microarray. The functions of the
package have expanded significantly in the differential expression
of RNA sequencing and the analysis of expression profiles.
According the gene expression differences of CAD and control group,
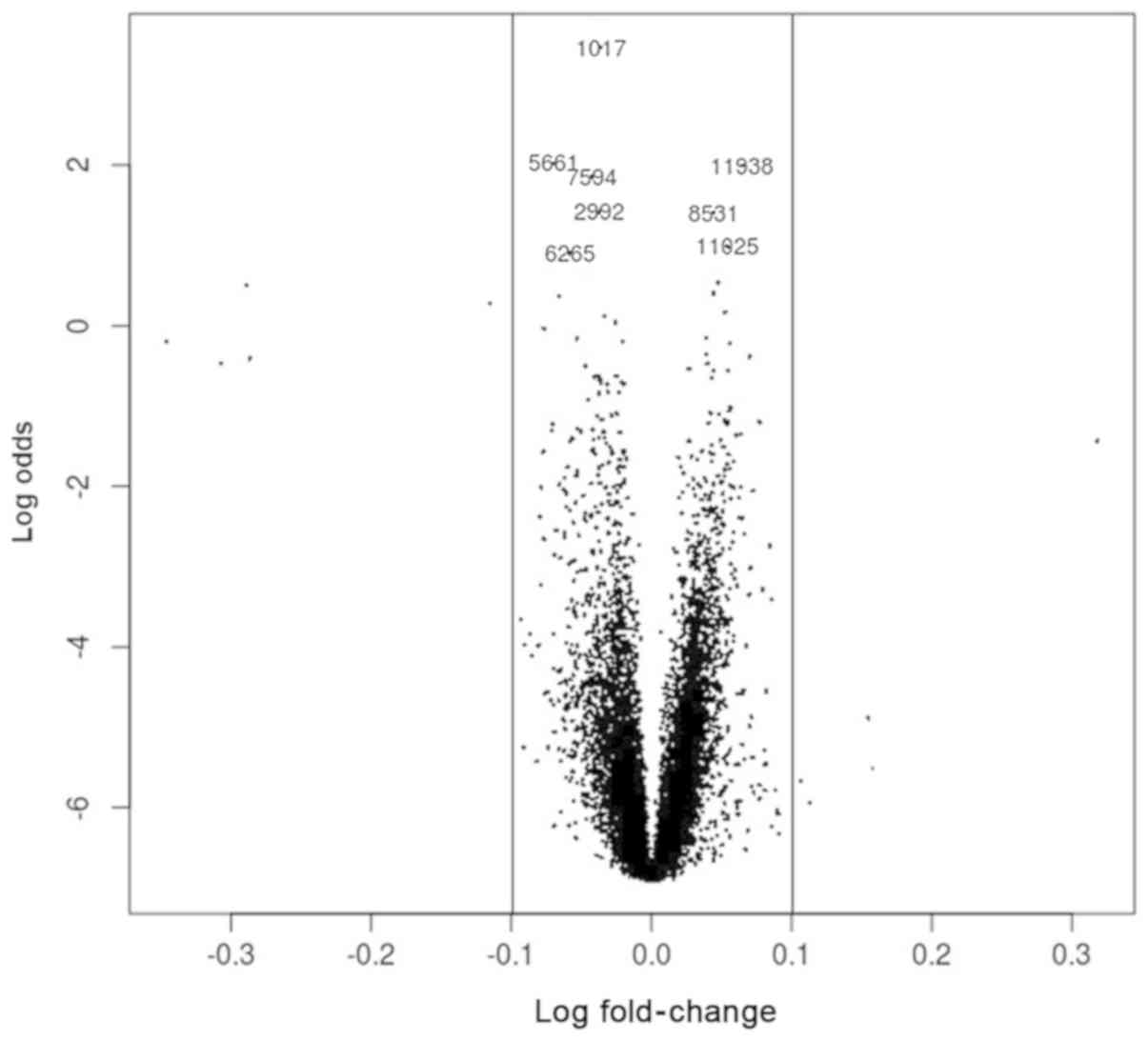
the logFC calculated by limma package was as a priori value
for the experiment (Fig. 1). From
TargetScan website we entered each miRNA to obtain the TargetScan
context score (TSCS) and PCT values of each gene. Combining the
data search in the website with the logFC obtained in the gene
expression profiling experiment, we used the variational Bayesian
Gaussian mixture model (VB-GMM) to calculate the targetscore of the
genes. The miRNA target genes were predicted by the targetscore
value.
Calculation of the targetscore. The VB-GMM model is
rearranged according to the Bayesian GMM model optimized by Li
et al (12). The target mRNAs
were predicted based on the sequence scores using the integrated
miRNA overexpression data and other predictive methods. The
miRNA-mRNA interaction is estimated by the posterior probability
distribution of the observed variables, and the targetscore value
is calculated as a weighted sigmoid-transformed fold-change.
Variational Bayesian EM means the variational Bayesian expectation
maximization. It is based on variational inference to find the edge
distribution of minimized Kullback-Leibler distance to approximate
the joint distribution, and use the mean field to reduce the
complexity of the joint estimation. Variational Bayesian is a
technique for approximating computational complexity in Bayesian
estimation and machine learning. It also used the maximum a
posteriori (MAP), that is, with the single most probable
parameter value instead of a complete Bayesian estimate. This is
why we consider it as an extension of the EM algorithm to some
extent. In addition, the variational Bayesian also obtains the
optimal solution through a series of mutually dependent equations.
We obtained the TSCS, PCT and logFC values of the genes in the
experimental data, and deduced the maximum likelihood (ML) value by
VB-GMM as the posterior probability of each gene. The targetscore
values were calculated by the following formula to quantify the
target genes of miRNA.
targetscore=11+ex×p¯
In the formula, × refers to the logFC value of the
gene, and p refers to the posterior value inferred.
Results
Preliminary screening with the
targetscore value
In the results, we predicted 362 miRNA-1-related
gene targetscore values >0.333, of which 47 genes were
validated. After the prediction of miRNA-155, there were 648 gene
targetscore values >0.333, of which 63 genes were validated. The
miRNA-146a had 428 target genes of the targetscore value >0.333
and only 1 gene was validated. According to the targetscore value
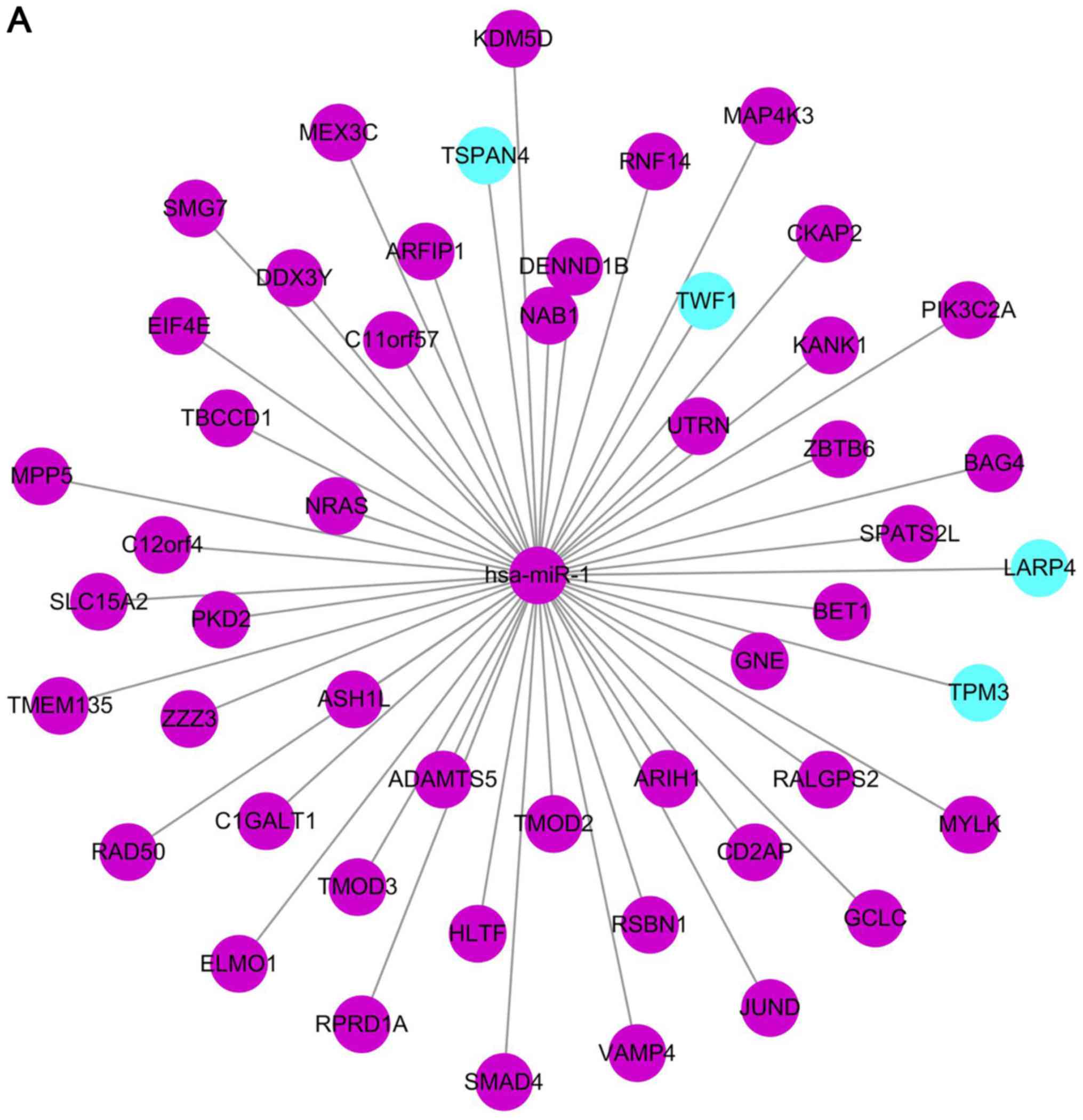
from high to low order, we extracted the top 50 genes. Of the first
50 genes of miRNA-1, the validated genes were LARP4, TSPAN4,
TWF1 and TPM3. In the first 50 genes of miRNA-155, the
validated genes are LPL, ZNF652, BET1 and TWF1. There
were only non-validated genes in the first 50 genes of miRNA146a
(Fig. 2).
Selection of the targetscore threshold
and target genes
To further increase the credibility of the target
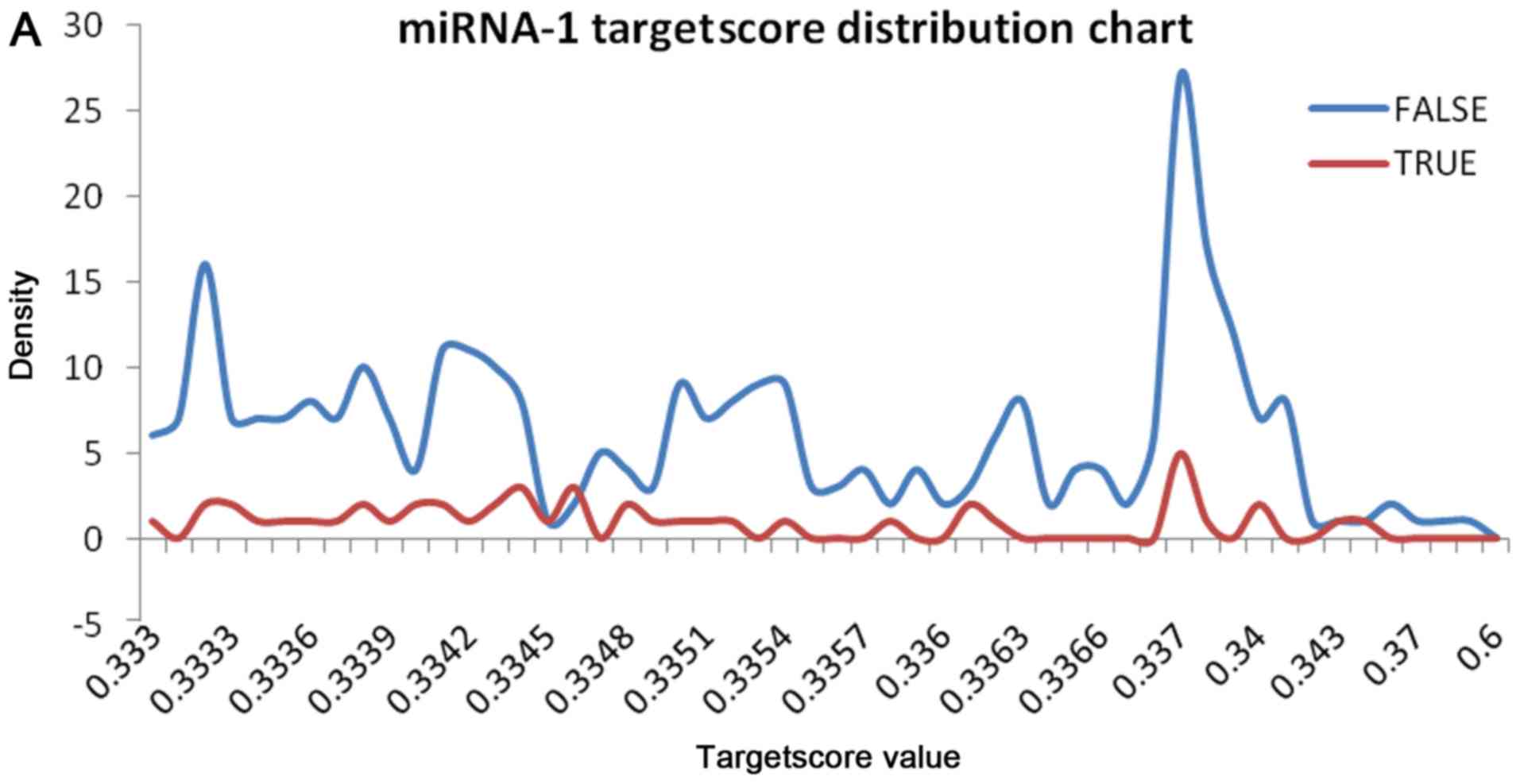
genes and make accurate screening results, we analyzed the density
overlap between the targetscore distribution of the validated and
the non-validated target genes to make the screening result more
exact. We counted the number of the validated and the non-validated
target genes in each targetscore value and drew a curve graph for
convenient observation (Fig. 3). In
the end, we chose the little overlap between the two distributions
as the targetscore threshold.
We observed the overlap on the miRNA-1 and found
that the distribution overlap decreases when targetscore = 0.341.
Therefore, we selected targetscore = 0.341 as the threshold for
screening the target genes for miRNA-1. We observed the miRNA-155
target gene overlap of targetscore distribution in the same way.
When targetscore is 0.344, the overlap was significantly reduced,
so we chose it as the standard for filtering inaccurate miRNA-155
target genes. We screened miRNA-1 and miRNA-155 target genes again
with the thresholds selected by analyzing the density distribution
of the genes. There were a total of 18 miRNA-1 target genes which
are DDX3Y, KDM5D, TMOD3, ZBTB6, MAP4K3, LARP4, RPRD1A, TSPAN4,
UTRN, CD2AP, CAGALT1, C12orf4, BET1, HLTF, SMAD4, MPP5, KANK1,
and CKAP2 (Table I).
miRNA-155 has 19 target genes, including PF4V1, LPL, DDX3Y,
LRIF1, LZTFL1, KCTD9, PRPF39, RNF146, FEM1C, MAP4K3, PAQR3,
C11orf71, FAM3C, BBS10, HMGCS1, TRMT2B, YTHDC2, UGDH, and
ZNF652 (Table II).
 | Table I.The miRNA-1 target genes. |
Table I.
The miRNA-1 target genes.
| External gene ID | LogFC | TSCS | TargetScan PCT | targetscore |
|---|
| DDX3Y | −0.28881 | −0.045 | −0.31 | 0.565146 |
| KDM5D | −0.28676 | −0.126 | 0 | 0.384165 |
| TMOD3 | −0.07866 | −0.046 | −0.06 | 0.370506 |
| ZBTB6 | −0.06244 | −0.15 | −0.63 | 0.347949 |
| MAP4K3 | −0.06196 | −0.298 | −0.56 | 0.347653 |
| LARP4 | −0.05549 | −0.11 | −0.8 | 0.344617 |
| RPRD1A | −0.05434 | −0.065 | −0.05 | 0.344199 |
| TSPAN4 | −0.05361 | −0.248 | −0.62 | 0.343959 |
| UTRN | −0.05173 | −0.207 | −0.6 | 0.343371 |
| CD2AP | −0.04958 | −0.332 | −0.89 | 0.342759 |
| C1GALT1 | −0.04657 | −0.219 | −0.4 | 0.341989 |
| C12orf4 | −0.04641 | −0.142 | −0.62 | 0.34195 |
| BET1 | −0.04592 | −0.223 | −0.77 | 0.341833 |
| HLTF | −0.04491 | −0.088 | −0.36 | 0.341596 |
| SMAD4 | −0.04413 | −0.088 | −0.7 | 0.34142 |
| MPP5 | −0.04341 | −0.273 | −0.7 | 0.341258 |
| KANK1 | −0.04295 | −0.077 | −0.18 | 0.341156 |
| CKAP2 | −0.04248 | −0.103 | −0.12 | 0.341054 |
 | Table II.The miRNA-155 target genes. |
Table II.
The miRNA-155 target genes.
| External gene ID | LogFC | TSCS | PCT | TargetScan
targetscore |
|---|
| PF4V1 | 0.113045 | −0.195 | −0.06 | 0.465517 |
| LPL | −0.08697 | −0.04 | −0.07 | 0.416066 |
| DDX3Y | −0.28881 | −0.215 | −0.01 | 0.412147 |
| LRIF1 | −0.07936 | −0.329 | −0.07 | 0.377031 |
| LZTFL1 | −0.07061 | −0.192 | −0.07 | 0.355845 |
| KCTD9 | −0.06963 | −0.111 | −0.14 | 0.354505 |
| PRPF39 | −0.06604 | −0.098 | −0.08 | 0.350639 |
| RNF146 | −0.06437 | −0.055 | −0.07 | 0.349273 |
| FEM1C | −0.06326 | −0.208 | −0.06 | 0.348484 |
| MAP4K3 | −0.06196 | −0.184 | −0.06 | 0.347653 |
| PAQR3 | −0.06076 | −0.056 | −0.06 | 0.346964 |
|
C11orf71 | −0.05819 | −0.147 | −0.07 | 0.345704 |
| FAM3C | −0.05812 | −0.074 | −0.07 | 0.345674 |
| BBS10 | −0.0573 | −0.099 | −0.07 | 0.345325 |
| HMGCS1 | −0.05637 | −0.265 | −0.07 | 0.344948 |
| TRMT2B | −0.05617 | −0.064 | −0.06 | 0.344872 |
| YTHDC2 | −0.05569 | −0.141 | −0.06 | 0.34469 |
| UGDH | −0.05498 | −0.089 | −0.07 | 0.34443 |
| ZNF652 | −0.05379 | −0.187 | −0.66 | 0.344021 |
Unlike miRNA-1 and miRNA-155, there is only one
validated gene in the miRNA-146a prediction result. So the
re-screening method chosen to predict miRNA-146a is different from
the other two miRNAs. We decided to re-screen the target genes of
miRNA-146a according to the value of the |logFC|. Based on past
experience, we regarded |logFC|≥0.05 as the standard for
re-screening target genes of miRNA-146a, and the targetscore
threshold is 0.343. The genes, including PTGS2, EIF1AY, GUF1,
TUBB1, PCYOX1, CHM, TRMT1L, ANKRD46, STEAP4, B3GNT2, TMEM185B,
QDPR, CDC37L1, PRMT3, ZNF652, and DCAF17, were selected
as target genes for miRNA-146a (Table
III).
 | Table III.The miRNA-146a target genes. |
Table III.
The miRNA-146a target genes.
| External gene
ID | LogFC | TSCS | PCT | TargetScan
targetscore |
|---|
| PTGS2 | −0.09076 | −0.163 | −0.14 | 0.443186 |
| EIF1AY | −0.307 | −0.2 | 0 | 0.384102 |
| GUF1 | −0.07941 | −0.114 | −0.16 | 0.37725 |
| TUBB1 | 0.089999 | −0.066 | −0.15 | 0.369688 |
| PCYOX1 | −0.06924 | −0.103 | −0.16 | 0.354002 |
| CHM | −0.06777 | −0.049 | −0.16 | 0.352296 |
| TRMT1L | −0.06562 | −0.267 | −0.15 | 0.350279 |
| ANKRD46 | −0.06506 | −0.129 | −0.14 | 0.349811 |
| STEAP4 | −0.06161 | −0.095 | −0.16 | 0.347444 |
| B3GNT2 | −0.06078 | −0.139 | −0.16 | 0.346977 |
|
TMEM185B | −0.0586 | −0.204 | −0.16 | 0.345892 |
| QDPR | −0.05846 | −0.115 | −0.14 | 0.345829 |
| CDC37L1 | −0.05836 | −0.148 | −0.13 | 0.345781 |
| PRMT3 | −0.05643 | −0.323 | −0.15 | 0.34497 |
| ZNF652 | −0.05379 | −0.295 | −0.14 | 0.344021 |
| DCAF17 | −0.05333 | −0.108 | −0.16 | 0.343869 |
Relevant references of validated genes. Some of the
predicted genes in the target gene were validated, but most of the
genes were not validated. We compiled the relevant literature
information of the validated genes into a Table. Since the first 50
target genes of miRNA-146a are non-validated, we listed the
literature information only of the validated gene in all the genes
(Table IV).
 | Table IV.The validated target gene
references. |
Table IV.
The validated target gene
references.
| miRNA | Target gene | Target gene (Entrez
ID) | Experiments | Refs. (PMID) |
|---|
| Hsa-miR-1 | LARP4 | 113251 | Luciferase reporter
assay | 20144220 |
| Hsa-miR-1 | TSPAN4 | 7106 | Microarray | 15685193 |
| Hsa-miR-1 | TWF1 | 5756 | Microarray | 15685193 |
| Hsa-miR-1 | TPM3 | 7170 | pSILAC | 18668040 |
| Hsa-miR-155 | LPL | 4023 | pSILAC | 18668040 |
| Hsa-miR-155 | ZNF652 | 22834 | luciferase reporter
assay | 18367535 |
| Hsa-miR-155 | BET1 | 10282 | pSILAC | 18668040 |
| Hsa-miR-155 | TWF1 | 5756 | pSILAC | 18668040 |
| Hsa-miR-146a | TRAF6 | 7189 | western
blotting/northern blotting | 18504431 |
Discussion
miRNAs have important effect because they could
regulate gene expression at the post-transcriptional level by
binding to the 3′-UTR of mRNAs. Several scientific gains have found
that miRNAs have the function of regulating specific genes. With
the increase of the research of CAD diagnosis and prediction by
miRNA, we found some meaningful results. Wang et al found
that the genetic variant of one of the miRNA-1 target genes binding
site was associated with the major adverse cardiovascular events of
CAD (2). In the study, researchers
compared the target gene COG6 rs9548934 C→T variant with the wide
genotype CC, and found that the genetic variant could significantly
reduce the risk of CAD. This means that a diminutive variation in
the gene miRNA-binding sites may have an influence on numerous
target mRNAs, and the polymorphism in miRNA target sites could be
modulated in miRNA-1 targeting. The scholars have utilized the
bioinformatics tool to confirm that MAP3K10 is the target gene of
miRNA-155, and the results proved that targeting MAP3K10 may be a
major mechanism for the anti-atherosclerosis effect of miR-155
(13). It has been confirmed that
miRNAs could control the main cell function to regulate the
progression of atherosclerosis. miRNA-146a was reported to be a
regulator of the inflammatory process and influence the pathology
of CAD (14). From the literature,
we found that miRNA-1, miRNA-155 and miRNA-146a were all closely
related to the pathological process of CAD. They can control the
expression of effective target genes to affect the probability of
the major adverse cardiovascular events of CAD. So we selected
these three miRNAs for target gene prediction in order to do the
preparatory work for clarifying the mechanism of CAD.
In this study, we chose the targetscore method to
predict the target genes of miRNAs, and the greatest advantage of
this method is that it could be associated with the selected
disease to make the results more specific. Screening the accurate
target genes not only can make the mechanism of miRNAs in CAD
clear, but also can help the future of medicine research and
development. After we made the prediction, we found that a large
number of the genes in results were validated thus indicating the
results were credible. Nevertheless the initial screening of the
results were too large, we used the targetscore value for
re-screening to make the consequence accurate. First we considered
the top 50 genes of the targetscore value as the precise target
genes and analyzed them. We found that LARP4, TSPAN4, TWF1
and TPM3 of miRNA-1 were validated. The LPL, ZNF652,
BET1 and TWF1 of miRNA-155 were also validated. But the
first 50 genes of miRNA146a were all non-validated. In order to
further refine the results, we used the density overlap between the
targetscore distribution of the validated and the non-validated
target genes to re-screen. By observing the density overlap, we
selected the thresholds of miRNA-1 as targetscore = 0.341, and the
thresholds of miRNA-155 as targetscore = 0.344. Unlike the other
two miRNAs, there was only one validated gene in the miRNA-146a
predicted results. Therefore the re-screening method cannot use the
density overlap, so we refined its prediction result based on the
|logFC| values (12). In the end, we
used the |logFC| value >0.05 as the standard of miRNA-146a exact
screening and the targetscore threshold is 0.343. According to the
targetscore threshold, we finally confirmed that the miRNA-1 target
genes are DDX3Y, KDM5D, TMOD3, ZBTB6, MAP4K3, LARP4, RPRD1A,
TSPAN4, UTRN, CD2AP, CAGALT1, C12orf4, BET1, HLTF, SMAD4, MPP5,
KANK1, and CKAP2. The miRNA-155 target genes are
PF4V1, LPL, DDX3Y, LRIF1, LZTFL1, KCTD9, PRPF39, RNF146, FEM1C,
MAP4K3, PAQR3, C11orf71, FAM3C, BBS10, HMGCS1, TRMT2B, YTHDC2,
UGDH, and ZNF652. The miRNA-146a target genes are
PTGS2, EIF1AY, GUF1, TUBB1, PCYOX1, CHM, TRMT1L, ANKRD46,
STEAP4, B3GNT2, TMEM185B, QDPR, CDC37L1, PRMT3, ZNF652, and
DCAF17. In the final result, the LARP4 and TSPAN4 of miRNA-1
as well as the LPL and ZNF652 of miRNA-155 were validated.
We retrieved the references of the validated genes,
including LARP4, TSPAN4, LPL and ZNF652, and
carefully analyzed them. Takane et al considered that
miRNA-1 is an evolutionarily conserved miRNA which controls the
expression of a large number of target genes and attempted to
explore miRNA-1-mediated gene regulatory mechanisms by analyzing
miRNA-target gene pairs (15). They
believe that the complementarily between miRNA and 3′-UTR was weak
when the method was chosen, so the phylogenetic profiling approach
was chosen as the research method. Five miRNA families, including
miRNA-1, were extracted from five species of Diptera animals.
Firstly, the potential target genes were extracted according to the
optimal free-energy information. Then, the sequence of the seed was
matched by RNAhybrid software, and the potential binding site of
the short RNA in the target sequence was predicted. Finally, the
target genes were screened according to the ‘enrichment’ index. The
results show that miRNA-1 downregulates the expression of LARP4. It
suggests that miRNAs can regulate their own miRNA pathway by
controlling the expression of significant translation factors. Lim
et al transfected a double-stranded piece of miRNA-1 into
HeLa cells and used microarray to select the mRNAs whose expression
profiles were different from the LocusLink database (16). They observed that miRNA-1 is
preferentially expressed in the heart and skeletal muscle of
mammals, and TSPAN4 is a target gene of miRNA-1. The predictive
results confirmed the hypothesis that vertebrate miRNAs have a
large number of target genes. Selbach et al indicated that
recognizing miRNA-target gene pairs is one of the most critical
aspects of understanding life (17).
They measured the protein difference between diverse transfected
HeLa cells by the pulsed SILAC (pSILAC) method, and finally
verified 16 pSILAC genes by western blotting. Yin et al
analyzed the gene expression of miRNA-155 by cell experiment, and
the target gene was predicted by TargetScan Bayesian method
(18). Seven transcriptional
regulatory genes including ZNF652 were regulated by miRNA-155.
Although the validated genes are only a small part
in the final prediction results, our results were obtained by the
targetscore method which is more accurate than the TargetScan
method. The targetscore method uses the limma package to calculate
the logFC value as the a priori value. After the gene
expression profiling, the logFC value was obtained, and it was
calculated as the posterior value by VB-GMM model with TSCS and
PCT. The targetscore method is more closely related to the disease
than the TargetScan method, and the predicted target genes are more
accurate. So we believe that genes with high targetscore values are
more likely to be the target genes that are regulated by miRNAs
during the development and progression of CAD. In this study, our
major contribution is to predict the target mRNAs of the chosen
miRNAs with the gene expression profiles, which can effectively
reduce the workload of screening. Most of the predicted results are
non-validated genes, which is a good sign for the research in the
future. Because the results predicted by our method could
effectively avoid the false-positive, and provide more direction
and ideas to research the mechanism of miRNA affecting CAD.
Acknowledgements
This article was completed in the careful guidance
and strong support of Director Jiupei Cheng. The rigorous academic
attitude, high professionalism, profound knowledge and innovative
spirit of Dean YiWen Wang have had a significant impact on me. I
would like to pay special tribute to Dr Zheng Huang for his
guidance on the experiments, and also express my heartfelt thanks
to Mr. CunMing Fang for his assistance in reference literature
search. (X-L Ma).
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
XLM and RF designed the study. XLM was involved in
data collection. XY performed the statistical analysis and prepared
the figures. XLM and XY drafted the report. RF contributed
substantially to its revision. All authors read and approved the
final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Eriksen A, Tillin T, O'Connor L, Brage S,
Hughes A, Mayet J, McKeigue P, Whincup P, Chaturvedi N and Forouhi
NG: The impact of health behaviours on incident cardiovascular
disease in Europeans and South Asians - a prospective analysis in
the UK SABRE study. PLoS One. 10:e01173642015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wang L, Zhi H, Li Y, Ma G, Ye X, Yu X,
Yang T, Jin H, Lu Z and Wei P: Polymorphism in miRNA-1 target site
and circulating miRNA-1 phenotype are associated with the decreased
risk and prognosis of coronary artery disease. Int J Clin Exp
Pathol. 7:5093–5102. 2014.PubMed/NCBI
|
|
3
|
Lai Y, He S, Ma L, Lin H, Ren B, Ma J, Zhu
X and Zhuang S: HOTAIR functions as a competing endogenous RNA to
regulate PTEN expression by inhibiting miR-19 in cardiac
hypertrophy. Mol Cell Biochem. 432:179–187. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Liu H, Li G, Zhao W and Hu Y: Inhibition
of miR-92a may protect endothelial cells after acute myocardial
infarction in rats: Role of KLF2/4. Med Sci Monit. 22:2451–2462.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Castoldi G, Di Gioia CR, Bombardi C,
Catalucci D, Corradi B, Gualazzi MG, Leopizzi M, Mancini M, Zerbini
G, Condorelli G, et al: MiR-133a regulates collagen 1A1: Potential
role of miR-133a in myocardial fibrosis in angiotensin II-dependent
hypertension. J Cell Physiol. 227:850–856. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wu ZW, Liu YF, Wang S and Li B: miRNA-146a
induces vascular smooth muscle cell apoptosis in a rat model of
coronary heart disease via NF-κB pathway. Genet Mol Res.
14:18703–18712. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kishore R, Verma SK, Mackie AR, Vaughan
EE, Abramova TV, Aiko I and Krishnamurthy P: Bone marrow progenitor
cell therapy-mediated paracrine regulation of cardiac miRNA-155
modulates fibrotic response in diabetic hearts. PLoS One.
8:e601612013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Baumgarten A, Bang C, Tschirner A,
Engelmann A, Adams V, von Haehling S, Doehner W, Pregla R, Anker
MS, Blecharz K, et al: TWIST1 regulates the activity of ubiquitin
proteasome system via the miR-199/214 cluster in human end-stage
dilated cardiomyopathy. Int J Cardiol. 168:1447–1452. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lewis BP, Shih IH, Jones-Rhoades MW,
Bartel DP and Burge CB: Prediction of mammalian microRNA targets.
Cell. 115:787–798. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Friedman RC, Farh KK, Burge CB and Bartel
DP: Most mammalian mRNAs are conserved targets of microRNAs. Genome
Res. 19:92–105. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Nam JW, Rissland OS, Koppstein D,
Abreu-Goodger C, Jan CH, Agarwal V, Yildirim MA, Rodriguez A and
Bartel DP: Global analyses of the effect of different cellular
contexts on microRNA targeting. Mol Cell. 53:1031–1043. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Li Y, Goldenberg A, Wong KC and Zhang Z: A
probabilistic approach to explore human miRNA targetome by
integrating miRNA-overexpression data and sequence information.
Bioinformatics. 30:621–628. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhu J, Chen T, Yang L, Li Z, Wong MM,
Zheng X, Pan X, Zhang L and Yan H: Regulation of microRNA-155 in
atherosclerotic inflammatory responses by targeting MAP3K10. PLoS
One. 7:e465512012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang Y, Wang X, Li Z, Chen L, Zhou L, Li C
and Ouyang DS: Two single nucleotide polymorphisms (rs2431697 and
rs2910164) of miR-146a are associated with risk of coronary artery
disease. Int J Environ Res Public Health. 14:142017.
|
|
15
|
Takane K, Fujishima K, Watanabe Y, Sato A,
Saito N, Tomita M and Kanai A: Computational prediction and
experimental validation of evolutionarily conserved microRNA target
genes in bilaterian animals. BMC Genomics. 11:1012010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lim LP, Lau NC, Garrett-Engele P, Grimson
A, Schelter JM, Castle J, Bartel DP, Linsley PS and Johnson JM:
Microarray analysis shows that some microRNAs downregulate large
numbers of target mRNAs. Nature. 433:769–773. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Selbach M, Schwanhäusser B, Thierfelder N,
Fang Z, Khanin R and Rajewsky N: Widespread changes in protein
synthesis induced by microRNAs. Nature. 455:58–63. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yin Q, McBride J, Fewell C, Lacey M, Wang
X, Lin Z, Cameron J and Flemington EK: MicroRNA-155 is an
Epstein-Barr virus-induced gene that modulates Epstein-Barr
virus-regulated gene expression pathways. J Virol. 82:5295–5306.
2008. View Article : Google Scholar : PubMed/NCBI
|