Introduction
Ulcerative colitis (UC), a chronic non-specific
inflammatory bowel disease, typically starts in the rectum and
spreads to the proximal colon in a continuous manner, involving the
area surrounding the appendix (1,2). UC is
characterized by acute pain, vomiting, weight loss, diffuse mucosal
inflammation, diarrhoea and bloody stools. The symptoms affect the
quality of life of patients due to a high recurrence rate and the
lack of effective therapeutic options (3,4).
Furthermore, the mechanisms underlying the pathogenesis of UC have
not been fully elucidated. A previous study indicated that UC
triggers are primarily associated with the immune system,
apoptosis, the physical environment, mental psychology, genetic
inheritance and infection (5). The
human gut contains >100 trillion microorganisms, including
commensals and pathogens (6). A
previous study demonstrated that dysregulated interactions between
the immune system and the intestinal microbiota might lead to
colitis (7). Illumina MiSeq
sequencing reports have revealed that the diversity of the gut
flora in patients with UC is decreased, with significant
differences in the abundance of Bacteroides and
Lactobacillus, compared with healthy controls (8,9). The
aforementioned studies indicated that alterations in the gut flora
are closely related to the pathogenesis of UC.
First identified in 2012, irisin is a myogenic
factor (10) and a soluble
polypeptide fragment consisting of 112 amino acids (11), which is cleaved from the precursor
protein fibronectin type III domain containing 5(12). A recent report revealed that irisin
serves a key role in the occurrence of metabolic, chronic kidney
and autoimmune diseases (13), and
that there is a correlation between irisin and the level of
inflammation (14). Furthermore,
irisin may display anti-inflammatory effects by regulating the
expression of receptors and proteins in macrophages, resulting in a
reduction in the release of key proinflammatory factors, including
tumour necrosis factor-α (TNF-α), interleukin (IL)-1β and monocyte
chemoattractant protein-1(15).
However, the mechanisms underlying the anti-inflammatory properties
of irisin are not completely understood. A previous study
demonstrated that low concentrations of irisin increase the
expression of Toll-like receptor 4 (TLR4) in macrophages, as well
as the ability of macrophages to identify potential pathogens, by
indirectly promoting anti-inflammatory effects (16). Furthermore, the protein expression
levels of TLR4 and myeloid differentiation factor 88 (MyD88), as
well as the phosphorylation level of NF-κB, in macrophages were
decreased by high irisin concentrations, leading to a reduction in
the release of key proinflammatory cytokines and induction of
anti-inflammatory effects (16).
Another study demonstrated that irisin can control the inflammatory
response and reduce pathological progress in the inflammatory
response in a concentration-dependent manner in acute lung injury
(17). However, to the best of our
knowledge, no previous studies have investigated the role of irisin
in UC pathogenesis.
A previous study revealed that plasma irisin levels
in sedentary mice with UC fed a high-fat diet were decreased
compared with those in mice fed a standard diet (18). Furthermore, our preliminary data
(data not shown) suggested that irisin may be negatively correlated
with IL-12 and IL-23 levels and the abundance of
Enterococci, but positively correlated with the abundance of
Lactobacilli. However, the association between irisin and
intestinal microbiota in UC is still not completely understood.
Therefore, the present study aimed to investigate the therapeutic
effect of exogenous irisin, as well as its underlying mechanisms,
in an experimental mouse model of UC.
Materials and methods
Chemicals and reagents
Dextran sodium sulphate (DSS) salt (colitis grade;
lot no. Q5756) was obtained from MP Biomedicals, LLC. Recombinant
irisin (cat. no. 067-29A) was obtained from Phoenix
Pharmaceuticals, Inc. The CD64 antibody (cat. no. B166615) was
obtained from BioLegend, Inc.
Establishment of animal models
Mice were purchased from the Henan Experimental
Animal Center [license no. SCXK (Yu) 2015-0004]. A total of 40
C57/BL mice (age, 6 weeks; average weight, 37.767±4.487 g) were
randomly divided into the following four groups (n=10 per group;
five males and five females): i) Control; ii) irisin; iii) UC; and
iv) UC + irisin. The mice in each group were kept in different
cages and administered the corresponding treatments. The control
and irisin groups had access to normal autoclaved water. The UC and
UC + irisin groups had access to 4% DSS (dissolved in autoclaved
water) via drinking water. From day 1, the irisin and the UC +
irisin groups received an intraperitoneal injection of irisin
(0.0075 µg/g; dissolved in PBS) once a day, the UC group and the
control group received an intraperitoneal injection of 0.2 ml of
PBS, as described by Asadi et al (19). During the study, mice were fed a
standard rodent diet and were housed at 25˚C with 12-h light/dark
cycles and 45% humidity. Each morning, the mice were weighed, and
their faeces, faecal characteristics and general states were
observed and recorded. On day 10, mice were humanely sacrificed by
cervical dislocation as there were signs that the humane endpoints
of the study of weight loss and behavioural changes had been
reached. The humane endpoints were: i) General appearance: Very
rough coat; ii) mental status: Nearly moribund or coma; iii)
behaviour: Extremely low in-cage activity level or violent response
to external stimuli; iv) clinical signs: Obvious weight change
(~80% of initial body weight or a reduction in weight of ~20%) or
rare food and water intake. All efforts were made to minimize
animal suffering. The animal experiments were approved by the
Medical Ethics Committee of Henan University and performed in
accordance with the Guidelines of the Institutional Animal Care and
Use Committee (20).
Body weight, spleen weight,
macroscopic score and histological score alterations
Mice were weighed daily and the spleen weight of
each mouse was compared at the end of the experiment. The mice were
evaluated using macroscopic and histological scoring systems. The
histological score was assessed by hematoxylin and eosin staining
of colon sections. Colon tissues were fixed in 4% paraformaldehyde
overnight at room temperature and embedded in paraffin. The
paraffin embedded colon tissues were cut into 5 µm thick slices.
From each colon sample 5 sections were evaluated and this was
repeated in 10 mice per group. The slices were stained at room
temperature for 5 min with hematoxylin and 1 min with eosin.
Stained sections were observed under a light microscope with a
magnification of x200 in four randomly selected fields of view to
evaluate inflammatory cell infiltration and mucosal and epithelial
destruction. Tissues were histologically scored by two experienced
pathologists independently in a blinded manner, from 0 (no change)
to 6 (extensive cell infiltration and tissue damage), according to
the Siegmund method (Table SII)
(21). The sub-scores of
inflammatory cell infiltration and epithelial damage were added and
divided by two to form a total histological score. Tissues were
macroscopically scored from 0.0 (healthy) to 4.0 (maximal activity
of colitis), according to the Murano method (Table SI) (22). The sub-scores of weight loss, stool
consistency and occult/gross bleeding were added and divided by
three, forming a total macroscopic score.
Immunofluorescence staining of colon
tissue
Colon tissues were fixed in 4% paraformaldehyde at
room temperature overnight, paraffin-embedded, sectioned into 5 µm
slices, and incubated for 2 h at 60˚C. The sections were dewaxed
with xylene, hydrated with a descending alcohol series and blocked
using 5% BSA (cat. no. 9048-46-8; Sigma-Aldrich; Merck KGaA) at
room temperature for 1 h. Antigen retrieval was performed by
microwaving the sections. Subsequently, the sections were incubated
with a PE-labelled CD64 primary antibody (cat. no. 139303,
BioLegend, Inc.; 0.2 mg/ml) at room temperature for 1.5 h, followed
by nuclear staining with DAPI for 5 min at room temperature. The
sections were sealed with glycerol and observed using a
fluorescence microscope with a magnification of x200. CD64
expression was quantified using ImageJ software (version 1.51;
National Institutes of Health) according to the following formula:
Fluorescence integrated density=region of interest area x mean
fluorescence intensity.
ELISA measurement of IL-12 and IL-23
levels in plasma
Prior to sacrifice on day 10, whole blood (0.5 ml)
was collected from the submandibular vein of each mouse. Blood
samples were centrifuged at 1,509.3 x g for 10 min at 4˚C to
separate the plasma. Subsequently, plasma levels of IL-12 and IL-23
were measured using Hermes Criterion Biotechnology IL-12 mouse
ELISA kits and Hermes Criterion Biotechnology IL-23 mouse ELISA
kits respectively (cat. nos. I085 and I114; Elixir Canada Medicine
Company Ltd.) according to the manufacturer's protocol.
Fecal DNA extraction
On day 10, fecal samples were collected from each
mouse. Fecal bacterial DNA was extracted using the TIANamp Stool
DNA kit (cat. no. DP328-02; Tiangen Biotech Co., Ltd.) according to
the manufacturer's instructions.
Library preparation and
sequencing
MetaVx™ library preparation and Illumina MiSeq
sequencing were conducted at Genewiz, Inc. DNA quantity of the
fecal bacterial DNA was determined using a Qubit 2.0 fluorometer
(Invitrogen; Thermo Fisher Scientific, Inc.) and a library was
prepared using the MetaVx™ Library Preparation kit (Genewiz, Inc.).
V3 and V4 are two hypervariable regions of the prokaryotic 16S rDNA
(23). The V3 and V4 regions were
amplified using the same following primers: Forward,
5'-CCTACGGRRBGCASCAGKVRVGAAT-3' and reverse,
5'-GGACTACNVGGGTWTCTAATCC-3'. PCR was performed using 30-50 ng
template DNA (EasyTaq® DNA Polymerase, cat. no.
AP111-01, TransGen Biotech). The first-round PCR products were used
as templates for a second round of amplicon enrichment by PCR (94˚C
for 3 min, followed by 24 cycles at 94˚C for 5 sec, 57˚C for 90 sec
and 72˚C for 10 sec, and a final extension at 72˚C for 5 min). In
addition, indexed adapters were added to the ends of the 16S rDNA
amplicons to generate indexed libraries that were ready for
downstream NGS on the MiSeq platform. The quality and concentration
of the library were tested using the Agilent 2100 bioanalyzer
(Agilent Technologies Inc.) and Qubit 2.0 fluorometer (Invitrogen;
Thermo Fisher Scientific, Inc.), respectively. DNA libraries were
multiplexed and loaded on an Illumina MiSeq instrument (Illumina,
Inc.) according to the manufacturer's instructions. Sequencing was
performed using a 2x300 paired-end configuration and the data were
analysed using Miseq Control software (version 2.5.0.5; Illumina,
Inc.). To process the raw data, all the forward and reverse reads
were assembled in pairs, the sequences containing N in the results
were filtered and sequences >200 bp in length were retained.
Subsequently, the spliced and filtered sequences were compared with
Basic Local Alignment Search Tool (BLAST, https://blast.ncbi.nlm.nih.gov/Blast.cgi) match
lengths (against reference sequences in the database)to remove the
chimera sequence (<90% match). In the operational taxonomic unit
(OTU) analysis, the 16S reads were clustered using the VSEARCH
software (version 1.9.6; https://github.com/torognes/vsearch) with a pair-wise
identity cutoff of 97%, and the 16S rRNA reference database was
Silva 132 (https://www.arb-silva.de/search/). Representative
sequences of each OTU were analyzed using the Ribosome Database
Program classifier (version 2.2, http://rdp.cme.msu.edu/classifier/classifier.jsp;jsessionid=D5D6C78C6C197C015E237D0FD7A85246.10.0.0.9).
Bayesian algorithm and the composition of each sample at different
species classification levels was calculated. According to the OTU
analysis results, the random sampling method was used to calculate
the α diversity index of Shannon and Chao1, and the dilution curve
was obtained. Unweighted unifrac analysis was applied to compare
whether there were significant differences in microbial communities
among samples. The Bray-Curtis distance matrix among samples was
used for PCoA (Principal Coordinate Analysis) to indicate β
diversity and the P-value was calculated using a non-parametric
MANOVA. To compare the hierarchical relationships among groups, the
UPGMA (Unweighted Pair Group Method for Arithmetic Mean) clustering
tree was constructed by the non-weighted mean method in
hierarchical clustering. Anosim analysis was confirmed according to
Bray-Curtis. Linear discriminant analysis effect size analysis was
performed by LDA (Linear Discriminant Analysis; huttenhower.sph.harvard.edu/galaxy) to
calculate the impact of species richness on the difference effect
and to identify the species with a significant difference. Based on
the β diversity distance matrix and environmental factor data, the
redundancy analysis was determined.
Statistical analysis
Statistical analyses were performed using SPSS
software (version 19.0; IBM Corp.). Data are presented as the mean
± SD. Values were obtained from 10 animals per group. For normally
distributed data, comparisons among multiple groups were analysed
by one-way ANOVA followed by Tukey's post hoc test. For
non-normally distributed data, comparisons among multiple groups
were analysed by the Kruskal-Wallis test followed by the
Dunn-Bonferroni post hoc test. Correlations between specific
bacteria and the histological score or IL-12/23 levels were
determined by Pearson correlation analysis. P<0.05 was
considered to indicate a statistically significant difference.
Results
Irisin improves the macroscopic and
histopathological scores of UC model mice
In the UC group, body weight was significantly
decreased from day 4-7 compared with the control group (P<0.01;
Fig. 1A). Furthermore, the
macroscopic (P<0.01; Fig. 1C)
and histological (P<0.01; Fig.
1D and E) scores were
significantly increased in the UC group compared with the control
group. Irisin did not significantly alter the colonic tissues in
normal mice; however, irisin decreased the macroscopic (P<0.05)
and histological (P<0.05) scores in UC model mice. Furthermore,
the spleen weight of the UC group was increased compared with the
control group, and irisin treatment decreased UC-mediated
alterations to the spleen weight; however, the differences were not
statistically significant (Fig.
1B).
Irisin downregulates the number of
CD64+ cells in colonic tissues in UC model mice
Compared with the control group, the number of
CD64+ cells in colonic tissues in the irisin group was
not significantly altered; however, the UC group displayed a
significantly increased number of CD64+ cells compared
with the control group (P<0.01; Fig.
2A and B). CD64 fluorescence
staining was significantly decreased in the irisin-treated UC model
mice compared with untreated UC model mice (P<0.05; Fig. 2A and B).
Irisin reduces plasma IL-12 and IL-23
levels in UC model mice
The levels of IL-12 and IL-23 in the plasma of
normal mice were 893.15±222.76 and 685.67±137.55 pg/ml,
respectively. There was no significant difference in IL-12 and
IL-23 levels between the control and irisin (911.63±129.57 and
698.69±163.15 pg/ml, respectively) groups. However, the levels of
IL-12 (1,147.37±233.92; P<0.05) and IL-23 (1,137.27±156.49
pg/ml; P<0.01) in the UC group were significantly increased
compared with the control group. Irisin significantly reduced
UC-mediated upregulation of IL-12 (911.04±122.78 pg/ml; P<0.05).
In addition, irisin decreased UC-mediated upregulation of IL-23
levels, but the decrease was not significant (Fig. 3). Collectively, the results
suggested that irisin displayed an anti-inflammatory effect in UC
model mice.
Alterations to the gut microbiota
structure of UC model mice following treatment with irisin
A total of 6,064,158 reads and 1,516,039,500 base
pairs were analysed in the four groups. Quality filtering on joined
sequences was performed, and sequences that did not fulfil the
criteria were discarded. Finally, a total of 2,409,318 reads with
an average length of 450.78 base pairs were obtained. The
multivariate analysis of variance of the PCoA matrix scores
indicated that primary coordinate 1 and primary coordinate 2
accounted for 22.94 and 9.84% of the total structural changes,
respectively (Fig. 4A). The
intestinal flora in control and irisin groups were significantly
different from that in UC and UC + irisin groups, which indicated
that changes in flora structure play an important role in the
pathogenesis of UC. In accordance with UPGMA clustering, the four
groups were divided into four different clustering trees (Fig. 4B). The dilution curve analysis
indicated that the curves of each group reached a plateau,
suggesting that the majority of the microbiota were identified
(Fig. 4C). According to the Shannon
and Chao1 indices (Fig. 4D and
E), the majority of the diversity
was identified in all samples. The results indicated that there was
a decrease in diversity in UC group compared with control group
(P<0.01) for both the Shannon and Chao1 indices. Analysis of the
microbial diversity in the stools of mice with active UC indicated
that the five bacteria that displayed the greatest alterations in
relative abundance were Alloprevotella, Bacteroides,
Lachnospiraceae-UCG-001, Prebotellaceae-UCG-001 and
Rikenellaceae-RCB-gut-group (Fig. S1).
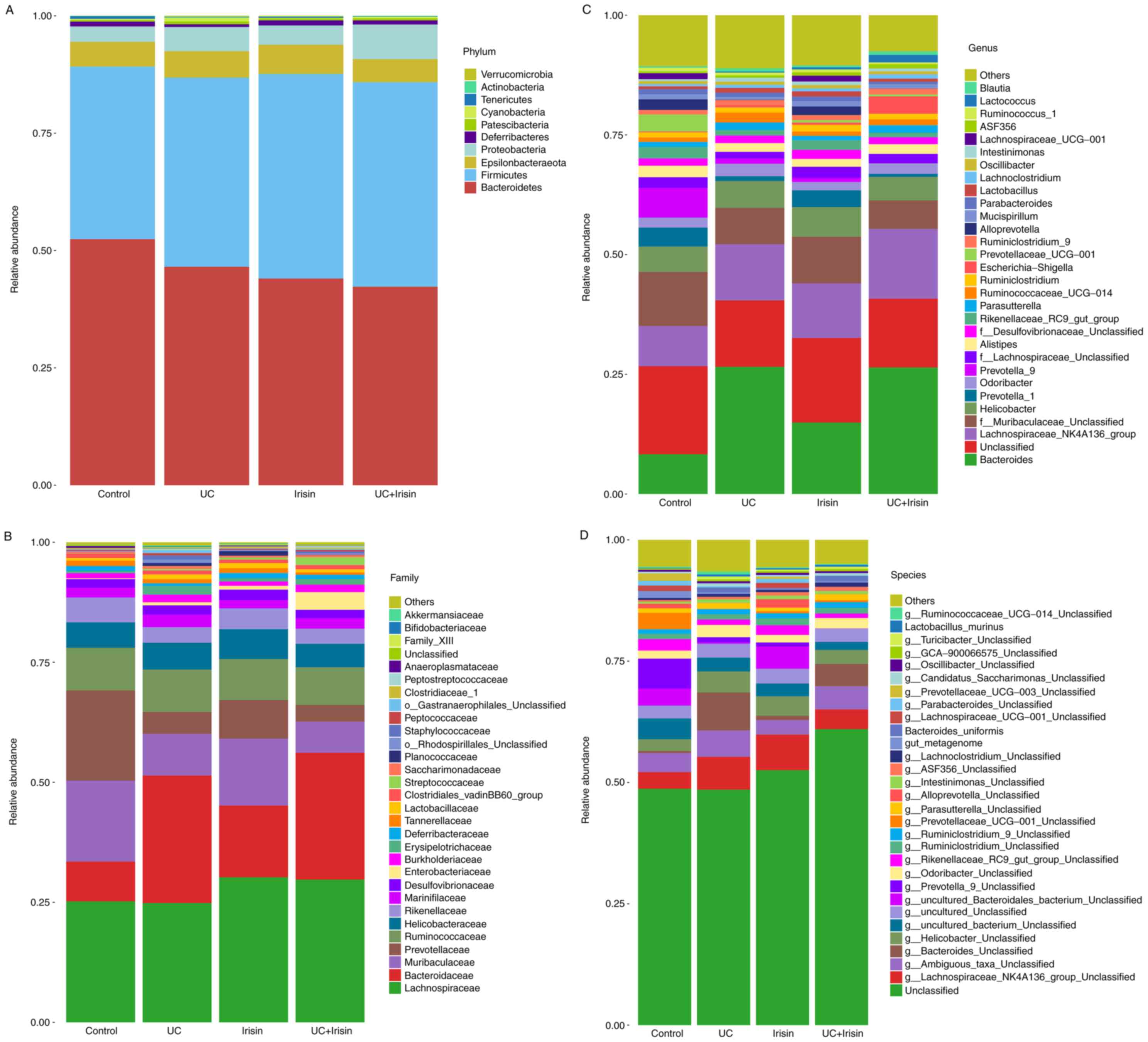
Irisin alters the gut microbiota
composition
The gut microbiota community structure histograms
indicated the microbial species present and their relative
abundance (Fig. 5). All samples
contained five main phyla: Bacteroidetes, Firmicutes,
Epsilonbacteraeota, Proteobacteria and Deferribacteres
(Fig. 5A). The composition of the
microbiota in the control and irisin groups was similar. The
abundance of Deferribacteres in the UC group was lower
compared with the control and irisin groups (0.58 vs. 1.03%,
1.10%), and slightly higher in the UC + irisin group (0.89%)
compared with the UC group. In addition, the abundance of
Cyanobacteria (0.52%) and Patescibacteria (0.62%) in
the UC group was highest among the four groups.
In all samples, 20 main families were identified,
including Lachnospiraceae, Bacteroidaceae, Muribaculaceae,
Prevotellaceae, Deferribacteres and Lactobacillaceae
(Fig. 5B). Following treatment with
irisin, the abundance of Deferribacteres in the UC model
mice was slightly upregulated (from 0.58 to 0.89%); however, the
abundance remained lower compared with the control and irisin
groups (1.03 and 1.09% respectively). In UC group, the relative
abundance of Lactobacillaceae (0.99%),
o-Rhodospirillales-Unclassified (0.67%) and
Staphylococcaceae (0.89%) were all increased compared with
control group. With the treatment of irisin, the abundance of
Lactobacillaceae (0.66%),
o-Rhodospirillales-Unclassified (0.31%) and
Staphylococcaceae (0.40%) were all decreased compared with
UC group.
Sequencing data identified 31 genera of microbial
flora. The abundance of Bacteroides,
Lachnospiraceae-NK4A136-group, f-Muribaculaceae-Unclassified,
Prevotella-1, Prevotella-9, Prevotellaceae-UCG-001 and
Lachnospiraceae-UCG-001 was changed most (>1% of total
composition in all groups). The abundance of Bacteroides
(8.29 vs. 26.57%) and Lachnospiraceae-NK4A136-group (8.38
vs. 11.71%) was higher in the UC group than in the control group.
By contrast, the abundance of f-Muribaculaceae-Unclassified
(11.29 vs. 7.59%), Prevotella-1 (3.99 vs. 1.00%),
Prevotella-9 (3.88% vs. 0.00%),
Prevotellaceae-UCG-001 (3.55 vs. 0.00%) and
Lachnospiraceae-UCG-001 (1.25 vs. 0.00%) was decreased.
Following treatment with irisin, the abundance of
Ruminococcaceae-UCG-014 decreased from 2.05 to 1.25%, which
was similar to the control group 0.93%. Lactobaillus
decreased from 0.99 to 0.66%, which was similar to the control
group 0.57% (Fig. 5C).
Furthermore, 31 main species were identified in all
four groups. For example, the relative abundance of
g-Bacteroides-Unclassified was higher in the UC group
compared with the control and irisin groups (7.82 vs. 0.38%,
0.90%). However, the relative abundance of
g-Bacteroides-Unclassified was decreased in the UC + irisin
group compared with the UC group (4.62 vs. 7.82%; Fig. 5D).
Heatmap at different taxonomic
levels
At the phylum level, Deferribacteres was
present at a lower level in the UC group compared with the other
three groups. By contrast, the abundance of Patescibacteria
and Cyanobacteria was higher in the UC group compared with
the control group (Fig. 6A). At the
class level, the abundance of Erysipelotrichia was increased
in the UC group compared with the control and irisin groups, and
slightly decreased in the UC + irisin group compared with the UC
group (Fig. 6B).
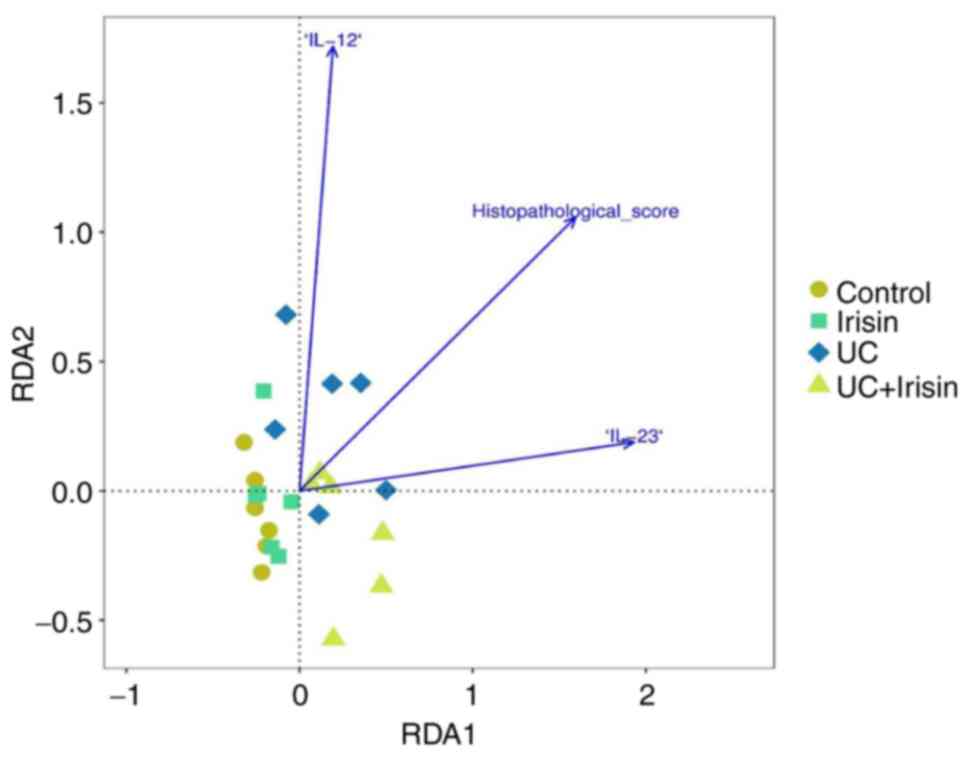
Correlation analysis between
alterations to the gut flora and the histopathological score, as
well as the plasma levels of IL-12 and IL-23 in the four
groups
A positive association between the histopathological
score and IL-12/23 levels was identified (Fig. 7). Compared with the other three
groups, the structure of the gut flora in the UC group was more
closely related to histopathological score and IL-12/23 levels. In
addition, IL-12 served a more important role in the correlation
compared with IL-23. Furthermore, the correlation between the top
five most variable flora (Fig. S1)
in the UC group and IL-12 and IL-23 levels, and histopathological
score were analysed (Table SIII).
Bactoroides was positively correlated with the
histopathological score (P=0.001; R=0.977) and IL-23 levels
(P=0.008; R=0.924). The other three types of microbiota,
Alloprevotella (P=0.001; R=-0.943),
Lachnospiraceae-UCG-001 (P=0.000; R=-0.973) and
Rikenollaceae-RC8-gut-group (P=0.001; R=-0.971), were
negatively correlated with the histopathological score.
Lachnospiraceae-UCG-001 (P=0.01; R=-0.873) and
Rikenollaceae-RC8-gut-group (P=0.049; R=-0.814) were also
negatively correlated with IL-23 levels.
Analysis of the gut flora structure in
the four groups
The composition of the gut flora was significantly
different among the four groups (P=0.001; R=0.515; Fig. 8). The LDA scores indicated that the
g-Ruminococcaceae-UCG-014 and g-Family-XIII-UCG-001
played important roles in the UC and irisin groups, respectively
(Fig. 9). In the control group, a
total of 12 different types of microorganisms (LDA score >3.0)
served a vital role in the gut flora structure. In particular,
f-Prevotellaceae displayed the most significant role
(Fig. 9). But the UC + irisin group
had no dominant bacteria.
Discussion
UC is a chronic non-specific inflammatory bowel
disease with a high recurrence rate, for which the current
therapies are largely ineffective (24). Previous studies have indicated the
environment, autoimmunity, psychology and genetic inheritance are
closely related to the occurrence of UC (25,26).
Therefore, UC is not a disease induced by a single factor. Previous
studies have also suggested that the interaction between disordered
intestinal flora and an abnormal immune response may lead to
intestinal mucosal dysfunction (27). Moreover, dysfunction of the
intestinal mucosal barrier may serve as a trigger for UC and is
associated with the incidence of disease (28).
Intestinal microorganisms consist of commensal
bacteria, probiotics and pathogenic bacteria (29). Under physiological conditions, all
types of microorganisms maintain symbiotic or antagonistic
relationships in the intestine to form a dynamic and balanced
micro-ecological system (30).
Healthy intestinal flora can protect the intestine, improve the
metabolism, regulate immunity, and help resist inflammation and
tumour growth (31,32). Previous studies have reported that
exercise optimizes the composition of the intestinal flora and
improves intestinal microecology (33,34).
Sufficient exercise, for example, 30 min of swimming a day, may
amplify the antibacterial effect of the intestine and relieve
intestinal barrier dysfunction induced by chronic inflammation
(35). A previous study revealed
that exercise increased the diversity of the intestinal microbiota
in professional rugby players and simultaneously reduced the level
of inflammatory markers (36). The
aforementioned studies indicated that the beneficial effect of
exercise is partly due to the increased intestinal microbial
diversity that occurs as a result of exercise. Additionally, Allen
et al (37) revealed that
voluntary physical activity alters the intestinal microbes present
in mice; however, the mechanisms underlying how movement can alter
the flora of the inflammatory intestine are not completely
understood.
In the present study, the symptoms of the UC model
mice were consistent with the clinical symptoms of patients with
UC, and the macroscopic and histological scores of UC model mice
were increased compared with the control mice. The relative
abundance of Bacteroides and
Lachnospiraceae-NK4A136-group were increased in UC model
mice. The levels of inflammatory factors in the plasma decreased in
UC model mice following the intraperitoneal injection of irisin,
and UC symptoms were also alleviated. Alterations to the relative
abundance of Ruminococcaceae-UCG-014 and Lactobaillus
in irisin treated mice were identified by sequencing. Furthermore,
a previous study reported that the symptoms of DSS-induced UC in
mice were improved following HuangQin treatment, and the abundance
of Deferribacteres in the microflora, which was reduced by
UC, was increased following treatment (38). Similarly, in the present study, the
abundance of Deferribacteres in the UC + irisin group was
higher compared with the UC group, and the symptoms of the UC model
mice were alleviated by irisin treatment. Therefore, irisin may
exert its anti-inflammatory effects by inducing alterations in the
intestinal flora; however, the mechanism by which irisin affects
intestinal microbiota needs further investigation.
Intestinal microbiota belong to a diverse anaerobic
species and extraction may be challenging (39). Consequently, the mechanism
underlying intestinal microbial alterations in UC is not completely
understood. Previous studies have indicated that certain
microorganisms produce a large number of metabolites that serve as
a major source of intestinal antigens, resulting in an IgA-based
antibody response (40,41). Frehn et al (42) observed a decrease in E.
coli-specific antibody IgA expression in the faeces of patients
with UC. In the present study, high-throughput sequencing
technology was used to analyse the composition and alterations to
the intestinal flora in different groups more accurately and
thoroughly compared with the detection of IgA antibody expression.
Furthermore, quantitative comparisons were made at different
taxonomic levels.
A number of studies have indicated that the
occurrence of UC is not only related to intestinal microbiota
alterations but is also related to alterations in the microbiota of
the intestinal mucosal (27,43).
The microbiota in the external and internal intestinal mucosal
layers of patients with UC are significantly different compared
with controls. Moreover, the compositional structure of the
microbiota in the external mucosal layer, in which
Clostridium is predominant, is significantly different
compared with controls (44).
Therefore, intestinal flora alterations may serve an important role
during the progression of UC.
Due to the differences in UC course, stages,
treatments and analytical methods in each patient, as well as the
complex structure and function of the intestinal flora, at present,
there is no consensus regarding alterations in intestinal aerobic
bacteria and common anaerobic bacteria in patients with UC. As a
result, the specific roles of bacteria in the progression of the
disease have not yet been fully elucidated, and alterations in the
gut flora and their relationship with pathological changes in the
intestinal mucosa remain unclear. To date, the majority of studies
have primarily focused on the relationship between a certain type
of intestinal flora and UC; however, a comprehensive comparison
between common intestinal flora and UC has not been performed. In
the present study, a relatively thorough analysis of the intestinal
microbiota in mice in each of the four groups was conducted.
To the best of our knowledge, a consensus regarding
the anti-inflammatory mechanism underlying irisin has not been
reached. Studies have indicated that irisin inhibits the
inflammatory response and reduces pathological alterations during
the progression of acute lung injury in a concentration-dependent
manner (45,46). Irisin may reduce the expression of
NF-κB (p65) in the nucleus and increase its expression level in the
cytosol by inhibiting apoptosis (17). Mazur-Bialy et al (16) reported that lower concentrations of
irisin can not only enhance the expression of TLR4 in macrophages,
but can also promote the ability of macrophages to recognize
potential pathogens, indirectly increasing anti-inflammatory
effects. Furthermore, irisin also significantly reduces the levels
of TLR4 and MyD88 protein in macrophages, as well as the
phosphorylation of NF-κB, leading to a decreased release of key
pro-inflammatory cytokines, which results in an anti-inflammatory
effect. Another study also reported that the anti-inflammatory
effect of irisin may be due an improvement in the levels of TNF-α
and TNF superfamily member 11, two factors that drive lymph node
hyperplasia (47). In the present
study, the macroscopic and histopathological scores, the abundance
of CD64+ cells and the level of proinflammatory
cytokines in the plasma, such as IL-12 and IL-23, were decreased in
irisin-treated UC model mice compared with untreated UC model
mice.
IL-12 is a heterodimer molecule composed of p40 and
p35 subunits connected by disulfide bonds. Under physiological
conditions, IL-12 is produced by mature dendritic cells and
activated monocyte macrophages and provides important signals that
promote the differentiation and proliferation of T helper (Th)1
cells, thereby participating in the inflammatory response (48). IL-23 is formed by p40 and p19
subunit chains and serves a key role in the differentiation and
expansion of Th cells and resident lymphocytes (49,50). A
previous study indicated that the expression level of IL-12 mRNA in
UC is significantly increased compared with normal colon tissue and
may serve an important role in the pathogenesis of UC (51). Another study reported that the p40
chain serves an important role in intestinal inflammation (52). Genome-wide association studies have
indicated that mutations in the gene encoding the IL-23 receptor
and the site encoding the p40 gene are genetic risks for the
development of UC (53,54). Monoclonal antibodies against the p40
chain blocked the activity of IL-12 and IL-23 and have displayed
initial success for the treatment of UC (55). In the present study, the plasma
levels of IL-12 and IL-23 in mice were significantly increased in
UC model mice compared with control mice. The macroscopic score and
histopathological score of UC model mice were significantly
decreased by irisin treatment. Furthermore, the number of
CD64+ cells, and the plasma levels of IL-12 and IL-23
were also decreased in UC model mice following treatment with
irisin.
During the present study, three mice died. The first
death occurred in the UC group on day 5 of the experiment, and on
day 6, two further deaths were recorded (one in the UC group and
one in the UC + Irisin group). According to previous study
(56), due to the mucosal damage
and anticoagulation effects of DSS, the modest initial effects are
followed by increasingly worsening symptoms, including increased
intestinal permeability, severe bleeding and mortality. DSS-induced
mortality can be variable dependent on the concentration and
duration of treatment (57).
Shimizu et al (58) reported
that in a 4% DSS model, 2 out of 10 rats died before the day 7 of
DSS administration, which was similar to the mortality rate of the
present study. In addition, Axelsson et al (59) demonstrated that neither T, B nor NK
cells were critical for the induction of DSS-induced colitis in
mice. Administration of 5% DSS to athymic nu/nu CD-I(BR) mice
resulted in a rapid decline in body weight and a short survival
time, with no animals surviving beyond day 9. Furthermore, the
administration of 5% DSS in SCID mice resulted in a 50% survival
rate at day 9 of DSS administration. In order to minimize animal
mortality in our preliminary experiments, an appropriate DSS load
for the reliable induction of colitis and comparable severity of
disease between animals was established according to the method
conducted by Vowinkel et al (60). The results of the preliminary
experiments indicated that the protocol that was used in the
present study led to lowest morality rate, but still represented a
classic UC model with obvious body weight loss and histological
damage. The death of the three mice in the present study was caused
by severe diarrhea, severe blood in the stool, dehydration,
continuous weight loss and damage to the liver coagulation
mechanism; however, the detailed mechanism underlying the death
observed in the present study requires further investigation.
The present study demonstrated the anti-inflammatory
effects of irisin, as well as the accompanied alterations to the
intestinal microbiome. In addition, a possible preliminary
relationship between the anti-inflammatory effect of irisin and
alterations to the intestinal flora was identified. Future studies
using specific order or gene knockout mice should be conducted to
support the results of the present study. A limitation of the
present study is that the relationship between alterations to the
intestinal microbiota and inflammation is controversial. For
example, colonic infusion of donor human intestinal flora can
reverse UC in selected patients, which suggests that differences in
the gut microbiota may be the cause of milder inflammation
(61). However, by investigating
the correlation between fecal flora alterations and inflammatory
indicators, other studies have indicated that alterations to the
gut microbiota may be a consequence of inflammation (62). Microbiota transferring artificial
colonization with divergent species or other specific gnotobiotic
techniques is a key strategy that could be used to explain the
relationship between alterations to the intestinal microbiota and
intestinal inflammation. Furthermore, only the anti-inflammatory
effect of irisin was investigated in the present study; therefore,
comparing irisin with other clinically used drugs requires further
investigation.
In conclusion, the present study suggested that
irisin ameliorated gut inflammation potentially by altering the gut
microbiota in UC model mice. Therefore, irisin may serve as a novel
therapeutic agent for UC.
Supplementary Material
Analysis of the difference in the
content of flora in UC group and normal group.
Disease activity index.
Pathologic score evaluation.
Correlation between specific bacteria
and histopathological score, IL-12 and IL-23 levels.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National Natural Science
Foundation of China (grant nos. 81500430 and U1304802 to XHL), the
Key Project of Science and Technology Research of the Education
Department of Henan (grant nos. 17A320019 to HCW), the Henan
Science and Technology Planning Project (grant nos. 182102310544
and 192102310045 to XHL; grant no. 182102310566 to HCW; grant no.
182102310567 to RLY) and the Henan Medical Science and Technology
Tackling Project (grant no. 201702136 to ZFD).
Availability of data and materials
The datasets used and analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
LXH and XTC performed the histopathological
examination and immunofluorescence experiment, and contributed
equally to this paper. XHL designed the research study. RLY
conducted IL-12 and IL-18 measurement, and the correlation
analysis. RLY and LXH confirmed the authenticity of the raw data.
JNY and HCW helped to interpret and analyze the data. ZFD and YXL
conducted high throughput sequencing experiments. XTC and XHL
revised the work critically for important intellectual content. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
The animal experiments were approved by the Medical
Ethics Committee of Henan University (approval no.
HUSOM2020-103).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Anzai H, Hata K, Kishikawa J, Ishii H,
Yasuda K, Otani K, Nishikawa T, Tanaka T, Kiyomatsu T, Kawai K, et
al: Appendiceal orifice inflammation is associated with proximal
extension of disease in patients with ulcerative colitis.
Colorectal Dis. 18:0278–0282. 2016.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Podolsky DK: Inflammatory bowel disease. N
Engl J Med. 347:417–429. 2002.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Grinspan A and Kornbluth A: Positioning
therapy for ulcerative colitis. Curr Gastroenterol.
17(29)2015.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Khor B, Gardet A and Xavier RJ: Genetics
and pathogenesis of inflammatory bowel disease. Nature.
474:307–317. 2011.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Legaki E and Gazouli M: Influence of
environmental factors in the development of inflammatory bowel
diseases. World J Gastrointest Pharmacol Ther. 7:112–125.
2016.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Frank DN, St Amand AL, Feldman RA,
Boedeker EC, Harpaz N and Pace NR: Molecular-phylogenetic
characterization of microbial community imbalances in human
inflammatory bowel diseases. Proc Natl Acad Sci USA.
104:13780–13785. 2007.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Ihara S, Hirata Y and Koike K: TGF-β in
inflammatory bowel disease: A key regulator of immune cells,
epithelium, and the intestinal microbiota. J Gastroenterol.
52:777–787. 2017.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Manichanh C, Rigottier-Gois L, Bonnaud E,
Gloux K, Pelletier E, Frangeul L, Nalin R, Jarrin C, Chardon P,
Marteau P, et al: Reduced diversity of faecal microbiota in Crohn's
disease revealed by a metagenomic approach. Gut. 55:205–211.
2006.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Schwab C, Berry D, Rauch I, Rennisch I,
Ramesmayer J, Hainzl E, Heider S, Decker T, Kenner L, Muller M, et
al: Longitudinal study of murine microbiota activity and
interactions with the host during acute inflammation and recovery.
ISME J. 8:1101–1114. 2014.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Boström P, Wu J, Jedrychowski MP, Korde A,
Ye L, Lo JC, Rasbach KA, Boström EA, Choi JH, Long JZ, et al: A
PGC1-alphadependent myokine that drives brown-fat-like development
of white fat and thermogenesis. Nature. 481:463–468.
2012.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Yi Z, Yi S and Shu-Zhe D: The novel
Myokine-Irisin. Chin J Biochem Mol Biol. 33:429–435. 2017.
|
|
12
|
Farmer SR: Boning up on Irisin. N Engl J
Med. 380:1480–1482. 2019.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Mahgoub MO, D'Souza C, Al Darmaki RSMH,
Baniyas MMYH and Adeghate E: An update on the role of irisin in the
regulation of endocrine and metabolic functions. Peptides.
104:15–23. 2018.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Buscemi S, Corleo D, Vasto S, Buscemi C,
Barile AM, Rosafio G, Randazzo C, Currenti W and Galvano F: Serum
Irisin concentrations in severely inflamed patients. Horm Metab
Res. 52:246–250. 2020.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Mazur-Bialy AI: Superiority of the
Non-glycosylated form over the glycosylated form of Irisin in the
attenuation of adipocytic meta-inflammation: A potential factor in
the fight against insulin resistance. Biomolecules.
9(394)2019.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Mazur-Bialy AI, Pocheć E and Zarawski M:
Anti-inflammatory properties of Irisin, mediator of physical
activity, are connected with TLR4/MyD88 signaling pathway
activation. Int J Mol Sci. 18:701–712. 2017.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Shao L, Meng D, Yang F, Haibo S and Dongqi
T: Irisin-mediated protective effect on LPS-induced acute lung
injury via suppressing inflammation and apoptosis of alveolar
epithelial cells. Biochem Biophys Res Commun. 487:194–200.
2017.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Mazur-Bialy AI, Bilski J, Wojcik D,
Brzozowski B, Surmiak M, Hubalewska-Mazgaj M, Chmura A, Magierowski
M, Magierowska K, Mach T and Brzozowski T: Beneficial effect of
voluntary exercise on experimental colitis in mice fed a High-Fat
Diet: The role of Irisin, adiponectin and proinflammatory
biomarkers. Nutrients. 9(410)2017.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Asadi Y, Gorjipour F, Behrouzifar S and
Vakili A: Irisin peptide protects brain against ischemic injury
through reducing apoptosis and enhancing BDNF in a rodent model of
stroke. Neurochem Res. 43:1549–1560. 2018.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Pitts M: Applied Research Ethics National
Association/Office of Laboratory Animal Welfare. Institutional
Animal Care and Use Committee. A Guide to the New ARENA/OLAW IACUC
Guidebook. Lab Anim (NY). 31:40–42. 2002.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Siegmund B, Lehr HA, Fantuzzi G and
Dinarello CA: IL-1β-converting enzyme (caspase-1) in intestinal
inflammation. Proc Natl Acad Sci USA. 98:13249–13254.
2001.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Murano M, Maemura K, Hirata I, Toshina K,
Nishikawa T, Hamamoto N, Sasaki S, Saitoh O and Katsu K:
Therapeutic effect of intracolonically administered nuclear factor
kappa B (p65) antisense oligonucleotide on mouse dextran sulphate
sodium (DSS)-induced colitis. Clin Exp Immunol. 120:51–58.
2000.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Jumpstart Consortium Human Microbiome
Project Data Generation Working Group. Evaluation of 16S rDNA-based
community profiling for human microbiome research. PLoS One.
7(e39315)2012.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Ng SC, Shi HY, Hamidi N, Underwood FE,
Tang W, Benchimol EI, Panaccione R, Ghosh S, Wu JCY, Chan FKL, et
al: Worldwide incidence and prevalence of inflammatory bowel
disease in the 21st century: A systematic review of
population-based studies. Lancet. 390:2769–2778. 2018.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Porter RJ, Kalla R and Ho GT: Ulcerative
colitis: Recent advances in the understanding of disease
pathogenesis. F1000Res. 9(F1000)2020.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Colquhoun C, Duncan M and Grant G:
Inflammatory bowel diseases: Host-microbial-environmental
interactions in dysbiosis. Diseases. 8(E13)2020.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Kaser A, Zeissig S and Blumberg RS:
Inflammatory bowel disease. Annu Rev Immunol. 28:573–621.
2010.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Martini E, Krug SM, Siegmund B, Neurath MF
and Becker C: Mend your fences: The epithelial barrier and its
relationship with mucosal immunity in inflammatory bowel disease.
Cell Mol Gastroenterol Hepatol. 4:33–46. 2017.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Qin J, Li R, Raes J, Arumugam M, Burgdorf
KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, et al: A
human gut microbial gene catalogue established by metagenomic
sequencing. Nature. 464:59–65. 2010.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Etienne-Mesmin L, Chassaing B, Desvaux M,
De Paepe K, Gresse R, Sauvaitre T, Forano E, de Wiele TV, Schüller
S, Juge N and Blanquet-Diot S: Experimental models to study
intestinal microbes-mucus interactions in health and disease. FEMS
Microbiol Rev. 43:457–489. 2019.PubMed/NCBI View Article : Google Scholar
|
|
31
|
O'Callaghan AA and Corr SC: Establishing
boundaries: The relationship that exists between intestinal
epithelial cells and gut-dwelling bacteria. Microorganisms 7:
Efficiency of cancer therapy. Front Microbiol.
10(1050)2019.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Ma W, Mao Q, Xia W, Dong G, Yu C and Jiang
F: Gut microbiota shapes the efficiency of cancer therapy. Front
Microbiol. 10(1050)2019.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Mika A, Van Treuren W, González A, Herrera
JJ, Knight R and Fleshner M: Exercise is more effective at altering
gut microbial composition and producing stable changes in lean mass
in juvenile versus adult male F344 rats. PLoS One.
10(e0125889)2015.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Mohr AE, Jäger R, Carpenter KC, Kerksick
CM, Purpura M, Townsend JR, West NP, Black K, Gleeson M, Pyne DB,
et al: The athletic gut microbiota. J Int Soc Sports Nutr.
17(24)2020.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Luo B, Xiang D, Nieman DC and Chen P: The
effects of moderate exercise on chronic stress-induced intestinal
barrier dysfunction and antimicrobial defense. Brain Behav
Immunity. 39:99–106. 2014.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Clarke SF, Murphy EF, O'Sullivan O, Lucey
AJ, Humphreys M, Hogan A, Hayes P, O'Reilly M, Jeffery IB,
Wood-Martin R, et al: Exercise and associated dietary extremes
impact on gut microbial diversity. Gut. 63:1913–1920.
2014.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Allen JM, Berg Miller ME, Pence BD,
Whitlock K, Nehra V, Gaskins HR, White BA, Fryer JD and Woods JA:
Voluntary and forced exercise differentially alters the gut
microbiome in C57BL/6J mice. J Appl Physiol. 118:1059–1066.
2015.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Yang Y, Gang C, Qian Y, Juan Y, Xueting C,
Pamo T, Xiaolan C, Chunping H, Shuangquan Z and Peng C: Gut
microbiota drives the attenuation of dextran sulphate
sodium-induced colitis by Huangqin decoction. Oncotarget.
8:48863–48874. 2017.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Liu JH, Chen ZL, Li Y and Yu DJ:
Development of denaturant gradient gel electrophoresis for analysis
of intestinal microflora community diversity. Chin J Veterinary Sci
Technol. 35:2006.
|
|
40
|
Donaldson GP, Ladinsky MS, Yu KB, Sanders
JG, Yoo BB, Chou WC, Conner ME, Earl AM, Knight R, Bjorkman PJ and
Mazmanian SK: Gut microbiota utilize immunoglobulin A for mucosal
colonization. Science. 360:795–800. 2018.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Moor K, Diard M, Sellin ME, Felmy B,
Wotzka SY, Toska A, Bakkeren E, Arnoldini M, Bansept F, Co AD, et
al: High-avidity IgA protects the intestine by enchaining growing
bacteria. Nature. 544:498–502. 2017.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Frehn L, Jansen A, Bennek E, Mandic AD,
Temizel I, Tischendorf S, Verdier J, Tacke F, Streetz K, Trautwein
C and Sellge G: Distinct patterns of IgG and IgA against food and
microbial antigens in serum and feces of patients with inflammatory
bowel diseases. PLoS One. 9(e106750)2014.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Kurashima Y and Kiyono H: Mucosal
ecological network of epithelium and immune cells for gut
homeostasis and tissue healing. Annu Rev Immunol. 35:119–147.
2017.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Pei LY, Ke YS, Zhao HH, Wang L, Jia C, Liu
WZ, Fu QH, Shi MN, Cui J and Li SC: Role of colonic microbiota in
the pathogenesis of ulcerative colitis. BMC Gastroenterol.
19(10)2019.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Li X, Jamal M, Guo P, Jin Z, Zheng F, Song
X, Zhan J and Wu H: Irisin alleviates pulmonary epithelial barrier
dysfunction in sepsis-induced acute lung injury via activation of
AMPK/SIRT1 pathways. Biomed Pharmacother.
118(109363)2019.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Chen K, Xu Z, Liu Y, Wang Z, Li Y, Xu X,
Chen C, Xia T, Liao Q, Yao Y, et al: Irisin protects mitochondria
function during pulmonary ischemia/reperfusion injury. Sci Transl
Med. 9(eaao6298)2017.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Narayanan SA, Metzger CE, Bloomfield SA
and Zawieja DC: Inflammation-induced lymphatic architecture and
bone turnover changes are ameliorated by Irisin treatment in
chronic inflammatory bowel disease. FASEB J. 32:4848–4861.
2018.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Murphy KM, Ouyang W and Farrar JD:
Signaling and transcription in T helper development. Annu Rev
Immunol. 18:451–494. 2000.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Fuss IJ, Becker C, Yang Z, Groden C,
Hornung RL and Heller F: Both IL-12p70 and IL-23 are synthesized
during active Crohn's disease and are down-regulated by treatment
with anti-IL-12 p40 monoclonal antibody. Inlamm Bowel Dis. 12:9–15.
2006.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Grivennikov DI, Wang K, Mucida D, Wtewart
C, Schnabl B and Jauch D: Adenoma-linked barrier defects and
microbial products drive IL-23/IL-17-mediated tumor growth. Nature.
491:254–258. 2012.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Wang BY, Liu FL, Hu Z, Yu CH, Xia XT, Fan
JY and Li X: Effects of Tongxieyaofang on serum levels of
interleukin-12 and neuropeptide Y in rats with ulcerative colitis.
Hun J Traditional Chin Med. 32:166–167. 2016.
|
|
52
|
Fiorino G, Allocca M, Correale C, Roda G,
Furfaro F, Loy L, Zilli A, Peyrin-Biroulet L and Danese S:
Positioning ustekinumab in moderate-to-severe ulcerative colitis:
New kid on the block. Expert Opin Biol Ther. 20:421–427.
2020.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Beaudoin M, Goyette P, Boucher G, Lo KS,
Rivas MA, Stevens C, Alikashani A, Ladouceur M, Ellinghaus D,
Törkvist L, et al: Deep resequencing of GWAS loci identifies rare
variants in CARD9, IL23R and RNF186 that are associated with
ulcerative colitis. PLoS Genet. 9(e1003723)2013.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Bianchi E and Rogge L: The IL-23/IL-17
pathway in human chronic inflammatory diseases-new insight from
genetics and targeted therapies. Microbes Infect. 21:246–253.
2019.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Niederreiter L, Adolph TE and Kaser A:
Anti-IL-12/23 in Crohn's disease: Bench and bedside. Curr Drug
Targets. 14:1379–1384. 2013.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Eichele DD and Kharbanda KK: Dextran
sodium sulfate colitis murine model: An indispensable tool for
advancing our understanding of inflammatory bowel diseases
pathogenesis. World J Gastroenterol. 23:6016–6029. 2017.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Chassaing B, Aitken JD, Malleshappa M and
Vijay-Kumar M: Dextran sulfate sodium (DSS)-induced colitis in
mice. Curr Protoc Immunol. 104:15.25.1–15.25.14. 2014.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Shimizu T, Suzuki M, Fujimura J, Hisada K,
Yoshikazu O, Obinata K and Yamashiro Y: The relationship between
the concentration of dextran sodium sulfate and the degree of
induced experimental colitis in weanling rats. Pediatr
Gastroenterol Nutr. 37:481–486. 2003.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Axelsson LG, Landström E, Goldschmidt TJ,
Grönberg A and Bylund-Fellenius AC: Dextran sulfate sodium (DSS)
induced experimental colitis in immunodeficient mice: Effects in
CD4(+) -cell depleted, athymic and NK-cell depleted SCID mice.
Inflamm Res. 45:181–191. 1996.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Vowinkel T, Kalogeris TJ, Mori M,
Krieglstein CF and Granger DN: Impact of dextran sulfate sodium
load on the severity of inflammation in experimental colitis. Dig
Dis Sci. 49:556–564. 2004.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Borody TJ, Warren EF, Leis S, Surace R and
Ashman O: Treatment of ulcerative colitis using fecal
bacteriotherapy. Clin Gastroenterol. 37:42–47. 2003.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Zhang T, Chen Y, Wang Z, Zhou Y, Zhang S,
Wang P, Xie S and Jiang B: Changes of fecal flora and its
correlation with inflammatory indicators in patients with
inflammatory bowel disease. Nan Fang Yi Ke Da Xue Xue Bao.
33:1474–1477. 2013.PubMed/NCBI(In Chinese).
|