Introduction
Nasopharyngeal carcinoma (NC) is a type of cancer
that arises from the nasopharynx epithelium. NC is an uncommon type
of cancer that has a variable geographical distribution (1). As of 2012, there were >86,500 cases
of NC globally, accounting for 0.6% of all diagnosed cancers
(2,3). Notably, >70% of new cases were
diagnosed in East and Southeast Asia (4). Furthermore, incidences of NC cases are
also common among the population of Southern China and other
Chinese populations in Hong Kong, Singapore and Taiwan, which may
be due to polygenic events and genetic factors (5). Therefore, identification of the genes
causing susceptibility to NC may aid in the identification of
individual communities and populations that have an increased risk
of developing NC. This could lead to further understanding of the
pathogenesis and malignancy of NC (6,7).
Previous linkage analysis has revealed several chromosomal regions
that contain various genes susceptible to NC (8). Studies from China have reported the
susceptibility of chromosome regions 6p22, 4p15.1-q12 and
3p21.31-21.2, and their involvement in human leukocyte antigens and
pathogenesis in NC (9,10). Despite advances in cancer research,
the genomic alleles that are responsible for cancer susceptibility
are not fully understood (11). In
the present study, it was investigated whether functional
polymorphisms in short palate lung and nasal epithelium clone 1
(SPLUNC1) and its interaction with myelodysplasia syndrome 1 (MDS1)
may have an influence on the risk or severity of nasopharyngeal
carcinoma among the Chinese population. The SPLUNC1 gene encodes a
secreted protein that is present at the surface of the
nasopharyngeal epithelium, which acts as an innate immunity
defensive molecule and has previously been identified to be a
potent risk factor for NC (12,13).
SPLUNC1 also acts as a potent tumor suppressor gene, which
increases the susceptibility to NC, however, its underlying
mechanism is not fully understood (14). MDS1-ectopic viral integration site 1
(EVI1) is encoded by the MDS1 and EVI1 Complex Locus (MECOM) gene
and is comprised of three different proteins: EVI1, MDS1 and
MDS1-EVI1(15). EVI1 is a
transcription factor that has the ability to suppress TGF-β,
thereby promoting tumor growth (16), and the imbalance of EVI1 and
MDS1-EVI1 proteins is considered to be an important factor for the
pathogenesis of NC (17). Indeed,
to the best of our knowledge, the gene-to-gene interaction between
SPLUNC1 and MDS1-EVI1 polymorphisms has not been previously
studied. The significant gene-to-gene interaction impact on other
types of cancer may affect the function of SPLUNC1 and MDS1-EVI1,
which may play a crucial role in the development of NC. Moreover,
these polymorphisms may cause individual vulnerability towards
carcinogenesis. The aim of the present study was to investigate the
relationship between polymorphisms in SPLUNC1 (rs2752903, T>C)
and MDS1-EVI1 (rs6774494, G>A), and the risk of developing NC
among the Chinese population.
Materials and methods
Ethical approval
All procedures were carried out in accordance with
the Declaration of Helsinki 1964 and its later amendments. Written
informed consent was provided by all patients and participants.
Consent was obtained from each subject regarding their personal
information on demographic factors, any history of medical
information, tobacco smoking and alcohol consumption using a
standard questionnaire. All protocols and procedures were approved
by the Hospital Ethical Research Committee of the Jining Medical
University (Jining, Shandong, China; approval no.
JMU/AH/OncDept/2017-34GA2).
Study participants
The study population consisted of 1,059 patients
(mean age, 42.3±0.26 years; age range, 31-72 years) with NC
enrolled at the Affiliated Hospital of Jining Medical University
(Jining, Shandong, China). All patients were of Han Chinese
ethnicity and residents in Jining and the surrounding regions.
Patients were newly diagnosed with NC, which was confirmed by
pathological examination between July 2014 and June 2019, with a
response rate of 94%, where 6% of the patients were either shifted
to other hospital or left the city and were lost during follow-up.
Patients treated with chemotherapy and radiotherapy with other
types of cancer prior to surgery were not included in the study.
Cancer staging was carried out based on the Tumor, Node, Metastasis
(TNM) staging system, according to the American Joint Committee on
Cancer system (AJCC) Cancer Staging Manual, 2009. The staging was
determined by one senior pathologist and one doctor of the hospital
based on pathological examination of the tissues (18). A total of 891 control participants
(mean age, 44.1±0.17 years; age range, 30-65 years) were randomly
selected from the same region within the Jining area from a cancer
screening community program carried out during the same period,
with a response rate of 90%, where and 10% of the patients moved
out of the city and were lost during the follow-up. Control
participants were selected based on those who had no episodes of
any type of cancer, regardless of the age or sex of the
participants. Furthermore, the association between a functional
polymorphism in SPLUNC1 and its interaction with MDS1, which may
influence NC susceptibility, were examined based on age, sex,
smoking and alcohol consumption status and TNM classification.
Polymorphism genotyping
In total, ~5 ml peripheral blood were drawn into a
tube from the study participants. Genomic DNA was then extracted
using the TIANamp Blood DNA kit (cat. no. DP318; Tiangen Biotech
Co., Ltd.) by following the manufacturer's protocol, and the
genotypes examined following a PCR-based protocol. Initially, the
genotyping was carried out without knowing the status of the case
subjects and the control subjects. The genotypes of SPLUNC1
(rs2752903, T>C) were analyzed using the PCR-restriction
fragment length polymorphism method, according to the protocol by
Ara et al (19). The forward
primer, 5'-TTGCCGTCCCAAGCAATGGATGA-3' and reverse primer,
5'-TCTGGGAAGGGACAGAAGATGAC-3', were used to amplify and sequence
the target region containing the T/C site (Qiagen, Inc.). PCR
amplification was carried out by mixing the reaction mixture, which
was comprised of the template DNA, 0.5 µM both the forward and
reverse primers. PCR amplification was carried out with a 25 µl
master mix that contained ~120 ng template DNA, both forward and
reverse primers (~0.5 µM), dNTP (0.4 mM), MgCl2 (1.7 mM)
and 2U Taq polymerase (Promega Corporation). The PCR thermocycling
conditions were as follows: Denaturation at 92˚C for 3 min; 32
cycles of annealing at 93˚C for 35 sec, 55˚C for 25 sec and 72˚C
for 25 sec; and a final elongation step at 72˚C for 5 min.
MDS1-EVI1 (rs6774494, G>A), genotypes were analyzed using
amplification-refractory mutation system-PCR, which is an
economical method for genotyping single-nucleotide polymorphism.
The amplification was carried out using the following primers:
MDS1-EVI1 forward, 5'-TTGAGGCCCGTTTAGATACCA-3' and reverse,
5'-CTTTCCTTGGAGCAATGTAGTT-3' for the MDS1-EVI1 G-allele, and
forward, 5'-ACTATCAGGACACGCC-3' and reverse, 5'-TAACCACCGAATGGTG-3'
for the MDS1-EVI1 A-allele (Qiagen, Inc.). PCR amplification was
carried out by mixing a 15 µl reaction mixture that contained ~20
ng template DNA, 0.5 µM of both the forward and reverse primers,
0.3 mM dNTP, 1.6 mM MgCl2 and 3U Taq polymerase
supplemented with Q-solution (Qiagen, Inc.). The PCR thermocycling
conditions were as follows: Denaturation at 95˚C for 10 min; 32
cycles of annealing at 93˚C for 32 sec, 57˚C for 25 sec and 72˚C
for 30 sec; and a final elongation step at 72˚C for 8 min.
Statistical analysis
The genotype frequencies and the distribution
between the patient and control groups were compared by departures
from Hardy-Weinberg equilibrium by gene counting and tested using a
two-sided χ2 test (20).
The parameters which constitute the departure from Hardy-Weinberg
include allele frequency and number of genotypes. Smokers were
defined as participants who smoked up to 1 year before the date of
a cancer diagnosis for patients, or the date of interview for
controls. Alcohol drinkers were defined as participants who drank
alcohol ≥ once per week for >6 months. Unconditional logistic
regression analysis was used for calculating the odds ratio (OR)
and 95% CI to estimate the association between the SPLUNC1 and
MDS1-EVI1 polymorphisms and the risk of developing NC. Gene-gene
interaction analysis was tested for its null hypotheses from the
multiplicative joint effect models based on the logistic regression
model. χ2 test was used to estimate the correlation
between the genotypes and their clinical parameters. P-values, ORs
and 95% CIs were calculated and adjusted for sex, age, alcohol and
tobacco use where appropriate. All statistical tests were two-sided
and P<0.05 was used as the criterion of statistical
significance. All analyses were performed using SPSS version 21
(IBM Corp.).
Results
In the present study, the effect of SPLUNC1 CC and
MDS1-EVI1 AA genotypes on the susceptibility of NC in the
investigated province, comprised of 1,059 patients and 891
controls, was investigated. As demonstrated in Table I, the NC and controls groups were
comparable in terms of the sex composition (P=0.43), they differed
significantly in comparing both age groups (P=0.014). The younger
age group (>40) has a lower number of NC patients compared with
the control (45.61 vs. 51.18%), where whereas the older age group
(≤40) has a higher number of NC patients compared with the control
(54.39 vs. 48.82%). The number of patients with NC was almost
comparable to the controls with respect to those cases who have a
history of smoking. However, for patients with NC, there was a
higher number of alcohol drinkers compared with the controls (43.72
vs. 30.64%). It was also observed that 23.51, 39.57, 33.90 and
3.02% patients with NC were classified as TNM stage I, II, III and
IV, respectively. Table II
presents the details of the investigated population based on
history of tobacco and alcohol consumption, including both controls
and patients (n=1,950). Table III
shows the genotyping results and frequencies of SPLUNC1 and
MDS1-EVI1 polymorphisms in patients with NC and controls. In both
cases, the genotypic frequency does not deviate from the expected
Hardy-Weinberg equilibrium. Furthermore, the distributions of the
genotypes that were compared with the NC cases and control are
presented in Table III and
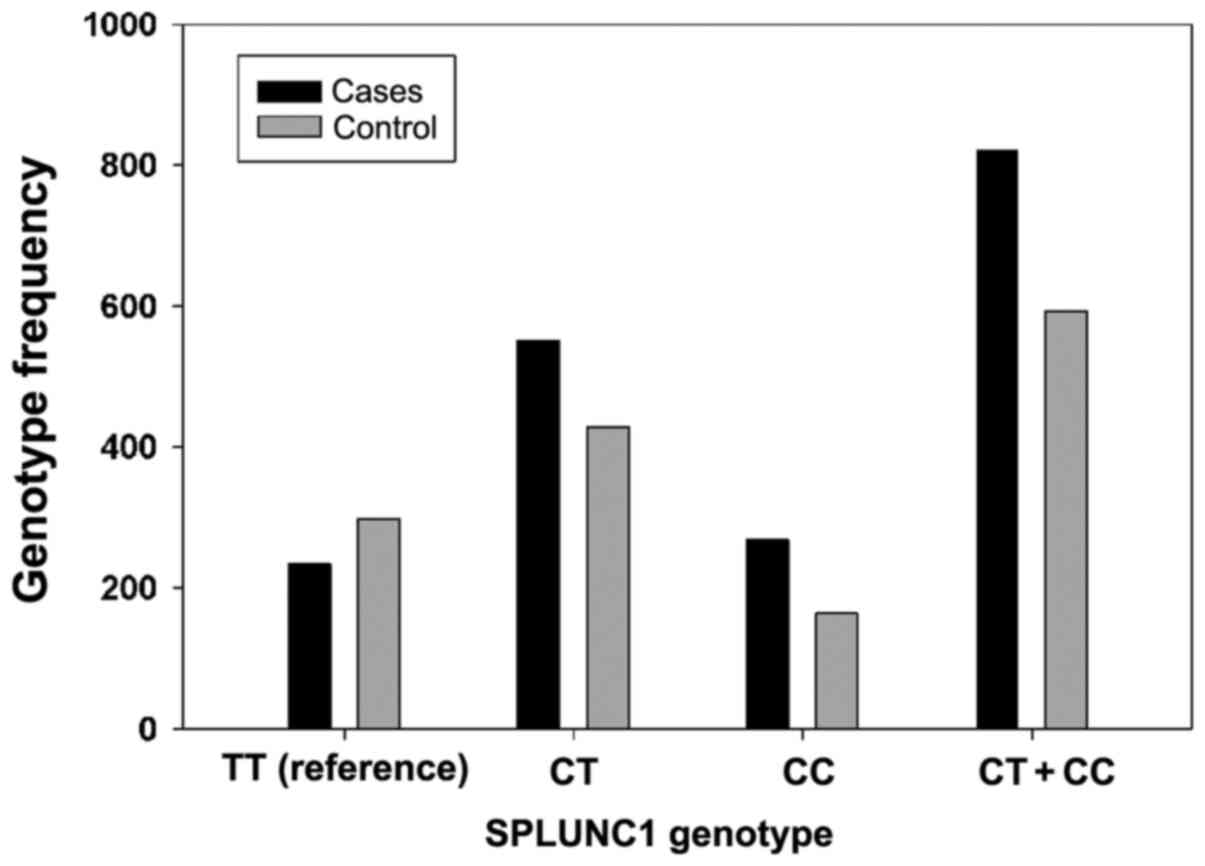
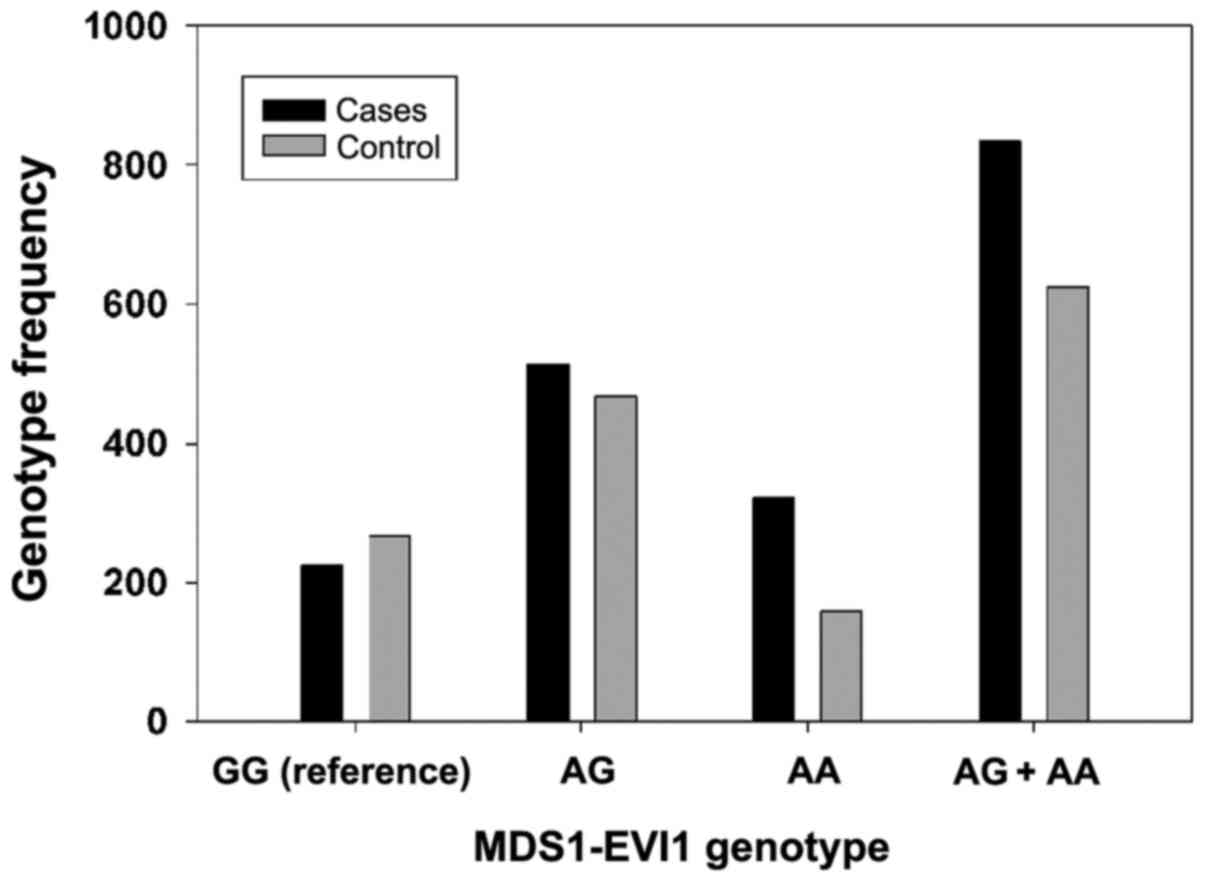
graphically presented in Figs. 1
and 2. It was observed that the
SPLUNC1 CT + CC genotypes may be a genetic risk factor for NC
(Fig. 1). By contrast, it was
observed that the MDS1-EVI1 AG + AA genotypes may be a potential
risk factor for NC among the studied population (Fig. 2). The frequencies of the SPLUNC1 TT,
CT and CC genotypes in patients with NC were 22.10, 52.12 and
25.78% respectively, which was significantly different compared
with the controls (Table III).
This difference was particularly observed in the CC genotype (25.78
patients vs. 18.52% controls). The CC homozygote was also the most
dominant genotype. Based on logistic regression analysis with
MDS1-EVI1, the AA allele was observed to have a more significant
risk for NC compared with patients carrying the AG allele (AA
allele; OR, 2.15; 95% CI, 1.62-3.15). Thus, indicating that the
MDS1-EVI1 AA allele is the higher-risk allele. The present study
also observed no direct relationship between the age group, tumor
invasion stage and lymph node involvement at the time of diagnosis
and polymorphisms of these genes. Moreover, the logistic regression
analysis also showed that subjects with SPLUNC1 CC genotypes had a
significantly higher susceptibility to NC compared with the TT
genotype (OR, 2.76; 95% CI, 1.96-3.81) when adjusted for sex, age,
smoking and drinking status. However, in the present analyses,
there was no correlation between sex, tobacco smoking and alcohol
consumption on the risk of lymph node related to the CT + CC
genotype. Furthermore, the gene-gene interaction between SPLUNC1
and MDS1-EVI1 polymorphisms was analyzed (Table IV). This analysis found that
patients with NC that have the SPLUNC1 CC genotype were more likely
to have the MDS1-EVI1 AA genotype compared with the controls (10.29
vs. 2.92%; P<0.002). Moreover, the SPLUNC1 CC genotype alone was
also associated with an increased risk in developing NC (OR, 2.76;
95% CI, 1.28-4.81; P<0.0078) when compared between patients with
NC and control individuals. In the case of control subjects, the
MDS1-EVI1 AA genotype did not show a higher risk compared with the
SPLUNC1 CC genotype (OR, 6.75; 95% CI, 3.41-12.11; P<0.002;
Table IV). However, the presence
of both SPLUNC1 CC and MDS1-EVI1 AA genotypes was associated with a
markedly higher risk for NC compared with others whom do not carry
both the genotypes (OR, 6.75; 95% CI, 3.41-12.11). Overall, this
analysis suggested that there was a multiplicative interaction
between the genotypes of SPLUNC1-CC and MDS1-EVI1-AA, which was
associated with an increased risk of developing nasopharyngeal
carcinoma.
 | Table ICharacteristics of patients with NC
(n=1,059) and controls (n=891) in the Chinese population. |
Table I
Characteristics of patients with NC
(n=1,059) and controls (n=891) in the Chinese population.
| Characteristics | Patients with NC, n
(%) | Controls, n (%) |
χ2a | P-value |
|---|
| Age, years | | | 6.01 | 0.014 |
|
>40 | 483 (45.61) | 456 (51.18) | | |
|
≤40 | 576 (54.39) | 435 (48.82) | | |
| Sex | | | 4.075 | 0.43 |
|
Male | 662 (62.51) | 517 (58.02) | | |
|
Female | 397 (37.49) | 374 (41.98) | | |
| Smoker | 359 (33.90) | 288 (32.32) | 0.54 | 0.46 |
| Alcohol drinker | 463 (43.72) | 273 (30.64) | 35.23 | <0.00001 |
| Clinical stage | | | | |
|
I | 249 (23.51) | | | |
|
II | 419 (39.57) | | | |
|
III | 359 (33.90 | | | |
|
IV | 32 (3.02) | | | |
| Local tumor invasion
(T-classification) | | | | |
|
T1 | 186 (17.56) | | | |
|
T2 | 539 (50.90) | | | |
|
T3 | 229 (21.62) | | | |
|
T4 | 105 (9.92) | | | |
| Lymph node
involvement (N-classification) | | | | |
|
N0 | 239 (22.57) | | | |
|
N1 | 512 (48.35) | | | |
|
N2 | 221 (20.87) | | | |
|
N3 | 87 (8.22) | | | |
| Presence of
metastasis | | | | |
|
Yes | 480 (54.67) | | | |
|
No | 579 (45.33) | | | |
 | Table IICharacterization of the study
population (n=1,950) based on history of tobacco and alcohol
consumption in patients with NC (n=1,059) and controls (n=891). |
Table II
Characterization of the study
population (n=1,950) based on history of tobacco and alcohol
consumption in patients with NC (n=1,059) and controls (n=891).
| Serial no. |
Characteristics | Population, n |
|---|
| 1 | Patients with NC
with no history of tobacco or alcohol use | 237 |
| 2 | Patients with NC
with history of tobacco or alcohol use | 822 |
| 3 | Controls with no
history of tobacco or alcohol use | 330 |
| 4 | Controls with
history of tobacco or alcohol use | 561 |
 | Table IIIGenotype frequencies of SPLUNC1 and
MDS1-EVI1 among patients with NC (n=1,059) and controls (n=891) and
their association with the risk of developing NC. |
Table III
Genotype frequencies of SPLUNC1 and
MDS1-EVI1 among patients with NC (n=1,059) and controls (n=891) and
their association with the risk of developing NC.
| Genotypes | Patients with NC, n
(%) | Controls, n
(%) | OR (95% CI) | χ2 | P-value |
|---|
| SPLUNC1 (rs2752903,
T>C) | | | | 39.884 | <0.00001 |
|
TT
(Reference) | 234 (22.10) | 298 (33.45) | 1.0 | | |
|
TT Smoker
(Reference) | 75 (7.08) | 59 (6.62) | | | |
|
TT Alcohol
drinker (Reference) | 103 (9.73) | 74 (8.31) | | | |
|
CT | 552 (52.12) | 428 (48.04) | 1.51
(1.22-2.14) | | 0.014 |
|
CC | 269 (25.78) | 165 (18.52) | 2.76
(1.96-3.81) | | 0.046 |
|
CT + CC | 821 (77.9) | 593 (66.56) | 1.43
(1.04-1.91) | | 0.021 |
|
CT + CC
Smoker | 284 (26.82) | 229 (25.70) | | | |
|
CT + CC
Alcohol drinker | 360 (33.99) | 199 (22.33) | | | |
| MDS1-EVI1
(rs6774494, G>A) | | | | 50.24 | <0.00001 |
|
GG
(Reference) | 224 (21.15) | 266 (29.85) | 1.0 | | |
|
GG Smoker
(Reference) | 69 (6.52) | 56 (6.29) | | | |
|
GG Alcohol
drinker (Reference) | 91 (8.59) | 71 (7.97) | | | |
|
AG | 514 (48.54) | 467 (52.41) | 1.37
(1.19-2.06) | | 0.21 |
|
AA | 321 (30.31) | 158 (17.73) | 2.15
(1.62-3.15) | | 0.018 |
|
AG + AA | 835 (78.85) | 625 (70.14) | 1.53
(1.00-2.36) | | 0.032 |
|
AG + AA
Smoker | 290 (27.38) | 232 (26.04) | | | |
|
AG + AA
Alcohol drinker | 372 (35.13) | 202 (22.67) | | | |
 | Table IVRisk of NC associated with SPLUNC1
and MDS1-EVI1 genotypes in patients with NC (n=1,059) and controls
(n=891). |
Table IV
Risk of NC associated with SPLUNC1
and MDS1-EVI1 genotypes in patients with NC (n=1,059) and controls
(n=891).
| Genotype | |
|---|
| SPLUNC1 | MDS1-EVI1 | Patients with NC, n
(%) | Controls, n
(%) | OR (95% CI) | P-value |
|---|
| TT | GG | 51 (4.82) | 79 (8.87) | 1.00 | |
| | AG | 135 (12.75) | 163 (18.29) | 1.28
(0.87-2.51) | 0.072 |
| | AA | 47 (4.44) | 54 (6.06) | 1.59
(0.93-3.16) | 0.063 |
| CT | GG (Reference) | 109 (10.29) | 148 (16.61) | 1.24
(0.83-2.42) | 0.002 |
| | AG | 254 (23.98) | 208 (23.34) | 2.24
(1.87-3.43) | 0.059 |
| | AA | 117 (11.05) | 81 (9.09) | 2.81
(1.42-5.27) | 0.017 |
| CC | GG (Reference) | 85 (8.03) | 47 (5.27) | 2.76
(1.28-4.81) | 0.0078 |
| | AG | 152 (14.35) | 85 (9.54) | 2.97
(1.54-5.69) | 0.087 |
| | AA | 109 (10.29) | 26 (2.92) | 6. 75
(3.41-12.11) | 0.002 |
Discussion
In the present investigation, it was observed that
SPLUNC1 and MDS1-EVI1 polymorphisms may trigger the development of
nasopharyngeal carcinoma among the Chinese population. By examining
1,059 patients with NC and 891 controls, the study showed that
SPLUNC1 CC genotype increases the risk for the development of NC.
Whereas MDS1-EVI1 AA genotypes were observed to be associated with
an increased risk of developing NC. Moreover, the gene-gene
interaction between SPLUNC1 and MDS1-EVI1 polymorphisms increased
the risk for NC in an additive manner. This suggests that the case
subjects carrying both of these genotypes are associated with
higher risks for the occurrence of NC. The results of the present
study implied the risk between the association of SPLUNC1 and
MDS1-EVI1 polymorphisms and NC. This increased risk may be due to
the importance of these genes in providing genomic integrity and
suppressing cancerous cells (21,22).
Several genes have been previously reported to be
associated with nasopharyngeal carcinoma, such as cytochrome P450
2E1, arylamine N-acetyltransferase 2, SPLUNC1, DNA repair protein
XRCC1, DNA repair protein RAD51 homolog 2, MDS1-EVI1 and
interleukins (23). Liu et
al (24) reported on PLUNC
proteins and deduced that the expression level of nasopharyngeal
carcinoma-related protein/SPLUNC1 was higher in nasopharynx cells
under chronic inflammation. They also observed that SPLUNC1 played
a major role in the differentiation of NC cells, and might be an
important antimicrobial protein involved in innate immunity defense
(24). SPLUNC1 was found to be
responsible for inhibiting the Toll-like receptor 9/NF-κB signaling
pathway and reduced the inflammatory microenvironment in
Epstein-Barr virus-associated NC (25). Whereas, Zhang et al (26) observed that von Ebner minor salivary
gland protein and SPLUNC1 genes were expressed in nasopharyngeal
epithelial tissue and the trachea, based on cDNA microarray
hybridization. In addition, it has been suggested that SPLUNC1 is
associated with the prognosis of NC and that the positive
expression of SPLUNC1 in NC cases infers an improved prognosis
(27). Yew et al (28) reported that the single nucleotide
polymorphism rs1407019, which lies within the intronic enhancer
region of SPLUNC1, was associated with susceptibility to NC, in
which the A allele was responsible for an increased risk for
NC.
The association between the mutation of MDS1-EVI1
and tumor susceptibility has also been reported by Heller et
al (29). Moreover, Wilson
et al (30) observed that
mice lacking an inactivating mutation in the MDS1-EVI1 allele
tended to develop a lower rate of tumor growth compared with mice
harboring the MDS-EVI1 allele. The overexpression of SPLUNC1 also
gives rise to the inactivity of MDS1-EVI1, which has been evidenced
in various tumor types (31).
Moreover, there are reports that have suggested that polymorphisms
among the SPLUNC1 and MDS1-EVI1 genes lead to functional
consequences (14). For example,
previous reports have suggested MDS1-EVI1 as a proto-oncogene, and
its overexpression was associated with leukemia and myelodysplastic
syndrome (32). In addition, a case
study on SPLUNC1 polymorphisms reported that two promoter single
nucleotide polymorphisms, rs2752903 and rs750064, were
significantly associated with NC in a Cantonese-speaking Chinese
population from Guangdong province in China (14). Moreover, the aberrant expression of
MDS1-EVI1 has been reported in various cancer studies (32,33).
However, its role in NC has not been extensively studied, and the
present study could not draw any significant relationship between
SPLUNC1 and MDS1-EVI1 genotypes on the prognostic status of
nasopharyngeal carcinoma (33).
These findings suggest that the polymorphisms between SPLUNC1 and
MDS1-EVI1 genes may not serve as an absolute marker for prognosis
of NC. Therefore, it is recommended that investigations on a large
number of patients with prospective follow-up clinical measures are
performed. One limitation of the present study was that the
participants were from one community admitted to one hospital. In
future studies, these findings should be verified in other
populations with high cases of patients with NC.
In conclusion, the association between SPLUNC1
(rs2752903, T>C) and MDS1-EVI1 (rs6774494, G>A) polymorphisms
exhibited a higher risk of developing NC. The present study
suggested a gene-to-gene interaction with MDS1-EVI1 (rs6774494,
G>A) polymorphisms. The study also observed that SPLUNC1 CT + CC
genotypes are a genetic risk factor for the development of NC among
the studied Chinese population. Therefore, the present study
provided useful information for the development of screening for
nasopharyngeal carcinoma and its treatment.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by the Shandong
Province Medical Health Scientific Research Investigation (grant
no. SJ2017RH289).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
ZG, DH, WC, HL and CZ performed the experiments, the
data collection and the statistical analysis. DH, WC, HL and CZ
interpreted the data and proofread the manuscript. All authors read
and approved the final manuscript.
Ethics approval and consent to
participate
All procedures were carried out in accordance with
the Declaration of Helsinki 1964 and its later amendments. Informed
consent was provided by all patients and participants. Consent was
obtained from each subject regarding their personal information on
demographic factors, any history of medical information, tobacco
smoking and alcohol consumption using a standard questionnaire. All
protocols and procedures were approved by the Hospital Ethical
Research Committee of the Jining Medical University (Jining,
Shandong, China; approval no. JMU/AH/OncDept/2017-34GA2).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Wei WI and Kwong DL: Current management
strategy of nasopharyngeal carcinoma. Clin Exp Otorhinolaryngol.
3:1–12. 2010.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Wang Y, Zhang Y and Ma S: Racial
differences in nasopharyngeal carcinoma in the United States.
Cancer Epidemiol. 37:793–802. 2013.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Lang J, Hu C, Lu T, Pan J and Lin T:
Chinese expert consensus on diagnosis and treatment of
nasopharyngeal carcinoma: Evidence from current practice and future
perspectives. Cancer Manag Res. 11:6365–6376. 2019.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Adham M, Kurniawan AN, Muhtadi AI, Roezin
A, Hermani B, Gondhowiardjo S, Tan IB and Middeldorp JM:
Nasopharyngeal carcinoma in Indonesia: Epidemiology, incidence,
signs, and symptoms at presentation. Chin J Cancer. 31:185–196.
2012.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Jain A, Chia WK and Toh HC: Immunotherapy
for nasopharyngeal cancer-a review. Chin Clin Oncol.
5(22)2016.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Yang QY, He YQ, Xue WQ, Zhou T, Liao Y,
Zheng MQ, Jia YJ, Yuan LL and Jia WH: Association between serum
cotinine level and serological markers of Epstein-Barr virus in
healthy subjects in south china where nasopharyngeal carcinoma is
endemic. Front Oncol. 9(865)2019.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Wang TM, Zhou T, He YQ, Xue WQ, Zhang JB,
Zheng XH, Li XZ, Zhang SD, Zeng YX and Jia WH: Fine-mapping of HLA
class I and II genes identified two independent novel variants
associated with nasopharyngeal carcinoma susceptibility. Cancer
Med. 7:6308–6316. 2018.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Bei JX, Zuo XY, Liu WS, Guo YM and Zeng
YX: Genetic susceptibility to the endemic form of NPC. Chin Clin
Oncol. 5(15)2016.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Guo XC, Scott K, Liu Y, Dean M, David V,
Nelson GW, Johnson RC, Dilks HH, Lautenberger J, Kessing B, et al:
Genetic factors leading to chronic Epstein-Barr virus infection and
nasopharyngeal carcinoma in South East China: Study design, methods
and feasibility. Hum Genomics. 2:365–375. 2006.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Zhang P, Zhang L, Liu H, Zhao L, Li Y,
Shen JX, Liu Q, Liu MZ and Xi M: Clinicopathologic characteristics
and prognosis of tongue squamous cell carcinoma in patients with
and without a history of radiation for nasopharyngeal carcinoma: A
matched case-control study. Cancer Res Treat. 49:695–705.
2017.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Guo XC, O'Brien SJ, Winkler C, Scott K,
Hutcheson H, David V, Kessing B, Zheng YM, Liao J, Lui Y, et al:
Association study of chromosome 4 STRs polymorphisms with
nasopharyngeal carcinoma. Yi Chuan. 28:783–790. 2006.PubMed/NCBI(In Chinese).
|
|
12
|
Zhou HD, Li XL, Li GY, Zhou M, Liu HY,
Yang YX, Deng T, Ma J and Sheng SR: Effect of SPLUNC1 protein on
the pseudomonas aeruginosa and Epstein-Barr virus. Mol Cell
Biochem. 309:191–197. 2008.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Zhou HD, Li GY, Yang YX, Li XL, Sheng SR,
Zhang WL and Zhao J: Intracellular co-localization of SPLUNC1
protein with nanobacteria in nasopharyngeal carcinoma epithelia
HNE1 cells depended on the bactericidal permeability increasing
protein domain. Mol Immunol. 43:1864–1871. 2006.PubMed/NCBI View Article : Google Scholar
|
|
14
|
He Y, Zhou G, Zhai Y, Dong X, Lv L, He F
and Yao K: Association of PLUNC gene polymorphisms with
susceptibility to nasopharyngeal carcinoma in a Chinese population.
J Med Genet. 42:172–176. 2005.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Métais JY and Dunbar CE: The MDS1-EVI1
gene complex as a retrovirus integration site: Impact on behavior
of hematopoietic cells and implications for gene therapy. Mol Ther.
16:439–449. 2008.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Kurokawa M, Mitani K, Yamagata T,
Takahashi T, Izutsu K, Ogawa S, Moriguchi T, Nishida E, Yazaki Y
and Hirai H: The evi-1 oncoprotein inhibits c-Jun N-terminal kinase
and prevents stress-induced cell death. EMBO J. 19:2958–2968.
2000.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Bei JX, Li Y, Jia WH, Feng BJ, Zhou G,
Chen LZ, Feng QS, Low HQ, Zhang H, He F, et al: A genome-wide
association study of nasopharyngeal carcinoma identifies three new
susceptibility loci. Nat Genet. 42:599–603. 2010.PubMed/NCBI View
Article : Google Scholar
|
|
18
|
Edge SB and Compton CC: The American joint
committee on cancer: The 7th edition of the AJCC cancer staging
manual and the future of TNM. Ann Surg Oncol. 17:1471–1484.
2010.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Ara S, Lee PS, Hansen MF and Saya H: Codon
72 polymorphism of the TP53 gene. Nucleic Acids Res.
18(4961)1990.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Emigh TH: A comparison of tests for
Hardy-Weinberg equilibrium. Biometrics. 36:627–642. 1980.PubMed/NCBI
|
|
21
|
Wu MY, Huang SJ, Yang F, Qin XT, Liu D,
Ding Y, Yang S and Wang XC: Detection of nasopharyngeal carcinoma
susceptibility with single nucleotide polymorphism analysis using
next-generation sequencing technology. Oncotarget. 8:52708–52723.
2017.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Chin C, Hahn WC, Getz G and Meyerson M:
Making sense of cancer genomic data. Genes Dev. 25:534–555.
2011.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Hildesheim A and Wang CP: Genetic
predisposition factors and nasopharyngeal carcinoma risk: A review
of epidemiological association studies, 2000-2011: Rosetta stone
for NPC: Genetics, viral infection, and other environmental
factors. Semin Cancer Biol. 22:107–116. 2012.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Liu H, Zhang X, Wu J, French SW and He Z:
New insights on the palate, lung, and nasal epithelium clone
(PLUNC) proteins: Based on molecular and functional analysis of its
homolog of YH1/SPLUNC1. Exp Mol Pathol. 100:363–369.
2016.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Ou C, Sun Z, Zhang H, Xiong W, Ma J, Zhou
M, Lu J, Zeng Z, Bo X, Chen P, et al: SPLUNC1 reduces the
inflammatory response of nasopharyngeal carcinoma cells infected
with the EB virus by inhibiting the TLR9/NF-κB pathway. Oncol Rep.
33:2779–2788. 2015.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Zhang B, Nie X, Xiao B, Xiang J, Shen S,
Gong J, Zhou M, Zhu S, Zhou J, Qian J, et al: Identification of
tissue-specific genes in nasopharyngeal epithelial tissue and
differentially expressed genes in nasopharyngeal carcinoma by
suppression subtractive hybridization and cDNA microarray. Genes
Chromosomes Cancer. 38:80–90. 2003.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Zhang W, Zeng Z, Wei F, Chen P, Schmitt
DC, Fan S, Guo X, Liang F, Shi L, Liu Z, et al: SPLUNC1 is
associated with nasopharyngeal carcinoma prognosis and plays an
important role in all-trans-retinoic acid-induced growth inhibition
and differentiation in nasopharyngeal cancer cells. FEBS J.
281:4815–4829. 2014.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Yew PY, Mushiroda T, Kiyotani K,
Govindasamy GK, Yap LF, Teo SH, Lim PV, Govindaraju S, Ratnavelu K,
Sam CK, et al: Identification of a functional variant in SPLUNC1
associated with nasopharyngeal carcinoma susceptibility among
Malaysian Chinese. Mol Carcinog. 51 (Suppl 1):E74–E82.
2012.PubMed/NCBI View
Article : Google Scholar
|
|
29
|
Heller G, Rommer A, Steinleitner K, Etzler
J, Hackl H, Heffeter P, Tomasich E, Filipits M, Steinmetz B,
Topakian T, et al: EVI1 promotes tumor growth via transcriptional
repression of MS4A3. J Hematol Oncol. 8(28)2015.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Wilson M, Tsakraklides V, Tran M, Xiao YY,
Zhang Y and Perkins AS: EVI1 interferes with myeloid maturation via
transcriptional repression of Cebpa, via binding to two far
downstream regulatory elements. J Biol Chem. 291:13591–13607.
2016.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Steinleitner K, Rampetsreiter P, Köffel R,
Ramanathan G, Mannhalter C, Strobl H and Wieser R: EVI1 and
MDS1/EVI1 expression during primary human hematopoietic progenitor
cell differentiation into various myeloid lineages. Anticancer Res.
32:4883–4889. 2012.PubMed/NCBI
|
|
32
|
Wieser R: The oncogene and developmental
regulator EVI1: Expression, biochemical properties, and biological
functions. Gene. 396:346–357. 2007.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Yuan X, Wang X, Bi K and Jiang G: The role
of EVI-1 in normal hematopoiesis and myeloid malignancies (Review).
Int J Oncol. 47:2028–2036. 2015.PubMed/NCBI View Article : Google Scholar
|