Introduction
Myeloid-derived suppressor cells (MDSCs) are
heterogeneous, immature cell populations of myeloid origin
including monocyte/macrophage, granulocyte and dendritic cells,
which are one of the major components of the tumor microenvironment
protecting cancer cells from the host immune system attack
(1-3).
MDSCs promote tumor growth by producing immunosuppressive molecules
in the TME such as interleukin-10(4), transforming growth factor-β (5) and reactive oxygen species, as well as
expressing cell surface receptors that inhibit T cell proliferation
and activation and producing molecules that directly promote tumor
growth and invasions such as vascular endothelial growth factor
(6) and matrix metalloproteinases
(7). There are two major subsets
of MDSCs, identified in both mice and humans: Monocytic MDSCs
(M-MDSCs) are morphologically and phenotypically similar to
monocytes whereas polymorphonuclear MDSCs (PMN-MDSCs) are similar
to neutrophils. In mice, M-MDSCs and PMN-MDSCs are defined as
CD11b+Ly6G-Ly6Chigh cells and
CD11b+Ly6G+Ly6Clow cells,
respectively (8). In humans,
M-MDSCs and PMN-MDSCs are defined as
CD11b+CD14+CD15-CD33+HLA-DR-/low
cells and
CD11b+CD14-CD15+CD33+
cells, respectively (9,10). It is reported that MDSCs accumulate
in patients with renal cell carcinoma (9,11),
breast cancer (12,13), ovarian cancer (14,15),
melanoma (16) and head and neck
cancer (17,18). In some clinical studies, MDSCs are
therapeutic targets alone (19-21)
or combined with other target drugs such as PD-1/PD-L1 and CTLA-4
immune checkpoint inhibitors (22-24).
In in vitro MDSCs studies, there are some
different methods to generate MDSCs in mice and humans. One of the
major methods is that mouse primary bone marrow cells or human
peripheral blood mononuclear cells are co-cultured with mouse or
human tumor cell lines or cytokines and then purified by magnetic
beads or flow sorting with the use of myeloid cell surface markers
such as CD33, CD11b or CD14 (25-29).
Another major method is to first obtain high purity myeloid cells,
such as monocytes, by magnetic beads or flow sorting and then
co-culturing with tumor cell lines or cytokines. Taking a
reductionist approach, the combination of GM-CSF and IL6 could
induce normal peripheral blood mononuclear cells (PBMCs) to become
CD33+ HLA-DR-/low MDSCs more consistent and
potency than combinations of tumor cell-secreted cytokines
(30). The present study
hypothesized that an obvious limitation of the studies isolating
induced MDSCs or freshly monocytes by magnetic beads or flow
sorting is the high cost of magnetic bead sorting kits or flow
cytometers with sorting function. To overcome this problem, the
present study showed that MDSCs were induced from monocytes by
plastic adhesive sorting with the cytokine combination of IL6 and
GM-CSF. This new model could significantly simplify the
manufacturing process of MDSCs and promote drug discovery with
MDSCs.
Materials and methods
Isolation of PBMCs
Peripheral blood mononuclear cells (PBMCs) were
isolated from anonymous healthy donors. Briefly, the buffy coats
were diluted 1:1 in 0.9% NaCl, then centrifuged at 400 x g for 30
min at room temperature, according to the protocol of density
gradient centrifugation (Ficoll-Paque). Following centrifugation,
PBMCs on the second layer were harvested and washed twice with 30
ml of cold PBS. All human blood samples (150-200 ml) were collected
between March 2018 and September 2020 and were performed under the
guidelines of the Medical Ethics Committee of the Affiliated
Tianyou Hospital, College of Life and Health Sciences, Wuhan
University of Science and Technology. (institutional review board
approval: WUSTLL-20180009). Each patient involved in the study was
asked to sign a written informed consent form and agreed to the use
of their samples in scientific research. All the specimens were
anonymized and handled according to accepted ethical and legal
standards.
Monocytes isolation with plastic
adhesion
For monocytes isolation, PBMCs were resuspended in
RPMI-1640 medium (Dalian Meilun Biotech Co., Ltd.) supplemented
with 10% Fetal Bovine Serum (ExCell Bio. ExCell Biotech Co., Ltd.),
seeded in a surface-treated 6-well plate at 2x107
cells/ml in 2 ml. After 1 h of adherence in a 5% CO2
container at 37˚C, non-adherent cells were removed by washing twice
with RPMI-1640 medium and the remaining adherent cells were mostly
monocytes.
T cell isolation and activation
For suppression assay, PBMCs isolated from volunteer
blood in the above steps were used to sort T cells. According to
the instructions (Pan T Cell Isolation kit, human, Miltenyi Biotec
GmbH), the PBMCs cell pellet was first resuspended in 40 µl of
buffer per 107 total cells; second, 10 µl of Pan T Cell
Biotin-Antibody Cocktail, 30 µl of buffer and 20 µl of Pan T Cell
MicroBead Cocktail was added per 107 total cells
successively. Then, the LS Column was placed in the magnetic field
of a suitable MACS Separator (MidiMACS; Miltenyi Biotec GmbH), the
cell suspension added into the column and the flow-through
containing unlabeled cells, representing the enriched T cells, was
collected. Finally, an appropriate amount of anti-CD3/CD28 magnetic
beads was added to activate the T cells (the ratio of magnetic
beads to T cells was 2:1).
IL6/GM-CSF induce monocytes to
M-MDSCs
For the induction of M-MDSCs, adherent cells were
kept cultured in a surface-treated 6-well plate in the complete
induction medium [RPMI-1640 with 10% FBS, 10 ng/ml IL6 (PeproTech,
Inc.) and 10 ng/ml GM-CSF (PeproTech, Inc.)] for 7 days. The medium
was refreshed on day 3 and day 5.
Flow cytometry
Briefly, adherent cells were harvested with 10 min
of dissociation with a gentle non-enzymatic cell dissociation
reagent Versene (Gibco; Thermo Fisher Scientific, Inc.) followed by
gently scraping. Antibody staining was performed in PBS
supplemented with 2% FBS for 30 min at 4˚C in the dark. For
intracellular antibody staining, cells were treated with the
True-Nuclear Transcription Factor Buffer Set. The antibodies used
were: CD3 (UCHT1, cat. no. 300458), CD22 (HIB22, cat. no. 302524),
CD14 (63D3, cat. no. 367104), CD15 (HI98, cat. no. 301908), CD33
(HIM3-4, cat. no. 303304), CD11b (ICRF44, cat. no. 301324), HLA-DR
(TU39, cat. no. 361707), phosphorylated (p)-STAT3 (A16089B, cat.
no. 698905), PD-L1 (29E.2A3, cat. no. 329705) (all from BioLegend,
Inc.) and two SinoBiological antibodies of Bcl-2 (cat. no.
100126-R204-F), Caspase3 (cat. no. 10050-MM02) and corresponding
isotype control. All the antibodies were diluted 1:2.
Suppression assay
The inhibitory function of the induced M-MDSCs were
evaluated by their ability to inhibit the proliferation of
allogeneic T cells in the following Suppression Assay: T cells were
labeled with carboxyfluorescein succinimidyl ester (CFSE; 5 µM;
MilliporeSigma) and seeded at 2x105 cells per well in
96-well plates with 100 µl RPMI-1640 medium at 37˚C for 4 days.
M-MDSCs from the above cytokine induction cultures were added to T
cells at ratios of 1:4. T cell proliferation was provided by
anti-CD3/CD28 stimulation beads (Gibco; Thermo Fisher Scientific,
Inc.) and IL2 (Beijing T&L Biotechnology Co., Ltd.).
Suppression Assay wells were measured by flow cytometry for T cell
proliferation after four days. T cells with or without stimulation
and activated T cells co-cultured with monocytes were used as
controls.
Reverse transcription-quantitative
(RT-q) PCR for suppression gene expression
For the suppression gene expression study, monocytes
and M-MDSCs were harvested, then total cellular RNA was extracted
from 5x106 cells using the RNeasy Mini kits (Qiagen
GmbH). RNA (1 µg) was reverse transcribed to cDNA with random
hexamer primers using the Transcriptor First Strand cDNA Synthesis
kit (Roche Molecular Diagnostics). Afterward, relative cDNA was
amplified with gene specific-primers using Hieff qPCR SYBR Green
Master Mix (Shanghai Yeasen Biotechnology Co., Ltd.) and run on a
Bio-Rad CFX Maestro with PCR cycling conditions: 95˚C 5 min; 95˚C
15 sec; 55˚C 15 sec; and 72˚C 30 sec for 30 cycles. (Bio-Rad
Laboratories, Inc.). Data were acquired and analyzed using Bio-Rad
CFX Maestro 1.1 software (Bio-Rad Laboratories, Inc.). Gene
expression was normalized to the house-keeping gene (GAPDH) and
fold change relative to monocytes was determined. Gene specific
primers for reverse transcription-quantitative PCR are shown in
Table I. All experiments were
performed in at least three independent experiments. RNA
extraction, cDNA synthesis, and qPCR were performed according to
the manufacturer's protocols.
 | Table IGene specific primers for reverse
transcription-quantitative PCR. |
Table I
Gene specific primers for reverse
transcription-quantitative PCR.
| Gene | Forward primer (5'
à 3') | Reverse primer (5'
à 3') |
|---|
| GAPDH |
TTAAAAGCAGCCCTGGTGAC |
CTCTGCTCCTCCTGTTCGAC |
| VEGF |
CACACAGGATGGCTTGAAG |
AGGGCAGAATCATCACGAAG |
| NOX2 |
TGCCAGTCTGTCGAAATCTGC |
ACTCGGGCATTCACACACC |
| TGF-β |
GCAGAAGTTGGCATGGTAGC |
CCCTGGACACCAACTATTGC |
| PDL1 |
TATGGTGGTGCCGACTACAA |
TGCTTGTCCAGATGACTTCG |
| ARG1 |
GTTTCTCAAGCAGACCAGCC |
GCTCAAGTGCAGCAAAGAGA |
| PDL2 |
ACCGTGAAAGAGCCACTTTG |
GCGACCCCATAGATGATTATGC |
IFN-γELISA assay
According to the protocol (Human IFN-γ ELISA kit,
U-CyTech biosciences), 50 µl of diluted coating antibody solution
and 100 µl PBS was added to each well of the ELISA plate, then
incubated overnight at 4˚C. Then, 100 µl of diluted
standard/blank/samples was added to the wells and the plate sealed
and incubated for 2 h at 37˚C. Diluted detection antibody solution
(100 µl) was added and incubated for 1 h at 37˚C. Finally 100 µl of
diluted SPP conjugate, 100 µl of TMB substrate solution and 100 µl
of stop solution was added into each well (resulting in a yellow
color) and the plate read at 450 nm within 30 min.
WP1066 treatment on induced
M-MDSCs
After 7 days of IL6/GM-CSF induction, M-MDSCs
induced in vitro were treated with different concentrations
of STAT3 inhibitor WP1066 (0, 5 and 10 µM for 24 h and DMSO without
WP1066 as a control. Flow cytometry of p-STAT3 expression levels
and suppression protein PD-L1 expression was determined. In
addition, the toxicity analysis of WP1066 on inducted M-MDSCs was
also determined by detecting the level of cell apoptosis and the
expression of apoptosis-relative protein Bcl-2 and Caspase3 by flow
cytometry.
Statistical analysis
Statistical analysis was performed using GraphPad
Prism 5 software (GraphPad Software, Inc.). The results were shown
as means ± SEM from at least three independent experiments. Single
comparison between two groups was analyzed by Student's t test.
Comparisons between multiple groups were determined using ANOVA
followed by the Newman-Keuls multiple-comparison test. P<0.05
was considered to indicate a statistically significant
difference.
Results
The purity of monocytes before and
after plastic adhesion
Isolating monocytes from PBMCs and co-culturing with
tumor cells or inducing with cytokines is a common way to obtain
MDSCs for in vitro studies. The present study wanted to
explore the effect of monocytes on cell yield, purity and monocyte
phenotype using the plastic adhesion isolation method.
Peripheral blood mononuclear cells were isolated
from healthy donors by density gradient centrifugation with a
median PBMCs count of 720.5x106 cells with a range
(540.0x106 to 790.0x106) and median viability
of 96.5% (90.0-97.8). The median monocytes percentage and cell
count in PBMC were 11% (9.8-21.6) and 77.6x106
(50.6x106-176.8x106) cells respectively.
After 1 h of plastic adhesion enrichment, the total number of cells
reduced by 76%, with an average of 165.0x106 cells and
the median viability was 98.4%. The median percentage of monocytes
increased to 21.8% as the number of 34.6x106 on average
(Fig. 1A and B). Compare with monocytes separated from
flow cytometry, monocytes isolated by plastic adhesion were mixed
with a proportion of lymphocytes. However, after 7 days of
induction with IL6 and GM-CSF, the mean of the total cells was
23.1x106 and the median percentage of monocytes
increased to 94% (82-98%).
The presence of non-monocytes during
MDSCs induction
Unlike the high purity of monocytes produced by
isolation methods such as flow cytometry cell sorting or magnetic
bead sorting, there were numerous non-monocytes such as
CD3+ T cells or CD22+ B cells during the
MDSCs induction process of the present study. As shown in Fig. 1A and B, the median percentage of
CD3+ T cells in 1 h of adherent cells was 52.6%
(34.8-68.0%). Meanwhile, median percentage of CD22+ B
cells in 1 h of adherent cells was 16.6% (9.7-23.5%); median
percentage of CD3-CD22-CD14- cells
was 10% (6.8-15.9%). However, after 3 PBS washes and medium changes
in the 7 days of the culture process, non-monocytes all decreased
to the lowest level ~2-5% (Fig. 1C
and D).
The phenotype of MDSCs
Similar to the common definition of human MDSCs
(3), the phenotype of in
vitro-generated MDSCs was evaluated for CD33, CD11b, HLA-DR,
CD14 and CD15 expression by flow cytometry in the present study. As
described above, human PMN-MDSCs were defined as
CD11b+CD14-CD15+CD33+cells
and M-MDSCs were defined as
CD11b+CD14+CD15-CD33+HLA-DR-/low
cells. As shown in Fig. 2A, MDSCs
show high expression of CD33, CD11b and CD14 and low expression of
HLA-DR. However, MDSCs did not show CD15 expression. The results
indicated that the method generated only a single subset of
CD11b+CD14+CD15-CD33+HLA-DR-/low
M-MDSCs but no
CD11b+CD14-CD15+CD33+
PMN-MDSCs. At the same time, the present study also revealed the
changes of above surface marker expression compared with 1 h
adherent monocytes (data not shown).
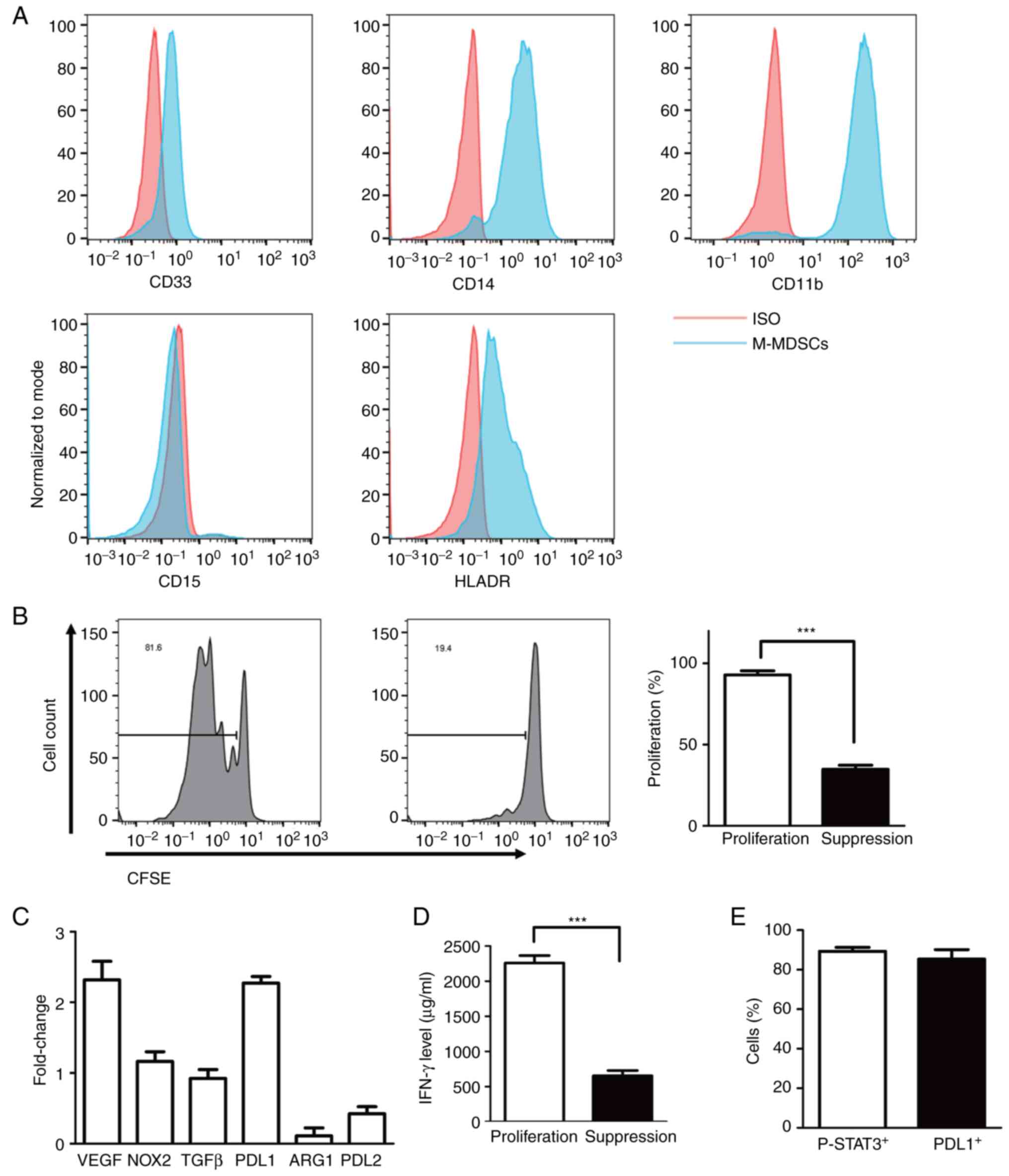 | Figure 2Phenotype of induced MDSCs and
suppressing ability. (A) Flow analysis of the cell-surface
expression of CD33, CD11b, CD14, CD15 and HLA-DR on in vitro
induced MDSCs with IL6/GM-CSF for 7 days. (B) and (D) The ability
of suppressing T cells proliferation in M-MDSCs at ratio of 1:4 as
determined by CFSE label assay and secretion of IFN-γ by ELISA. (C)
Gene expression of relative suppressive mechanisms in MDSCs such as
VEGF, NOX2, TGF-β, PD-L1, ARG1, PD-L2 in induced M-MDSCs as
determined by reverse transcription-quantitative PCR. (E)
Percentage of positive p-STAT3 and PDL1 cells in induced M-MDSCs.
***P<0.0001. MDSCs, myeloid-derived suppressor cells;
HLA, human leukocyte antigen; GM-CSF, granulocyte-macrophage
colony-stimulating factors; M-MDSCs, monocytic MDSCs; CFSE,
carboxyfluorescein succinimidyl ester; NOX2, NADPH oxidase 2; PD-L,
programmed death ligand; ARG1, arginase 1; p-, phosphorylated. |
The suppressive ability of MDSCs and
suppress gene expression
The inhibitory effect of MDSCs generated from
adherent monocytes was evaluated by their ability to inhibit
allogeneic T cell proliferation and IFN-γ secretion. As only
M-MDSCs were produced, high-purity MDSCs were separated with
Versene for 10 min and carefully scraped off the surface of the
treated 6-well plate. MDSCs were then mixed with CFSE-labeled
allogeneic T cells at MDSCs: T cells ratios of 1:4, followed by
stimulation with anti-CD3/CD28 beads. T cell proliferation was
evaluated by flow cytometry after 4 days. Notably, the
proliferation of T cells was significantly inhibited with 98% of
proliferation T cells with control compared with 36% of
proliferation T cells with MDSCs co-culture (Fig. 2B). To reveal the molecular
mechanism of suppressing T cell proliferation, some putative
suppressor genes including VEGF, ARG1, PD-L1, PD-L2, NOX2 and TGFβ
were tested by RT-qPCR. As shown in Fig. 2C, the suppressor genes of PD-L1 and
VEGF were shown to be upregulated, which may affect the inhibitory
ability of MDSCs. Last, the percentage of p-STAT3 and PD-L1
positive cells both reached ~90% (Fig.
2E). As shown in Fig. 2D, the
level of IFN-γ was significantly decreased in suppressed T
cells.
WP1066 treatment leads to
downregulation of p-STAT3 and PD-L1
As an important component of the immunosuppressive
microenvironment of solid tumors, MDSCs have been extensively
studied as therapeutic targets (31). WP1066 is a mature and widely used
inhibitor of JAK and STAT3 signaling pathways. However, there are
no reports of WP1066 targeting MDSCs. In vitro generated
MDSCs here were treated with dimethyl sulfoxide (DMSO) plus 0, 5
and 10 µM WP1066 for 24 h to evaluate the inhibitory effect of
WP1066 on MDSCs. As shown in Fig.
3A, MDSCs treated with 10 µM WP1066 revealed more effectiveness
compared with 0 and 5 µM WP1066 in the proliferation of T cells.
Similarly, the expression of p-STAT3 (Fig. 3B) and PD-L1 (Fig. 3C) were significantly reduced after
10 µM WP1066 treatment. Finally, with the high-intensity inhibitory
effect of WP1066 on M-MDSC cells, the ability of T cells to release
IFN-γ was also effectively restored, as shown in Fig. 3D.
WP1066 treatment leads to upregulation
of MDSCs apoptosis
In addition to reducing the ability of MDSCs to
inhibit T cell proliferation, WP1066 treatment also resulted in an
upregulation of cell apoptosis in MDSCs. As shown in Fig. 4A, induced MDSCs treated with 5 µM
WP1066 revealed almost the same level of upregulation of early
apoptosis and late apoptosis after 24 h of WP1066 treatment,
whereas MDSCs treated with 10 µM WP1066 showed more cells with late
apoptosis than early apoptosis. Despite the detection of cell
viability, classical proteins associated with cell death such as
Bcl-2 and Caspase3 were also examined. As shown in Fig. 4B, 5 µM WP1066 did not affect Bcl-2
protein expression in the resulting MDSCs, however, 10 µM WP1066
showed a significant reduction of Bcl-2+ cells.
Similarly, induced MDSCs treated with 10 µM WP1066 showed more
decrease in Caspase3+ cells implying more late apoptosis
or dead cells (Fig. 4C).
Discussion
The present study established a simple in
vitro model to generate MDSCs from monocytes isolated from
fresh PBMCs by plastic adhesion isolation. Plastic adhesion
isolation is a simple and inexpensive widely used technique for the
isolation of human monocytes from PBMCs. Evidently, compared with
monocytes isolated by magnetic beads or flow sorting, the yield of
adherence monocytes was significantly lower and mixed with a high
proportion of lymphocytes. However, only M-MDSCs-like cells were
successfully generated from adherence monocytes by co-treatment
with IL6 and GM-CSF for 7 days similar to
CD11b+CD14+CD15-CD33+HLA-DR-/low
cells. However, it is not claimed to be the best model to generate
MDSCs from human bone marrow or PBMCs. Nevertheless, it is
suggested that this model will significantly remove the barriers of
research funding and high-tech equipment and allow more researchers
to study MDSCs-related problems. In addition, it is hypothesized
that the feasibility of this technique in diagnosis aspects is not
high because diagnosis requires reliable results and high
reproducibility. Although this method is the simplest and cheapest
method to obtain monocytes, the reliability and consistency of the
conclusions using this method are poor and therefore is not
suitable for diagnostic applications.
A limitation of the present study is that it only
focused on the most commonly used combination of IL6 and GM-CSF to
induce MDSCs with greater inhibitory capacity. Further studies will
investigate whether MDSCs can be induced from adherence monocytes
with the treatment of other different combinations of cytokines or
conditional medium of human cell lines. Based on CD14 and CD16
expression, monocytes could be further divided into three main
subsets (32), therefore, another
limitation of the present study is that it did not explore the
proportion of each subset in adherent monocytes.
The first notable phenomenon is that the presence of
no-monocytes in the process of early induction did not affect the
successful induction of final MDSCs. It was hypothesized that the
powerful function of the cytokine combination of IL6 and GM-CSF
masked the influence of other lymphocytes early in the process. As
in other studies, the STAT3 signaling pathway was activated in
MDSCs induced by IL6/GM-CSF, but the characteristics, phenotype and
the mechanisms by which MDSCs inhibited T cell proliferation were
different. For example, in Casacuberta-Serra et al (33), MDSCs express low levels of CD14,
but high PD-L1 expression. Bian et al (34), show that arginase-1 is neither
inherently expressed in MDSC nor required for MDSC-mediated
inhibition. Zhan et al (35), reveal that the suppressive function
of CD33+HLA-DRlow MDSCs is dependent on the programmed death
ligand-1/programmed death ligand-2 pathway. However, the IL6/GM-CSF
induced MDSCs in the present study lacked ARG1 and NOX2 expression
but upregulated VEGF and PD-L1 expression, suggesting that this
model is not a classic MDSCs that suppressed T cells proliferation
through L-arginine depletion or RNS production. Furthermore, the
expression of PD-L1 was upregulated to suppress T cell
proliferation. Therefore, this model may be used for therapeutic
targets of PD-L1 on MDSCs, but not for other conventional
mechanisms of intracellular regulatory molecules in MDSCs isolated
from cancer patients.
STAT3 activation is involved in the generation and
function of MDSCs and has been identified as a promising
therapeutic target for anticancer drugs (36). The present study demonstrated that
the STAT3 inhibitor WP1066 had the potential to reduce the level of
immunosuppression and upregulate cell apoptosis in MDSCs. It was
observed that MDSCs treated with 10 µM WP1066 for 24 h
significantly reduced p-STAT3 levels and PD-L1 expression,
suggesting that WP1066 could directly inhibit the function of
MDSCs. Furthermore, downregulation of Bcl-2 and Caspase3 in MDSCs
indicated that WP1066 could promote MDSCs apoptosis to change the
tumor microenvironment.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by the Department of
Science and Technology of Hubei Province (project number
2020BCB048) and Hubei Province Technology Innovation Special Major
Project (project number 2019ACA168).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
XL and TZ participated in research design. HH, YX,
QY and TL performed the experiments, analyzed the data and were
major contributors in writing the manuscript. LG provided technical
guidance and participated in data acquisition and analysis. HH and
YX confirm the authenticity of all the raw data. All the authors
read and approved the final version of the manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of the Affiliated Tianyou Hospital, College of Life and
Health Sciences, Wuhan University of Science and Technology. Each
patient involved in the study was asked to sign a written informed
consent form and agreed to the use of their samples in scientific
research.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Kumar V, Patel S, Tcyganov E and
Gabrilovich DI: The nature of myeloid-derived suppressor cells in
the tumor microenvironment. Trends Immunol. 37:208–220.
2016.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Gabrilovich DI: Myeloid-derived suppressor
cells. Cancer Immunol Res. 5:3–8. 2017.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Li K, Shi H, Zhang B, Ou X, Ma Q, Chen Y,
Shu P, Li D and Wang Y: Myeloid-derived suppressor cells as
immunosuppressive regulators and therapeutic targets in cancer.
Signal Transduct Target Ther. 6(362)2021.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Huang B, Pan PY, Li Q, Sato AI, Levy DE,
Bromberg J, Divino CM and Chen SH: Gr-1+CD115+ immature myeloid
suppressor cells mediate the development of tumor-induced T
regulatory cells and T-cell anergy in tumor-bearing host. Cancer
Res. 66:1123–1131. 2006.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Lee CR, Kwak Y, Yang T, Han JH, Park SH,
Ye MB, Lee W, Sim KY, Kang JA, Kim YC, et al: Myeloid-derived
suppressor cells are controlled by regulatory T cells via TGF-β
during murine colitis. Cell Rep. 17:3219–3232. 2016.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Chen JY, Lai YS, Chu PY, Chan SH, Wang LH
and Hung WC: Cancer-derived VEGF-C increases chemokine production
in lymphatic endothelial cells to promote CXCR2-dependent cancer
invasion and MDSC recruitment. Cancers (Basel).
11(1120)2019.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Yan HH, Pickup M, Pang Y, Gorska AE, Li Z,
Chytil A, Geng Y, Gray JW, Moses HL and Yang L: Gr-1+CD11b+ myeloid
cells tip the balance of immune protection to tumor promotion in
the premetastatic lung. Cancer Res. 70:6139–6149. 2010.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Kusmartsev S, Nefedova Y, Yoder D and
Gabrilovich DI: Antigen-specific inhibition of CD8+ T cell response
by immature myeloid cells in cancer is mediated by reactive oxygen
species. J Immunol. 172:989–999. 2004.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Ochoa AC, Zea AH, Hernandez C and
Rodriguez PC: Arginase, prostaglandins, and myeloid-derived
suppressor cells in renal cell carcinoma. Clin Cancer Res.
13:721s–726s. 2007.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Almand B, Clark JI, Nikitina E, van Beynen
J, English NR, Knight SC, Carbone DP and Gabrilovich DI: Increased
production of immature myeloid cells in cancer patients: A
mechanism of immunosuppression in cancer. J Immunol. 166:678–689.
2001.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Mirza N, Fishman M, Fricke I, Dunn M,
Neuger AM, Frost TJ, Lush RM, Antonia S and Gabrilovich DI:
All-trans-retinoic acid improves differentiation of myeloid cells
and immune response in cancer patients. Cancer Res. 66:9299–9307.
2006.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Alshetaiwi H, Pervolarakis N, McIntyre LL,
Ma D, Nguyen Q, Rath JA, Nee K, Hernandez G, Evans K, Torosian L,
et al: Defining the emergence of myeloid-derived suppressor cells
in breast cancer using single-cell transcriptomics. Sci Immunol.
5(eaay6017)2020.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Shou D, Wen L, Song Z, Yin J, Sun Q and
Gong W: Suppressive role of myeloid-derived suppressor cells
(MDSCs) in the microenvironment of breast cancer and targeted
immunotherapies. Oncotarget. 7:64505–64511. 2016.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Baert T, Vankerckhoven A, Riva M, Van
Hoylandt A, Thirion G, Holger G, Mathivet T, Vergote I and
Coosemans A: Myeloid derived suppressor cells: Key drivers of
immunosuppression in ovarian cancer. Front Immunol.
10(1273)2019.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Taki M, Abiko K, Baba T, Hamanishi J,
Yamaguchi K, Murakami R, Yamanoi K, Horikawa N, Hosoe Y, Nakamura
E, et al: Snail promotes ovarian cancer progression by recruiting
myeloid-derived suppressor cells via CXCR2 ligand upregulation. Nat
Commun. 9(1685)2018.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Filipazzi P, Valenti R, Huber V, Pilla L,
Canese P, Iero M, Castelli C, Mariani L, Parmiani G and Rivoltini
L: Identification of a new subset of myeloid suppressor cells in
peripheral blood of melanoma patients with modulation by a
granulocyte-macrophage colony-stimulation factor-based antitumor
vaccine. J Clin Oncol. 25:2546–2553. 2007.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Pandit R, Lathers DM, Beal NM, Garrity T
and Young MR: CD34+ immune suppressive cells in the peripheral
blood of patients with head and neck cancer. Ann Otol Rhinol
Laryngol. 109:749–754. 2000.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Lathers DM, Achille N, Kolesiak K, Hulett
K, Sparano A, Petruzzelli GJ and Young MR: Increased levels of
immune inhibitory CD34+ progenitor cells in the peripheral blood of
patients with node positive head and neck squamous cell carcinomas
and the ability of these CD34+ cells to differentiate into immune
stimulatory dendritic cells. Otolaryngol Head Neck Surg.
125:205–212. 2001.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Suzuki E, Kapoor V, Jassar AS, Kaiser LR
and Albelda SM: Gemcitabine selectively eliminates splenic
Gr-1+/CD11b+ myeloid suppressor cells in tumor-bearing animals and
enhances antitumor immune activity. Clin Cancer Res. 11:6713–6721.
2005.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Blattner C, Fleming V, Weber R, Himmelhan
B, Altevogt P, Gebhardt C, Schulze TJ, Razon H, Hawila E, Wildbaum
G, et al: CCR5+ myeloid-derived suppressor cells are
enriched and activated in melanoma lesions. Cancer Res. 78:157–167.
2018.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Lin S, Wang J, Wang L, Wen J, Guo Y, Qiao
W, Zhou J, Xu G and Zhi F: Phosphodiesterase-5 inhibition
suppresses colonic inflammation-induced tumorigenesis via blocking
the recruitment of MDSC. Am J Cancer Res. 7:41–52. 2017.PubMed/NCBI
|
|
22
|
Kim K, Skora AD, Li Z, Liu Q, Tam AJ,
Blosser RL, Diaz LA Jr, Papadopoulos N, Kinzler KW, Vogelstein B
and Zhou S: Eradication of metastatic mouse cancers resistant to
immune checkpoint blockade by suppression of myeloid-derived cells.
Proc Natl Acad Sci USA. 111:11774–11779. 2014.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Steele CW, Karim SA, Leach JDG, Bailey P,
Upstill-Goddard R, Rishi L, Foth M, Bryson S, McDaid K, Wilson Z,
et al: CXCR2 inhibition profoundly suppresses metastases and
augments immunotherapy in pancreatic ductal adenocarcinoma. Cancer
Cell. 29:832–845. 2016.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Sun L, Clavijo PE, Robbins Y, Patel P,
Friedman J, Greene S, Das R, Silvin C, Van Waes C, Horn LA, et al:
Inhibiting myeloid-derived suppressor cell trafficking enhances T
cell immunotherapy. JCI Insight. 4(e126853)2019.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Youn JI, Collazo M, Shalova IN, Biswas SK
and Gabrilovich DI: Characterization of the nature of granulocytic
myeloid-derived suppressor cells in tumor-bearing mice. J Leukoc
Biol. 91:167–181. 2012.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Dufait I, Schwarze JK, Liechtenstein T,
Leonard W, Jiang H, Escors D, De Ridder M and Breckpot K: Ex vivo
generation of myeloid-derived suppressor cells that model the tumor
immunosuppressive environment in colorectal cancer. Oncotarget.
6:12369–12382. 2015.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Schröder M, Loos S, Naumann SK, Bachran C,
Krötschel M, Umansky V, Helming L and Swee LK: Identification of
inhibitors of myeloid-derived suppressor cells activity through
phenotypic chemical screening. Oncoimmunology.
6(e1258503)2016.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Marigo I, Bosio E, Solito S, Mesa C,
Fernandez A, Dolcetti L, Ugel S, Sonda N, Bicciato S, Falisi E, et
al: Tumor-induced tolerance and immune suppression depend on the
C/EBPbeta transcription factor. Immunity. 32:790–802.
2010.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Lechner MG, Megiel C, Russell SM, Bingham
B, Arger N, Woo T and Epstein AL: Functional characterization of
human Cd33+ and Cd11b+ myeloid-derived suppressor cell subsets
induced from peripheral blood mononuclear cells co-cultured with a
diverse set of human tumor cell lines. J Transl Med.
9(90)2011.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Zhang Y, Wilt E and Lu X: Human isogenic
cell line models for neutrophils and myeloid-derived suppressor
cells. Int J Mol Sci. 21(7709)2020.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Fultang L, Panetti S, Ng M, Collins P,
Graef S, Rizkalla N, Booth S, Lenton R, Noyvert B, Shannon-Lowe C,
et al: MDSC targeting with gemtuzumab ozogamicin restores T cell
immunity and immunotherapy against cancers. EBioMedicine.
47:235–246. 2019.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Sheng Y, Duan X, Liu Y, Li F, Ma S, Shang
X, Wang X, Liu Y, Xue R and Qin Z: Tie2-expressing
monocytes/macrophages promote cerebral revascularization in
peri-infarct lesions upon ischemic insult. Signal Transduct Target
Ther. 6(295)2021.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Casacuberta-Serra S, Parés M, Golbano A,
Coves E, Espejo C and Barquinero J: Myeloid-derived suppressor
cells can be efficiently generated from human hematopoietic
progenitors and peripheral blood monocytes. Immunol Cell Biol.
95:538–548. 2017.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Bian Z, Abdelaal AM, Shi L, Liang H, Xiong
L, Kidder K, Venkataramani M, Culpepper C, Zen K and Liu Y:
Arginase-1 is neither constitutively expressed in nor required for
myeloid-derived suppressor cell-mediated inhibition of T-cell
proliferation. Eur J Immunol. 48:1046–1058. 2018.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Zhan X, Hu S, Wu Y, Li M, Liu T, Ming S,
Wu M, Liu M and Huang X: IFN-γ decreased the suppressive function
of CD33+HLA-DRlow myeloid cells through
down-regulation of PD-1/PD-L2 signaling pathway. Mol Immunol.
94:107–120. 2018.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Weber R, Riester Z, Hüser L, Sticht C,
Siebenmorgen A, Groth C, Hu X, Altevogt P, Utikal JS and Umansky V:
IL-6 regulates CCR5 expression and immunosuppressive capacity of
MDSC in murine melanoma. J Immunother Cancer.
8(e000949)2020.PubMed/NCBI View Article : Google Scholar
|


















