Introduction
Among all intracranial tumors, the incidence of
glioma is relatively high, accounting for ~80% of the cases with
this disease (1). In addition,
glioma is a challenging disease to be treated by surgery (total
removal) and the prognosis is relatively poor with common
recurrence of the tumor. Due to differences in age, gender and
other reasons, the incidence of glioma also differs (2). In spite of the use of multimodal
therapy, such as surgical excision, complete resection is
considered to be impossible since a large amount of brain tissue is
involved. In addition, due to the strong resistance of glioma to
chemotherapy, its aggressiveness and the unique pathophysiological
characteristics of the central nervous system (3), the recurrence rate of this disease
approaches 100%. Therefore, screening influential prognostic
factors is considered to be an effective solution to the
aforementioned problems.
In addition, accumulating evidence has revealed the
importance of the tumor microenvironment (TME) in the trend of
tumor growth (4,5). The synergistic interaction between
cancer and their supporting cells leads to the immortal status of
cancer cells resulting in uncontrolled proliferation, resistance to
apoptosis and evasion of immune surveillance, all of which comprise
the malignant phenotypes of cancer. Therefore, the treatment
response and clinical outcome of patients with cancer are
significantly affected by the TME (6,7). The
structural components of the TME mainly consist of host stromal
cells and recruited immune cells. By studying the interaction of
the TME with its various components it is possible to combine the
traits of tumor cells with the TME mechanism, leading to the
identification of novel biomarkers and potential drug targets for
biotherapy (7). The TME mesenchyma
is a genetically stable therapeutic target, unlike tumor cells that
constantly mutate. The infiltrating immune cells are mainly
composed of macrophages and monocytes and are deemed to exhibit
tumor-promoting and immunosuppressive functions (8). The tumor-infiltrating immune cells
(TICs) are markedly related to glioma cells. A previous study has
indicated that the TICs of TME are a prospective indicator of a
curative effect (9).
Based on bioinformatics analysis, an optimal
biomarker for biological prediction was developed as a prognostic
indicator. This biomarker was termed spleen tyrosine kinase (SYK),
a non-receptor type of protein tyrosine kinases in the Src family
(10). SYK has been widely
reported in various hematopoietic malignancies and certain primary
epithelial tumors as a pro-survival factor; it has been detected in
several cell types, such as melanocytes, human nasal fibroblasts
and liver cells (11-13).
However, a limited number of reports have investigated the role of
SYK in glioma (14). In the
present study, it was hypothesized that SYK was also involved in
the pathogenesis of glioma and that SYK intervention may be pivotal
in delaying the deterioration of glioma and improve the prognosis
of these patients. Therefore, an in-depth study was conducted on
the differential genes between the immune and the matrix components
in glioma specimens in order to explore the potential impact of
this biomarker in the TME of glioma.
Materials and methods
Data preparation
The transcriptome RNA-sequencing data were available
in the Genomic Data Commons (GDC) portal of The Cancer Genome Atlas
(TCGA) GDC official website (https://portal.gdc.cancer.gov/). The data
corresponding to 698 patients with glioma and their mRNA expression
levels were obtained.
Acquisition of immune and stromal
scores and Estimation of STromal and Immune cells in MAlignant
Tumor tissues using Expression data (ESTIMATE) score
The ratio of the immune-stromal component of each
sample in the TME was computed by the R language (version 3.6.2;
r-project.org/) using ESTIMATE (15). The ratio was expressed in three
different forms, including the immune score, the stromal score and
the ESTIMATE score, which were positively associated with the
proportion of immune and stromal cells and the sum of the two,
respectively; this indicated that as the score for each component
increased, their corresponding component accounted for a more
significant proportion of the TME.
Survival analysis
Survival analysis was performed using the R language
(version 4.0.1; https://www.r-project.org/) survival and survminer
packages. The ggsurvplot function was used to plot the survival
curve. A total of 510 out of 698 tumor samples had detailed
survival records ranging from 0-12 years that could be selected to
conduct survival analysis. The Kaplan-Meier method was used to
depict the survival spline and the logarithmic rank was utilized to
assess the statistical significance. P<0.05 was considered to
indicate a statistically significant difference.
Discrepancy analysis of scores with
clinicopathological stages
The clinical data received from the glioma samples
were obtained from the TCGA database. The data were analyzed by R.
In addition, Wilcoxon rank sum or Kruskal-Wallis's rank sum tests
were regarded as the important tests according to the count of the
clinical stages for comparison.
Heatmaps
The heatmaps of the differentially expressed genes
(DEGs) were generated using the R language with package pheatmap.
In this way, the results could be divided into high-expression and
low-expression groups.
Analysis of DEGs in the immune and
stromal scores between high-score and low-score groups
The immune and stromal scores were compared with the
median scores and 698 tumor samples were marked with high or low
scores. The R language with the Limma package (16) (http://www.bioconductor.org/packages/release/bioc/html/limma.html)
was applied to analyze gene expression differentiation; the DEGs
were obtained by comparing the high-score with the low-score
samples. The log2 values obtained from the high-score compared with
the low-score groups of DEGs were converted into decimals and fold
change (FC). A result >1 and a false discovery rate (FDR)
<0.05 were considered meaningful.
Gene Ontology (GO) and Kyoto
Encyclopedia of Genes and Genomes (KEGG) enrichment analyses
The Database for Annotation, Visualization and
Integrated Discovery database (http://david.ncifcrf.gov) merges biological data and
analysis tools that can be used for genetic difference analysis as
well as pathway enrichment. A total of 1,438 DEGs were outlined
with GO and KEGG (genome.jp/) enrichment analyses using cluster
profile, enrichplot and ggplot2 package in the R language. Only the
results with P-value and q-value of <0.05 were considered to be
markedly enriched.
Protein-protein interaction (PPI)
network and Cox regression analyses
The Search Tool for the Retrieval of Interacting
Genes/Proteins (STRING) database (http://string-db.org/) was employed to establish the
PPI network and subsequently the Cytoscape version 3.7.2 was used
to reestablish the network. The network was constructed by using
nodes with the confidence of interaction >0.95. In addition, the
intersection of the previous 30 genes in the PPI network was
selected as an example and the univariate Cox regression study was
analyzed using the survival package in R language.
Gene set enrichment analysis
(GSEA)
GSEA is a reliable and practical gene analysis
method (17), which confirms the
correlation between the target gene required and the signal
transduction pathway by ranking the target gene's high- and
low-expression status. GSEA analysis included the total
transcriptome of the entire tumor samples and only the gene sets
with NOM P<0.05 and FDR q<0.06 were regarded as
exceptional.
Cell culture
The present study used U87 as tumor cells and HEB as
normal cells; HEB and U87 cells (ATCC HTB-14) were purchased from
the American Type Culture Collection. The U87 cell line has been
analyzed and verified by STR (Haixing Biosciences Co., Ltd). The
cells were cultured at 37˚C in 5% CO2 in a mixture of
DMEM (Gibco; Thermo Fisher Scientific, Inc.) with 10% FBS (Gibco;
Thermo Fisher Scientific, Inc.) and cultured in an incubator WH-400
(Wiggens GmbH).
Reverse transcription-quantitative
(RT)-qPCR
A total of 1x106 cells used for RT-qPCR,
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) was used for total RNA extraction, RNA reversed transcription
using Takara PrimeScript RT reagent kit (Takara Biotechnology Co.,
Ltd.) and analyzed by RT-qPCR using Takara TB Green Premix Ex Taq
II (Takara Biotechnology Co., Ltd.) in Bio-Rad iQ5 system (Bio-Rad
Laboratories, Inc.). The following thermocycling conditions were
used for qPCR: Initial denaturation at 95˚C for 1 min, 40 cycles at
95˚C for 5 sec, 60˚C for 30 sec and 72˚C for 20 sec. All samples
used β-actin as an internal reference and the results were
quantified for relative expression by the 2-ΔΔCq method
(18). RNA extraction, cDNA
synthesis and qPCR were performed according to the manufacturer's
protocols; these experiments replicated three times. The primers
were as follows: SYK forward, 5'-CTGTCGGTGGCTGCCTTTGAC-3' and
reverse, 5'-TGTGGAGGGTGAGTCCTGGG-3'; β-actin forward,
5'-TGGCACCCAGCACAATGAA-3' and reverse,
5'-CTAAGTCAGAGTCCGCCTAGAAGCA-3'.
Western blotting
HEB and U87 were collected, the total cellular
protein was extracted by adding RIPA lysis buffer (Beijing Solarbio
Science & Technology Co., Ltd.) and the protein concentration
was determined by the BCA method (Beijing Solarbio Science &
Technology Co., Ltd.). The total protein (50 µg/per lane) from each
sample was separated using 10% SDS-gel. The proteins were separated
by sodium dodecyl sulfate-polyacrylamide gel electrophoresis
(SDS-PAGE) and then transferred onto a PVDF (Roche Diagnostics
GmbH). After being blocked with 5% (w/v) skimmed milk for 2 h at
room temperature, followed by incubation with the primary anti-SYK
antibodies (1:1,000; cat. no. A2123; ABclonal Biotech Co., Ltd.)
and anti-β-actin antibodies (1:4,000; cat. no. AY0573; Shanghai
Abways Biotechnology Co., Ltd.) overnight at 4˚C. Following
incubation with goat anti-rabbit IgG/HRP (1:1,000; cat. no.
ab131368; Abcam) and goat anti-mouse IgG/HRP conjugated secondary
antibodies (1:1,000; cat. no. SE131; Beijing Solarbio Science &
Technology Co., Ltd.) for 2 h at room temperature, targeted bands
were developed using the BeyoECL Plus kit (MilliporeSigma) and the
grayscale values of each band were analyzed by Image J software
(version 2.0, National Institutes of Health) and assessed
semiquantitatively.
Statistical analysis
Experiments were statistically evaluated using
GraphPad Prism 8.0 (GraphPad Software, Inc.) and the results are
presented as mean ± standard deviation (SD). Unpaired Student's
t-test was used to evaluate differences between groups. Pearson's
correlation coefficient was utilized to analyze the correlation.
P<0.05 was considered to indicate a statistically significant
difference.
Results
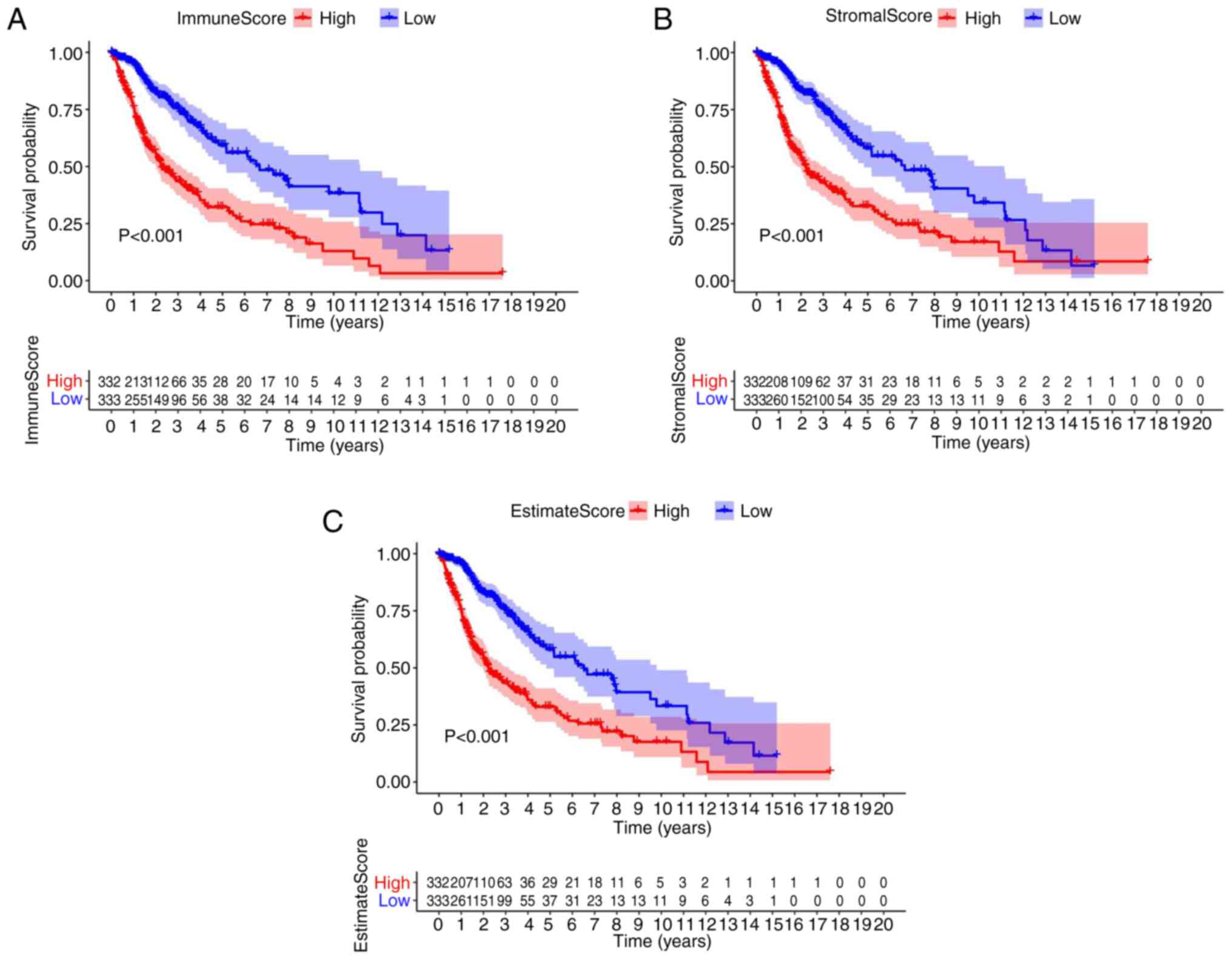
Association of survival with the score
of patients with glioma
To ascertain the association between immune and
stromal estimate ratio and the survival rate, survival analysis was
performed for the immune score, stromal score, as well as for the
ESTIMATE score, using the Kaplan-Meier survival analysis. The
counts of the immune or stromal components of the TME were
expressed as estimates in the immune score or stromal score. In
comparison with the median, patients with glioma were segmented
into high and low groups. The results indicated that the scores of
the immune and stromal cell contents were markedly associated with
the survival of patients with glioma (Fig. 1A-C). In brief, the data revealed
that the immune, stromal and estimate components of the TME were
more appropriate indicators of the prognosis in patients with
glioma (P<0.01).
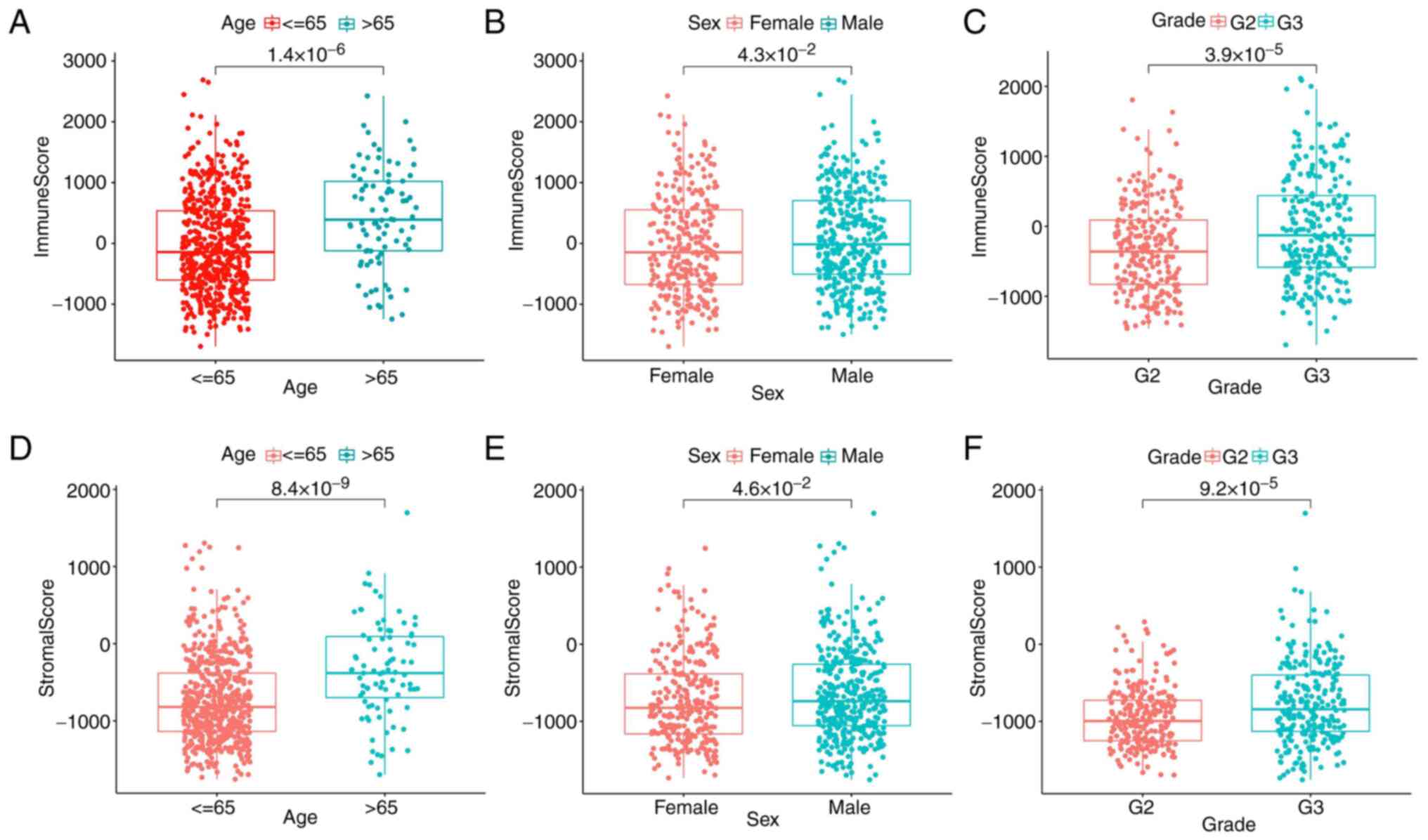
Association of the score with the
clinicopathological stage of patients with glioma
The clinical information of patients with glioma in
the TCGA database was obtained to ascertain the association between
immune and stromal components and the clinicopathological traits.
The results found that the immune scores of patients >65 years
were significantly higher than those of patients <65 years
(Fig. 2A) and the immune scores of
patients with G3 grade were significantly higher than those of G2
grade (Fig. 2C). It was noteworthy
that there was little association with sex. However, stromal scores
were positively associated with tumor age and grade (Fig. 2D and F), sex had less influence on stromal
scores (Fig. 2E). All the results
further indicated that the quantity of immune and stromal
components was relevant to glioma evolution, such as age and grade.
However, there was little association with sex.
The DEGs shared by the immune and
stromal scores are principally enriched in immune-related
genes
Comparative analyses of high- and low-score samples
were performed to ensure the substantial variation in the gene
profiles of the immune and stromal components of the TME. Compared
with the median, 1,655 DEGs were selected in the high- and
low-score samples of the immune score. A total of 1,026 upregulated
and 629 downregulated genes were identified in all gene sets
(Fig. 3A, C and D).
By analogy, 1,813 DEGs were acquired in the stromal score, with
1,204 upregulated and 609 downregulated genes (Fig. 3B and D). The intersecting point analysis of the
Venn plot indicated that 950 upregulated genes overlapped in the
aggregate in the immune score and stromal score analysis, whereas
488 downregulated genes overlapped in the same type of analysis.
These genes (a total of 1,438 genes) may influence the status of
the TME. During KEGG enrichment analysis, neuroactive
ligand-receptor interaction, cytokine-cytokine receptor interaction
and tuberculosis enrichment were shown (Fig. 3E). From GO enrichment analysis, it
was concluded that the DEGs identified were mainly consistent with
the GO terms regarding immunity. For example, the terms neutrophil
activation, neutrophil-mediated immunity and leukocyte migration
were highlighted (Fig. 3F).
Therefore, the entire function in DEGs was seemingly connected with
immune-related activities, which confirmed that the existence of
immune factors had an influence on the TME of patients with
glioma.
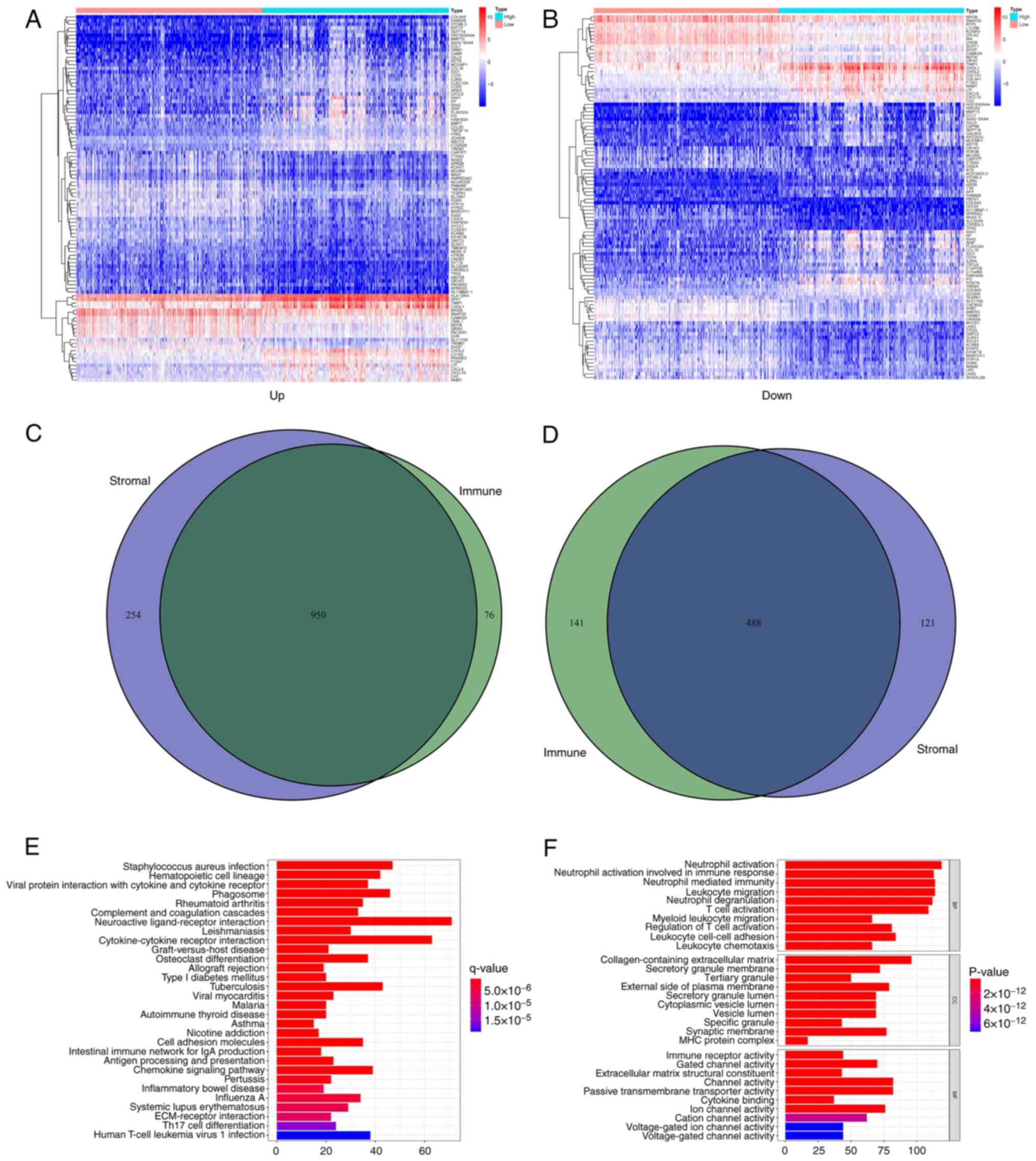 | Figure 3Heatmaps, Venn plots and enrichment
analysis of GO and KEGG of DEGs. (A) Heatmap of DEGs obtained by
comparing the high and low score group in the immune score. The
abscissa is the name with 100 genes and the vertical is the sample
ID (not shown in the figure). DEGs were obtained by Wilcoxon rank
sum test (q=0.05 and FC >1) following log2 conversion as the
importance threshold. (B) Heatmap of DEGs obtained by comparing the
high and low score group in the stromal score. (C) Venn plots for
upregulated DEGs both to the immune score and the stromal score
with q<0.05 and FC >1 following log2 conversion as the DEG
importance filtering threshold. (D) Venn plots for downregulated
DEGs both to the immune score and the stromal score with q<0.05
and FC >1 following log2 conversion as the DEGs importance
filtering threshold. (E) The barplot obtained by GO enrichment
analysis, included BP, CC and MF. The abscissa represents the
number of gene enrichments, whereas the color represents the
significance of each gene enrichment. (F) The barplot obtained by
KEGG enrichment analysis. GO, Gene enrichment; KEGG, Kyoto
Encyclopedia of Genes and Genomes; DEGs, differentially expressed
genes; ID, identity; FC, fold change; BP, Biological Process; CC,
Cellular Component; MF, Molecular Function. |
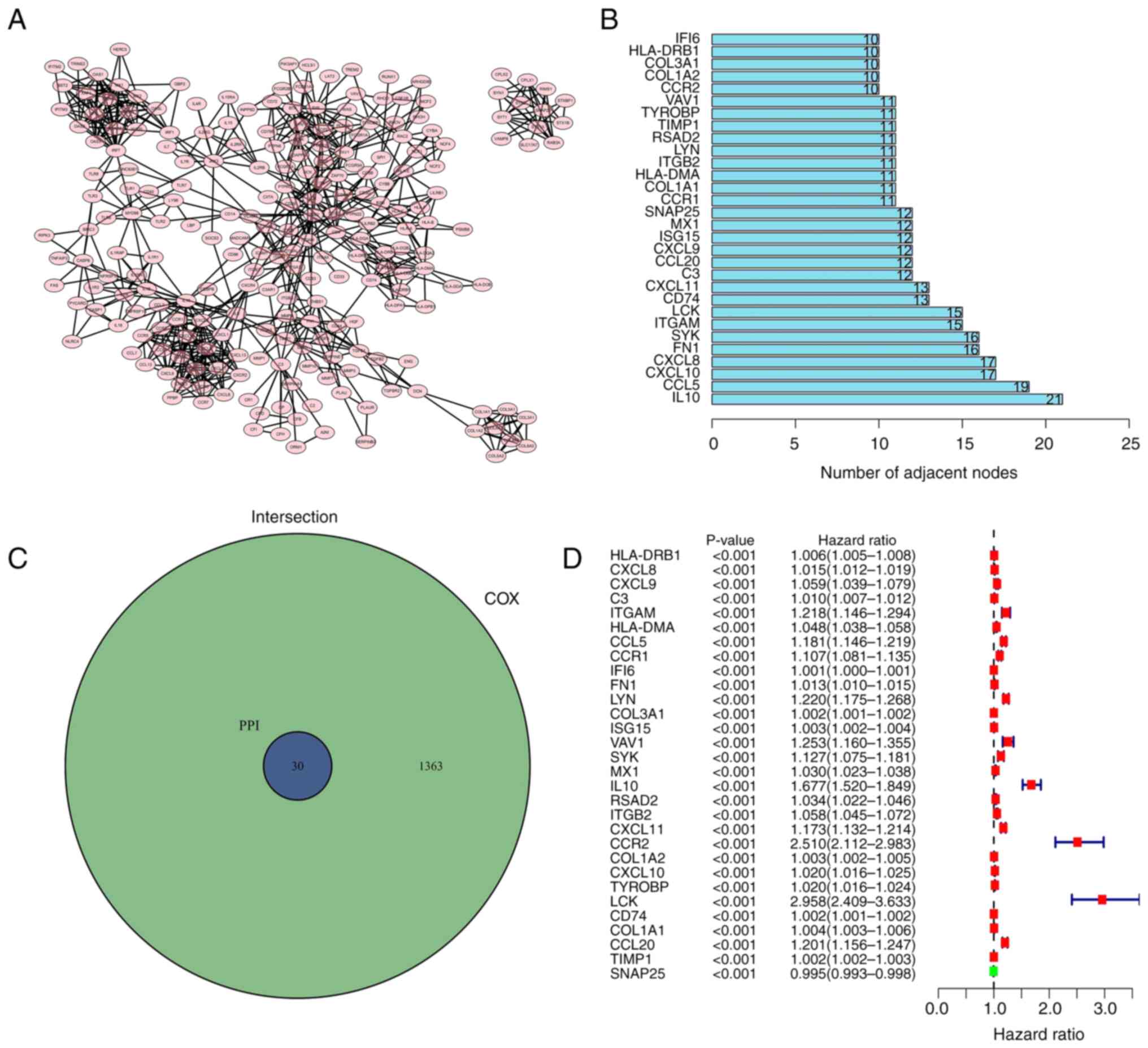
Intersection analysis of the PPI
network and univariate Cox regression analysis
To investigate whether protein interactions
/Users/e.kouneni/Downloads/ETM-19384-290776/FIG.3.tif
were present among the 1,438 DEGs, the
PPI network was established using Cytoscape software (3.7.2)
according to the STRING database
Fig. 4A displays
the interaction between genes (confidence interval 0.95); the bar
chart indicates the previous 30 genes permutated by the count of
nodes (Fig. 4B). A total of 30
genes were intersected and screened in the PPI and the genes with a
P-value of <0.05 were selected by the univariate Cox regression
analysis (Fig. 4C). To determine
the risk value among the 30 factors, a univariate Cox regression
analysis was performed on patients with glioma. Fig. 4D indicated that only a limited
number of genes exhibited low-risk values.
Association between survival analysis
and SYK expression in patients with glioma
The risk genes (ITGAM; CCL5; CCR1; LYN; SYK; VAV1;
IL10; CXCL11; CCR2; LCK and CCL20) were selected through
intersection analysis of the PPI network and univariate Cox
regression analysis. SYK ranked in the top five of genes identified
in the intersection analysis of the PPI network. SYK demonstrated
significance in the univariate Cox regression analysis. The genes
were queried one by one using gene card (https://www.genecards.org). The analysis indicated
that SYK demonstrated a significant association with the occurrence
and development of tumors (19).
SYK has been extensively studied in the occurrence and development
of breast, pancreatic and colon cancers, as well as in the
development of other types of tumor. However, the studies that have
examined its role in glioma are not in-depth analyses (20-22).
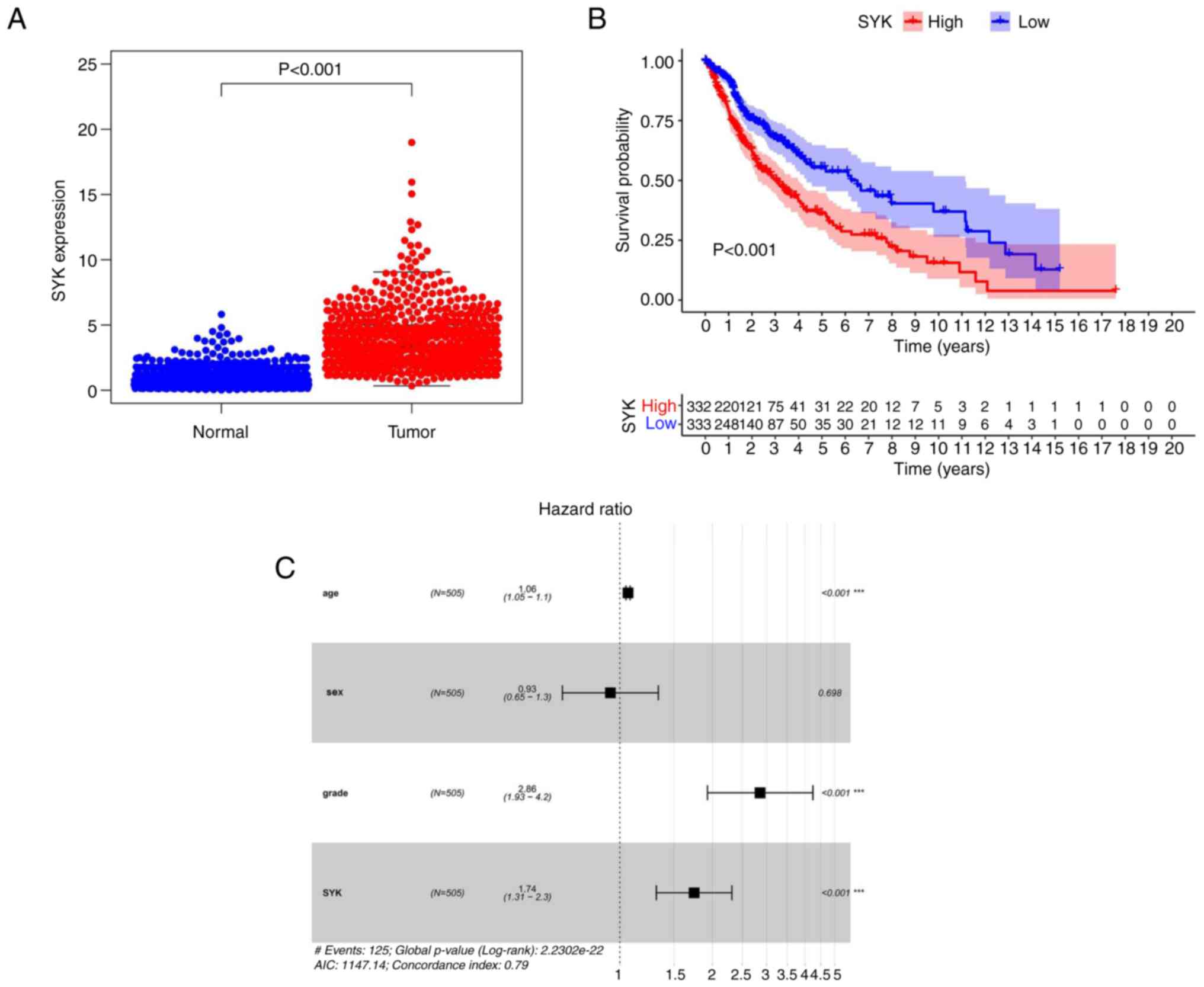
SYK was finally selected. The data obtained from the TCGA database
indicated that SYK expression was significantly increased in the
tumor group compared with that noted in the normal tissue group
(P<0.001; Fig. 5A). Survival
analysis of SYK indicated that the survival rate of patients with
glioma and low SYK expression was apparently higher compared with
that noted in patients with glioma and high SYK expression.
Moreover, the entire survival of tumor patients was apparently
reduced (Fig. 5B). Multivariate
Cox survival analysis confirmed that a high SYK expression level
was an autocephalous predictor of undesirable prognosis in patients
with glioma (P<0.001). The results indicated that SYK was a
high-risk gene in glioma (Fig.
5C). In summary, these results indicated that SYK exhibited a
significantly higher expression in glioma samples compared with
that noted in standard samples; its expression was an autocephalous
predictor of undesirable prognosis in patients with glioma.
SYK may be an indicator of TME
regulation
Since SYK levels were inversely associated with the
survival of patients with glioma, their data were divided into
high-expression and low-expression groups and subsequently
severally contrasted to the median level of SYK expression in GSEA.
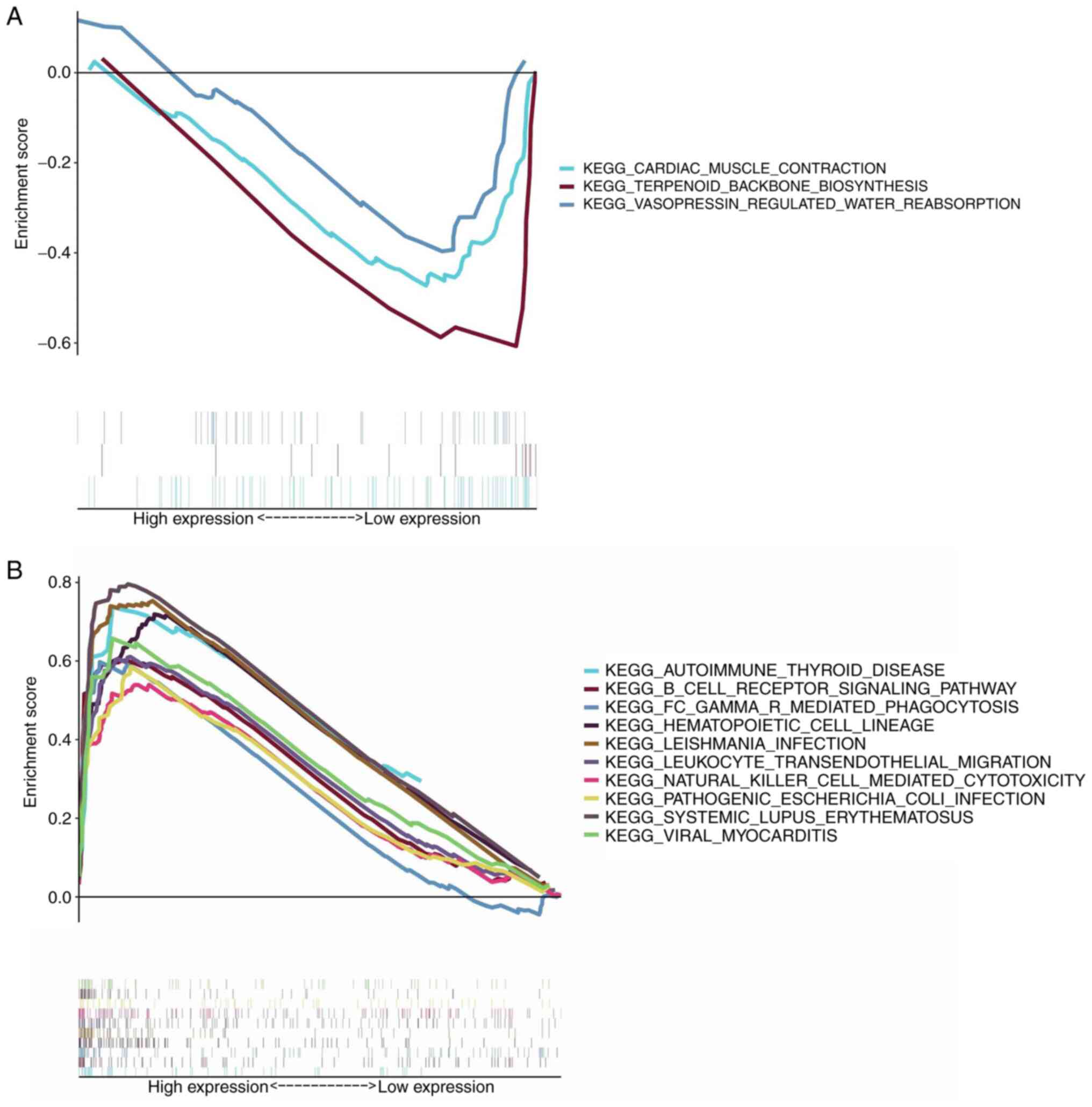
As shown in Fig. 6A, almost no
gene set enrichment was noted in the low-expression group of SYK.
The SYK high-expression group was principally enriched in
immune-related activities. For example, the B cell receptor
signaling pathway, the hematopoietic cell lineage and the
autoimmune thyroid disease were the main enriched pathways
(Fig. 6B). The aforementioned
results revealed that SYK could be a potential indicator of the TME
status.
Correlation between SYK expression and
the ratio of TICs
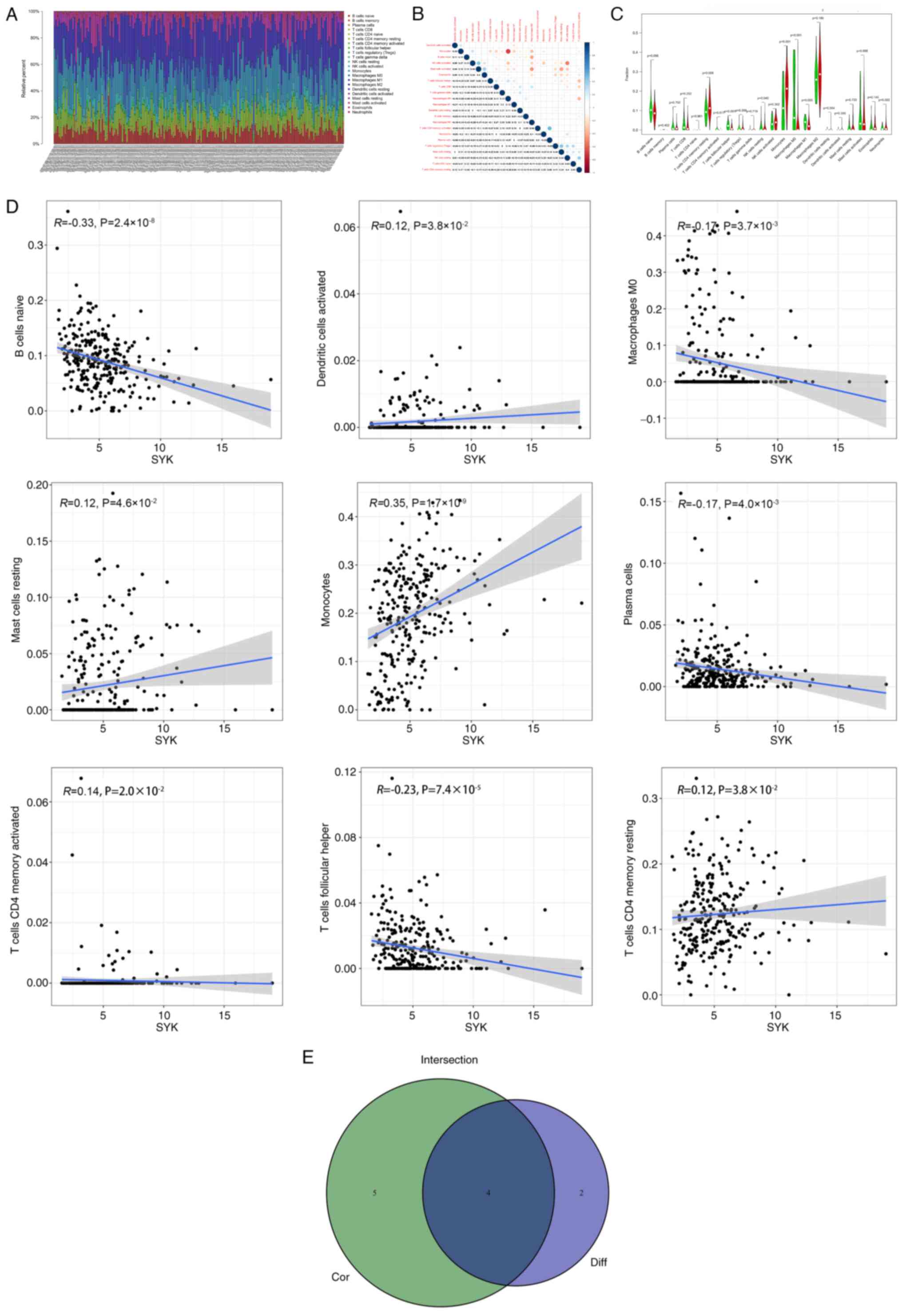
To further verify the association between the
expression of SYK and the immune microenvironment, the ratio of
tumor-infiltrating immune subsets was analyzed by utilizing the
CIBERSORT algorithm. A spectrum of 22 immune cells was established
in glioma samples (Fig. 7A and
B). The immune cell difference
analysis results indicated that six different immune cells were
associated with the expression of SYK (Fig. 7C). The immune cell correlation
analysis results indicated that nine different immune cells were
associated with the expression of SYK (Fig. 7D). The difference and correlation
analyses indicated that four different immune cells were associated
with SYK expression. Among them, two types of immune cells
positively correlated with SYK expression, which comprised cluster
of differentiation (CD) 4 memory resting T cells and monocytes and
two different types of immune cells were inversely associated with
SYK expression, including T follicular helper cells and macrophages
M0 (Fig. 7E). These results
demonstrated the contribution of SYK in modulating the immune
response and playing a crucial role in the development of the
TME.
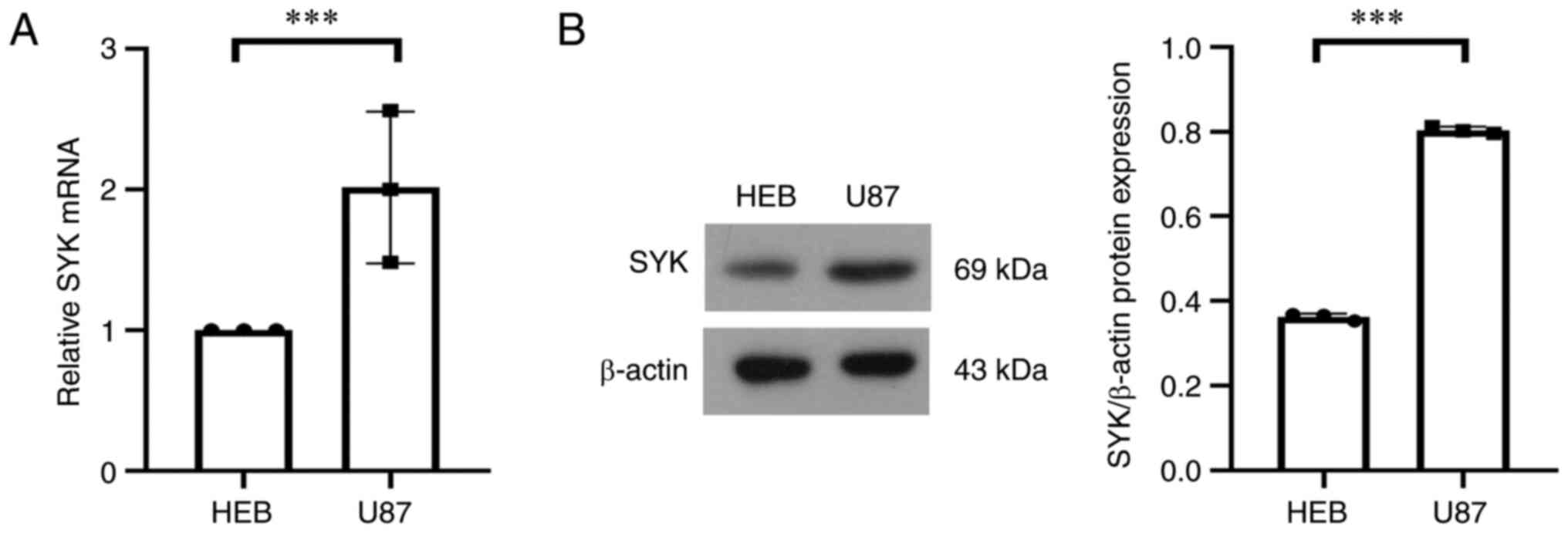
The expression of SYK is associated
with glioma
According to the results of the present study, the
expression of SYK was closely associated with glioma. The
expression of SYK in tumor cells and normal cells was verified by
using western blot and RT-qPCR and the results showed that the
expression level of SYK in tumor cells was significantly higher
compared with that in normal cells (Fig. 8A and B). These results further proved that the
high expression of SYK was closely associated with the development
of glioma in terms of protein expression and gene expression.
Discussion
In the present study, the data derived from the TCGA
database were used to determine the TME genes associated with
survival in patients with glioma. By using Cox regression analysis,
SYK was finally identified from the list of DEGs to be closely
associated with the survival of patients with glioma. SYK was shown
to participate in immunization processes. Eventually, SYK was
confirmed to be a preponderant target of the TME status for
patients with glioma by a series of bioinformatics analyses.
During tumor occurrence and development, the TME is
always in effect. Therefore, the exploration of the latent remedial
targets of TME refactoring and the further induction of the
TME-related changes from a tumor-friendly to a tumor-suppressive
phenotype is required for conducting valuable research. Previous
studies have shown that the occurrence of tumors is markedly
associated with the immune microenvironment (23,24).
In the transcriptional analysis of glioma, the analysis of the data
downloaded from the TCGA database indicated that the immune
component of the TME exhibited a particular effect on the
postoperative predictive outcomes of the patients. In particular,
the evolution in glioma (such as intrusion and transfer) was
associated with the quantity of immune and stromal components in
the TME. The aforementioned analysis indicated the importance of
investigating the mutual effect of tumor and immune cells, which
can provide new directions for developing more promising treatment
options.
Following systematic analysis, the most significant
gene, SYK, was identified to be responsible for the survival of
patients with glioma. SYK is a pivotal member in the immune cell
signaling pathways as it modulates proliferation, differentiation
and cell survival by activating a series of signal transduction
pathways, such as an element of immune receptor signal transduction
(25,26). In addition, SYK is a crucial part
of the B lymphocyte signaling receptor (27) and it can regulate a variety of
biological functions of B lymphocytes; it is also closely connected
with the activation and maturation of B cells (12). The lack of SYK expression, which is
involved in the TME, leads to impaired development and maturation
of immune cells and, in severe cases, to severe combined
immunodeficiency disease. Therefore, in the presence of mutations,
abnormal proliferation of cells occurs, which leads to the loss of
immunity and eventually to tumor development (28). Several studies have investigated
the function of SYK inhibitors. Entospletinib, an inhibitor of SYK,
has shown promising results in clinical trials performed for the
treatment of B cell malignancies (29,30).
In addition, several oral SYK inhibitors have already been
evaluated in clinical trials, including fostamatinib (R788),
entospletinib (GS-9973) and TAK-659 (31-33).
It has been shown that SYK inhibition can block the propagation and
migration of glioma cells in vitro (14). Furthermore, SYK may be involved in
the regulation of macrophage polarization in TME (34). Therefore, a deeper analysis of the
relationship in SYK expression and the TME was performed. Certain
immune-related signaling pathways were identified by GSEA analysis.
For example, FC gamma R-mediated phagocytosis, the B cell receptor
signaling pathway and the hematopoietic cell lineage pathway were
markedly enriched in the SYK high-expression group. The violin plot
indicated that the number of T cell CD4 memory resting, monocytes
and M2 macrophages in the SYK high-expression group was higher
compared with that of the SYK low-expression group, which revealed
that the expression of SYK was closely associated with the number
of immune cells in the TME. Macrophages are roughly segmented into
M1 and M2 genres according to their functions. The M1 macrophages
participate in the inflammatory response, pathogen clearance and
antitumor immunity. Nevertheless, the M2 macrophages are different
from M1 and they influence the anti-inflammatory response, the
wound healing process and the pro-tumorigenic properties (35,36).
As depicted in the violin diagram, the number of M1 macrophages was
lower and that of M2 was higher in the high-expression group of
SYK, further supporting the possibility that SYK may take part in
the tumor-promoting properties of glioma.
The current study suggested that SYK may affect the
proliferation and migration of glioma cells by affecting the B cell
receptor signaling pathways and the hematopoietic cell lineage. In
summary, SYK is a potential cancer-promoting gene and it is
expected to be a new target for the treatment of glioma; it may
also provide new ideas for the therapy of glioma. Therefore, the
accuracy of the combined analysis requires further investigation to
define SYK expression, tumor-infiltrating B cell subtypes and
mutagen-driven patterns prior to the treatment of patients with
glioma with SYK inhibitors. However, the present study lacks
glioblastoma tumor tissues and the immunological role and specific
mechanisms of SYK in glioma have not been explored in depth. The
role of SYK and its mechanism in glioma will continue to be
explored through cell experiments and animal experiments in the
future.
Acknowledgements
Not applicable.
Funding
Funding: This study was supported by the Natural Science
Foundation of Shandong Province, China (grant no.
ZR202102190696).
Availability of data and materials
The datasets generated and/or analyzed during the
current study are available in the TCGA database (https://portal.gdc.cancer.gov).
Authors' contributions
CW, PL and YS performed the experiments and wrote
the manuscript. TL, XX and JG searched the literature and analyzed
the data. HS and ZG searched the literature and revised the
manuscript. RX designed the experiments and revised the manuscript.
All authors read and approved the final manuscript. CW and RX
confirm the authenticity of all the raw data.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ostrom QT, Bauchet L, Davis FG, Deltour I,
Fisher JL, Langer CE, Pekmezci M, Schwartzbaum JA, Turner MC, Walsh
KM, et al: The epidemiology of glioma in adults: A ‘state of the
science’ review. Neuro Oncol. 16:896–913. 2014.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Gigineishvili D, Shengelia N, Shalashvili
G, Rohrmann S, Tsiskaridze A and Shakarishvili R: Primary brain
tumour epidemiology in Georgia: First-year results of a
population-based study. J Neurooncol. 112:241–246. 2013.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Lee E, Yong RL, Paddison P and Zhu J:
Comparison of glioblastoma (GBM) molecular classification methods.
Semin Cancer Biol. 53:201–211. 2018.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Hinshaw DC and Shevde LA: The tumor
microenvironment innately modulates cancer progression. Cancer Res.
79:4557–4566. 2019.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Roma-Rodrigues C, Mendes R, Baptista PV
and Fernandes AR: Targeting tumor microenvironment for cancer
therapy. Int J Mol Sci. 20(840)2019.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Quail DF and Joyce JA: Microenvironmental
regulation of tumor progression and metastasis. Nat Med.
19:1423–1437. 2013.PubMed/NCBI View
Article : Google Scholar
|
|
7
|
Wood SL, Pernemalm M, Crosbie PA and
Whetton AD: The role of the tumor-microenvironment in lung
cancer-metastasis and its relationship to potential therapeutic
targets. Cancer Treat Rev. 40:558–566. 2014.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Tomaszewski W, Sanchez-Perez L, Gajewski
TF and Sampson JH: Brain tumor microenvironment and host state:
Implications for immunotherapy. Clin Cancer Res. 25:4202–4210.
2019.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Mollaoglu G, Jones A, Wait SJ,
Mukhopadhyay A, Jeong S, Arya R, Camolotto SA, Mosbruger TL,
Stubben CJ, Conley CJ, et al: The lineage-defining transcription
factors SOX2 and NKX2-1 determine lung cancer cell fate and shape
the tumor immune microenvironment. Immunity. 49:764–779.
2018.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Liu P, Guo J, Xu X, Sun H and Gong Z:
Prognostic biomarker SYK and its correlation with immune
infiltrates in glioma. https://doi.org/10.21203/rs.3.rs-839283/v1.
|
|
11
|
Deng GM, Kyttaris VC and Tsokos GC:
Targeting syk in autoimmune rheumatic diseases. Front Immunol.
7(78)2016.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Mócsai A, Ruland J and Tybulewicz VL: The
SYK tyrosine kinase: A crucial player in diverse biological
functions. Nat Rev Immunol. 10:387–402. 2010.PubMed/NCBI View
Article : Google Scholar
|
|
13
|
Krisenko MO and Geahlen RL: (2015) Calling
in SYK: SYK's dual role as a tumor promoter and tumor suppressor in
cancer. Biochim Biophys Acta. 1853:254–263. 2010.
|
|
14
|
Moncayo G, Grzmil M, Smirnova T, Zmarz P,
Huber RM, Hynx D, Kohler H, Wang Y, Hotz HR, Hynes NE, et al: SYK
inhibition blocks proliferation and migration of glioma cells and
modifies the tumor microenvironment. Neuro Oncol. 20:621–631.
2018.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Robin X, Turck N, Hainard A, Tiberti N,
Lisacek F, Sanchez JC and Müller M: pROC: An open-source package
for R and S+ to analyze and compare ROC curves. BMC Bioinformatics.
12(77)2011.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: Limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43(e47)2015.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Subramanian A, Kuehn H, Gould J, Tamayo P
and Mesirov JP: GSEA-P: A desktop application for gene set
enrichment analysis. Bioinformatics. 23:3251–3253. 2007.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Yu Y, Rahmanto YS, Lee MH, Wu PH, Phillip
JM, Huang CH, Vitolo MI, Gaillard S, Martin SS, Wirtz D, et al:
Inhibition of ovarian tumor cell invasiveness by targeting SYK in
the tyrosine kinase signaling pathway. Oncogene. 37:3778–3789.
2018.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Repana K, Papazisis K, Foukas P, Valeri R,
Kortsaris A, Deligiorgi E and Kyriakidis D: Expression of Syk in
invasive breast cancer: Correlation to proliferation and
invasiveness. Anticancer Res. 26:4949–4954. 2006.PubMed/NCBI
|
|
21
|
Layton T, Stalens C, Gunderson F, Goodison
S and Silletti S: Syk tyrosine kinase acts as a pancreatic
adenocarcinoma tumor suppressor by regulating cellular growth and
invasion. Am J Pathol. 175:2625–2636. 2009.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Shakeel S, Mahjabeen I, Kayani MA and
Faryal R: Association of SYK genetic variations with breast cancer
pathogenesis. Asian Pac J Cancer Prev. 14:3309–3314.
2013.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Qian Y, Zhai E, Chen S, Liu Y, Ma Y, Chen
J, Liu J, Qin C, Cao Q, Chen J and Cai S: Single-cell RNA-seq
dissecting heterogeneity of tumor cells and comprehensive dynamics
in tumor microenvironment during lymph nodes metastasis in gastric
cancer. Int J Cancer. 151:1367–1381. 2022.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Xiao Y and Yu D: Tumor microenvironment as
a therapeutic target in cancer. Pharmacol Ther.
221(107753)2021.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Taniguchi T, Kobayashi T, Kondo J,
Takahashi K, Nakamura H, Suzuki J, Nagai K, Yamada T, Nakamura S
and Yamamura H: Molecular cloning of a porcine gene syk that
encodes a 72-kDa protein-tyrosine kinase showing high
susceptibility to proteolysis. J Biol Chem. 266:15790–15796.
1991.PubMed/NCBI
|
|
26
|
Qu C, Zheng D, Li S, Liu Y, Lidofsky A,
Holmes JA, Chen J, He L, Wei L, Liao Y, et al: Tyrosine kinase SYK
is a potential therapeutic target for liver fibrosis. Hepatology.
68:1125–1139. 2018.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Feng G and Wang X: Role of spleen tyrosine
kinase in the pathogenesis of chronic lymphocytic leukemia. Leuk
Lymphoma. 55:2699–2705. 2014.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Sada K, Takano T, Yanagi S and Yamamura H:
Structure and function of Syk protein-tyrosine kinase. J Biochem.
130:177–186. 2001.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Wang X, Guo J, Ning Z and Wu X: Discovery
of a natural syk inhibitor from chinese medicine through a
docking-based virtual screening and biological assay study.
Molecules. 23(3114)2018.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Poe JC, Jia W, Di Paolo JA, Reyes NJ, Kim
JY, Su H, Sundy JS, Cardones AR, Perez VL, Chen BJ, et al: (2018)
SYK inhibitor entospletinib prevents ocular and skin GVHD in mice.
JCI Insight. 3(122430)2018.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Singh R, Masuda ES and Payan DG: Discovery
and development of spleen tyrosine kinase (SYK) inhibitors. J Med
Chem. 55:3614–3643. 2012.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Chen L, Monti S, Juszczynski P, Daley J,
Chen W, Witzig TE, Habermann TM, Kutok JL and Shipp MA:
SYK-dependent tonic B-cell receptor signaling is a rational
treatment target in diffuse large B-cell lymphoma. Blood.
111:2230–2237. 2008.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Koerber RM, Held SAE, Heine A, Kotthoff P,
Daecke SN, Bringmann A and Brossart P: Analysis of the
anti-proliferative and the pro-apoptotic efficacy of Syk inhibition
in multiple myeloma. Exp Hematol Oncol. 4(21)2015.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Joshi S, Liu KX, Zulcic M, Singh AR, Skola
D, Glass CK, Sanders PD, Sharabi AB, Pham TV, Tamayo P, et al:
Macrophage Syk-PI3Kγ inhibits antitumor immunity: SRX3207, a novel
dual Syk-PI3K inhibitory chemotype relieves tumor
immunosuppression. Mol Cancer Ther. 19:755–764. 2020.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Mills CD, Kincaid K, Alt JM, Heilman MJ
and Hill AM: M-1/M-2 macrophages and the Th1/Th2 paradigm. J
Immunol. 164:6166–6173. 2000.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Chanmee T, Ontong P, Konno K and Itano N:
Tumor-associated macrophages as major players in the tumor
microenvironment. Cancers (Basel). 6:1670–1690. 2000.PubMed/NCBI View Article : Google Scholar
|