Introduction
Hypopharyngeal carcinoma is a primary malignant
tumor of the hypopharynx, accounting for 3-5% of the malignancies
in the upper aerodigestive tract. Early diagnosis of hypopharyngeal
cancer is hard because the early stages of hypopharyngeal carcinoma
have no specific symptoms. Studies have reported that 60-80% of
these patients had ipsilateral lymph node metastases and ≤40% of
these patients have contralateral occult lymph node tumor deposits
(1-3). Thus, the majority of patients with
hypopharyngeal cancer have a poor prognosis and low survival rate
(4). Therefore, identifying early
stage indicators or biomarkers to improve patient survival is
urgent.
Unlike normal linear RNA, the 3′ and 5′ ends of
circular RNAs (circRNAs) are linked by covalent bonds and lack
polarities or polyadenylated tails, thereby rendering them stable
in tissues, serum and urine (5).
Owing to this characteristic, the potential of circRNAs as
biomarkers for human cancer has attracted significant focus. In
addition, circRNAs are widely involved in cancer; ciRS-7 in HeLa
cells (6), Hsa_ circ_001569 in
colorectal cancer (7), circHIPK3
in several types of cancer (8),
f-circM9, f-circPR in hematological malignancy (9), and circTCF25 in urinary bladder
carcinoma (10). Previous studies
have demonstrated that the main function of circRNAs is that they
can function as a microRNA (miRNA) sponge, binding to miRNAs and
regulating them and their downstream gene targets, through a
competing endogenous (ce) RNA mechanism (11).
The present study comprehensively investigated the
expression profile of circRNAs in HCa patients. The results
identified a circRNA signature in HCa and suggested that a core
miRNA-ceRNA network, regulating both the ErbB and Hippo signaling
pathways, may have important roles in HCa progression.
Materials and methods
Patients and specimens
The study included three patients with HCa who
underwent partial or radical cystectomies at the First Affiliated
Hospital of Kunming Medical University (Kunming, China); samples
were collected from March 2017 to October 2017. All three patients
were male and their ages were 44, 54 and 56. Following surgery, the
matched specimens were immediately preserved in liquid nitrogen
until use. All patient samples were confirmed by pathological
examination and none of the patients received neoadjuvant therapy.
The study was approved by the Second Department of Otolaryngology
Head and Neck Surgery of the First Affiliated Hospital of Kunming
Medical University (Kunming, China). Written informed consent was
obtained from all the participants in the study.
Total RNA isolation and quality
control
Total RNA was isolated from samples using TRIzol
reagent (Thermo Fisher Scientific, Inc., Waltham, MA, USA)
following the manufacturer’s protocol. The quantity and quality of
total RNA samples were measured using NanoDrop ND-1000 (Thermo
Fisher Scientific, Inc.). RNA integrity was assessed and confirmed
via electrophoresis using denaturing agarose gels. Isolated RNA
samples were stored at −80°C prior to use.
Library preparation and sequencing
Total RNA from three matched HCa samples and
adjacent normal tissues were treated with Epicenter Ribo-Zero rRNA
Removal kit (Illumina, Inc., San Diego, CA, USA) and RNase R
(Epicenter; Illumina, Inc.) to remove ribosomal and linear RNA.
Then, the RNA-seq libraries were constructed using TruSeq Stranded
Total RNA HT/LT Sample Prep kit (Illumina, Inc.). Sequencing was
determined on Illumina Hiseq 2500 instrument with 2×150 bp paired
reads.
Computational analysis of circRNAs
The clean reads were obtained after the raw reads
were preprocessed with the FastQC quality control tool (12). CircRNAs were identified using CIRI
(v.1.2) pipeline with default parameters (13). Genomic circRNAs were mapped to the
human reference genome (GRCh37) by BWA (14). All circRNAs were annotated for
circRNA-hosting genes with the application of GENCODE v24 (15). The identified circRNAs were
converted to circRNA ID with web server circBase (16).
Principal component analysis (PCA)
PCA was performed as previously described (17). A total of 4,634 distinct circRNAs
with non-zero raw counts across the six samples were isolated and
expressions of circRNAs were normalized with the reads per Million
mapped reads (RPM) method and the expression matrix (each row
represented a gene, each column represented a sample) were used for
PCA. The prcomp package from R was used to perform PCA and the
default parameters were used (18). The ggplot2 package from R was used
to draw the scatter plot (19).
Normalization and differential expression
analysis of circRNAs
Two steps were performed to normalize circRNA
expression for depth. Firstly, the total back-spliced reads in a
sample were counted and that number was divided by 1,000,000. This
resulted in the ‘per million’ scaling factor. Secondly, the read
counts were divided by the ‘per million’ scaling factor. This
method normalized for sequencing depth, giving RPM. CircRNAs were
isolated with RPM>0 across 6 samples and Mann-Whitney U test
(20) (paired=T) followed by
Benjamini-Hochberg multiple testing correction (21) were applied to identify the
differentially expressed (DE) circRNAs. FDR<0.05 and a fold
change of >2.0 or <0.5 were the selection criteria for
significant DE circRNAs.
Functional enrichment analysis
Gene ontology (GO) term enrichment analysis and
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment
analysis were conducted with web server DAVID 6.8 (22). P<0.05 was considered as
statistically significant.
CeRNA network
The top 20 upregulated circRNAs and the top 20
downregulated circRNAs were used to survey miRNA targets with the
web tool CircInteractome (23).
Specifically, CircInteractome downloads the mature sequences of
circRNAs from the UCSC browser mirror (http://genome.mdc-berlin.de) (24) and predicts miRNAs that target
circRNA by surveying for 7-mer or 8-mer complementarity to the seed
region, as well as the 3′end of each miRNA using the TargetScan
algorithm (25). The complete
miRNA list and sequences were taken from the miRBase (http://www.mirbase.org/) (16). miRNA downstream targets were
isolated with mirPath 3.0 (26)
which was also used for miRNA KEGG pathway analysis. The ceRNA
network was displayed by Cytoscape (v3.5.1) (27).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted from pooled normal and tumor
tissue samples using TRIzol (Thermo Fisher Scientific, Inc.), and 1
µg of total RNA was reverse transcribed into first-strand
cDNA using a PrimeScript RT Reagent kit (Takara Bio, Inc., Otsu,
Japan), according to the manufacturer’s protocols. qPCR was
performed with a SYBR-Green real-time PCR kit (Thermo Fisher
Scientific, Inc.) using the ABI StepOnePlus Real-Time PCR system
(Applied Biosystems; Thermo Fisher Scientific, Inc.). CircRNAs were
analyzed with 18s rRNA as the internal standard and miRNA was
analyzed with U6 as the internal standard. The reactions were
prepared as follows: 7.5 µl SYBR Premixm Ex Taq II, 0.25
µl ROX Reference Dye II, 0.125 µl forward primer,
0.125 µl reverse primer, 5 µl RNase-free water, and 2
µl cDNA. The thermocycling conditions were: one step at 95°C
for 30 sec, followed by 40 cycles of 95°C for 5 sec and 60°C for 30
sec, and a final step of 95°C for 15 sec, 60°C for 15 sec and 95°C
for 15 sec. Primer sequences are listed in Table I; expression levels were
quantified via the 2−ΔΔCq method (28).
 | Table IPrimer sequences used for reverse
transcription-quantitative polymerase chain reaction analysis. |
Table I
Primer sequences used for reverse
transcription-quantitative polymerase chain reaction analysis.
| Gene | Forward primer
(5′-3′) | Reverse primer
(5′-3′) |
|---|
|
hsa_circ_0004670 |
GCTCCCAAGCAAAAGAGAAG |
CTGCTTTGTGCTTCCGTATTC |
|
hsa_circ_0005703 |
TGGAGGAGAGGATCGAGTTC |
GTTCTGGATGGTCTGCTTGG |
|
hsa_circ_0003214 |
TGTGTTTGGAACTGCTACCG |
ATCAGCCAGGGCACTCAATA |
|
hsa_circ_0003146 |
CAACGACCTGGTGAAGAGG |
GTCCGAGATCTCCAGCTTGT |
|
hsa_circ_0002059 |
GCCGAGTTAATGGTGGGTTT |
ACCAAATTCAGCCAGAATGC |
|
hsa_circ_0002617 |
TTCCCCAGGAGTGTCAAGAT |
TGGCAAAGATGAAAAGCTGA |
|
hsa_circ_0000660 |
CTTCCAGTGGGAATCCACAT |
AGACATTCTTCCCTTCCAACAA |
|
hsa_circ_0054309 |
CTCTCAGCATGGGACCTTTT |
CGATTTGGTTCTCCCATATCA |
|
hsa_circ_0003279 |
CTCTGTGCACGACTCTCAGG |
TCCCTTCTTCGCTCTTCTCA |
|
hsa_circ_0000239 |
GAATGTTCAAATTGCTGCCATA |
CCAGCAGCCCAACAATTACT |
|
hsa_circ_0091382 |
CTGCAGGGTCTGTTTTTACCA |
CCCATCCAGATCAAGAGAGC |
|
hsa_circ_0004811 |
GGATCCAAAGGCACGTTTTA |
AGAACTTCAGGCGCCAAGTA |
|
hsa_circ_0007480 |
GTTGGAGGAAGGGAAAGAGC |
ATGGCCACATCCCTAAATGT |
|
hsa_circ_0013084 |
GGATGCTGCAAAAACGAGA |
TGGGTTGTTTATACGACTTGGA |
|
hsa_circ_0008836 |
CCTTTTGAGCTGGGAAAACT |
TCTGAAGGAATTCGGGACAG |
|
hsa_circ_0059060 |
GTGGAAGTGGAGAACCCAGA |
ATGGGATGCTAGCCTTGAGA |
|
hsa_circ_0001312 |
TCAGTACTCTGGGGGAAAGG |
GCTGGGACAGATGAAACCAT |
|
hsa_circ_0005027 |
TGTTGAGTTCGGCAGCATAC |
ACACACCTCTGCAACCACAA |
|
hsa_circ_0008287 |
CCGAGCCACCTAAACAACAG |
TCTGGGAGCGTCAGAAAGTT |
|
has-miR-548c-3p |
CATTGGCATCTATTAGGTTG |
GTATTAAGTTGGTGCAAAAG |
| 18s rRNA |
ACCTGGTTGATCCTGCCAG |
TCCAAGTAGGAGAGGAGCG |
| U6 |
GTGCTCGCTTCGGCAGCA |
TGGAACGCTTCACGAATTTG |
Expression analysis of miR-548c-3p
Two methods were used to investigate the expression
of miR-548c-3p among normal and tumor samples. The first was
RT-qPCR, as detailed above. The second was in-silico analysis. The
miRNA dataset of the esophageal carcinoma cohort from The Cancer
Genome Atlas (TCGA) project (29)
was exploited. There were 13 normal samples and 184 tumor samples
in this dataset. Normalized miRNA expressions of miR-548c-3p were
compared between normal and tumor samples. Mann-Whitney U test was
applied to test the significance.
Survival analysis
A Kaplan-Meier curve was used to examine the
clinical relevance of miR-548c-3p levels in the patients’ outcomes
(30). Patients were separated
into two groups according to the median expression of
hsa-miR-548c-3p using TCGA clinical and expression dataset.
Differences between groups were analyzed using log-rank test
(31) and two-tailed P-values
<0.05 were considered statistically significant. Statistical
analyses were performed using the survival package (version 2.39-5)
in R (version 3.4.3).
Expression correlation of hsa-miR-548c-3p
and its targeted genes
The miRNA and mRNA datasets of the esophageal
carcinoma cohort from TCGA (29)
were used for the correlation analysis. Common samples were
isolated according to the sample barcodes. The Pearson correlation
method was used to assess the expression association between
hsa-miR-548c-3p and the targeted genes. Significance of association
was determined by the R package cor.test (alternative=‘two.sided’,
method=‘pearson’). Then, P-values were corrected with
Benjamini-Hochberg procedure for multiple testing.
Statistical analysis
All statistical analyses were generated using R
(32). The Pearson correlation
method was used to assess the expression association. Significances
of associations were determined by the R package cor.test.
Mann-Whitney U test was used for comparisons between two groups.
Benjamini-Hochberg procedure was applied for multiple testing.
Log-rank test was used for Kaplan-Meier survival curves. P<0.05
was considered to indicate a statistically significant
difference.
Results
Identification of DE circRNAs in HCa
To identify DE circRNAs in HCa, circRNA sequencing
(Seq) was performed using three matched normal and HCa tissue
samples, and an average of 90 million reads was achieved for each
sample. A total of 4,634 distinct circRNAs with at least two unique
back-spliced reads across six samples using CIRI pipeline (13) were identified and the expressions
of circRNAs were normalized and represented by reads per million
mapped reads (RPM) values. Genetic distances across 6 samples were
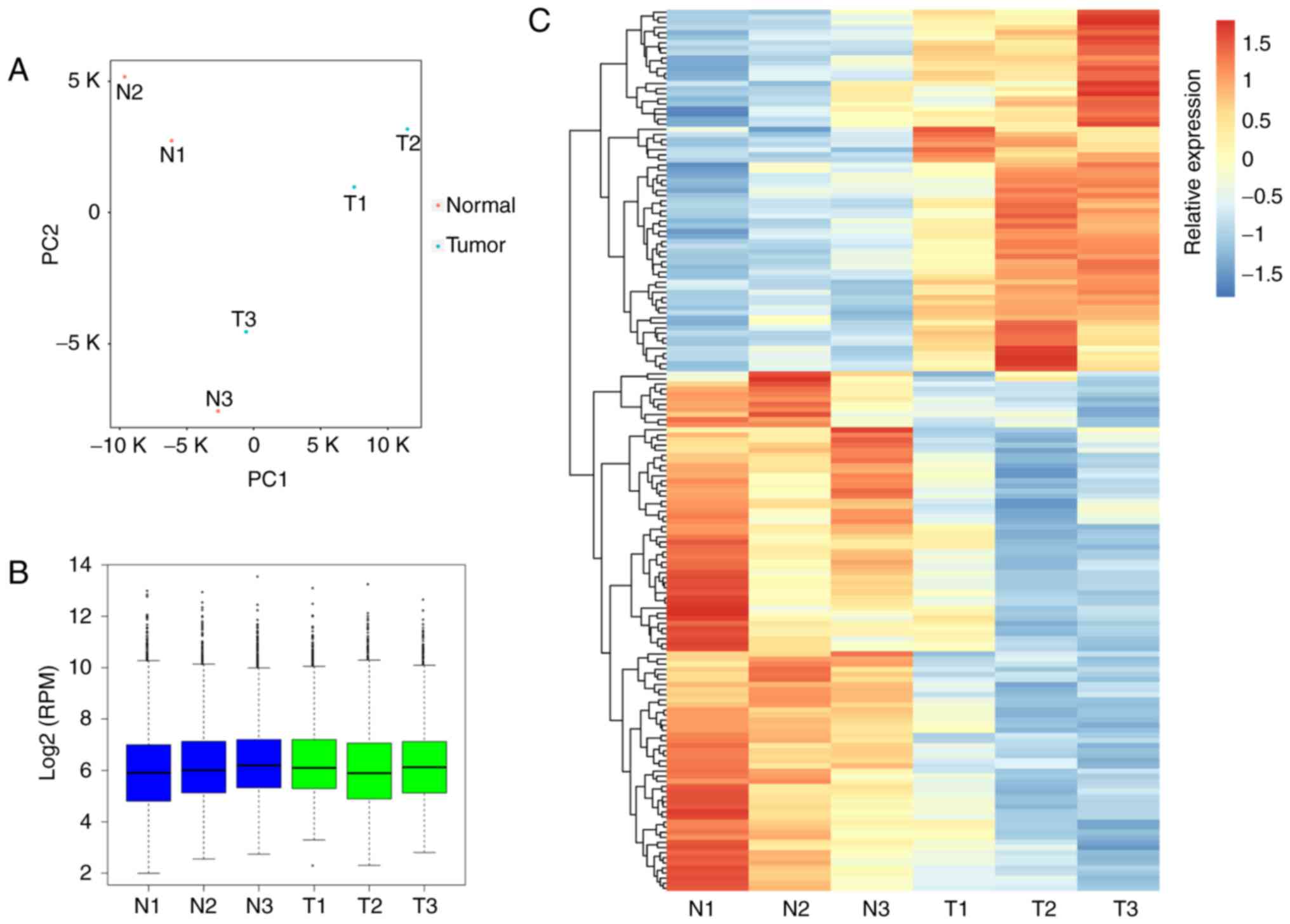
evaluated using PCA (Fig. 1A),
and the normalized expression level (RPM) of circRNAs across the
six samples is illustrated in Fig.
1B. Following statistical analysis, 71 and 102 circRNAs were
determined to be significantly upregulated and downregulated,
respectively (Table II). The DE
circRNAs between tumor and adjacent normal samples were presented
in a heatmap (Fig. 1C). To
confirm the circRNA-Seq results, RT-qPCR was performed to assess
the expression of 19 of the above DE circRNAs in both normal and
tumor samples. The results confirmed that 12 of them were
consistently upregulated or downregulated with the circRNA-Seq
results (Fig. 2).
 | Table IIDifferentially expressed
circRNAs. |
Table II
Differentially expressed
circRNAs.
| circRNA ID
(CIRI) | circRNA ID
(circBase) | Adjusted
P-value | FC | Gene |
|---|
|
chr16:21973780-21987564 |
hsa_circ_0005690 | 0.022002929 | 9.346453412 | UQCRC2 |
|
chr2:242343242-242357524 |
hsa_circ_0004924 | 0.042106003 | 4.237176918 | FARP2 |
|
chr7:72873865-72884813 |
hsa_circ_0004670 | 0.003579475 | 4.136199328 | BAZ1B |
|
chr5:133871547-133887899 |
hsa_circ_0005608 | 0.036132329 | 3.805820016 | PHF15 |
|
chr22:41979962-41980607 |
hsa_circ_0005703 | 0.030406606 | 3.76275756 | PMM1 |
|
chr3:48019354-48040369 |
hsa_circ_0005255 | 0.039183919 | 3.736266721 | MAP4 |
|
chr12:27521194-27523163 |
hsa_circ_0009009 | 0.01165016 | 3.553331882 | ARNTL2 |
|
chr9:117399269-117401006 |
hsa_circ_0002318 | 0.041159198 | 3.381349856 C | 9orf91 |
|
chr1:165859440-165860559 |
hsa_circ_0006758 | 0.041758958 | 3.367866028 | UCK2 |
|
chr16:50321822-50322261 |
hsa_circ_0000699 | 0.043912028 | 3.314878392 | ADCY7 |
|
chr16:89484691-89497734 |
hsa_circ_0000727 | 0.035411987 | 3.287797218 | ANKRD11 |
|
chr8:98817580-98837381 |
hsa_circ_0003214 | 0.018483558 | 3.272900498 | LAPTM4B |
|
chr19:48229068-48229481 |
hsa_circ_0003146 | 0.006019062 | 3.268823068 | EHD2 |
|
chr1:118003110-118045592 |
hsa_circ_0002059 | 0.010572316 | 3.169435071 | MAN1A2 |
|
chr14:92264128-92268765 |
hsa_circ_0032969 | 0.040149042 | 3.130356466 | TC2N |
|
chr2:210968827-211019335 |
hsa_circ_0002617 | 0.020968937 | 3.121028088 C | 2orf67 |
|
chr15:94899365-94945248 |
hsa_circ_0000660 | 0.032292149 | 3.092238966 | MCTP2 |
|
chr2:43655238-43657441 |
hsa_circ_0054309 | 0.002782953 | 3.091209609 | THADA |
|
chr2:110321942-110323436 |
hsa_circ_0009020 | 0.03917314 | 3.05906561 | SEPT10 |
|
chr19:2137009-2138713 |
hsa_circ_0048344 | 0.040805597 | 3.048246172 | AP3D1 |
|
chr16:4311779-4312702 |
hsa_circ_0002439 | 0.046240525 | 2.987845154 | TFAP4 |
|
chr11:128993340-128997200 |
hsa_circ_0005027 | 0.001764073 | 2.945490868 | ARHGAP32 |
|
chr8:62460629-62479877 |
hsa_circ_0084604 | 0.018794062 | 2.926235997 | ASPH |
|
chr3:155628480-155643155 |
hsa_circ_0008184 | 0.037854875 | 2.918397576 | GMPS |
|
chr16:47531309-47581459 |
hsa_circ_0004791 | 0.048387875 | 2.9019822 | PHKB |
|
chr20:35457456-35467844 |
hsa_circ_0060219 | 0.015622827 | 2.89815993 | KIAA0889 |
|
chr3:195101737-195112876 |
hsa_circ_0007331 | 0.048715375 | 2.872843159 | ACAP2 |
|
chr22:29517344-29521404 |
hsa_circ_0004547 | 0.044771757 | 2.802768657 | KREMEN1 |
|
chr2:122260742-122287901 |
hsa_circ_0002374 | 0.037227807 | 2.731455995 | CLASP1 |
|
chr3:128514202-128526514 |
hsa_circ_0006346 | 0.046003237 | 2.690296373 | RAB7A |
|
chr4:83793096-83796975 |
hsa_circ_0003549 | 0.044620973 | 2.679827252 | SEC31A |
|
chr12:1399017-1481143 |
hsa_circ_0024997 | 0.011882022 | 2.668639886 | ERC1 |
|
chr13:96409897-96416207 | #N/A | 0.005392448 | 2.660521497 | #N/A |
|
chr16:30715384-30715636 |
hsa_circ_0039076 | 0.030665031 | 2.628113834 | SRCAP |
|
chr7:2400344-2404164 |
hsa_circ_0004869 | 0.033796697 | 2.611610945 | EIF3B |
|
chr2:55209650-55214834 |
hsa_circ_0001006 | 0.045899911 | 2.579018715 | RTN4 |
|
chr12:42768664-42792796 |
hsa_circ_0003961 | 0.010866489 | 2.568055885 | PPHLN1 |
|
chr7:2404006-2406083 |
hsa_circ_0001671 | 0.022907729 | 2.551488457 | EIF3B |
|
chr4:87685745-87689129 |
hsa_circ_0007948 | 0.022945704 | 2.547028768 | PTPN13 |
|
chr1:176085759-176105683 |
hsa_circ_0015373 | 0.029930944 | 2.536801963 | RFWD2 |
|
chr22:36737414-36745300 |
hsa_circ_0004470 | 5.2192E-05 | 2.529021894 | MYH9 |
|
chr2:32396355-32409407 |
hsa_circ_0053423 | 0.031086982 | 2.514457353 | SLC30A6 |
|
chr7:138951078-138957186 |
hsa_circ_0005594 | 0.027733961 | 2.504492053 | UBN2 |
|
chr9:95030455-95032265 |
hsa_circ_0008367 | 0.016513149 | 2.486273648 | IARS |
|
chr7:65705311-65751696 |
hsa_circ_0006041 | 0.005514578 | 2.410252357 | TPST1 |
|
chr4:75040222-75067087 |
hsa_circ_0069981 | 0.032657959 | 2.370951734 | MTHFD2L |
|
chr1:246021797-246093239 |
hsa_circ_0017289 | 0.010842885 | 2.366575966 | SMYD3 |
|
chr22:29090019-29091861 |
hsa_circ_0004811 | 0.001831625 | 2.363098777 C | HEK2 |
|
chr16:3900297-3901010 |
hsa_circ_0007637 | 0.009154577 | 2.351311388 C | REBBP |
|
chr2:168920009-168931741 |
hsa_circ_0003279 | 0.000393597 | 2.342009076 | STK39 |
|
chr10:101728871-101731891 |
hsa_circ_0008393 | 0.049564434 | 2.328443205 D | NMBP |
|
chr9:140646782-140652463 |
hsa_circ_0001904 | 0.031745459 | 2.316860866 | EHMT1 |
|
chr22:46125304-46136418 |
hsa_circ_0001247 | 0.013526942 | 2.31185877 | ATXN10 |
|
chr14:23419522-23421892 |
hsa_circ_0005663 | 0.04538212 | 2.257072167 | HAUS4 |
|
chr12:122773035-122801402 | #N/A | 0.015647694 | 2.25006903 | #N/A |
|
chr4:3188323-3190820 | #N/A | 0.025850456 | 2.249708035 | #N/A |
|
chr3:172363412-172365904 |
hsa_circ_0007042 | 0.026989758 | 2.238835674 | NCEH1 |
|
chr2:10799297-10808849 |
hsa_circ_0008511 | 0.042758286 | 2.221921485 | NOL10 |
|
chr10:70696691-70703013 |
hsa_circ_0007097 | 0.040819924 | 2.219586601 DD | X50 |
|
chrX:14868626-14877456 |
hsa_circ_0006971 | 0.009211245 | 2.215797457 | FANCB |
|
chr1:23356961-23385660 |
hsa_circ_0007822 | 0.027240129 | 2.206406433 | KDM1A |
|
chr20:13539654-13561628 |
hsa_circ_0002001 | 0.017808498 | 2.188222168 | TASP1 |
|
chr7:139741443-139757834 |
hsa_circ_0004684 | 0.026813206 | 2.168759032 | PARP12 |
|
chr7:72883846-72884813 |
hsa_circ_0003866 | 0.012904268 | 2.108744306 | BAZ1B |
|
chr1:247319707-247323115 | #N/A | 0.039522611 | 2.102327859 | #N/A |
|
chr18:21644103-21663045 |
hsa_circ_0047270 | 0.020649137 | 2.07972359 | TTC39C |
|
chr1:31532050-31532424 |
hsa_circ_0000045 | 0.009259898 | 2.078191163 | PUM1 |
|
chr2:63660878-63667005 |
hsa_circ_0003497 | 0.033526319 | 2.072706608 | WDPCP |
|
chr10:128859931-128908618 |
hsa_circ_0020462 | 0.014021298 | 2.068064026 D | OCK1 |
|
chr1:44773981-44804994 |
hsa_circ_0007693 | 0.022406409 | 2.066977275 | ERI3 |
|
chr7:99621041-99621930 |
hsa_circ_0001727 | 0.043987221 | 2.025308683 | ZKSCAN1 |
|
chr16:11873021-11876244 |
hsa_circ_0005420 | 0.045974717 | 0.499738449 | ZC3H7A |
|
chr20:13539654-13561628 |
hsa_circ_0002001 | 0.03937226 | 0.497281358 | TASP1 |
|
chr3:43341245-43345284 |
hsa_circ_0004089 | 0.007985302 | 0.491228173 | SNRK |
|
chrX:77084527-77086392 | #N/A | 0.020792399 | 0.489952895 | #N/A |
|
chr2:11905658-11907984 |
hsa_circ_0002229 | 0.013904514 | 0.48799699 | LPIN1 |
|
chr17:60111147-60112969 |
hsa_circ_0004273 | 0.010727519 | 0.487251947 | MED13 |
|
chr2:168920009-168986268 |
hsa_circ_0005882 | 0.018253559 | 0.481253188 | STK39 |
|
chr16:53532302-53534241 |
hsa_circ_0004072 | 0.021867418 | 0.480875236 | AKTIP |
|
chr9:95030455-95032265 |
hsa_circ_0008367 | 0.024608207 | 0.480576493 | IARS |
|
chr9:14146687-14179779 |
hsa_circ_0086376 | 0.025017135 | 0.480402794 | NFIB |
|
chr12:122773035-122801402 | #N/A | 0.027697748 | 0.475833919 | #N/A |
|
chr20:35457456-35467844 |
hsa_circ_0060219 | 0.01965515 | 0.472752875 | KIAA0889 |
|
chr15:80412669-80415142 |
hsa_circ_0000643 | 0.019656594 | 0.47081423 | ZFAND6 |
|
chr4:144449020-144451679 | #N/A | 0.002041818 | 0.470785346 | #N/A |
|
chr15:62299506-62306191 |
hsa_circ_0000607 | 0.01862274 | 0.47022119 | VPS13C |
|
chr21:40578033-40584633 | #N/A | 0.038815333 | 0.466807062 | #N/A |
|
chr10:128859931-128908618 |
hsa_circ_0020462 | 0.048526313 | 0.46671725 D | OCK1 |
|
chr1:32381495-32385259 |
hsa_circ_0007364 | 0.012904962 | 0.464916349 | PTP4A2 |
|
chr2:148811959-148990964 | #N/A | 0.018584205 | 0.463574046 | #N/A |
|
chr8:37734626-37735069 |
hsa_circ_0001789 | 0.019846769 | 0.462223466 | RAB11FIP1 |
|
chr13:28748408-28752072 |
hsa_circ_0004372 | 0.023765765 | 0.461563778 | PAN3 |
|
chr18:9931806-9937063 |
hsa_circ_0006990 | 0.039623112 | 0.461171429 | VAPA |
|
chr17:34910660-34923615 |
hsa_circ_0003930 | 0.021748331 | 0.458221861 | GGNBP2 |
|
chr22:46125304-46136418 |
hsa_circ_0001247 | 0.008045429 | 0.453521935 | ATXN10 |
|
chr2:206992520-206994966 |
hsa_circ_0002431 | 0.036437006 | 0.452588631 | NDUFS1 |
|
chr2:62100136-62103369 |
hsa_circ_0001018 | 0.029799501 | 0.451702188 CC | T4 |
|
chr11:119144577-119145663 |
hsa_circ_0000362 | 0.025621724 | 0.450849769 C | BL |
|
chr4:39734978-39747430 | #N/A | 0.03460933 | 0.450272258 | #N/A |
|
chr22:29682911-29683123 |
hsa_circ_0008044 | 0.010096395 | 0.44308523 | EWSR1 |
|
chr14:21971315-21972024 |
hsa_circ_0000523 | 0.047512563 | 0.44023344 | METTL3 |
|
chr14:50615002-50616948 | #N/A | 0.013896814 | 0.437352627 | #N/A |
|
chr8:101232506-101243516 | #N/A | 0.028204026 | 0.430720792 | #N/A |
|
chr7:72883846-72884813 |
hsa_circ_0003866 | 0.004390368 | 0.424421462 | BAZ1B |
|
chr18:46858233-46906128 |
hsa_circ_0002501 | 0.03573514 | 0.420126159 D | YM |
|
chr1:246784730-246797889 |
hsa_circ_0017311 | 0.028462249 | 0.41970008 | CNST |
|
chr12:1399017-1481143 |
hsa_circ_0024997 | 0.040174171 | 0.417295868 | ERC1 |
|
chr12:27521194-27523163 |
hsa_circ_0009009 | 0.024711114 | 0.414722924 | ARNTL2 |
|
chr1:52959282-52975384 |
hsa_circ_0003632 | 0.045708435 | 0.414593811 | ZCCHC11 |
|
chr5:50055476-50059076 |
hsa_circ_0006787 | 0.018488474 | 0.412395338 | PARP8 |
|
chr3:179096128-179104417 |
hsa_circ_0002219 | 0.029224282 | 0.403006229 | MFN1 |
|
chr10:88203031-88206206 | #N/A | 0.022884862 | 0.399217754 | #N/A |
|
chr17:26490568-26499644 |
hsa_circ_0003638 | 0.013335258 | 0.397922038 | NLK |
|
chr16:8952206-8953192 |
hsa_circ_0000669 | 0.000715169 | 0.397901388 | CARHSP1 |
|
chr2:186946056-186964557 | #N/A | 0.017444656 | 0.397798992 | #N/A |
|
chr11:85685750-85695016 |
hsa_circ_0006629 | 0.01991069 | 0.396956177 | PICALM |
|
chr7:91980263-91991587 | #N/A | 0.035701238 | 0.389282593 | #N/A |
|
chr14:35519989-35522657 |
hsa_circ_0006424 | 0.04322634 | 0.387507944 | FAM177A1 |
|
chr12:42768664-42792796 |
hsa_circ_0003961 | 0.014465456 | 0.386034041 | PPHLN1 |
|
chr1:246021797-246093239 |
hsa_circ_0017289 | 0.011181456 | 0.38452595 | SMYD3 |
|
chr10:27431315-27434519 |
hsa_circ_0005633 | 0.009131314 | 0.384171219 | YME1L1 |
|
chr12:129299319-129299615 |
hsa_circ_0000462 | 0.006941025 | 0.380095649 | SLC15A4 |
|
chr1:118003110-118045592 |
hsa_circ_0002059 | 0.002220417 | 0.379882753 | MAN1A2 |
|
chr6:55966269-56006781 | #N/A | 0.022261578 | 0.375691727 | #N/A |
|
chr8:17123415-17126465 |
hsa_circ_0008592 | 0.040012263 | 0.373149729 | VPS37A |
|
chr10:99915849-99923154 |
hsa_circ_0004419 | 0.032103617 | 0.372519687 C | 10orf28 |
|
chr20:17933230-17934761 |
hsa_circ_0006704 | 0.020560715 | 0.371748201 | SNX5 |
|
chr1:31532050-31532424 |
hsa_circ_0000045 | 0.040901772 | 0.368955101 | PUM1 |
|
chrX:14868626-14877456 |
hsa_circ_0006971 | 0.032386045 | 0.36887067 | FANCB |
|
chr16:3900297-3901010 |
hsa_circ_0007637 | 0.020340132 | 0.36603697 C | REBBP |
|
chr4:103644027-103647840 |
hsa_circ_0006007 | 0.02570568 | 0.359726565 | MANBA |
|
chr1:62907158-62907970 | #N/A | 0.047427014 | 0.35583852 | #N/A |
|
chr18:18619432-18624147 |
hsa_circ_0006733 | 0.037595523 | 0.353388689 | ROCK1 |
|
chr14:52977957-53011089 |
hsa_circ_0031939 | 0.01966747 | 0.351937505 | TXNDC16 |
|
chr21:37711076-37717005 |
hsa_circ_0001189 | 0.010082196 | 0.351308477 | MORC3 |
|
chr1:94685813-94697199 |
hsa_circ_0003310 | 0.0259611 | 0.351119971 | ARHGAP29 |
|
chr3:47103652-47108608 |
hsa_circ_0065159 | 0.020521933 | 0.350064342 | SETD2 |
|
chr8:71126137-71128999 | #N/A | 0.048201309 | 0.344144613 | #N/A |
|
chr16:71779046-71779517 |
hsa_circ_0002505 | 0.046185136 | 0.343484211 | AP1G1 |
|
chr5:31421378-31424578 |
hsa_circ_0005524 | 0.044966353 | 0.34303411 | DROSHA |
|
chr11:1307231-1317024 |
hsa_circ_0008301 | 0.018942131 | 0.342187543 | TOLLIP |
|
chr6:108242132-108243113 | #N/A | 0.012934139 | 0.340934595 | #N/A |
|
chr19:48229068-48229481 |
hsa_circ_0003146 | 0.020562164 | 0.340720207 | EHD2 |
|
chr3:37170553-37190529 |
hsa_circ_0003264 | 0.013154932 | 0.336803278 | LRRFIP2 |
|
chr7:65705311-65751696 |
hsa_circ_0006041 | 0.039282961 | 0.334902033 | TPST1 |
|
chr13:96409897-96416207 | #N/A | 0.0177537 | 0.332540513 | #N/A |
|
chr8:68200189-68214701 | #N/A | 0.011526157 | 0.328702879 | #N/A |
|
chr7:27668989-27689252 |
hsa_circ_0006773 | 0.003935998 | 0.328463848 | HIBADH |
|
chr10:32308785-32310215 |
hsa_circ_0006408 | 0.043640924 | 0.325878207 | KIF5B |
|
chr7:77407654-77408131 | #N/A | 0.023396605 | 0.323146621 | #N/A |
|
chr3:56600621-56601081 |
hsa_circ_0001312 | 0.029755856 | 0.321652942
CCDC | 66 |
|
chr2:234271722-234299129 | #N/A | 0.02313964 | 0.321131598 | #N/A |
|
chr7:73100965-73101425 |
hsa_circ_0005588 | 0.042073587 | 0.318524548 | WBSCR22 |
|
chr7:72873865-72884813 |
hsa_circ_0004670 | 0.043342921 | 0.316315928 | BAZ1B |
|
chr2:242282406-242283312 |
hsa_circ_0059060 | 0.007810999 | 0.315723287 | SEPT2 |
|
chr3:47139444-47144913 |
hsa_circ_0001289 | 0.039274732 | 0.313502465 | SETD2 |
|
chr2:43655238-43657441 |
hsa_circ_0054309 | 0.038540875 | 0.309703944 | THADA |
|
chr21:46275124-46281186 |
hsa_circ_0001200 | 0.0474882 | 0.302025085 | PTTG1IP |
|
chr5:179976930-179980471 |
hsa_circ_0008836 | 0.028790905 | 0.292442462 C | NOT6 |
|
chr1:87185189-87190088 |
hsa_circ_0013084 | 0.01567065 | 0.280991627 | SH3GLB1 |
|
chr19:53577392-53578436 |
hsa_circ_0007480 | 0.031784533 | 0.275005495 | ZNF160 |
|
chr16:53289511-53297009 | #N/A | 0.003705027 | 0.260024152 | #N/A |
|
chr22:29090019-29091861 |
hsa_circ_0004811 | 0.008438851 | 0.252361922 C | HEK2 |
|
chrX:117718697-117724265 |
hsa_circ_0091382 | 0.032765014 | 0.247214419 D | OCK11 |
|
chr11:128993340-128997200 |
hsa_circ_0005027 | 0.041341275 | 0.246885645 | ARHGAP32 |
|
chr15:34542498-34543258 |
hsa_circ_0034346 | 0.039707643 | 0.246690583 | SLC12A6 |
|
chr10:70152894-70154208 |
hsa_circ_0000239 | 0.008512372 | 0.236281903 | RUFY2 |
|
chr1:236966727-236979843 | #N/A | 0.036146024 | 0.222019071 | #N/A |
|
chr19:33604672-33605325 |
hsa_circ_0008287 | 0.042162087 | 0.207707068 | GPATCH1 |
|
chr2:168920009-168931741 |
hsa_circ_0003279 | 0.001767352 | 0.196582978 | STK39 |
|
chr1:179087721-179091002 | #N/A | 0.019369988 | 0.169662407 | #N/A |
|
chr18:9524591-9525849 |
hsa_circ_0005158 | 0.043392731 | 0.166193525 | RALBP1 |
|
chr22:36737414-36745300 |
hsa_circ_0004470 | 0.042494836 | 0.147382721 | MYH9 |
Next, the distribution of circRNAs in different DNA
elements and chromosomes was examined. The bar diagram of Fig. 3A demonstrates the % of
back-spliced junction reads on intron, intergenic, and exon areas.
The majority of circRNAs belonged to exonic, followed by intronic
and intergenic elements (Fig.
3B). These dysregulated circRNAs are widely distributed in all
chromosomes, including sex chromosomes X (Fig. 3C).
Functional enrichment analysis of genes
producing DE circRNAs
To reveal the dysregulated pathways underlying HCa,
first KEGG pathway enrichment analyses were performed for genes
that matched DE circRNAs. The results demonstrated that genes
containing downregulated circRNAs were enriched in endocytosis,
ubiquitin-mediated proteolysis, and Janus kinase (JAK)/signal
transducer and activator of transcription (STAT) signaling pathways
(Fig. 4A), whereas there were no
KEGG pathways enriched with genes producing upregulated
circRNAs.
Next, GO term enrichment analyies was performed for
genes that produced aberrantly expressed circRNAs. Biological
processes, such as the establishment of spindle orientation,
response to fungicide, positive regulation of transcription, cell
division were significantly enriched (Fig. 4B), whereas genes producing
downregulated circRNAs were related to autophagy, mitochondrion
organization actin cytoskeleton organization, membrane fission, and
cell-cell adhesion pathways (Fig.
4C). These results suggested that multiple pathways may
contribute to HCa pathogenesis and progression.
CircRNAs regulate the ErbB and Hippo
pathways through a miRNA-CeRNA network
The role of circRNAs as a miRNA sponge is the main
mechanism of circRNA function in tumor cells (11,33). Therefore, we further investigated
the roles of circRNAs in HCa progression through establishing a
ceRNA network. Firstly, the top 20 upregulated and top 20
downregulated circRNAs were isolated and were converted to circRNA
ID using circBase database (34).
Secondly, miRNAs targeting DE-circRNAs were isolated with the web
server CircInteractome (23).
Specifically, CircInteractome downloaded the mature sequences of
all of the reported circRNAs from the UCSC browser, then to
characterize miRNA-circRNA interactions, CircInteractome
incorporated the ability to search using the TargetScan algorithm,
which predicts miRNAs that target circRNA by surveying for 7-mer or
8-mer complementarity to the seed region as well as the 3′end of
each miRNA (23). A total of 191
and 182 miRNAs were putatively identified as the targets of
upregulated and downregulated circRNAs, respectively. Networks
consisted of circRNAs and miRNAs were displayed using Cytoscape
software (27). The results
demonstrated extensive interactions between miRNAs and upregulated
(Fig. 5A), and downregulated
circRNAs (Fig. 5B). Then, KEGG
pathway enrichment analysis was performed for the miRNAs targeted
by the top 40 DE circRNAs, in order to explore the altered
biological processes using mirPath 3.0 (26). Genes targeted by miRNAs were
significantly enriched in multiple signaling pathways, including
the ErbB, the Hippo, the Ras, the transforming growth factor
(TGF)-β, the phosphoinositide 3-kinase/AKT serine/threonine kinase
and the Wnt signaling pathways (Fig.
5C).
To get further insight into the function of circRNAs
in the ErbB and Hippo signaling pathways, miRNA-ceRNA networks were
constructed corresponding to the two pathways using Cytoscape. For
the miRNA-ceRNA network regulating the ErbB pathway, there were 33
circRNAs, 43 miRNAs and 74 ErbB pathway genes (Fig. 6A). In the ErbB miRNA-ceRNA
network, we isolated a subnetwork consisting of circRNAs
(hsa_circ_0008287 and hsa_circ_0005027), miRNAs (hsa-miR-548c-3p)
and 38 ErbB pathway genes which had the most interaction between
miRNAs and targeted genes (Fig.
6B). Hsa_circ_0008287 and hsa_circ_0005027 were significantly
downregulated in tumor samples compared with normal (Figs. 2 and 6C). In a similar manner, the miRNA-ceRNA
network regulating the Hippo pathway was constructed, consisting of
33 circRNAs, 43 miRNAs and 110 Hippo pathway genes (Fig. 7A). In the Hippo miRNA-ceRNA
network, we also isolated a subnetwork consisting of circRNAs
(hsa_circ_0008287 and hsa_circ_0005027), miRNAs (hsa-miR-548c-3p)
and 61 Hippo pathway genes, which had the most interaction between
miRNAs and targeted genes (Fig.
7B).
To further investigate the important role of this
subnet-work in tumor progression, the miRNA and mRNA datasets of
the esophageal carcinoma cohort from TCGA (29) were exploited. The esophageal
carcinoma cohort contains 13 normal samples and 184 tumor samples.
In this cohort, the miRNA hsa-miR-548c-3p expression between normal
and tumor samples was detected, and its clinical relevance to
patient survival was analyzed. The results suggested that
hsa-miR-548c-3p was highly expressed in tumor samples compared with
normal samples (Fig. 8A and B),
and its high expression was significantly associated with lower
survival in patients with esophageal carcinoma (Fig. 8C). These findings suggested that
hsa-miR-548c-3p is an oncogenic miRNA, which is consistent with the
hypothesis that in tumor samples circRNAs were downregulated
resulting in more oncogenic hsa-miR-548c-3p being released, and
highly expressed hsa-miR-548c-3p may promote HCa progression
through downstream target genes. To confirm the negative regulation
of hsa-miR-548c-3p on the ErbB and Hippo pathway genes, the
expression correlation of hsa-miR-548c-3p and its targeted genes
were also analyzed. Many of the targeted genes were negatively
correlated with hsa-miR-548c-3p levels, which supported a negative
regulatory role of hsa-miR-548c-3p on the ErbB and Hippo pathways
(Table III). The present
results demonstrated that circRNAs regulate HCa progression through
multiple pathways and identifying a miRNA-ceRNA network that
regulated the ErbB and Hippo signaling pathways.
 | Table IIIExpression correlation of
hsa-miR-548c-3p and its targeted genes in The Cancer Genome Atlas
esophageal carcinoma cohort. |
Table III
Expression correlation of
hsa-miR-548c-3p and its targeted genes in The Cancer Genome Atlas
esophageal carcinoma cohort.
| miRNA | Gene | Correlation
coefficient | FDR |
|---|
|
hsa-miR-548c-3p | PIK3CA | −0.179442021 | 0.011633 |
|
hsa-miR-548c-3p | ABL1 | −0.16430793 | 0.021044 |
|
hsa-miR-548c-3p | PIK3R1 | −0.159510982 | 0.025159 |
|
hsa-miR-548c-3p | AKT3 | −0.152832805 | 0.032027 |
| hsa-miR-548c-3p
C | TGF | −0.139434991 | 0.050682 |
| hsa-miR-548c-3p
D | LG2 | −0.136001942 | 0.056703 |
|
hsa-miR-548c-3p | FRMD6 | −0.128579884 | 0.07175 |
|
hsa-miR-548c-3p | GAB1 | −0.119111167 | 0.095493 |
|
hsa-miR-548c-3p | GDF6 | −0.114236586 | 0.109949 |
|
hsa-miR-548c-3p | BTC | −0.114017567 | 0.110637 |
| hsa-miR-548c-3p
CC | ND2 | −0.113880294 | 0.11107 |
|
hsa-miR-548c-3p | SOS2 | −0.106454244 | 0.136518 |
|
hsa-miR-548c-3p | ABL2 | −0.103988246 | 0.14589 |
|
hsa-miR-548c-3p | STAT5B | −0.103173372 | 0.149092 |
|
hsa-miR-548c-3p | ERBB4 | −0.099703628 | 0.163324 |
| hsa-miR-548c-3p
C | BLB | −0.083838426 | 0.241477 |
| hsa-miR-548c-3p
D | LG4 | −0.083217471 | 0.244993 |
|
hsa-miR-548c-3p | BMPR1A | −0.082135258 | 0.251205 |
|
hsa-miR-548c-3p | PAK7 | −0.079722196 | 0.265448 |
| hsa-miR-548c-3p
D | LG1 | −0.072417936 | 0.311876 |
|
hsa-miR-548c-3p | CAMK2D | −0.070087122 | 0.327746 |
|
hsa-miR-548c-3p | LEF1 | −0.068131773 | 0.341454 |
| hsa-miR-548c-3p
C | TNNA3 | −0.06669216 | 0.351776 |
|
hsa-miR-548c-3p | PAK2 | −0.066482615 | 0.353295 |
|
hsa-miR-548c-3p | PTK2 | −0.064644262 | 0.366794 |
|
hsa-miR-548c-3p | FZD1 | −0.063827484 | 0.372892 |
|
hsa-miR-548c-3p | NRG3 | −0.053532783 | 0.454989 |
|
hsa-miR-548c-3p | LATS2 | −0.052315837 | 0.46532 |
|
hsa-miR-548c-3p | RPS6KB1 | −0.049387268 | 0.490702 |
|
hsa-miR-548c-3p | NCK1 | −0.047228889 | 0.509871 |
|
hsa-miR-548c-3p | PRKCB | −0.045059635 | 0.529521 |
|
hsa-miR-548c-3p | FZD4 | −0.041762665 | 0.560101 |
|
hsa-miR-548c-3p | LLGL1 | −0.037399843 | 0.601828 |
|
hsa-miR-548c-3p | BMPR2 | −0.036025864 | 0.615251 |
|
hsa-miR-548c-3p | GSK3B | −0.022579459 | 0.752806 |
|
hsa-miR-548c-3p | GSK3B | −0.022579459 | 0.752806 |
|
hsa-miR-548c-3p | EGFR | −0.019225787 | 0.788581 |
|
hsa-miR-548c-3p | PRKCA | −0.017068489 | 0.811834 |
|
hsa-miR-548c-3p | LATS1 | −0.014844376 | 0.835982 |
|
hsa-miR-548c-3p | FZD7 | −0.012434723 | 0.862317 |
|
hsa-miR-548c-3p | PAK3 | −0.012235276 | 0.864504 |
|
hsa-miR-548c-3p | BRAF | −0.011521175 | 0.872343 |
|
hsa-miR-548c-3p | SOS1 | −0.007706896 | 0.914404 |
| hsa-miR-548c-3p
C | BL | −0.006374831 | 0.929156 |
| hsa-miR-548c-3p
C | RKL | −0.005591422 | 0.937844 |
|
hsa-miR-548c-3p | BMP5 | −0.004257302 | 0.952654 |
|
hsa-miR-548c-3p | EGF | 0.000395288 | 0.995601 |
|
hsa-miR-548c-3p | PIK3CB | 0.002839251 | 0.968414 |
|
hsa-miR-548c-3p | MAPK8 | 0.021309033 | 0.766301 |
| hsa-miR-548c-3p
C | RK | 0.031385585 | 0.661515 |
|
hsa-miR-548c-3p | LIMD1 | 0.031613315 | 0.659213 |
| hsa-miR-548c-3p
C | RB1 | 0.032804274 | 0.647223 |
|
hsa-miR-548c-3p | FZD3 | 0.040937544 | 0.567885 |
|
hsa-miR-548c-3p | PIK3R3 | 0.041780747 | 0.559931 |
|
hsa-miR-548c-3p | KRAS | 0.056284445 | 0.43211 |
| hsa-miR-548c-3p
C | TNNB1 | 0.057967563 | 0.418448 |
|
hsa-miR-548c-3p | NRAS | 0.065278707 | 0.362099 |
|
hsa-miR-548c-3p | CDKN1B | 0.0732999 | 0.306004 |
|
hsa-miR-548c-3p | MAPK1 | 0.083215756 | 0.245003 |
|
hsa-miR-548c-3p | EREG | 0.086515941 | 0.226721 |
|
hsa-miR-548c-3p | BBC3 | 0.102224529 | 0.152888 |
|
hsa-miR-548c-3p | ELK1 | 0.115802515 | 0.105129 |
|
hsa-miR-548c-3p | CSNK1E | 0.137981222 | 0.053163 |
|
hsa-miR-548c-3p | AXIN2 | 0.158253961 | 0.026346 |
|
hsa-miR-548c-3p | FZD5 | 0.169685512 | 0.017135 |
|
hsa-miR-548c-3p | ID2 | 0.202596669 | 0.004302 |
|
hsa-miR-548c-3p | CTNNA2 | 0.246858375 | 0.00047 |
Discussion
HCa is clinically difficult to diagnose and has a
poor prognosis, therefore, identifying early stage molecular
biomarkers has become urgent. CircRNAs, which are stable and easier
to extract and detect, are considered ideal candidates for
early-stage biomarkers. This is the first report on the expression
profile of circRNAs in HCa. In the present study, a number of
aberrantly expressed circRNAs in HCa samples were identified.
Pathway enrichment results revealed that circRNAs may regulate HCa
progression through multiple signaling pathways, especially the
ErbB and Hippo signaling pathways. These results provided several
potential biomarkers and therapeutic targets for HCa.
The ceRNA hypothesis was described as a way that
RNAs communicate with each other, via competing for binding to
miRNAs and regulating the expression of each other to construct a
complex post-transcriptional regulatory network (35,36). mRNAs and long non-coding (lnc)
RNAs may all serve as ceRNAs (37). It has been demonstrated that
circRNAs can also function as miRNA sponges (6,11).
The present study demonstrated that aberrantly expressed circRNAs
have extensive interactions with miRNAs, and those miRNAs exerted
their effect on multiple cancer-related pathways. These data
indicated that the circRNA-associated ceRNA network may have
crucial roles in HCa progression.
The activation of ErbB oncogenes has been described
in various types of human tumors, including hypopharynx carcinomas,
and it has been correlated with a poor prognosis. For example, one
study describing the molecular alterations in hypopharynx
carcinomas demonstrated that ErbB1 was amplified in 29% of patients
with hypopharyngeal squamous cell carcinomas (38). In addition, ErbB1 amplification is
correlated with a hypopharyngeal primary site (39). Another study reported that v-erbB
stained positively in 62.5% of hypopharyngeal squamous cell
carcinomas samples but negatively in normal mucosa (40). The present ceRNA network analysis
demonstrated that a circRNA (hsa_circ_0008287 and
hsa_circ_0005027)/miRNA (hsa-miR-548c-3p) axis may have important
roles in ErbB-mediated tumor progression (Fig. 6).
Another pathway that is likely to be associated with
hypopharynx carcinomas is the Hippo signaling pathway. The Hippo
pathway has generated considerable interest in recent years because
of its involvement in several key hallmarks of cancer progression
and metastasis (41). Regulation
of Hippo signaling can be an attractive alternative strategy for
cancer treatment (42-44). Previously, ACTL6A and p63 were
demonstrated to cooperatively promote head and neck squamous cell
carcinoma, through activation of the Hippo/Yes-associated protein 1
(YAP) pathway and YAP activation can predict poor patient survival
(45). The present ceRNA network
analysis demonstrated that a circRNA (hsa_circ_0008287 and
hsa_circ_0005027)/miRNA (hsa-miR-548c-3p) axis may have important
roles in Hippo-mediated tumor progression (Fig. 7).
Extensive evidence has suggested that miRNAs have
important roles in breast cancer. The miR-548 family has been
demonstrated to be involved in the pathogenesis of several cancers.
For example, miR-548-3p was significantly downregulated in breast
cancer and overexpression of miR-548-3p inhibited the proliferation
and promoted the apoptosis of breast cancer cells (46). Overexpression of miR-548c-3p was
also confirmed in prostate epithelial stem cells and in
castration-resistant prostate cancer cells (45). Overexpression of miR-548c-3p in
differentiated cells induced stem-like properties and
radio-resistance (45).
Re-analyses of published studies further revealed that miR-548c-3p
is significantly overexpressed in castration-resistant prostate
cancer cells and is associated with poor recurrence-free survival,
suggesting that miR-548c-3p is a functional biomarker for prostate
cancer aggressiveness (47). The
present results demonstrated that miR-548c-3p may have important
roles in HCa progression through modulating the ErbB and Hippo
pathways. Due to the crucial roles of miR-548c-3p in multiple types
of cancer, development of novel gene therapies based on miR-548c-3p
might be encouraged.
Taken together, the present study indicated that
hsa_ circ_0008287 and hsa_circ_0005027 were downregulated in HCa
and competitively bound miR-548c-3p with ErbB and Hippo signaling
pathway genes. Further studies are warranted on the roles of
hsa_circ_0008287, hsa_circ_0005027, and miR-548c-3p as potential
diagnostic biomarkers and therapeutic targets for HCa.
Acknowledgments
Not applicable.
Funding
This work was funded by Yunnan Applied Basic
Research Projects (grant no. 2016FB038).
Availability of data and materials
The sequencing data have been deposited in the Gene
Expression Omnibus (GEO) database under the accession number
GSE111423.
Authors’ contributions
CF designed experiments and helped analyze the data.
YaL, YuL and XC collected the samples. DL analyzed data. HZ
interpreted the results and wrote the manuscript. XH designed
experiments and interpreted the results. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
The study was approved by the Ethics Committee of
the First Affiliated Hospital of Kunming Medical University
(Kunming, China). Written informed consent was obtained from all
the participants in the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Abbreviations:
|
HCa
|
hypopharyngeal cancer
|
|
DE
|
differentially expressed
|
|
ceRNAs
|
competing endogenous RNA
|
|
RPM
|
reads per Million mapped reads
|
|
GO
|
gene ontology
|
|
KEGG
|
Kyoto Encyclopedia of Genes and
Genomes
|
|
PCA
|
principal component analysis
|
References
|
1
|
Pingree TF, Davis RK, Reichman O and
Derrick L: Treatment of hypopharyngeal carcinoma: A 10-year review
of 1,362 cases. Laryngoscope. 97:901–904. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chan JY and Wei WI: Current management
strategy of hypopharyngeal carcinoma. Auris Nasus Larynx. 40:2–6.
2013. View Article : Google Scholar
|
|
3
|
Lagha A, Chraiet N, Labidi S, Rifi H,
Ayadi M, Krimi S, Allani B, Raies H, Touati S and Boussen H: Larynx
preservation: What is the best non-surgical strategy. Crit Rev
Oncol Hematol. 88:447–458. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Carrasco Llatas M, Lopez Molla C, Balaguer
Garcia R, Ferrer Ramírez MJ, Guallart Doménech F, Estellés Ferriol
JE, Fernández Martínez S and Dalmau Galofre J: Hypopharyngeal
cancer: Analysis of the evolution and treatment results. Acta
Otorrinolaringol Esp. 60:3–8. 2009.In Spanish. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chen LL and Yang L: Regulation of circRNA
biogenesis. RNA Biol. 12:381–388. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Memczak S, Jens M, Elefsinioti A, Torti F,
Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer
M, et al: Circular RNAs are a large class of animal RNAs with
regulatory potency. Nature. 495:333–338. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Xie H, Ren X, Xin S, Lan X, Lu G, Lin Y,
Yang S, Zeng Z, Liao W, Ding YQ and Liang L: Emerging roles of
circRNA_001569 targeting miR-145 in the proliferation and invasion
of colorectal cancer. Oncotarget. 7:26680–26691. 2016.PubMed/NCBI
|
|
8
|
Zheng Q, Bao C, Guo W, Li S, Chen J, Chen
B, Luo Y, Lyu D, Li Y, Shi G, et al: Circular RNA profiling reveals
an abundant circHIPK3 that regulates cell growth by sponging
multiple miRNAs. Nat Commun. 7:112152016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Guarnerio J, Bezzi M, Jeong JC, Paffenholz
SV, Berry K, Naldini MM, Lo-Coco F, Tay Y, Beck AH and Pandolfi PP:
Oncogenic role of fusion-circRNAs derived from cancer-associated
chromosomal translocations. Cell. 165:289–302. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhong ZY, Lv MX and Chen JX: Screening
differential circular RNA expression profiles reveals the
regulatory role of circTCF25-miR-103a-3p/miR-107-CDK6 pathway in
bladder carcinoma. Sci Rep. 6:309192016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hansen TB, Jensen TI, Clausen BH, Bramsen
JB, Finsen B, Damgaard CK and Kjems J: Natural RNA circles function
as efficient microRNA sponges. Nature. 495:384–388. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Brown J, Pirrung M and McCue LA: FQC
Dashboard: Integrates FastQC results into a web-based, interactive,
and extensible FASTQ quality control tool. Bioinformatics.
33:3137–3139. 2017. View Article : Google Scholar
|
|
13
|
Gao Y, Wang J and Zhao F: CIRI: An
efficient and unbiased algorithm for de novo circular RNA
identification. Genome Biol. 16:42015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li H and Durbin R: Fast and accurate short
read alignment with Burrows-Wheeler transform. Bioinformatics.
25:1754–1760. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Harrow J, Frankish A, Gonzalez JM,
Tapanari E, Diekhans M, Kokocinski F, Aken BL, Barrell D, Zadissa
A, Searle S, et al: GENCODE: The reference human genome annotation
for the ENCODE project. Genome Res. 22:1760–1774. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kozomara A and Griffiths-Jones S: miRBase:
Integrating microRNA annotation and deep-sequencing data. Nucleic
Acids Res. 39:D152–D157. 2011. View Article : Google Scholar :
|
|
17
|
Lever J, Krzywinski M and Atman N: Points
of Significance: principal component analysis. Nat Methods.
14:641–642. 2017. View Article : Google Scholar
|
|
18
|
Hotelling H: Analysis of a complex of
statistical variables into principal components. J Educ Psychol.
24:417–441. 1933. View
Article : Google Scholar
|
|
19
|
Ginestet C: ggplot2: Elegant graphics for
data analysis. J R Stat Soc Ser A. 174:245. 2011. View Article : Google Scholar
|
|
20
|
Harris T and Hardin JW: Exact Wilcoxon
signed-rank and Wilcoxon Mann-Whitney ranksum tests. Stata J.
13:337–343. 2013.
|
|
21
|
Benjamini Y and Hochberg Y: Controlling
the false discovery rate: A practical and powerful approach to
multiple testing. J Roy Stat Soc B Met. 57:289–300. 1995.
|
|
22
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Dudekulay DB, Panda AC, Grammatikakis I,
De S, Abdelmohsen K and Gorospe M: CircInteractome: A web tool for
exploring circular RNAs and their interacting proteins and
microRNAs. RNA Biol. 13:34–42. 2016. View Article : Google Scholar
|
|
24
|
Casper J, Zweig AS, Villarreal C, Tyner C,
Speir ML, Rosenbloom KR, Raney BJ, Lee CM, Lee BT, Karolchik D, et
al: The UCSC genome browser database: 2018 update. Nucleic Acids
Res. 46:D762–D769. 2018.
|
|
25
|
Agarwal V, Bell GW, Nam JW and Bartel DP:
Predicting effective microRNA target sites in mammalian mRNAs.
Elife. 4:75542015. View Article : Google Scholar
|
|
26
|
Vlachos IS, Zagganas K, Paraskevopoulou
MD, Georgakilas G, Karagkouni D, Vergoulis T, Dalamagas T and
Hatzigeorgiou AG: DIANA-miRPath v3.0: Deciphering microRNA function
with experimental support. Nucleic Acids Res. 43:W460–W466. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(T)(−Delta Delta C) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
29
|
Cancer Genome Atlas Research Network
Analysis Working Group, Asan University, BC Cancer Agency; et al:
Integrated genomic characterization of oesophageal carcinoma.
Nature. 541:169–175. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lancar R and Funck-Brentano C: Survival
analysis example based on an event history model from a clinical
trial in cardiology. Rev Epidemiol Sante Publique. 47:613–618.
1999.In French.
|
|
31
|
Mantel N: Evaluation of survival data and
two new rank order statistics arising in its consideration. Cancer
Chemother Rep. 50:163–170. 1966.PubMed/NCBI
|
|
32
|
R Development Core Team: R: A language and
environment for statistical computing Vienna, Austria: The. R
Foundation for Statistical Computing; 2011
|
|
33
|
Guo JU, Agarwal V, Guo H and Bartel DP:
Expanded identification and characterization of mammalian circular
RNAs. Genome Biol. 15:4092014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Glazar P, Papavasileiou P and Rajewsky N:
circBase: A database for circular RNAs. RNA. 20:1666–1670. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Tay Y, Rinn J and Pandolfi PP: The
multilayered complexity of ceRNA crosstalk and competition. Nature.
505:344–352. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Salmena L, Poliseno L, Tay Y, Kats L and
Pandolfi PP: A ceRNA hypothesis: The rosetta stone of a hidden RNA
language. Cell. 146:353–358. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Karreth FA and Pandolfi PP: ceRNA
cross-talk in cancer: When ce-bling rivalries go awry. Cancer
Discov. 3:1113–1121. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Rodrigo JP, González MV, Lazo PS, Ramos S,
Coto E, Alvarez I, García LA and Suárez C: Genetic alterations in
squamous cell carcinomas of the hypopharynx with correlations to
clinico-pathological features. Oral Oncol. 38:357–363. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Rodrigo JP, Ramos S, Lazo PS, Alvarez I
and Suarez C: Amplification of ERBB oncogenes in squamous cell
carcinomas of the head and neck. Eur J Cancer. 32A:2004–2010. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Otsu M, Hayashi Y, Amatsu M and Itoh H:
Immunohistochemical study of p53, EGF, EGF-receptor, v-erb B and
ras p21 in squamous cell carcinoma of hypopharynx. Kobe J Med Sci.
40:139–153. 1994.PubMed/NCBI
|
|
41
|
Ma Y, Yang Y, Wang F, Wei Q and Qin H:
Hippo-YAP signaling pathway: A new paradigm for cancer therapy. Int
J Cancer. 137:2275–2286. 2015. View Article : Google Scholar
|
|
42
|
Santucci M, Vignudelli T, Ferrari S, Mor
M, Scalvini L, Bolognesi ML, Uliassi E and Costi MP: The Hippo
pathway and YAP/TAZ-TEAD protein-protein interaction as targets for
regenerative medicine and cancer treatment. J Med Chem.
58:4857–4873. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Guo L and Teng L: YAP/TAZ for cancer
therapy: Opportunities and challenges (Review). Int J Oncol.
46:1444–1452. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Liu AM, Xu MZ, Chen J, Poon RT and Luk JM:
Targeting YAP and Hippo signaling pathway in liver cancer. Expert
Opin Ther Targets. 14:855–868. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Saladi SV, Ross K, Karaayvaz M, Tata PR,
Mou H, Rajagopal J, Ramaswamy S and Ellisen LW: ACTL6A Is
Co-amplified with p63 in squamous cell carcinoma to drive YAP
activation, regenerative proliferation, and poor prognosis. Cancer
Cell. 31:35–49. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Shi Y, Qiu M, Wu Y and Hai L: MiR-548-3p
functions as an anti-oncogenic regulator in breast cancer. Biomed
Pharmacother. 75:111–116. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Rane JK, Scaravilli M, Ylipää A, Pellacani
D, Mann VM, Simms MS, Nykter M, Collins AT, Visakorpi T and
Maitland NJ: MicroRNA expression profile of primary prostate cancer
stem cells as a source of biomarkers and therapeutic targets. Eur
Urol. 67:7–10. 2015. View Article : Google Scholar
|