Introduction
In recent 15 years, the overall survival of patients
with multiple myeloma (MM) has been significantly prolonged with
the clinical application of new agents, including immunomodulatory
drugs, proteasome inhibitors and monoclonal antibodies (1-5).
Thalidomide was the first of these drugs to exhibit its clinical
activity and has been shown to be effective against the disease
throughout its stages.
Angiogenesis is an important factor for cancer
progression, which promotes tumor growth and metastasis; tumors
become necrotic or apoptotic in the absence of vascular support.
Vascular endothelial growth factor (VEGF) family members are key
factors of angiogenesis, and VEGF expression has been described to
play a role in the prognosis of a number of types of cancer, such
as colorectal cancer (6), breast
cancer (7), non-small cell lung
carcinoma (8) and Kaposi's
sarcoma (9). However, MM was the
first hematological malignancy in which the prognostic relevance of
angiogenesis was shown (10).
Thalidomide has been proven to exert an anti-angiogenic effect;
however, the detailed underlying mechanisms remain unclear.
DEPTOR, an inhibitor of the mammalian target of
rapamycin (mTOR), has been found to be overexpressed in MM cells
and is essential for their survival (11,12). It has been reported that patients
with MM with a high DEPTOR expression have good responses to
thalidomide-based therapy (13,14). Vascular endothelial cell-expressed
DEPTOR regulates endothelial cell activation and angiogenic
responses through extracellular signal-regulated kinase (ERK)1 and
signal transducer and activator of transcription (STAT)-mediated
chemokine expression (15). In
addition, endothelial cell-specific DEPTOR deficiency increases
CD31, hypoxia-inducible factor (HIF)-1α and VEGF expression, which
promotes angiogenesis (16).
However, whether DEPTOR can regulate the angiogenesis of MM cells
and the associated underlying mechanisms have not yet been
elucidated.
A previous study by the authors demonstrated that
the knockdown of DEPTOR by siRNA also inhibited autophagy in MM
cells (17). Basal autophagy is
necessary for cellular; housekeeping; to eliminate damaged
organelles, such as depolarized mitochondria due to reactive oxygen
species (ROS) (13,18,19). Mitochondrial ROS (mtROS) are
involved in the signal transduction pathways leading to nuclear
factor κB (NF-κB) activation (20-22). NF-κB activation plays a dominant
role in the secretion of interleukin (IL)-6, which stimulates the
secretion of VEGF (21). As
mentioned above, it was hypothesized that DEPTOR-mediated cellular
autophagy and mitROS may play pivotal regulatory roles in
angiogenesis in MM. The analysis described herein may be helpful
for the further understanding of the mechanisms underlying the
regulatory effects of DEPTOR on angiogenesis and thalidomide for
the treatment of MM.
Materials and methods
Validation of DEPTOR expression in MM
based on the Oncomine database
Oncomine (https://www.oncomine.org) is an online database
containing previously published and publicly available microarray
data. The Oncomine database Wwase used to validate the mRNA
expression of DEPTOR in MM cell lines. The DEPTOR gene was queried
in the database, and the results were filtered by selecting MM and
Cancer vs. Cancer Analysis. Statistical analysis was conducted
using Oncomine algorithms.
Overview of the potential pathways of
DEPTOR in regulating the biological role of MM cells using
microarray profiles from patients with MM
The microarray dataset GSE24080 of 559 patients with
MM (23) (Affymetrix Human Genome
U133 2.0 Array) was obtained from the Gene Expression Omnibus (GEO)
database. The raw array data (.CEL files) were pre-processed using
the Robust Multichip Average (RMA) algorithm (24). Microarray annotation information
was used to match probes with the corresponding genes. If multiple
probes matched a single gene, the probes with the highest quartile
range were selected, as previously reported (25). To further investigate the possible
pathways of DEPTOR involved in regulating the biological
characteristics of myeloma cells, KEGG-related Gene Set Enrichment
Analysis (GSEA) analysis was conducted on the on GSE24080 dataset
by the gseKEGG function of the clusterProfiler package in R
(26).
Cell lines and primary cells
The human MM cell lines, RPMI8226, MM.1S and U226,
were obtained from the American Type Culture Collection (ATCC) and
grown in RPMI-1640 medium (HyClone; Thermo Fisher Scientific, Inc.)
containing 10% heat-inactivated fetal calf serum (Biological
Industries). Human umbilical vein endothelial cells (HUVECs) and
the 293-FT cell line were obtained from the China Center for Type
Culture Collection and grown in Dulbecco's modified Eagle's medium
(DMEM) (HyClone; Thermo Fisher Scientific, Inc.) containing 10%
heat-inactivated fetal calf serum (Biological Industries). Bone
marrow (BM) aspiration and biopsy specimens were obtained from
patients newly diagnosed with MM according to diagnostic criteria
(27). Primary plasma cells were
isolated from BM specimens using CD138+
magnetic-activated cell sorting (MACS; Miltenyi Biotec GmbH). A
total of 22 patients were used in the present study and samples
were collected at the First Affiliated Hospital of Fujian Medical
University from October, 2015 to June, 2018. Approval for the study
was obtained from the Ethics Committee of the First Affiliated
Hospital of Fujian Medical University. Informed consent was
obtained in accordance with the Declaration of Helsinki.
Assessment of BM microvessel density
(MVD)
BM MVD was estimated as previously described
(28). Immunostaining for CD34
was used to evaluate the BM MVD. BM specimens used in the present
study were fixed in 10% formaldehyde overnight and embedded in
paraffin. Thin slides of 4 µm thickness were cut and
subjected to immunohistochemistry (IHC) for CD34 (clone My10; 1:50;
ab245689, Abcam) for 32 min at room temperature. Immunoreactivity
was detected with goat anti-rabbit immunoglobulins H&L (HRP)
(1:200; ab6721, Abcam). The reaction was revealed with
diaminobenzidine (DAB) and counterstained with Harris' hematoxylin
(Dako; Agilent Technologies, Inc.) for 30 min at room
temperature.
The staining was evaluated by 2 independent
pathologists under a light microscope (Olympus Corporation). Each
slide was scanned at ×100 magnification to determine 3 'hotspots',
defined as areas with the maximum number of microvessels.
Microvessels were counted in each of the 3 hotspots at ×400
magnification. Large vessels and vessels in the periosteum or bone
were excluded. Areas of staining with no discrete breaks were
counted as a single vessel. The presence of a lumen was not
required. MVD was estimated by determining the average number of
vessels in each of the 3 hotspots.
Cell treatment
Rapamycin (100 nM, Shanghai Shenggong Bioengineering
Service Co., Ltd.) was added to the RPMI8226 and MM.1S cells for 24
h. 3-MA (20 µM, Sigma-Aldrich; Merck KGaA) was added to the
U226 cells followed by culture for 24 h. SMER28 (10 µM,
Beyotime Institute of Biotechnology) and Mito-TEMPO (10 µM,
Sigma-Aldrich; Merck KGaA) were added to the RPMI8226 and MM.1S
cells followed by culture for 24 h with DEPTOR inhibition.
Thalidomide (Sigma-Aldrich; Merck KGaA) was added to the RPMI8226
and MM.1S cells and cultured for 24 h, with or without DEPTOR
inhibition.
Recombinant lentiviral vector
construction and infection of cells
Lentiviral vectors harboring Cas9 and sgRNA
(Shanghai GeneChem Co., Ltd.) were constructed for the transfection
of the MM cells. The sgRNAs targeting the DEPTOR gene were designed
using an online tool (http://crispr.mit.edu/). The sequences of the sgRNAs
were as follows: sgRNA1, GAG TGG CGG GGC GCA GCA AA; sgRNA2, CAA
GTA TGA GCG CAC CTT CA; sgRNA3, TCA GAA TGA ACT TCC GGC GG; and
control sgRNA, CGC TTC CGC GGC CCG TTC AA. The sgRNAs were packaged
into lentiviruses, which were named DEP1, DEP2, DEP3 and GFP,
respectively.
According to the manufacturer's protocol,
lentiviruses were transduced into RPMI8226 (multiplication of
infection, MOI=100) and MM.1S (MOI=66) cells with HitransGP (cat.
no. REVG005; Shanghai Genechem Co., Ltd.) for 5 days. The cells
were then tested for the DEPTOR mutation using the Cruiser™ Enzyme
(Genloci Biotechnologies Inc.) and tested for DEPTOR protein
disruption by western blot analysis. Sanger sequencing was
performed to confirm the presence of a mutation in the desired part
of the genome using the forward primer for DEPTOR sgRNA1, 5′-CTG
CCT ACC CAT AGG GAT TCC -3′; sgRNA2, 5′-CTT TGC TGT AGT ACT TCA TGG
-3′; and sgRNA3, 5′-TTT CTC CCC AGT GTC CAA CAA G-3′. The U226
cells were transfected with a DEPTOR overexpression vector
(GV143-DEPTOR, Genechem Co., Ltd.) at the concentration of 20
µg/ml in serum-free medium by electroporation. After 48 h,
the protein levels of the corresponding genes in transfected U226
cells were detected by western blot analysis. The condition medium
derived from the above-mentioned MM cell lines with different
expression levels of DEPTOR were collected and centrifuged at 360 ×
g for 5 min at room temperature and stored at −80°C.
Cell growth assay
A Cell Counting kit (CCK)-8 assay (Beyotime
Institute of Biotechnology, Inc.) was used to assess cell growth,
according to the manufacturer's instructions. Briefly,
1×104 cells/100 µl were seeded into 96-well
plates and cultured for different periods of time (0, 24, 48, 72,
96, 120, 144 and 168 h). For the last 4 h of culture, the cells
were incubated with 10 µl of CCK-8 at 37°C for 45 min. The
absorbance at 450 nm was measured using an automated microplate
reader (Tecan Infinite F50; Tecan Group, Ltd.) to detect
metabolically intact cells.
Tube formation assay
Matrigel (100 µl; BD Biosciences) was poured
into 96-well plates and then incubated at 37°C for 45 min. HUVECs
were collected and resuspended in the previously collected
conditioned medium at 2×105/ml, and 100 µl was
seeded on the Matrigel and cultured for 8 h at 37°C. The
morphological changes of the cells from 3 randomly chosen fields
were observed and photographed using a light microscope (Olympus
Corporation) at ×100 magnification. The number of tube-like
structures was measured using ImageJ software version 1.46
(National Institutes of Health).
Dual-luciferase reporter assay
RPMI8226 and MM.1S cells were seeded in 24-well
plates at 5×105/ml. The cells were transfected with the
pNFκB-TA-luc reporter plasmid (Beyotime Institute of Biotechnology)
at the concentration of 2 µg/ml and the Renilla
luciferase-expressing plasmid as an internal control using
Lipofectamine 2000 for 48 h (Lipo2000, Invitrogen Inc.; Thermo
Fisher Scientific, Inc.), according to the manufacturer's
instructions. A total of 4 groups of cells were prepared: The
negative control group, positive control group (TNF-α was used as a
NF-κB activator), and test groups treated with lentivirus GFP and
DEP3 for 48 h before harvesting. Firefly luciferase activities were
analyzed using the dual-luciferase reporter assay kit (Beyotime
Institute of Biotechnology).
Enzyme-linked immunosorbent assay
(ELISA)
The quantita-tive evaluation of IL-6 and VEGF in
cell culture supernatants was carried out using ELISA. The
cell-free supernatants were collected from cell cultures and
centrifuged at 360 × g for 5 min at 20°C. The concentration of
secreted VEGF proteins was determined using an ELISA kit
(Neobioscience), according to the manufacturer's instructions.
DAPI and MitoSOX staining
The cells were incubated with DAPI (1 µg/ml,
Beyotime Institute of Biotechnology) for 12 h, stained with the
MitoSOX dye (5 µM; MitoSOX Red Reagent, Invitrogen Inc.;
Thermo Fisher Scientific, Inc.) at 37°C for 30 min, and fixed with
4% paraformaldehyde for 20 min. After washing with PBS, the nuclear
morphology of the cells and mtROS staining were examined using a
fluorescence microscope (Olympus Corporation).
Flow cytometry
For mtROS measurements, the cells were stained with
the MitoSOX Red Reagent (5 µM; Invitrogen Inc.; Thermo
Fisher Scientific, Inc.) at 37°C for 30 min. The stained cells were
analyzed for fluorescence using a flow cytometer (FACSCalibur, BD
Biosciences). The data were analyzed using Flowjo software (Flowjo
10.0).
Cell apoptosis assay was performed using flow
cytometry with propidium iodide (PI) and Annexin V-APC staining
(APC Annexin V Apoptosis Detection kit with PI; 640932, BioLegend,
Inc.). The cells were harvested, washed twice with PBS, resuspended
in staining buffer, and stained with Annexin V-APC/PI, according to
the manufacturer's instructions. The cells were analyzed using a
flow cytometer (FACSCalibur, BD Biosciences).
Monodansylcadaverine (MDC) staining
The autofluorescent compound, MDC [Sangon Biotech
(Shanghai) Co., Ltd.] is a commonly used selective fluorescent
marker for autophagic vacuoles. The cells were plated on glass
coverslips and probed with a 1:1,000 dilution of MDC in staining
buffer (Cell-Based Assay Buffer Tablet dissolved in water).
Following 10 min of incubation at 37°C, the cells were washed twice
with assay buffer in the dark. A fluorescence microscope (Olympus
Corporation) was used immediately after MDC staining.
Western blot analysis
For protein extraction, the cells were lysed in RIPA
buffer (Beyotime Biosciences) containing the protease inhibitor
PMSF. The isolation of mitochondrial and cytosolic proteins was
performed using the Mitochondria/Cytosol Fractionation kit
(Beyotime Institute of Biotechnology). Cytoplasmic and nuclear
protein extracts were prepared using the NE-PER™ Nuclear and
Cytoplasmic Extraction Reagents (Thermo Fisher Scientific, Inc.),
according to the manufacturer's instructions.
Protein quantification were detected using
spectrophotometer (DS-11, DeNovix). A total of 80 µg
protein/lane were separated by SDS-PAGE on 8-16% Tris-glycine gels
(Beyotime Institute of Biotechnology), transferred to
polyvinylidene difluoride membranes (Millipore corp., Billerica,
MA, USA). The membranes were then blocked for 1 h at room
temperature with non-fat dry milk in TBST (Bio-Rad Laboratories,
Inc.). The membranes were incubated with antibodies against LC3
(1:1,000, ab51520, Abcam), Beclin 1 (1:1,000, ab62557, Abcam),
DEPTOR (1:1,000, ab244394, Abcam), VEGF (1:1,000, AV202, Beyotime
Institute of Biotechnology), phospho-p65 (p-p65, 1:1,000, ab109458,
Abcam), p65 (1:1,000, ab16502, Abcam), phospho-p50 (p-p50, 1:1,000,
710460, Invitrogen; Thermo Fisher Scientific, Inc.), p50 (1:1,000,
710450, Invitrogen; Thermo Fisher Scientific, Inc.), IκB-α
(1:1,000, ab7217, Abcam), cytochrome c (Cyt c;
1:1,000, ab90529, Abcam), cytochrome c oxidase subunit IV
(COX IV; 1:1,000, ab16056, Abcam), CRBN (1:2,000, ab98992, Abcam),
Lamin B (1:1,000, 66095-1, ProteinTech Group, Inc.) and β-actin
(1:2,000, ab8226, Abcam) at 4°C overnight. Following 3 washes with
TBS-T for 10 min, a peroxidase-conjugated secondary antibody
(1:10,000, A0216/A0208, Beyotime Institute of Biotechnology) was
incubated with the membranes for 2 h at room temperature. Signals
were detected by the ECL detection system (Beyotime Institute of
Biotechnology). The results were analyzed quantitatively by
densitometry using software ImageJ software version 1.46 (National
Institutes of Health).
Purpose proteins and loading control with similar
molecular weight were produced from the same western blot membrane.
Purpose protein detected by the ECL detection system, incubated
with stripping buffer for approximately 30 min under constant
agitation; following 3 washes with TBS-T for 10 min and then
blotted with antibodies against loading control (β-actin and Lamin
B). Following 3 washes with TBS-T for 10 min, a
peroxidase-conjugated secondary antibody (Beyotime Institute of
Biotechnology) was incubated for 2 h at room temperature. Signals
were detected by the ECL detection system (Beyotime Institute of
Biotechnology). The results were analyzed quantitatively by
densitometry using ImageJ software version 1.46 (National
Institutes of Health).
Statistical analysis
All experiments were performed in triplicate. The
data are presented as the means ± standard deviation (SD).
Differences were analyzed using the Student's t-test or one-way
ANOVA with the Tukey's HSD post hoc test, as appropriate. The
associations between the expression of DEPTOR and the expression of
VEGF and BM MVD and in patients with MM were assessed by Pearson's
correlation analysis. GraphPad 5.0 (GraphPad Software Inc.) was
used for statistical analyses. A P-value <0.05 was considered to
indicate a statistically significant difference.
Results
Validation of DEPTOR expression in MM
cell lines using the Oncomine database
The mRNA expression levels of DEPTOR in MM were
explored using the Oncomine database. According to the results of
the Cancer vs. Cancer Analysis, 3 datasets (Barretina CellLine,
Garnett CellLine and Wooster CellLine) were identified that
contained DEPTOR mRNA expression data obtained from different tumor
cell lines. As shown in Fig.
1A-C, 2 datasets (Barretina CellLine and Garnett CellLine)
demonstrated that DEPTOR mRNA expression was significantly higher
in MM than in the majority of other cancers (including bladder
cancer, brain and central nervous system cancer, breast cancer,
cervical cancer, colorectal cancer, esophageal cancer, gastric
cancer, head and neck cancer, kidney cancer, leukemia, liver
cancer, lung cancer, lymphoma, melanoma, ovarian cancer, pancreatic
cancer, prostate cancer and sarcoma); however, one dataset (Wooster
CellLine) revealed no differences between MM and other types of
cancer. The meta-analysis of these 3 datasets still suggested that
DEPTOR was overexpressed in MM compared with other types of cancer
(Fig. 1D).
Overview of potential pathways of DEPTOR
involved in regulating the biological pathway of MM cells using
microarray profiles
To investigate the possible pathways of DEPTOR
involved in the regulation of MM cells, KEGG-related GSEA analysis
was conducted on the GSE24080 dataset using the gseKEGG function of
the clusterProfiler package in R. As shown in Fig. 1E, the main enriched pathways
closely related to cancer development included protein processing
in the endoplasmic reticulum, oxidative phosphorylation, autophagy,
ubiquitin-mediated proteolysis, the mTOR signaling pathway and the
VEGF signaling pathway.
DEPTOR expression is negatively related
to VEGF expression and BM MVD in MM
The mRNA expression levels of VEGFA in MM were also
explored using the Oncomine database. As shown in Fig. S1A-C, VEGFA mRNA expression was
significantly higher in MM than in the majority of other types of
cancer in the Barretina CellLine and Garnett CellLine databases;
however, the Wooster CellLine database did not reveal any marked
differences between MM and other types of cancer. The meta-analysis
of these 3 datasets still suggested that VEGFA was overexpressed in
MM compared with other types of cancer (Fig. S1D).
Western blot analysis was used to examine the
expression levels of DEPTOR and VEGF in different MM cell lines
(RPMI8226, MM.1S and U226). The results revealed that the RPMI8226
cells had the highest expression of DEPTOR and the lowest
expression of VEGF, while the MM.1S cells had an intermediate
expression of DEPTOR and VEGF, and the U266 cells had the lowest
expression of DEPTOR and the highest expression of VEGF (Fig. 1F and G).
In the cohort of 22 patients newly diagnosed with
MM, the expression patterns of DEPTOR and VEGF in primary MM cells
isolated by MACS were validated by western blot analysis. The 22
patients with MM included 1 patient with the International Staging
System (ISS) I, 7 with ISS II and 14 with ISS III. The median age
was 65 years (range, 46-89 years). In total 12 patients were male
and 10 were female. Western blot analysis and Pearson's correlation
analysis revealed that the expression of DEPTOR negatively
correlated with the expression of VEGF (Pearson's r=−0.78, Fig. 1H and I). The MVD in BM specimens
of corresponding patients was investigated by IHC with an anti-CD34
antibody (Fig. S2). The median
BM MVD was 167 (range, 9-412 vessels/mm2). Using the
median relative expression among the 22 patients (0.12, which
corresponds to the average value between the 11 and 12th patients)
of DEPTOR as a cut-off value, the patients were divided into 2
groups, those with a high and low DEPTOR expression. The MVD in the
high expression DEPTOR group was significantly lower than in the
low expression DEPTOR group (96.9±22.0 vessels/mm2 vs.
281.2±28.6 vessels/mm2, respectively; P<0.001;
Fig. 1J). The expression of
DEPTOR negatively correlated with MVD (Pearson's r=−0.76) (Fig. 1K).
CRISPR/Cas9-mediated efficient DEPTOR
disruption in MM
The CRISPR/Cas9 system was used to disrupt the
protein expression of DEPTOR in the MM cell line, RPMI8226.
Lentiviruses (DEP1, DEP2 and DEP3) expressing sgRNA targeting
DEPTOR were transfected into RPMI8226 cells (Fig. S3A). At 5 days following
transduction, the fragments of the 3 different target sites
relevant to DEP1, DEP2 and DEP3 were amplified bu PCR and then
digested with Cruiser™ Enzyme (Fig.
S3B). The cleaved bands were observed in the DEP1, DEP2 and
DEP3 groups, but not in the control and GFP groups (Fig. S3C), indicating that insertion or
deletion mutations (indel) were introduced in the genomes. The
levels of DEPTOR protein expression were detected by western blot
analysis. The results revealed that there were significant
disruptive effects on the DEPTOR protein levels in each sgRNA group
compared with the GFP group. Among these, DEP3 displayed the most
potent disruptive effect. To further validate DEPTOR gene
disruption, the nucleotide sequences of the above PCR products of
target DNA in the DEP3 group were analyzed and it was confirmed
that the indel mutation was introduced into the genome (Fig. S3D). Therefore, lentivirus DEP3
was used in the subsequent experiments. After the RPMI8226 and
MM.1S cells were transfected with lentivirus DEP3, the suppression
of DEPTOR abolished cell proliferation over a 7-day culture period
(Fig. S3E and F). In addition,
the suppression of DEPTOR in RPMI8226 cells induced cell lysis and
necrosis from the 5th day (data not shown). This strongly suggests
that the high expression of DEPTOR is essential for maintaining
myeloma cell survival.
Reduction of DEPTOR expression in MM
cells attenuates the effects of thalidomide on proliferation,
autophagy and VEGF expression
Based on its antitumor and anti-angiogenic effects,
thalidomide was used to treat the MM cells and was proven to be
effective. As shown in Fig. 2A and
C, the rate of proliferation of the RPMI8226 and MM.1S cells
decreased with the increasing thalidomide concentration and
treatment duration. In addition, the expression of DEPTOR was
increased with the increasing thalidomide concentration, whereas
the expression of VEGF decreased (Fig. 2B and D). These results indicated
that DEPTOR expression was negatively associated with the
expression of VEGF. The RPMI8226 cells in which DEPTOR expression
was suppressed exhibited a marked anti-proliferative response to
thalidomide (Fig. 2E). Of note,
thalidomide failed to induce the death of DEPTOR-disrupted MM cells
(Fig. 2F). Since cell death is
not only related to apoptosis, but also to autophagy, the effects
of thalidomide on the autophagy of MM cells were further analyzed.
MDC staining demonstrated that thalidomide significantly increased
the level of autophagy in MM cells, and its effect was suppressed
by the autophagy inhibitor, 3-MA (Fig. 2G). Western blot analysis revealed
that thalidomide increased DEPTOR and CRBN expression, as well as
the expression of autophagy-related proteins (Beclin 1 and LC3
II/I), whereas it inhibited VEGF expression. These effects were
reversed by 3-MA (Fig. 2H). In
the RPMI8226 cells in which DEPTOR expression was disrupted
(suppressed), thalidomide had no marked effects on the levels of
CRBN, VEGF and autophagy-related proteins (Beclin 1 and LC3 II/I).
Furthermore, after the DEPTOR-disrupted RPMI8226 cells were treated
with thalidomide and 3-MA, the levels of autophagy-related proteins
were further inhibited, while VEGF protein levels and VEGF levels
in the culture media were increased (Fig. 2H and I). These data suggested that
DEPTOR-regulated autophagy played an important role in the
antitumor and anti-angiogenic effects of thalidomide.
Effects of DEPTOR on autophagy and
angiogenesis of MM cells
The effects of DEPTOR on angiogenesis in myeloma
cell lines were further validated by disrupting or overexpressing
DEPTOR. Based on the above-mentioned results of DEPTOR expression
levels in MM cell lines, the expression of DEPTOR was
suppressed/disrupted in RPMI8226 and MM.1S cells, and overexpressed
in U266 cells. The tube formation assay of HUVECs revealed that the
RPMI8226 and MM.1S cells in which DEPTOR was inhibited exhibited an
increased angiogenesis, while the DEPTOR-overexpressing U266 cells
exhibited a suppressed angiogenesis (Fig. 3A). To clarify the effects of
DEPTOR on angiogenesis and autophagy, the association between
angiogenesis and autophagy in MM was first investigated. As shown
in Fig. 3B, when the RPMI8226 and
MM.1S cells with a higher autophagic activity were treated with the
autophagy inhibitor, 3-MA, their VEGF expression levels increased
as autophagic activity decreased. On the other hand, when the U266
cells with a lower autophagic activity were treated with the
autophagy inducer, rapamycin, their VEGF expression levels
decreased as autophagy activity increased. Furthermore, DEPTOR
disruption in RPMI8226 and MM.1S cells decreased the expression of
autophagy-related proteins (Beclin-1 and LC3II/LCI) and enhanced
VEGF expression (Fig. 3C). The
overexpression of DEPTOR in the U266 cells led to the opposite
results (Fig. 3C). IL-6 and VEGF
levels in the cell supernatant were increased in the RPMI8226 and
MM.1S cells following the inhibition of autophagy or the disruption
of DEPTOR expression, and were decreased in the U266 cells
following the induction of autophagy or the overexpression of
DEPTOR (Fig. 3D).
Inhibition of autophagy by DEPTOR
disruption activates the NF-κB pathway by damaging mitochondria and
promoting the accumulation of mtROS
The present tudy then investigated the mechanisms
through which autophagy inhibition influences angiogenesis in MM
cells. Autophagy defects led to mitochondrial damage and subsequent
ROS accumulation. As shown in Fig. 4A
and B, staining with Mito-SOX, a mitochondrial superoxide
indicator, revealed that DEPTOR inhibition prominently increased
the mtROS levels in the RPMI8226 and MM.1S cells. As ROS can
directly activate the NF-κB pathway (20), the role of DEPTOR in regulating
the NF-κB pathway was also validated. As shown in Fig. 4C, DEPTOR disruption in RPMI8226
and MM.1S cells stimulated NF-κB activation, as detected by
luciferase assays. Western blot analysis demonstrated that DEPTOR
inhibition decreased Cyt c expression in the mitochondria
and increased Cyt c expression in the cytoplasm, which
indicated the potential damage to the mitochondrial membrane of
RPMI8226 and MM.1S cells (Fig.
4D-G). Further analyses suggested that DEPTOR disruption
decreased IκB-α expression in the cytoplasm and increased p-p50 and
p-p65 expression in the nucleus (Fig.
4H-O). These data revealed that DEPTOR disruption induced
mitochondrial damage and the accumulation of mtROS in MM cells,
which led to the activation of the NF-κB pathway.
 | Figure 4Mitochondrial damage signals in
DEPTOR-inhibited RPMI8226 and MM.1S cells. RPMI8226 and MM.1s cells
were infected with lentiviral GFP and DEP3 for 5 days. (A and B)
The mtROS levels of the RPMI8226 and MM.1S cells in the indicated
groups stained with the MitoSOX probe was evaluated using a
fluorescence microscope (scale bar, 10 µm) and flow
cytometric analysis, respectively. (C) The NF-κB activity of
RPMI8226 or MM.1S cells in the indicated groups was detected by
luciferase assays. Cells were co-transfected with pNF-κB-TA-luc
with Renilla luciferase reporter (as an internal control)
for 24 h and treated with GFP or DEP3 for 48 h. (D-G) The
mitochondrial and cytoplasmic fractions of the cultured cells were
separated using the Mitochondria/Cytosol Fractionation kit. Protein
expression of Cyt c in the (D) mitochondria and (E)
cytoplasm of the indicated groups for RPMI8226 cells was examined
by western blot analysis. Primary antibodies with COXIV and β-actin
was used as the mitochondrial and cytosolic loading control,
respectively. Protein expression of Cyt c in the (F)
mitochondria and (G) cytoplasm of the indicated groups for MM.1S
cells was examined by western blot analysis. Primary antibodies
with COXIV and β-actin was used as the mitochondrial and cytosolic
loading control, respectively. Protein expression levels of DEPTOR
and IκB-α in the cytoplasm of the indicated groups for (H) RPMI8226
and (I) MM.1S cells were examined by western blot analysis. (J-O).
The nuclear and cytoplasmic protein fractions of the cultured cells
were isolated using the NE-PER™ Nuclear and Cytoplasmic Extraction
Reagents. Protein expressions of (J) p-p50 and p-p65 in the
nuclear, (K) p50 and p65 in the whole cell lysates of the indicated
groups for RPMI8226 cells was examined by western blot analysis.
LaminB and β-actin was used as the loading control for the nuclear
extracts and the whole cell lysates, respectively. (L)
Quantification of p-p50 subunit and p-p65 subunit was normalized
for total p50 protein and total p65 protein (J and K),
respectively. Protein expressions of (M) p-p50 and p-p65 in the
nuclear, (N) p50 and p65 in the whole cell lysates of the indicated
groups for MM.1S was examined by western blot analysis,
respectively. LaminB and β-actin was used as the loading control
for the nuclear extracts and the whole cell lysates, respectively.
(O) Quantification of p-p50 subunit and p-p65 subunit was
normalized for total p50 protein and total p65 protein (M and N),
respectively. Error bars indicate the standard deviation from 3
independent experiments. *P<0.05 compared with GFP
group, **P<0.01 compared with GFP group. Cyt
c, cytochrome c; mtROS, mitochondrial reactive oxygen
species; p-p50, phospho-p50; p-p65, phospho-p65. |
Autophagy inducer and
mitochondrial-specific antioxidant reverse the effects of DEPTOR
disruption on autophagy and VEGF expression in MM cells
To confirm the findings described above, the
RPMI8226 and MM.1S cells in which DEPTOR was inhibited were treated
with SMER28 (an mTOR-independent autophagy inducer) or Mito-TEMPO
(a mitochondrial-specific antioxidant) for 24 h. As shown in
Fig. 5A, both SMER28 and
Mito-TEMPO reversed the effects of DEPTOR disruption on autophagic
activity and VEGF expression, but had no effect on DEPTOR
expression. When the RPMI8226 and MM.1s cells in which DEPTOR was
suppressed were treated with SMER28 or Mito-TEMPO, the detection of
the cell supernatant by ELISA revealed that IL-6 and VEGF levels
were decreased (Fig. 5B). In
addition, SMER28 and Mito-TEMPO reversed the mitochondrial damage,
mtROS accumulation and subsequent NF-κB pathway activation induced
by DEPTOR disruption (Figs. 5C
and 6). These results demonstrate
that DEPTOR disruption regulates VEGF by promoting the accumulation
of mtROS and activating the NF-κB pathway in an autophagy-dependent
manner rather than in an mTOR-dependent manner.
 | Figure 6Autophagy inducer and
mitochondrial-specific antioxidants reversed the effects of DEPTOR
on the angiogenesis of MM cells. RPMI 8226 and MM.1S cells were
transfected with lentivirus DEP3 for 5 days and exposed to SMER28
(10 µM) or Mito-TEMPO (10 µM) for 24 h. (A-D) The
mitochondrial and cytoplasmic fractions of the cultured cells were
separated using the Mitochondria/Cytosol Fractionation kit. Protein
expression of Cyt c in the (A) mitochondria and (B)
cytoplasm of the indicated groups for RPMI8226 cells was examined
by western blot analysis, respectively. Primary antibodies with
COXIV and β-actin was used as the mitochondrial and cytosolic
loading control, respectively. Protein expression of Cyt c
in the (C) mitochondria and (D) cytoplasm of the indicated groups
for MM.1S was examined by western blot analysis, respectively.
Primary antibodies with COXIV and β-actin was used as the
mitochondrial and cytosolic loading control, respectively. Protein
expression levels of DEPTOR and IκB-α in the cytoplasm of the
indicated groups for (E) RPMI8226 and (F) MM.1S cells were examined
by western blot analysis, respectively. (G-L) The nuclear and
cytoplasmic protein fractions of the cultured cells were isolated
using the NE-PER™ Nuclear and Cytoplasmic Extraction Reagents.
Protein expressions of (G) p-p50 and p-p65 in the nuclear, (H) p50
and p65 in the whole cell lysates of the indicated groups for
RPMI8226 cells was examined by western blot analysis, respectively.
LaminB and β-actin was used as the loading control for the nuclear
extracts and the whole cell lysates, respectively. (I)
Quantification of p-p50 subunit and p-p65 subunit was normalized
for total p50 protein and total p65 protein (G and H),
respectively. Protein expression levels of (J) p-p50 and p-p65 in
the nuclear, (K) p50 and p65 in the whole cell lysates of the
indicated groups for MM.1S was examined by western blot analysis,
respectively. Lamin B and β-actin was used as the loading controls
for the nuclear extracts and the whole cell lysates, respectively.
(L) Quantification of p-p50 subunit and p-p65 subunit was
normalized for total p50 protein and total p65 protein (J and K),
respectively. Error bars indicate the standard deviation from 3
independent experiments. *P<0.05 compared with GFP
group, **P<0.01 compared with GFP group. MM, multiple
myeloma; VEGF, vascular endothelial growth factor; Cyt C,
cytochrome C; mtROS, mitochondrial reactive oxygen species; p-p50,
phospho-p50; p-p65, phospho-p65. |
Discussion
DEPTOR is an mTOR-interacting protein and mau be
involved in the proliferation, apoptosis, autophagy and
differentiation of MM cells (11,12,17). However, whether DEPTOR can
regulate the angiogenesis of MM cells has not yet been elucidated.
The present study first validated the significance of the DEPTOR
mRNA expression level in MM using the Oncomine database. As shown
in Fig. 1, compared to the
majority of other common cancers, the expression of DEPTOR was
significantly increased in MM, which confirmed that DEPTOR plays an
important role in the pathogenesis of MM as previously reported
(11,29). Based on 3 MM cell lines and
primary BM specimens from patients with MM, it was verified that
DEPTOR protein expression may negatively regulate the expression of
VEGF and MVD in MM. DEPTOR is differentially expressed among
patients with MM, and it is required to maintain the
differentiation of myeloma cells. A higher expression of DEPTOR may
be associated with a better prognosis of patients with MM (29). The overexpression of DEPTOR in MM
is associated with the translocation of the MAF transcription
factors and the CCND1 and CCND3 genes (11), as well as with copy number gains
of 8q24 (30). It was
hypothesized that the differences in DEPTOR expression may be
related to genetic differences among the 3 cell lines. The present
study reveals for the first time, to the best of our knowledge,
that DEPTOR as an anti-angiogenic factor, negatively correlated
with VEGF expression and angiogenesis in patients with MM.
Therefore, the potential underlying mechanisms warrant further
investigation. The present study used the KEGG-related GSEA
analysis to confirm that DEPTOR may be involved in the regulation
of MM cells by autophagy, the mTOR signaling pathway and VEGF
signaling pathway.
Previous studies have reported that MM patients with
a high DEPTOR expression could benefit more from the administration
of thalidomide-based therapy than patients with a low DEPTOR
expression (13,14). Thalidomide exerts antitumor and
anti-angiogenic effects and is currently an important drug for the
treatment of MM (31).
Accordingly, exploring the mechanisms of DEPTOR in the thalidomide
treatment of MM is essential. In the present study, the results
revealed that thalidomide promoted autophagy and DEPTOR expression,
but inhibited VEGF expression in MM cells. However, the reduction
of DEPTOR expression attenuated the effects of thalidomide on
proliferation, apoptosis, autophagy and VEGF expression in MM
cells. Notably, in the DEPTOR-disrupted RPMI8226 cells, thalidomide
exerted no effects on CRBN, VEGF and autophagy-related proteins.
Furthermore, following treatment with thalidomide and 3-MA, the
levels of autophagy-related proteins were further inhibited, while
VEGF expression was increased. These data suggested that
DEPTOR-regulated autophagy may be essential for the antitumor and
anti-angiogenic effects of thalidomide, which was independent of
the target of thalidomide CRBN. Of note, DEPTOR seems to be
associated with a poor prognosis in patients with hepatocellular
carcinoma (32) and in some other
solid tumors (33), while it is
associated with a good prognosis in MM following thalidomide
treatment (29). Hence, DEPTOR
may be used as a predictor of the efficacy of thalidomide
treatment.
Thalidomide treatment can boost the dendritic cell-
(34), T cell- (35) and natural killer cell- (36) mediated immune response in the
tumor microenvironment, and inhibit pro-inflammatory cytokine TNF-α
production (37). In addition,
thalidomide plays an apoptotic role by downregulating Akt
activation and triggering caspase3 activation (38), and induces cell cycle arrest via
the upregulation of CDK inhibitor p21 (39). DEPTOR is important for cell
proliferation and survival (40).
Its upregulation may be the feedback response to thalidomide
induced cell apoptosis and cell cycle arrest.
Autophagy is a complex process regulated by
different multistep signaling pathways to maintain intracellular
homeostasis (41). In the present
study, the autophagic regulatory drugs, rapamycin, 3-MA and SMER28,
were used to treat MM cells. Rapamycin induces autophagy through
the mTOR signaling pathway, while SMER28 activates autophagy in
mTOR-independent manners (42).
In the present study, both rapamycin and SMER28 reduced the
expression and secretion of IL-6 and VEGF, which proved that
autophagy inducers inhibit angiogenesis. The inhibition of
autophagy by 3-MA increased the expression, and the secretion of
IL-6 and VEGF in MM. Therefore, MM cells can regulate autophagy to
modulate angiogenesis via multiple mechanisms. The association of
angiogenesis with DEPTOR-mediated autophagy shown in the present
study suggests that autophagy plays a role in angiogenesis iin
MM.
mtROS, the main source of endogenous ROS, have
emerged as essential signal transducers that mediate autophagy
(43), which can lead to NF-κB
signal activation and the secretion of IL-6 and VEGF (20,21). The data presented herein
demonstrated that DEPTOR inhibition prominently increased mtROS
levels and promoted the production of IL-6 and VEGF expression
through the NF-κB-associated pathway. As DEPTOR is an inhibitor of
mTOR, the majority of previous studies are based on the
mTOR-dependent pathway (16,29,44,45); however, whether DEPTOR can work
independently of mTOR remains unclear (46). In the present study, both
non-mTOR-dependent autophagy enhancer (SMER28) and mitochondrial
ROS scavenger (Mito-TEMPO) reversed the effects of DEPTOR. These
results indicated that DEPTOR can affect angiogenesis through the
non-mTOR-dependent autophagy/mtROS pathway rather than through
mTOR-dependent pathways. The possible mechanism of angiogenesis
regulated by DEPTOR in MM may be that autophagy activation
regulated by a high expression of DEPTOR can eliminate excessive
ROS produced by the mitochondria, which has effects on cell
survival, thereby blocking the NF-κB signal and downstream
molecules IL-6 and VEGF, and inhibiting subsequent angiogenesis.
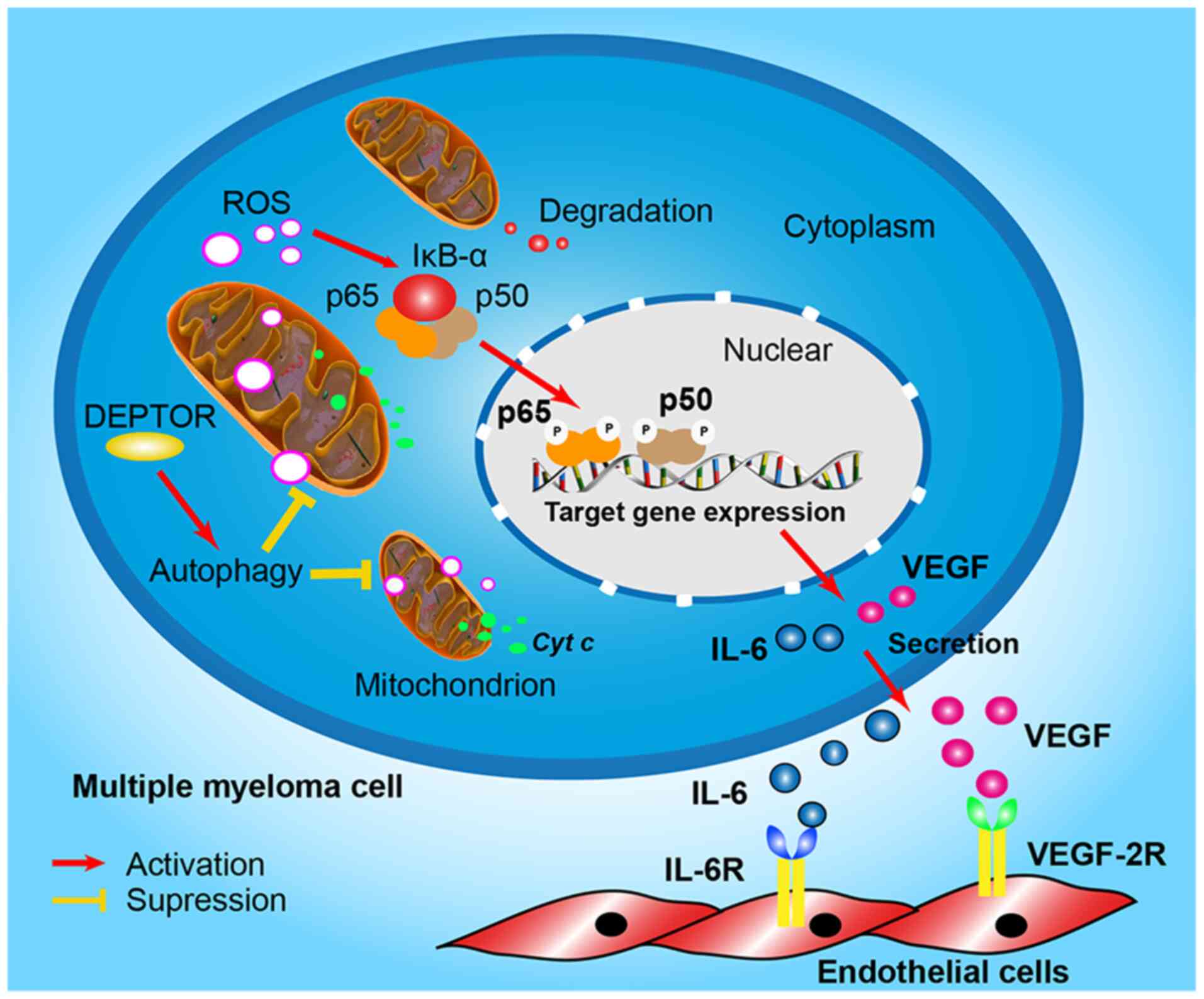
Fig. 7 proposes the pathway of
DEPTOR regulation of angiogenesis in MM.
Since DEPTOR is essential for the survival of MM
cells, the data of the present study demonstrated that almost all
MM cells died following the suppression of DEPTOR, and the effect
of DEPTOR cannot be validated in vivo, which was in
accordance with previous reports in the literature (11,29). Of note, it seems paradoxical that
the knockdown of DEPTOR can lead to death, but increase VEGF
expression and angiogenesis, which may promote the development of
cancer (47,48). Therefore, the MM cells may
preserve their proliferative potential by sacrificing angiogenesis
via the high expression of DEPTOR to maintain their own survival.
This phenomenon is similar to the role of E-cadherin in breast
cancer cells reported in recent literature, in which E-cadherin is
also essential for breast cancer cell survival, but at the expense
of dissemination and metastasis (49).
The present study is limited by the fact that no
immunohistochemistry of DEPTOR was performed. Indeed, MM is not a
solid tumor, and the identification of MM cells from bone marrow
specimens is difficult. In addition, whether thalidomide treatment
can increase the expression of DEPTOR needs to be further confirmed
in future clinical studies.
In conclusion, the present study demonstrates that
DEPTOR regulates cellular autophagy and mtROS in MM. Angiogenesis
may be involved in the process; however, it requires additional
study. These results are considered to be important for the further
understanding of the novel molecular mechanisms of thalidomide in
the treatment of MM.
Supplementary Data
Abbreviations:
|
MM
|
multiple myeloma
|
|
BM
|
bone marrow
|
|
DMEM
|
Dulbecco's modified Eagle's medium
|
|
ELISA
|
enzyme-linked immunosorbent assay
|
|
GEO
|
Gene Expression Omnibus
|
|
GSEA
|
Gene Set Enrichment Analysis
|
|
HUVECs
|
human umbilical vein endothelial
cells
|
|
IHC
|
immunohistochemistry
|
|
IL-6
|
interleukin 6
|
|
ISS
|
International Staging System
|
|
mTOR
|
mammalian target of rapamycin
|
|
MVD
|
microvessel density
|
|
NF-κB
|
nuclear factor-κB
|
|
PI
|
propidium iodide
|
|
RMA
|
robust multichip average
|
|
ROS
|
reactive oxygen species
|
|
sgRNAs
|
single guided RNAs
|
|
VEGF
|
vascular endothelial growth factor
|
Funding
The present study was supported by grants from the
National Natural Science Foundation of China (no. 81400160 and no.
82070218), the Natural Science Foundation of Fujian Province (no.
2017J01185), the Training Program of Outstanding Young Scientific
Researchers of Fujian Universities (no. 2016B027), the Key
Personnel Training Project for Young and Middle-Aged Person of
Health System of Health and Family Planning Commission of Fujian
Province (no. 2016-ZQN-50), and Education and Scientific Research
Project for Middle-aged and Young Teachers in Fujian Province (no.
JT180185).
Availability of data and materials
The gene expression data in this study can be found
online at the Gene Expression Omnibus under accession numbers
GSE24080 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE24080).
Data generated or analyzed during this study are available from the
corresponding author upon reasonable request.
Authors' contributions
ZZ and JC conceived the experiment and analyzed
data. JW performed the experiments, and generated and collected the
data. DQ interpreted the data. JW and ZZ wrote the manuscript. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
All experiments described in the present study were
approved by the Ethics Committee of the First Affiliated Hospital
of Fujian Medical University. Informed consent was obtained in
accordance with the Declaration of Helsinki.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
References
|
1
|
Singhal S, Mehta J, Desikan R, Ayers D,
Roberson P, Eddlemon P, Munshi N, Anaissie E, Wilson C, Dhodapkar
M, et al: Antitumor activity of thalidomide in refractory multiple
myeloma. N Engl J Med. 341:1565–1571. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Scott K, Hayden PJ, Will A, Wheatley K and
Coyne I: Bortezomib for the treatment of multiple myeloma. Cochrane
Database Syst Rev. 4:CD0108162016.PubMed/NCBI
|
|
3
|
Dimopoulos MA, San-Miguel J, Belch A,
White D, Benboubker L, Cook G, Leiba M, Morton J, Ho PJ, Kim K, et
al: Daratumumab plus lenalidomide and dexamethasone versus
lenalidomide and dexa-methasone in relapsed or refractory multiple
myeloma: Updated analysis of POLLUX. Haematologica. 103:2088–2096.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Cao Y, Wan N, Liang Z, Xie J, Wang S, Lin
T, Zhang T and Jiang J: Treatment outcomes in patients with newly
diagnosed multiple myeloma who are ineligible for stem-cell
transplantation: Systematic review and network meta-analysis. Clin
Lymphoma Myeloma Leuk. 19:e478–e488. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zeng Z, Lin J and Chen J: Bortezomib for
patients with previously untreated multiple myeloma: A systematic
review and meta-analysis of randomized controlled trials. Ann
Hematol. 92:935–943. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
André T, Kotelevets L, Vaillant JC,
Coudray AM, Weber L, Prévot S, Parc R, Gespach C and Chastre E:
Vegf, Vegf-B, Vegf -C and their receptors KDR, FLT-1 and FLT-4
during the neoplastic progression of human colonic mucosa. Int J
Cancer. 86:174–181. 2000. View Article : Google Scholar
|
|
7
|
Kurebayashi J, Otsuki T, Kunisue H, Mikami
Y, Tanaka K, Yamamoto S and Sonoo H: Expression of vascular
endothelial growth factor (VEGF) family members in breast cancer.
Jpn J Cancer Res. 90:977–981. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Decaussin M, Sartelet H, Robert C, Moro D,
Claraz C, Brambilla C and Brambilla E: Expression of vascular
endothelial growth factor (VEGF) and its two receptors
(VEGF-R1-Flt1 and VEGF-R2-Flk1/KDR) in non-small cell lung
carcinomas (NSCLCs): Correlation with angiogenesis and survival. J
Pathol. 188:369–377. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jussila L, Valtola R, Partanen TA, Salven
P, Heikkilä P, Matikainen MT, Renkonen R, Kaipainen A, Detmar M,
Tschachler E, et al: Lymphatic endothelium and Kaposi's sarcoma
spindle cells detected by antibodies against the vascular
endothelial growth factor receptor-3. Cancer Res. 58:1599–1604.
1998.PubMed/NCBI
|
|
10
|
Vacca A, Ribatti D, Roncali L, Ranieri G,
Serio G, Silvestris F and Dammacco F: Bone marrow angiogenesis and
progression in multiple myeloma. Br J Haematol. 87:503–508. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Peterson TR, Laplante M, Thoreen CC,
Sancak Y, Kang SA, Kuehl WM, Gray NS and Sabatini DM: DEPTOR is an
mTOR inhibitor frequently overexpressed in multiple myeloma cells
and required for their survival. Cell. 137:873–886. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhao Y, Xiong X and Sun Y: DEPTOR, an mTOR
inhibitor, is a physiological substrate of SCF(βTrCP) E3 ubiquitin
ligase and regulates survival and autophagy. Mol Cell. 44:304–316.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
de la Rubia J and Such E: DEPTOR
expression and response to thalidomide: Toward a new therapeutic
target in multiple myeloma? Leuk Lymphoma. 51:1960–1961. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Boyd KD, Walker BA, Wardell CP, Ross FM,
Gregory WM, Davies FE and Morgan GJ: High expression levels of the
mammalian target of rapamycin inhibitor DEPTOR are predictive of
response to thalidomide in myeloma. Leuk Lymphoma. 51:2126–2129.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bruneau S, Nakayama H, Woda CB, Flynn EA
and Briscoe DM: DEPTOR regulates vascular endothelial cell
activation and proinflammatory and angiogenic responses. Blood.
122:1833–1842. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ding Y, Shan L, Nai W, Lin X, Zhou L, Dong
X, Wu H, Xiao M, Zhou X, Wang L, et al: DEPTOR deficiency-mediated
mTORc1 hyperactivation in vascular endothelial cells promotes
angiogenesis. Cell Physiol Biochem. 46:520–531. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhang H, Chen J, Zeng Z, Que W and Zhou L:
Knockdown of DEPTOR induces apoptosis, increases chemosensitivity
to doxorubicin and suppresses autophagy in RPMI-8226 human multiple
myeloma cells in vitro. Int J Mol Med. 31:1127–1134. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang HR, Chen JM, Zeng ZY and Que WZ:
Knockdown of DEPTOR inhibits cell proliferation and increases
chemosensitivity to melphalan in human multiple myeloma RPMI-8226
cells via inhibiting PI3K/AKT activity. J Int Med Res. 41:584–595.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Cury PCC, Higashi F, Zacchi FFS, Palhares
RB, Quero AA, Dias ALMS, Crusoé EQ and Hungria VTM: Effect of
thalidomide on bone marrow angiogenesis in multiple myeloma
patients. Hematol Transfus Cell Ther. 42:159–163. 2020. View Article : Google Scholar :
|
|
20
|
Redza-Dutordoir M and Averill-Bates DA:
Activation of apoptosis signalling pathways by reactive oxygen
species. Biochim Biophys Acta. 1863:2977–2992. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Liu T, Zhang L, Joo D and Sun SC: NF-κB
signaling in inflammation. Signal Transduct Target Ther.
2:170232017. View Article : Google Scholar
|
|
22
|
Sun K, Xu L, Jing Y, Han Z, Chen X, Cai C,
Zhao P, Zhao X, Yang L and Wei L: Autophagy-deficient Kupffer cells
promote tumorigenesis by enhancing mtROS-NF-κB-IL1α/β-dependent
inflammation and fibrosis during the preneoplastic stage of
hepatocarcinogenesis. Cancer Lett. 388:198–207. 2017. View Article : Google Scholar
|
|
23
|
Shi L, Campbell G, Jones WD, Campagne F,
Wen Z, Walker SJ, Su Z, Chu TM, Goodsaid FM, Pusztai L, et al: The
MicroArray Quality Control (MAQC)-II study of common practices for
the development and validation of microarray-based predictive
models. Nat Biotechnol. 28:827–838. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Irizarry RA, Hobbs B, Collin F,
Beazer-Barclay YD, Antonellis KJ, Scherf U and Speed TP:
Exploration, normalization, and summaries of high density
oligonucleotide array probe level data. Biostatistics. 4:249–264.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang X, Lin Y, Song C, Sibille E and Tseng
GC: Detecting disease-associated genes with confounding variable
adjustment and the impact on genomic meta-analysis: With
application to major depressive disorder. BMC Bioinformatics.
13:522012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
He W, Fu L, Yan Q, Zhou Q, Yuan K, Chen L
and Han Y: Gene set enrichment analysis and meta-analysis
identified 12 key genes regulating and controlling the prognosis of
lung adenocarcinoma. Oncol Lett. 17:5608–5618. 2019.PubMed/NCBI
|
|
27
|
Rajkumar SV, Dimopoulos MA, Palumbo A,
Blade J, Merlini G, Mateos MV, Kumar S, Hillengass J, Kastritis E,
Richardson P, et al: International myeloma working group updated
criteria for the diagnosis of multiple myeloma. Lancet Oncol.
15:e538–e548. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lee N, Lee H, Moon SY, Sohn JY, Hwang SM,
Yoon OJ, Youn HS, Eom HS and Kong SY: Adverse prognostic impact of
bone marrow microvessel density in multiple myeloma. Ann Lab Med.
35:563–569. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Quwaider D, Corchete LA,
Misiewicz-Krzeminska I, Sarasquete ME, Pérez JJ, Krzeminski P, Puig
N, Mateos MV, García-Sanz R, Herrero AB and Gutiérrez NC: DEPTOR
maintains plasma cell differentiation and favorably affects
prognosis in multiple myeloma. J Hematol Oncol. 10:922017.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Carrasco DR, Tonon G, Huang Y, Zhang Y,
Sinha R, Feng B, Stewart JP, Zhan F, Khatry D, Protopopova M, et
al: High-resolution genomic profiles define distinct
clinicopathogenetic subgroups of multiple myeloma patients. Cancer
Cell. 9:313–325. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Mercurio A, Adriani G, Catalano A, Carocci
A, Rao L, Lentini G, Cavalluzzi MM, Franchini C, Vacca A and Corbo
F: A mini-review on thalidomide: Chemistry, mechanisms of action,
therapeutic potential and anti-angiogenic properties in multiple
myeloma. Curr Med Chem. 24:2736–2744. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chen J, Zhu H, Liu Q, Ning D, Zhang Z,
Zhang L, Mo J, Du P, Liu X, Song S, et al: DEPTOR induces a partial
epithelial-to-mesenchymal transition and metastasis via autocrine
TGFβ1 signaling and is associated with poor prognosis in
hepatocellular carcinoma. J Exp Clin Cancer Res. 38:2732019.
View Article : Google Scholar
|
|
33
|
Hu B, Shi D, Lv X, Wu F, Chen S and Shao
Z: Prognostic and clinicopathological significance of DEPTOR
expression in cancer patients: A meta-analysis. Onco Targets Ther.
11:5083–5092. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Reddy N, Hernandez-Ilizaliturri FJ, Deeb
G, Roth M, Vaughn M, Knight J, Wallace P and Czuczman MS:
Immunomodulatory drugs stimulate natural killer-cell function,
alter cytokine production by dendritic cells, and inhibit
angiogenesis enhancing the anti-tumour activity of rituximab in
vivo. Br J Haematol. 140:36–45. 2008.
|
|
35
|
Haslett PA, Corral LG, Albert M and Kaplan
G: Thalidomide costimulates primary human T lymphocytes,
preferentially inducing proliferation, cytokine production, and
cytotoxic responses in the CD8+ subset. J Exp Med. 187:1885–1892.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Chang DH, Liu N, Klimek V, Hassoun H,
Mazumder A, Nimer SD, Jagannath S and Dhodapkar MV: Enhancement of
ligand-dependent activation of human natural killer T cells by
lenalidomide: Therapeutic implications. Blood. 108:618–621. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Sampaio EP, Sarno EN, Galilly R, Cohn ZA
and Kaplan G: Thalidomide selectively inhibits tumor necrosis
factor alpha production by stimulated human monocytes. J Exp Med.
173:699–703. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Mitsiades N, Mitsiades CS, Poulaki V,
Chauhan D, Richardson PG, Hideshima T, Munshi NC, Treon SP and
Anderson KC: Apoptotic signaling induced by immunomodulatory
thalidomide analogs in human multiple myeloma cells: Therapeutic
implications. Blood. 99:4525–4530. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Escoubet-Lozach L, Lin IL, Jensen-Pergakes
K, Brady HA, Gandhi AK, Schafer PH, Muller GW, Worland PJ, Chan KW
and Verhelle D: Pomalidomide and lenalidomide induce p21 WAF-1
expression in both lymphoma and multiple myeloma through a
LSD1-mediated epigenetic mechanism. Cancer Res. 69:7347–7356. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Hu Y, Su H, Liu C, Wang Z, Huang L, Wang
Q, Liu S, Chen S, Zhou J, Li P, et al: DEPTOR is a direct NOTCH1
target that promotes cell proliferation and survival in T-cell
leukemia. Oncogene. 36:1038–1047. 2017. View Article : Google Scholar
|
|
41
|
Khandia R, Dadar M, Munjal A, Dhama K,
Karthik K, Tiwari R, Yatoo MI, Iqbal HMN, Singh KP, Joshi SK and
Chaicumpa W: A comprehensive review of autophagy and its various
roles in infectious, non-infectious, and lifestyle diseases:
Current knowledge and prospects for disease prevention, novel drug
design, and therapy. Cells. 8:6742019. View Article : Google Scholar :
|
|
42
|
Koukourakis MI, Giatromanolaki A,
Fylaktakidou K, Sivridis E, Zois CE, Kalamida D, Mitrakas A,
Pouliliou S, Karagounis IV, Simopoulos K, et al: SMER28 is a
mTOR-independent small molecule enhancer of autophagy that protects
mouse bone marrow and liver against radiotherapy. Invest New Drugs.
36:773–781. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Roca-Agujetas V, de Dios C, Lestón L, Mari
M, Morales A and Colell A: Recent insights into the mitochondrial
role in autophagy and its regulation by oxidative stress. Oxid Med
Cell Longev. 2019:38093082019. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Zhao L, Wang X, Yu Y, Deng L, Chen L, Peng
X, Jiao C, Gao G, Tan X, Pan W, et al: OTUB1 protein suppresses
mTOR complex 1 (mTORC1) activity by deubiquitinating the mTORC1
inhibitor DEPTOR. J Biol Chem. 293:4883–4892. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Wang Q, Zhou Y, Rychahou P, Harris JW,
Zaytseva YY, Liu J, Wang C, Weiss HL, Liu C, Lee EY and Evers BM:
Deptor is a novel target of Wnt/β-catenin/c-Myc and contributes to
colorectal cancer cell growth. Cancer Res. 78:3163–3175. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Catena V and Fanciulli M: Deptor: Not only
a mTOR inhibitor. J Exp Clin Cancer Res. 36:122017. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Jakob C, Sterz J, Zavrski I, Heider U,
Kleeberg L, Fleissner C, Kaiser M and Sezer O: Angiogenesis in
multiple myeloma. Eur J Cancer. 42:1581–1590. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Khan MA, Assiri AM and Broering DC:
Complement and macrophage crosstalk during process of angiogenesis
in tumor progression. J Biomed Sci. 22:582015. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Padmanaban V, Krol I, Suhail Y, Szczerba
BM, Aceto N, Bader JS and Ewald AJ: E-cadherin is required for
metastasis in multiple models of breast cancer. Nature.
573:439–444. 2019. View Article : Google Scholar : PubMed/NCBI
|