Introduction
Doxorubicin, a broad-spectrum antitumor drug, has
been reported to effectively treat various types of cancer,
including hepatocellular carcinoma, lung cancer and gastric cancer
(1-3). However, the accumulation of
doxorubicin in the body may lead to cardiotoxicity, which results
in cardiomyocyte apoptosis and collagen deposition, ultimately
leading to dilated cardiomyopathy and heart failure (4-7).
According to previous studies, a cumulative amount of 450-500
mg/m2 doxorubicin resulted in a ~5% incidence of
cardiotoxicity (8,9). Furthermore, the incidence of
cardiotoxicity was up to 18% at 550-660 mg/m2
doxorubicin (8,9). The degree of coronary artery
disease was a strong predictor for renal artery stenosis in
multivariate analysis (10).
However, the mechanism of doxorubicin-induced myocardial injury has
not been fully elucidated.
MicroRNAs (miRNAs/miRs) are short, single-stranded,
non-coding RNAs (11). Previous
studies proposed that miRNAs can regulate ~1/3 of human genes and
thereby exert their crucial biological functions in cellular
behavior or tumorigenesis (11-13). Additionally, numerous miRNAs have
been identified to be dysregulated in doxorubicin-treated
cardiomyocytes or animal hearts, and these dysregulated miRNAs may
influence several cellular processes, including oxidative damage
and apoptosis (14,15).
The miR-133 family consists of miR-133a-1,
miR-133a-2 and miR-133b, and serves an important role in the
pathophysiological processes of heart diseases (16,17). miR-133b, located on chromosome 6,
has been reported to be dysregulated in human myocardial infarction
and to regulate cardiac fibrosis (18,19). Additionally, miR-133b has been
widely reported to have antitumor potential (20-22) and to be closely associated with
cardiotoxicity (23-25). miR-133b serves a cardioprotective
role in morphine-preconditioned rat cardiomyocytes by inhibiting
cardiomyocyte apoptosis (26)
and can be used as a serum biomarker for cardiac fibrosis (27). Moreover, miR-133b expression is
downregulated in doxorubicin-treated rat ventricular cardiomyocytes
and rat cardiac tissues (28).
The present study focused on the role of miR-133b in
doxorubicin-induced myocardial injury.
In the current study, HL-1 cardiomyocytes and
C57BL/6 mice were treated with doxorubicin to establish a cell
model and animal model of myocardial injury, respectively. The
effects of miR-133b on fibrosis in cardiac tissues of
doxorubicin-treated mice were investigated. Subsequently, the role
of miR-133b in apoptosis and collagen deposition of
doxorubicin-preconditioned cardiomyocytes was investigated,
followed by exploration of the downstream targets of miR-133b.
Materials and methods
Ethics statement
All animal operations were in accordance with the
guidelines on the use and care of laboratory animals for biomedical
research (National Institutes of Health; no. 85-23; revised 1996).
The experimental protocol was approved by the Ethics Committee of
The First Affiliated Hospital of Nanjing Medical University
(Nanjing, China; approval no. 2019-041).
Cell culture and treatment
Mouse HL-1 cardiomyocytes (The Cell Bank of Type
Culture Collection of The Chinese Academy of Sciences) were
cultured at 37°C in DMEM supplemented with 10% FBS (Gibco; Thermo
Fisher Scientific, Inc.) in a humidified atmosphere containing 5%
CO2. For induction of apoptosis, different
concentrations of doxorubicin (2.5, 5 and 10 µM; Teva UK
Ltd.) were added to the medium for 6 h of culture at 37°C. HL-1
cardiomyocytes in the control group (Con) were treated with the
same dose of saline (Gibco; Thermo Fisher Scientific, Inc.).
Cell transfection
Full-length polypyrimidine tract binding protein 1
(PTBP1) or transgelin 2 (TAGLN2) was subcloned into pcDNA3.1 to
overexpress PTBP1 or TAGLN2, respectively, and empty pcDNA3.1 was
used as a control. Similarly, Bax was subcloned into pcDNA3.1 to
overexpress Bax to detect if fibrotic events were independent of
apoptosis. miR-133b mimics (miR-133b) and a negative control
(miR-NC) were used to overexpress miR-133b. The sequence of the
miR-133b mimics was 5′-UUU GGU CCC CUU CAA CCA GCU A-3′, and the
sequence of the scrambled miR-NC was 5′-UCA UUG CUA UGC ACU GUC CAC
C-3′. All pcDNA3.1 vectors (1 µg) and miR-133b mimics (50
nM) were purchased from Shanghai GenePharma Co., Ltd., and
transfected in HL-1 cardiomyocytes using Lipofectamine®
3000 (Invitrogen; Thermo Fisher Scientific, Inc.) at room
temperature for 48 h. After 48 h from transfection, subsequent
experiments were conducted.
Flow cytometry assay
HL-1 cardiomyocyte apoptosis was tested using a
PI/FITC Annexin V Apoptosis Detection kit (BD Pharmingen; BD
Biosciences). Briefly, HL-1 cardiomyocytes (5×105
cells/ml) were washed with cold PBS (Gibco; Thermo Fisher
Scientific, Inc.) twice and then resuspended in 1X binding buffer.
Subsequently, cell solution (3×104 cells; 100 µl)
was added to a culture tube. Then, 5 µl FITC Annexin V and 5
µl of PI were added at room temperature in the dark for 15
min. Finally, HL-1 cardiomyocytes were analyzed using an Attune NxT
flow cytometer (Invitrogen; Thermo Fisher Scientific, Inc.), and
apoptotic cardiomyocytes were calculated using FlowJo software
(version 10.0; FlowJo LLC). The apoptotic rate was calculated as
follows: (Cell number in Q3/cell number in all quadrants) ×100%,
with cells in Q3 representing early apoptotic cells.
Reverse transcription-quantitative PCR
(RT-qPCR)
Following the manufacturer's instructions, a
miRNeasy Mini kit (Qiagen, Inc.) was used to extract RNA from HL-1
cardiomyocyte and mouse heart samples. The iScript™ cDNA Synthesis
kit (Bio-Rad Laboratories, Inc.) was used to reverse transcribe RNA
(400 ng) to cDNA according to the manufacturer's protocol. The
relative mRNA expression was determined using SYBR qPCR (Bio-Rad
Laboratories, Inc.) with an ABI-7900 Real-Time PCR Detection System
(7900HT; Applied Biosystems; Thermo Fisher Scientific, Inc.). The
relative miR-133b expression was analyzed using Bulge-Loop™ miRNA
qPCR Primer Set (Guangzhou RiboBio Co., Ltd.) with an ABI-7900
Real-Time PCR Detection System. qPCR was performed with the
following thermocycling conditions: 95°C for 30 sec, followed by 40
cycles at 95°C for 5 sec and 60°C for 30 sec, and a melting curve
analysis protocol (60-95°C with temperature increments of 0.2°C
every 10 sec). GAPDH and U6 were used as internal controls for mRNA
and miRNA, respectively. The relative gene expression level was
calculated using the 2−ΔΔCq method (29). The primer sequences were as
follows: PTBP1 forward, 5′-GAT CCG ACG AGC TCT TCT C-3′ and
reverse, 5′-CTA TCG TTT CCA TTG GCT GC-3′; TAGLN2 forward, 5′-CCA
ACT GGT TTC CTA AGA AAT CC-3′ and reverse, 5′-CAA TCA CGT TCT TGC
CCT C-3′; LHFP forward, 5′-TTT GAC TTG GGC AAG TGT G-3′ and
reverse, 5′-CAT GTA CAC AGC AGC ATG G-3′; SEC61B forward, 5′-TTG
CTC TTC CCA GCT TCT C-3′ and reverse, 5′-TGT AGA ATC GCC ACA TCC
C-3′; CETN3 forward, 5′-TTA GCT CTG AGA GGT GAG C-3′ and reverse,
5′-TTC TTG CTT CTG TTC TTC AGA G-3′; FTL forward, 5′-CCG TGC ACT
CTT CCA GGA TGT-3′ and reverse, 5′-CCT TAT CCA GAT AGT GGC TTT CCA
G-3′; LDLRAP1 forward, 5′-TGA CAC TCA AGG TGT CAC C-3′ and reverse,
5′-TAG GAG ATC CTG TAA ATG GAC AC-3′; CLTA forward, 5′-ATA CTA CCAG
GAG AGC AAT GG-3′ and reverse, 5′-TAC TTT CAG GCT CTG ACT GC-3′;
GAPDH forward, 5′-CAT CTTC TTGT GCAG TGC C-3′ and reverse, 5′-CAA
ATC CGT TCA CACC GAC-3′; miR-133b forward, 5′-TTG GTC CCC TTCA ACCA
GCT A-3′ and reverse, 5′-CAG TGC GTG TCG TGG AGT-3′; and U6
forward, 5′-CTC GCT TCG GCA GCA CA-3′ and reverse, 5′-ACG CTT CAC
GAA TTT GCG T-3′.
Western blotting
HL-1 cardiomyocytes and mouse cardiac tissues were
lysed using RIPA lysis buffer (Beyotime Institute of
Biotechnology). A Bicinchoninic Acid Protein Assay kit (Thermo
Fisher Scientific, Inc.) was used to evaluate the concentration of
protein samples (1 µl). Subsequently, equal amounts of
protein samples (50 µg/lane) were subjected to 12% SDS-PAGE
and transferred to PVDF membranes, which were then blocked with 5%
skimmed milk at 4°C overnight. Primary antibodies (1:1,000) against
cleaved caspase-3 (cat. no. ab2302), Bcl-2 (cat. no. ab32124), Bax
(cat. no. ab32503), collagen I (cat. no. ab34710), collagen III
(cat. no. ab7778), fibronectin (cat. no. ab2413), collagen IV (cat.
no. ab214417), PTBP1 (cat. no. ab133734), TAGLN2 (cat. no.
ab121146) and GAPDH (cat. no. ab181602) were then incubated with
the membranes at 4°C overnight. Collagen I, III and IV, and
fibronectin are extracellular matrix (ECM) proteins. Finally,
HRP-conjugated secondary antibodies (cat. no. ab6721; 1:5,000) were
added to the membranes in the dark at room temperature for 1 h. All
antibodies were obtained from Abcam. Proteins were visualized using
an ECL Chemiluminescence kit (Thermo Fisher Scientific, Inc.) and
quantified using ImageJ software (version 1.46; National Institutes
of Health). GAPDH served as a loading control. The gray ratio of
the target bands to the internal reference bands was the relative
expression of the target proteins.
RNA immunoprecipitation (RIP) assay
The RIP assay was performed using a Magna RIP™
RNA-Binding Protein Immunoprecipitation kit (EMD Millipore)
according to the manufacturer's protocol. After centrifugation at
1,000 × g at 4°C for 5 min, HL-1 cardiomyocytes were lysed in
complete RIPA lysis buffer (cat. no. P0013B; Beyotime Institute of
Biotechnology). Subsequently, 100 µl supernatant was
collected and incubated with magnetic beads (1 mg) conjugated with
PTBP1 antibody (cat. no. ab133734; 1:10,000; Abcam) or control IgG
(cat. no. 12-370; 1:5,000; EMD Millipore) at 4°C for 6 h. After the
beads were washed, the complexes were treated with Proteinase K to
remove proteins. Beads were isolated from the supernatant after
centrifugation at 2,500 × g for 5 min at 4°C and washed with
washing buffer (10 mM Tris-HCl pH 7.5, 1 mM EDTA, 2 M NaCl and 0.1%
Tween-20), followed by another centrifugation step at 2,500 × g for
5 min at 4°C. Finally, the purified RNA level was measured by
RT-qPCR, as aforementioned.
Bioinformatics analysis
Based on the TargetScan version 7.2 website
(http://www.targetscan.org/vert_72/),
703 mRNAs were predicted to harbor binding site(s) for miR-133b
(data not shown). According to the starBase v2.0 website
(http://starbase.sysu.edu.cn/), PTBP1 was
predicted to bind to the 3′-untranslated region (UTR) of TAGLN2
(data not shown). According to the RNA-Protein Interaction
Prediction (http://pridb.gdcb.iastate.edu) based on the random
forest (RF) and support vector machine (SVM) algorithm, scores of
the combination between PTBP1 and TAGLN2 3′-UTR, and between PTBP1
and TAGLN2 were evaluated.
Luciferase reporter assay
A fragment of the PTBP1 or TAGLN2 3′-UTR containing
the predicted binding site of miR-133b was cloned into the pmirGLO
vector (Promega Corporation) to construct the wild-type
pmirGLO-PTBP1-3′-UTR or pmirGLO-TAGLN2-3′-UTR vectors. The mutant
pmirGLO-PTBP1-3′-UTR or pmirGLO-TAGLN2-3′-UTR vectors were also
generated by subcloning the mutated PTBP1 or TAGLN2 sequences in
the 3′-UTR complementary to miR-133b into the pmirGLO vectors. The
vectors were commercially obtained from Shanghai GenePharma Co.,
Ltd., and then separately transfected with miR-NC or miR-133b
mimics using Lipofectamine 3000, as aforementioned. The luciferase
activity was evaluated using a dual luciferase reporter assay
system (Promega Corporation) 48 h after transfection, and was
normalized to Renilla luciferase activity.
RNA pulldown assay
A total of 6×106 cells were cultured in a
100-mm culture dish. After discarding the culture medium, cells
were washed twice with PBS. Next, cells were digested using 0.5 ml
trypsin followed by addition of 2 ml PBS. Cells were transferred to
EP tubes followed by centrifugation at 1,000 × g at 4°C for 2 min
and were subsequently treated with 100 µl buffer solution
for 15 min. After another centrifugation at 1,000 × g at 4°C for 5
min, the supernatant was collected. The TAGLN2 sense or antisense
sequence was transcribed using T7 RNA polymerase (cat. no. AM2718;
Ambion; Thermo Fisher Scientific) and then purified using the
RNeasy Plus Mini kit (cat. no. 74134; Qiagen GmbH) according to the
manufacturer's protocol. Subsequently, the TAGLN2 sense or
antisense sequence was labeled with Biotin RNA Labeling Mix (Ambio
Life). Subsequently, a Magnetic RNA-Protein Pull-Down kit (cat. no.
20164; Pierce; Thermo Fisher Scientific, Inc.) was used for RNA
pulldown assays according to the manufacturer's instructions. Beads
were isolated from the supernatant after boiling, and the PTBP1
enrichment was analyzed by western blotting.
Mouse model and adeno-associated virus
(AAV) injection
A total of 50 C57BL/6 mice (20-25 g; 8 weeks old;
male; Beijing Vital River Laboratory Animal Technology Co., Ltd.)
were used in the present study and were grouped as follows: Sham
group (n=15), DOX+AAV-NC group (n=15) and DOX+AAV-miR-133b group
(n=20). Mice were raised with ad libitum access to water and
food in a temperature-controlled room (22±2°C) with a 12-h
light/dark cycle and a relative humidity of 40-60%. A cardiac
injury mouse model was generated by chronic intraperitoneal
injections (on days 0, 2, 4 and 6) of 4 mg/kg doxorubicin. The mice
in the sham group were treated with the same dose of PBS
(Invitrogen; Thermo Fisher Scientific, Inc.). All mice were
euthanized 4 weeks after the first injection of doxorubicin or PBS.
For AAV (serotype 9) injection, mice received a single-bolus
injection of AAV-miR-133b or empty AAV (AAV-NC) resuspended in PBS
via the tail vein at 1×1011 viral genomes per animal.
AAV vectors were purchased from Han Heng Biotechnology (Shanghai)
Co., Ltd. One week after injection of AAVs, the mice were treated
with doxorubicin or PBS. The mice were anesthetized with 1.5%
pentobarbital sodium (60 mg/kg) by intraperitoneal injection and
then sacrificed by cervical dislocation under anesthesia. The whole
experiment lasted 5 weeks, beginning from AAV injection and ending
up with the euthanasia of mice.
Echocardiography
Four weeks after the first injection of doxorubicin
or PBS, the mice were anesthetized with 2% isoflurane and placed in
the supine position. The left ventricular end-systolic diameter
(LVESD) and left ventricular end-diastolic diameter (LVEDD) values
were analyzed using a VEVO 770 high-resolution system
(VisualSonics, Inc.) with a RMV 704 probe at a frequency of 40 MHz.
Left ventricular ejection fraction (LVEF) and left ventricular
ejection shorting (LVES) were automatically calculated using an
echocardiographic instrument.
Masson's trichrome staining
Cardiac tissue samples were immobilized in 4%
paraformaldehyde for 48 h at 4°C, embedded in paraffin and sliced
into sections (5-µm-thick). Subsequently, sections were
subjected to Masson's trichrome staining as previously described
(30). The collagen fibers were
blue, the muscles and elastic fibers were red, cellulose was
purple-red, and the nucleus was blue-brown. Images were taken using
a light microscope Nikon model with a Spot Insight camera
(magnification, ×200). The fibrotic region was quantified using
ImageJ software (version 1.46; National Institutes of Health). The
percentage of fibrosis was calculated as fibrosis areas/total left
ventricular areas ×100%.
Measurement of collagen content
The collagen content was determined using a
quantitative dye-binding method, as previously described (30). A Sircol assay (Biocolor Ltd.) was
used to analyze cardiac tissues following the manufacturer's
instructions. The mouse heart was weighed and homogenized with
pepsin, and collagen content in each sample was quantified using
the Gen5™ BioTek software (Agilent Technologies, Inc.).
Statistical analysis
The data are displayed as the mean ± SD of 3
independent experiments. Statistical analysis was performed using
SPSS software (version 21.0; IBM Corp.). All data were assessed for
normal distribution (Shapiro-Wilk test) and homogeneity of variance
(Bartlett's test). Statistical analysis was performed using
unpaired Student's t-test for comparisons between two groups or
one-way ANOVA followed by Dunnett's or Tukey's post-hoc tests for
comparisons among multiple groups. P<0.05 was considered to
indicate a statistically significant difference.
Results
miR-133b expression is decreased by
doxorubicin treatment in HL-1 cardiomyocytes and cardiac
tissues
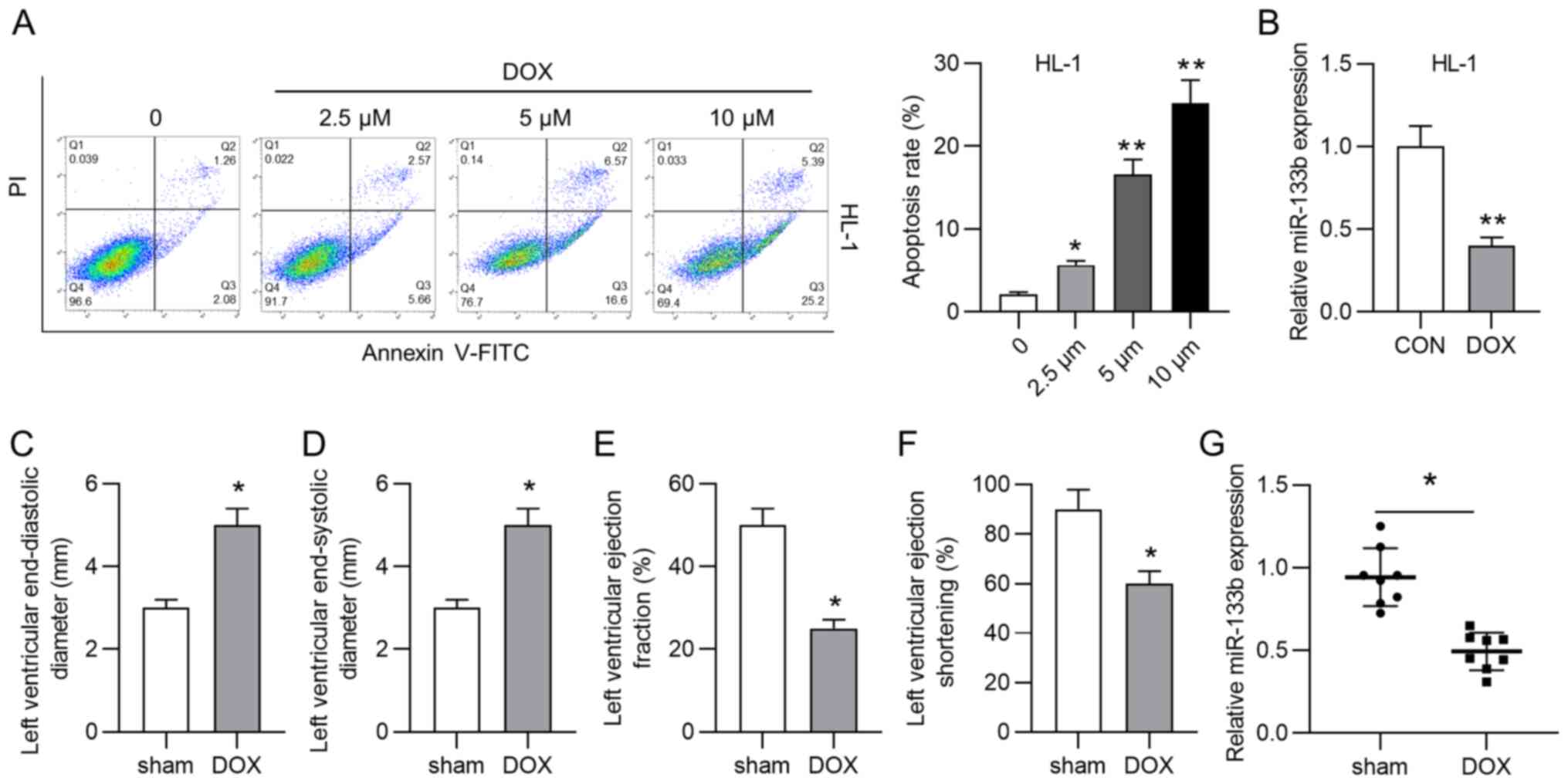
As shown in Fig.
1A, the apoptosis rate of HL-1 cardiomyocytes was significantly
increased by treatment with doxorubicin (2.5, 5 and 10 µM).
Considering that 10 µM doxorubicin resulted in the highest
apoptosis rate of HL-1 cardiomyocytes, 10 µM doxorubicin was
selected for subsequent assays. However, the role of miR-133b in
doxorubicin-induced cardiac injury remains unclear. According to
the RT-qPCR results, doxorubicin treatment significantly decreased
miR-133b expression in HL-1 cells (Fig. 1B). A mouse model of cardiac
injury was then established by injection of doxorubicin. Data from
echocardiography analysis demonstrated that the LVEDD and LVESD
were significantly increased by doxorubicin injection (Fig. 1C and D), while LVEF and LVES were
significantly decreased by doxorubicin injection (Fig. 1E and F). Additionally, miR-133b
expression was significantly downregulated in the DOX group
compared with in the sham group (Fig. 1G). All these experimental results
suggested that miR-133b expression was decreased by doxorubicin
treatment in vitro and in vivo.
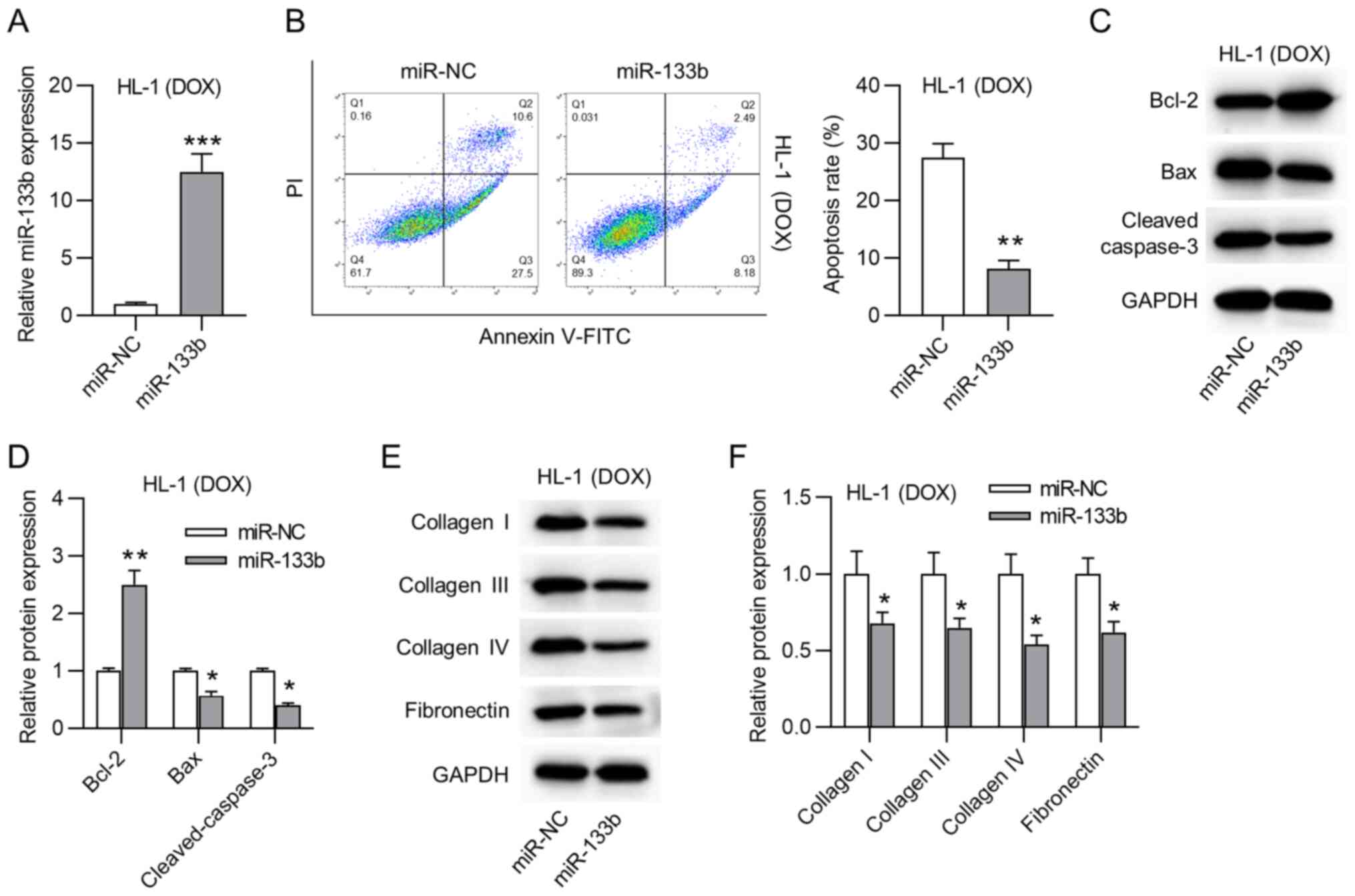
Overexpression of miR-133b inhibits the
apoptosis and collagen accumulation in doxorubicin-treated HL-1
cardiomyocytes
The biological function of miR-133b in
doxorubicin-treated HL-1 cardiomyocytes was then explored. First,
miR-133b was overexpressed by transfection using miR-133b mimics in
doxorubicin-treated HL-1 cardiomyocytes (Fig. 2A). miR-133b overexpression
significantly decreased the apoptosis rate of doxorubicin-treated
HL-1 cardiomyocytes (Fig. 2B).
Moreover, western blot assays revealed that miR-133b overexpression
triggered a significant increase in Bcl-2 protein expression, but a
significant decrease in Bax and cleaved caspase-3 protein
expression (Fig. 2C and D). As
shown in Fig. 2E and F,
overexpression of miR-133b significantly decreased the protein
expression levels of collagen I, III and IV, and fibronectin in the
doxorubicin-treated HL-1 cardiomyocytes. Moreover, it was revealed
that BAX was successfully overexpressed and that BAX overexpression
had no significant effects on the expression levels of ECM proteins
(Fig. S1A and B). Overall,
miR-133b overexpression inhibited the apoptosis and collagen
accumulation in the doxorubicin-treated HL-1 cardiomyocytes.
Overexpression of miR-133b alleviates
apoptosis and cardiac fibrosis in vivo
To explore the role of miR-133b in vivo, a
mouse model of doxorubicin-induced heart failure was established.
First, it was observed that miR-133b expression in the AAV-miR-NC
group was significantly decreased compared with in the sham group,
and miR-133b expression was significantly increased in cardiac
tissues by AAV-miR-133b injection (Fig. 3A). In addition, the injection of
doxorubicin induced a significant decrease in Bcl-2 protein
expression, but a significant increase in Bax and cleaved caspase-3
protein expression, and miR-133b overexpression reversed these
effects (Fig. 3B). Furthermore,
Masson's trichrome staining indicated that miR-133b overexpression
significantly attenuated doxorubicin-induced cardiac fibrosis and
collagen deposition (Fig. 3C-F).
Similarly, western blot assays suggested that the
doxorubicin-induced augmentation of the protein expression levels
of collagen I, III and IV, and fibronectin were also antagonized by
miR-133b overexpression (Fig.
3G). In conclusion, overexpression of miR-133b alleviated
apoptosis and cardiac fibrosis in vivo.
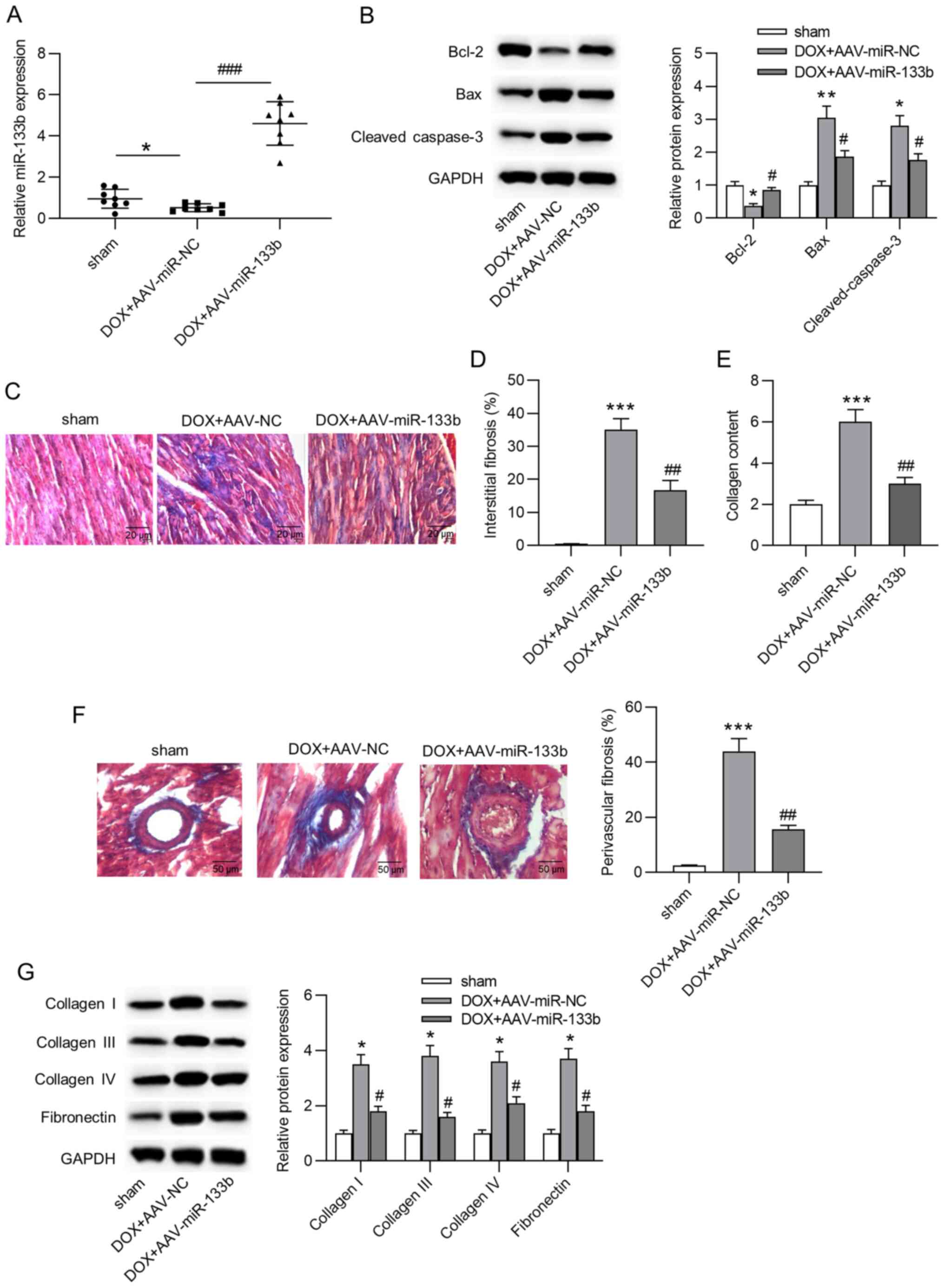 | Figure 3Overexpression of miR-133b alleviates
apoptosis and cardiac fibrosis in vivo. (A) Overexpression
efficacy of AAV-miR-133b was confirmed by reverse
transcription-quantitative PCR (n=8/group). (B) Protein expression
levels of Bcl-2, Bax and cleaved caspase-3 in cardiac tissues were
determined by western blot assays (n=8/group). (C and D) Masson's
trichrome staining was used to evaluate interstitial fibrosis in
the mouse heart (n=8/group). Scale bar, 20 µm. (E) Collagen
content in cardiac tissues (n=8/group). (F) Masson's staining of
myocardial peripheral vessels and the percentage of perivascular
fibrosis (n=8/group). Scale bar, 50 µm. (G) Protein
expression levels of collagen I, III and IV, and fibronectin in
cardiac tissues were determined by western blot assays (n=8/group).
*P<0.05, **P<0.01 and
***P<0.001 vs. sham; #P<0.05,
##P<0.01 and ###P<0.001 vs.
DOX+AAV-miR-NC. miR, microRNA; DOX, doxorubicin; NC, negative
control; AAV, adeno-associated virus. |
PTBP1 and TAGLN2 are targets of
miR-133b
Previously, miR-133b has been widely reported to
bind to the 3′-UTR of mRNAs to exert its biological functions
(26,31). Hence, it was hypothesized that
miR-133b functioned in the same way in HL-1 cardiomyocytes.
According to the TargetScan website, 703 mRNAs were predicted to
harbor binding site(s) for miR-133b (data not shown). The top 8
mRNAs (LHFP, SEC61B, CETN3, PTBP1, FTL, LDLRAP1, CLTA and TAGLN2)
with the highest total context score were chosen for subsequent
experiments. According to RT-qPCR, PTBP1 and TAGLN2 expression was
significantly upregulated in the doxorubicin-treated HL-1
cardiomyocytes compared with in control cells, suggesting that
PTBP1 and TAGLN2 may be target mRNAs of miR-133b (Fig. 4A). Subsequently, the binding site
between miR-133b and PTBP1 or TAGLN2 was predicted using the
TargetScan website (Fig. 4B).
Luciferase reporter assays demonstrated that the luciferase
activity of wild-type pmirGLO-PTBP1-3′-UTR or pmirGLO-TAGLN2-3′-UTR
was significantly inhibited by miR-133b mimics, while the mutant
pmirGLO-PTBP1-3′-UTR or pmirGLO-TAGLN2-3′-UTR displayed no changes
(Fig. 4C). Additionally, the
expression levels of PTBP1 and TAGLN2 were significantly
upregulated in the DOX group compared with in the sham group
(Fig. 4D). In addition, miR-133b
overexpression significantly decreased TAGLN2 and PTBP1 protein
expression in HL-1 cardiomyocytes (Fig. 4E). Coincidently, according to the
starBase website, PTBP1 was predicted to bind to the 3′-UTR of
TAGLN2 (data not shown). The interaction between the PTBP1 protein
and the TAGLN2 3′-UTR was then evaluated. Data from RNA-Protein
Interaction Prediction revealed that the RF classifier score and
SVM classifier score of the combination between PTBP1 and TAGLN2
3′-UTR were 0.7 and 0.29, respectively, while the scores of the
combination between PTBP1 and TAGLN2 were 0.8 and 0.26,
respectively (Fig. 4F).
Subsequently, an RNA pulldown assay indicated that PTBP1 was
enriched only in the input group (Fig. 4G), and a RIP assay demonstrated
that neither anti-IgG nor anti-PTBP1 immunoprecipitated TAGLN2
(Fig. 4H), indicating that PTBP1
was unable to bind to the TAGLN2 3′-UTR. Thus, PTBP1 and TAGLN2
served as targets of miR-133b.
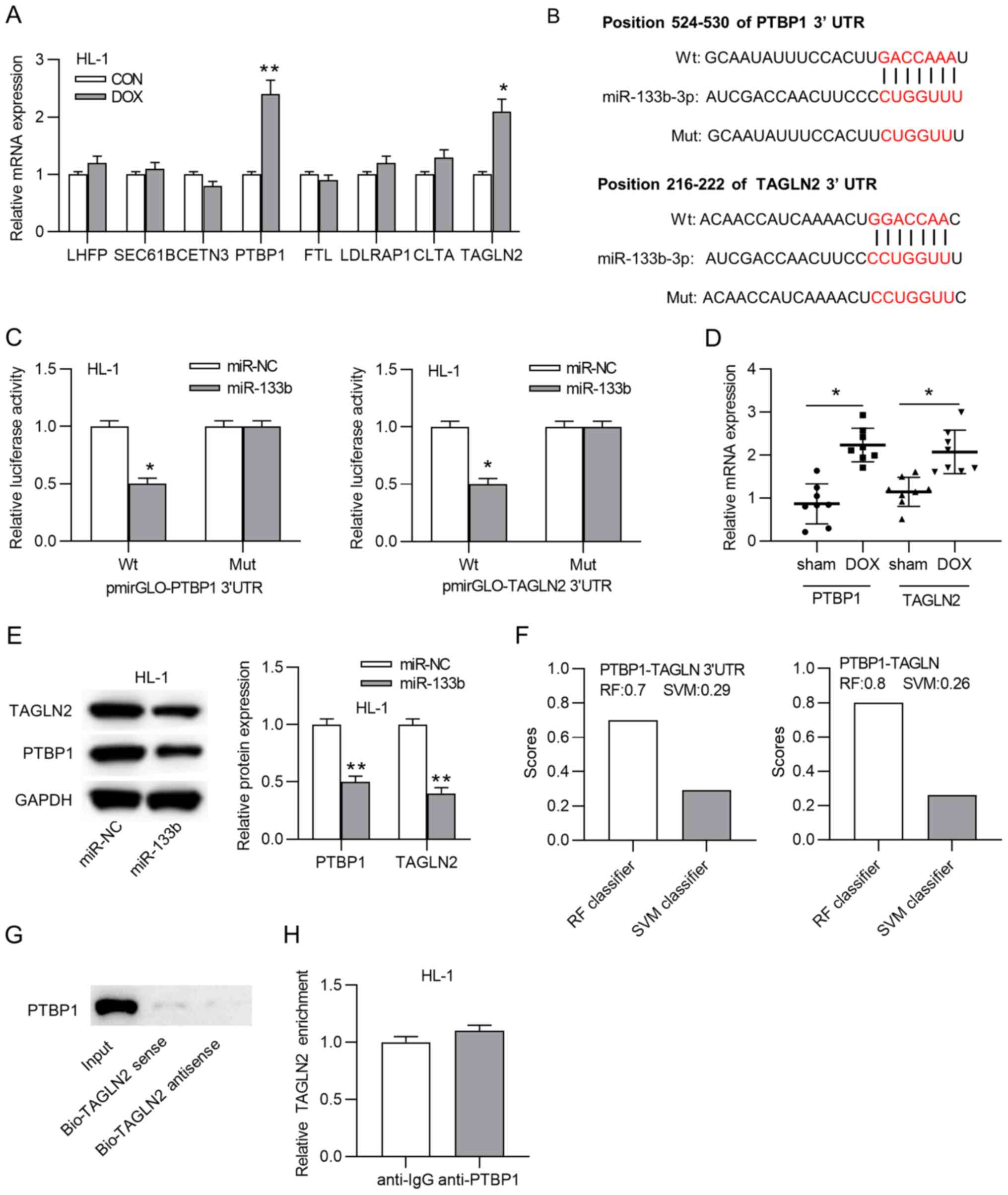 | Figure 4PTBP1 and TAGLN2 serve as targets of
miR-133b. (A) mRNA expression levels of predicted mRNAs in HL-1
cardiomyocytes treated with or without doxorubicin.
*P<0.05 and **P<0.01 vs. CON. (B)
Binding sites between miR-133b and PTBP1 or TAGLN2. (C) Luciferase
reporter assay was adopted to confirm the interaction between
miR-133b and PTBP1 or TAGLN2. *P<0.05 vs. miR-NC. (D)
mRNA expression levels of PTBP1 and TAGLN2 in the sham and DOX
groups. *P<0.05. (E) Western blot assays of the
changes in the PTBP1 and TAGLN2 protein expression levels in the
doxorubicin-treated HL-1 cardiomyocytes transfected with miR-NC or
miR-133b mimics. **P<0.01 vs. miR-NC. (F) RF
classifier score and SVM classifier score of the combination of
PTBP1 and the TAGLN2 3′-UTR or TAGLN2. (G) RNA pulldown and (H) RNA
immunoprecipitation assays were utilized to evaluate the
interaction between PTBP1 and TAGLN2. miR, microRNA; DOX,
doxorubicin; CON, control; NC, negative control; UTR, untranslated
region; PTBP1, polypyrimidine tract binding protein 1; TAGLN2,
transgelin 2; Wt, wild-type; Mut, mutant; Bio, biotin; RF, random
forest; SVM, support vector machine. |
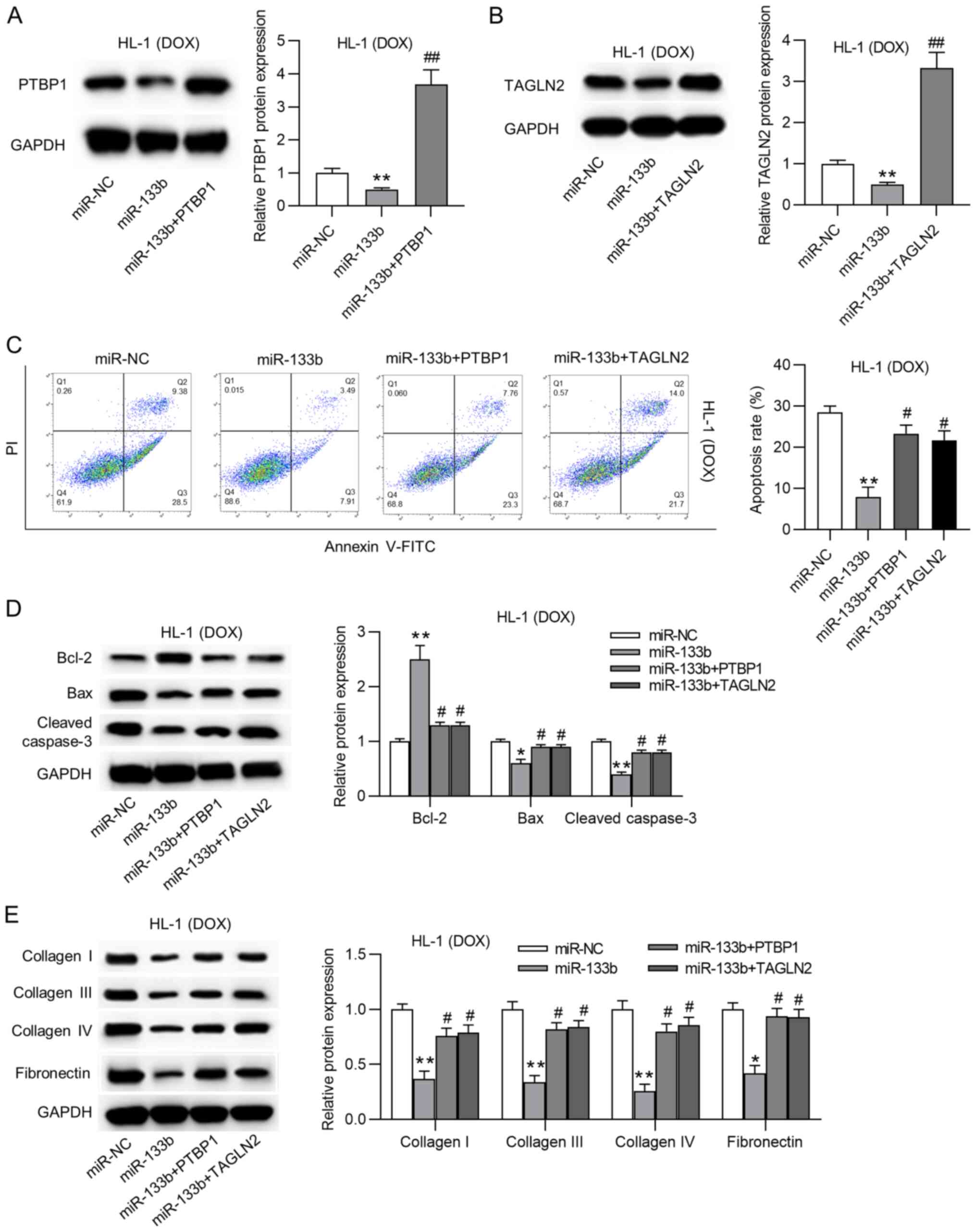
Overexpression of PTBP1 or TAGLN2
reverses the effects of miR-133b on apoptosis and collagen
accumulation
To validate whether miR-133b modulated apoptosis and
collagen deposition by targeting PTBP1 and TAGLN2 in the
doxorubicin-treated HL-1 cardiomyocytes, rescue assays were
performed via PTBP1 and TAGLN2 overexpression. PTBP1 and TAGLN2
overexpression efficiency was verified by RT-qPCR (Fig. S1C and D). The protein expression
levels of PTBP1 and TAGLN2 were significantly increased by
overexpression of PTBP1 and TAGLN2, respectively, following
miR-133b overexpression in the doxorubicin-treated HL-1
cardiomyocytes (Fig. 5A and B).
In addition, overexpression of either PTBP1 or TAGLN2 rescued the
miR-133b mimic-induced decreased apoptosis rate of the
doxorubicin-treated HL-1 cardiomyocytes (Fig. 5C). Moreover, the increase in
Bcl-2 expression and the decrease in Bax and cleaved caspase-3
expression resulting from miR-133b overexpression were reversed by
the overexpression of PTBP1 or TAGLN2 (Fig. 5D). Additionally, the suppressive
effects of miR-133b on the protein expression levels of collagen I,
III and IV, and fibronectin were abrogated by PTBP1 or TAGLN2
overexpression (Fig. 5E). All
these results confirmed that overexpression of PTBP1 or TAGLN2
reversed the effects of miR-133b on apoptosis and collagen
accumulation.
Discussion
Doxorubicin is one of the most important
chemotherapeutic drugs for the treatment of malignant tumors,
although the cardiotoxicity of doxorubicin severely limits its
clinical application (32,33). Previously, doxorubicin has been
reported to induce apoptosis and impair cardiac function in heart
failure (34,35). In the current study, doxorubicin
was used to treat HL-1 cardiomyocytes and mice to mimic
cardiomyocyte injury in vitro and heart failure in
vivo, respectively. The experimental results revealed that
doxorubicin strongly promoted apoptosis and impaired cardiac
function. Overexpression of Bax had no significant effects on the
expression levels of ECM proteins, indicating that fibrotic events
were independent of apoptosis.
Currently, numerous miRNAs have been shown to be
dysregulated in patients with myocardial diseases and to modulate
cardiomyocyte cellular processes and cardiac function (13,25,36). In addition, miRNAs serve crucial
roles in doxorubicin-induced myocardial diseases (15,37,38). Previously, relatively lower
expression levels of miR-133b have been identified in
doxorubicin-treated cells and rats (23,24,26). Similarly, miR-133b expression in
the present study was also downregulated in the doxorubicin-treated
HL-1 cardiomyocytes and mouse hearts. It has been previously
demonstrated that miR-133b decreases myocardial injuries by
inhibiting cardiomyocyte proliferation and the release of cytokines
(31). In addition, miR-133b is
involved in the regulation of cardiac fibrosis and cardioprotection
of morphine preconditioning in cardiomyocytes (26). Similarly, overexpression of
miR-133b in the current study inhibited apoptosis of HL-1
cardiomyocytes and suppressed collagen accumulation under
doxorubicin treatment. Additionally, AAV-induced overexpression of
miR-133b alleviated cardiac fibrosis in mice by decreasing collagen
deposition.
Notably, miR-133b exerts its biological function by
binding to the 3′-UTRs of mRNAs (26,31). miR-133b improves myocardial
injuries by targeting Rab27B in children with viral myocarditis
(31). Moreover, miR-133b
restricts cell proliferation, migration and invasion by targeting
EGFR in esophageal squamous cell carcinoma (39). Hence, the present study
hypothesized that miR-133b also functioned in the same way in HL-1
cardiomyocytes. After prediction and screening, PTBP1 and TAGLN2
were confirmed to serve as target mRNAs of miR-133b. miR-133b
targeted the 3′-UTR of PTBP1 and TAGLN2, and decreased the protein
expression levels of PTBP1 and TAGLN2, which is consistent with
previous studies (40,41).
PTBP1, also known as heterogeneous nuclear
ribonucleoprotein 1, belongs to a subfamily of ubiquitously
expressed heterogeneous nuclear ribonucleoproteins (42,43). These proteins have been reported
to potentially influence pre-mRNA processing or other aspects of
mRNA metabolism and transport (44). Previously, PTBP1 has been
reported to induce cardiomyocyte apoptosis by enhancing caspase
activity and caspase-dependent DNA fragmentation to activate
apoptotic signaling (45). PTBP1
regulates pulmonary arterial hypertension by splicing pyruvate
kinase muscle isoforms 1 and 2 (46). PTBP1 expression is upregulated in
diabetic heart disease, and the top categories for PTBP1-regulated
genes involve RNA transport, MAPK signaling and endocytosis,
according to Kyoto Encyclopedia of Genes and Genomes pathway
analysis (47). In addition,
TAGLN2 promotes hypoxia-induced apoptosis of rat cardiomyocytes by
increasing the expression levels of the apoptotic proteins
caspase-8, caspase-9 and caspase-3, and decreasing Bcl-2 expression
(48). Similarly, in the present
study, the rescue assays indicated that overexpression of PTBP1 or
TAGLN2 reversed the effects of miR-133b on apoptosis and collagen
accumulation.
In conclusion, miR-133b alleviated
doxorubicin-induced cardiac fibrosis in mice by decreasing collagen
deposition and suppressed apoptosis of doxorubicin-preconditioned
cardiomyocytes. The protective effects of miR-133b on cardiomyocyte
apoptosis and cardiac fibrosis were mediated by inhibition of PTBP1
and TAGLN2 expression. Therefore, miR-133b may be used as a
biomarker to identify patients who may develop dilated
cardiomyopathy after chemotherapy in the future.
Supplementary Data
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
ZL designed the experiments. ZL, JM and XG conducted
the experiments. ZL, ZY, QG, JT and XG analyzed the data and made
the graphs. ZL and XG wrote the manuscript. ZL, JM and XG confirmed
the authenticity of all the raw data. All authors have read and
approved the final manuscript.
Ethics approval and consent to
participate
The animal experimental protocol was approved by the
ethical committee of The First Affiliated Hospital of Nanjing
Medical University (Nanjing, China; approval no. 2019-041).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
Funding
The present study was supported by the Nanjing Medical Science
and Technology Development Special Fund (grant no. N2903-302).
References
|
1
|
Rivankar S: An overview of doxorubicin
formulations in cancer therapy. J Cancer Res Ther. 10:853–858.
2014. View Article : Google Scholar
|
|
2
|
Genovese I, Fiorillo A, Ilari A,
Masciarelli S, Fazi F and Colotti G: Binding of doxorubicin to
Sorcin impairs cell death and increases drug resistance in cancer
cells. Cell Death Dis. 8:e29502017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Borlle L, Dergham A, Wund Z, Zumbo B,
Southard T and Hume KR: Salinomycin decreases feline sarcoma and
carcinoma cell viability when combined with doxorubicin. BMC Vet
Res. 15:362019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Von Hoff DD, Layard MW, Basa P, Davis HL
Jr, Von Hoff AL, Rozencweig M and Muggia FM: Risk factors for
doxorubicin-induced congestive heart failure. Ann Intern Med.
91:710–717. 1979. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
du Pré BC, Dierickx P, Crnko S, Doevendans
PA, Vos MA, Geijsen N, Neutel D, van Veen TAB and van Laake LW:
Neonatal rat cardiomyocytes as an in vitro model for circadian
rhythms in the heart. J Mol Cell Cardiol. 112:58–63. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hole LD, Larsen TH, Fossan KO, Limé F and
Schjøtt J: Diazoxide protects against doxorubicin-induced
cardiotoxicity in the rat. BMC Pharmacol Toxicol. 15:282014.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
De Beer EL, Bottone AE and Voest EE:
Doxorubicin and mechanical performance of cardiac trabeculae after
acute and chronic treatment: A review. Eur J Pharmacol. 415:1–11.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Unverferth DV, Magorien RD, Leier CV and
Balcerzak SP: Doxorubicin cardiotoxicity. Cancer Treat Rev.
9:149–164. 1982. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Cecen E, Dost T, Culhaci N, Karul A, Ergur
B and Birincioglu M: Protective effects of silymarin against
doxorubicin-induced toxicity. Asian Pac J Cancer Prev.
12:2697–2704. 2011.
|
|
10
|
Burlacu A, Siriopol D, Voroneanu L, Nistor
I, Hogas S, Nicolae A, Nedelciuc I, Tinica G and Covic A:
Atherosclerotic renal artery stenosis prevalence and correlations
in acute myocardial infarction patients undergoing primary
percutaneous coronary interventions: Data from Nonrandomized
Single-Center Study (REN-ACS)-A single center, prospective,
observational study. J Am Heart Assoc. 4:e0023792015. View Article : Google Scholar
|
|
11
|
Lu TX and Rothenberg ME: MicroRNA. J
Allergy Clin Immunol. 141:1202–1207. 2018. View Article : Google Scholar :
|
|
12
|
Bernardo BC, Ooi JY, Lin RC and McMullen
JR: MiRNA therapeutics: A new class of drugs with potential
therapeutic applications in the heart. Future Med Chem.
7:1771–1792. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Rupaimoole R and Slack FJ: MicroRNA
therapeutics: Towards a new era for the management of cancer and
other diseases. Nat Rev Drug Discov. 16:203–222. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhao L, Qi Y, Xu L, Tao X, Han X, Yin L
and Peng J: MicroRNA-140-5p aggravates doxorubicin-induced
cardiotoxicity by promoting myocardial oxidative stress via
targeting Nrf2 and Sirt2. Redox Biol. 15:284–296. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gupta SK, Garg A, Avramopoulos P,
Engelhardt S, Streckfuss-Bömeke K, Batkai S and Thum T: MiR-212/132
cluster modulation prevents Doxorubicin-Mediated atrophy and
cardiotoxicity. Mol Ther. 27:17–28. 2019. View Article : Google Scholar :
|
|
16
|
Li N, Zhou H and Tang Q: MiR-133: A
suppressor of cardiac remodeling? Front Pharmacol. 9:9032018.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang Y, Li M, Xu L, Liu J, Wang D, Li Q,
Wang L, Li P, Chen S and Liu T: Expression of Bcl-2 and microRNAs
in cardiac tissues of patients with dilated cardiomyopathy. Mol Med
Rep. 15:359–365. 2017. View Article : Google Scholar
|
|
18
|
Cortez-Dias N, Costa MC, Carrilho-Ferreira
P, Silva D, Jorge C, Calisto C, Pessoa T, Robalo Martins S, de
Sousa JC, da Silva PC, et al: Circulating miR-122-5p/miR-133b Ratio
is a specific early prognostic biomarker in acute myocardial
infarction. Circ J. 80:2183–2191. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhang L and Wang H: Long Non-coding RNA in
CNS injuries: A new target for therapeutic intervention. Mol Ther
Nucleic Acids. 17:754–766. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Atef MM, Amer AI, Hafez YM, Elsebaey MA,
Saber SA and Abd El-Khalik SR: Long non-coding RNA EGFR-AS1 in
colorectal cancer: A potential factor in tumorigenesis and survival
via miRNA-133b sponge and EGFR/STAT3 axis regulation. Br J Biomed
Sci. 2020.Epub ahead of print.
|
|
21
|
Zhao N, Liu H, Zhang A and Wang M:
Expression levels and clinical significance of miR-203 and miR-133b
in laryngeal carcinoma. Oncol Lett. 20:2132020. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chen J, Li Y, Li Z and Cao L: LncRNA
MST1P2/miR-133b axis affects the chemoresistance of bladder cancer
to cisplatin-based therapy via Sirt1/p53 signaling. J Biochem Mol
Toxicol. 34:e224522020. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Sandhu H, Cooper S, Hussain A, Mee C and
Maddock H: Attenuation of Sunitinib-induced cardiotoxicity through
the A3 adenosine receptor activation. Eur J Pharmacol. 814:95–105.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Cooper SL, Sandhu H, Hussain A, Mee C and
Maddock H: Involvement of mitogen activated kinase kinase 7
intracellular signalling pathway in Sunitinib-induced
cardiotoxicity. Toxicology. 394:72–83. 2018. View Article : Google Scholar
|
|
25
|
Hanousková B, Skála M, Brynychová V,
Zárybnický T, Skarková V, Kazimírová P, Vernerová A, Souček P,
Skálová L, Pudil R and Matoušková P: Imatinib-induced changes in
the expression profile of microRNA in the plasma and heart of
mice-A comparison with doxorubicin. Biomed Pharmacother.
115:1088832019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
He SF, Zhu HJ, Han ZY, Wu H, Jin SY, Irwin
MG and Zhang Y: MicroRNA-133b-5p is involved in cardioprotection of
morphine preconditioning in rat cardiomyocytes by targeting fas.
Can J Cardiol. 32:996–1007. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Panizo S, Carrillo-López N, Naves-Díaz M,
Solache-Berrocal G, Martínez-Arias L, Rodrigues-Díez RR,
Fernández-Vázquez A, Martínez-Salgado C, Ruiz-Ortega M, Dusso A, et
al: Regulation of miR-29b and miR-30c by vitamin D receptor
activators contributes to attenuate uraemia-induced cardiac
fibrosis. Nephrol Dial Transplant. 32:1831–1840. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Roca-Alonso L, Castellano L, Mills A,
Dabrowska AF, Sikkel MB, Pellegrino L, Jacob J, Frampton AE, Krell
J, Coombes RC, et al: Myocardial MiR-30 downregulation triggered by
doxorubicin drives alterations in β-adrenergic signaling and
enhances apoptosis. Cell Death Dis. 6:e17542015. View Article : Google Scholar
|
|
29
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
30
|
Tao L, Bei Y, Chen P, Lei Z, Fu S, Zhang
H, Xu J, Che L, Chen X, Sluijter JP, et al: Crucial role of miR-433
in regulating cardiac fibrosis. Theranostics. 6:2068–2083. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhang Y, Sun L, Sun H, Liu X, Luo X, Li C,
Sun D and Li T: Overexpression of microRNA-133b reduces myocardial
injuries in children with viral myocarditis by targeting Rab27B
gene. Cell Mol Biol (Noisy-le-grand). 63:80–86. 2017. View Article : Google Scholar
|
|
32
|
Balli E, Mete UO, Tuli A, Tap O and Kaya
M: Effect of melatonin on the cardiotoxicity of doxorubicin. Histol
Histopathol. 19:1101–1108. 2004.PubMed/NCBI
|
|
33
|
Ganey PE, Carter LS, Mueller RA and
Thurman RG: Doxorubicin toxicity in perfused rat heart. Decreased
cell death at low oxygen tension. Circ Res. 68:1610–1613. 1991.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Saltiel E and McGuire W: Doxorubicin
(adriamycin) cardiomyopathy. West J Med. 139:332–341.
1983.PubMed/NCBI
|
|
35
|
Mitani I, Jain D, Joska TM, Burtness B and
Zaret BL: Doxorubicin cardiotoxicity: Prevention of congestive
heart failure with serial cardiac function monitoring with
equilibrium radionuclide angiocardiography in the current era. J
Nucl Cardiol. 10:132–139. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Li J, Zhang S, Zou Y, Wu L, Pei M and
Jiang Y: MiR-145 promotes miR-133b expression through c-myc and
DNMT3A-mediated methylation in ovarian cancer cells. J Cell
Physiol. 235:4291–4301. 2020. View Article : Google Scholar
|
|
37
|
Ruggeri C, Gioffré S, Achilli F, Colombo
GI and D'Alessandra Y: Role of microRNAs in doxorubicin-induced
cardiotoxicity: An overview of preclinical models and cancer
patients. Heart Fail Rev. 23:109–122. 2018. View Article : Google Scholar :
|
|
38
|
Lu Q, Huo J, Liu P, Bai L and Ma A: lncRNA
HOXB-AS3 protects doxorubicin-induced cardiotoxicity by targeting
miRNA-875-3p. Exp Ther Med. 19:1388–1392. 2020.PubMed/NCBI
|
|
39
|
Zeng W, Zhu JF, Liu JY, Li YL, Dong X,
Huang H and Shan L: MiR-133b inhibits cell proliferation, migration
and invasion of esophageal squamous cell carcinoma by targeting
EGFR. Biomed Pharmacother. 111:476–484. 2019. View Article : Google Scholar
|
|
40
|
Sugiyama T, Taniguchi K, Matsuhashi N,
Tajirika T, Futamura M, Takai T, Akao Y and Yoshida K: MiR-133b
inhibits growth of human gastric cancer cells by silencing pyruvate
kinase muscle-splicer polypyrimidine tract-binding protein 1.
Cancer Sci. 107:1767–1775. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhao F, Zhou LH, Ge YZ, Ping WW, Wu X, Xu
ZL, Wang M, Sha ZL and Jia RP: MicroRNA-133b suppresses bladder
cancer malignancy by targeting TAGLN2-mediated cell cycle. J Cell
Physiol. 234:4910–4923. 2019. View Article : Google Scholar
|
|
42
|
Coelho MB, Ascher DB, Gooding C, Lang E,
Maude H, Turner D, Llorian M, Pires DE, Attig J and Smith CW:
Functional interactions between polypyrimidine tract binding
protein and PRI peptide ligand containing proteins. Biochem Soc
Trans. 44:1058–1065. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zhang H, Wang D, Li M, Plecitá-Hlavatá L,
D'Alessandro A, Tauber J, Riddle S, Kumar S, Flockton A, McKeon BA,
et al: Metabolic and proliferative state of vascular adventitial
fibroblasts in pulmonary hypertension is regulated through a
MicroRNA-124/PTBP1 (polypyrimidine tract binding protein
1)/pyruvate kinase muscle axis. Circulation. 136:2468–2485. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Pina JM, Reynaga JM, Truong AAM and
Keppetipola NM: Post-Translational modifications in polypyrimidine
tract binding proteins PTBP1 and PTBP2. Biochemistry. 57:3873–3882.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Zhang J, Bahi N, Llovera M, Comella JX and
Sanchis D: Polypyrimidine tract binding proteins (PTB) regulate the
expression of apoptotic genes and susceptibility to
caspase-dependent apoptosis in differentiating cardiomyocytes. Cell
Death Differ. 16:1460–1468. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Caruso P, Dunmore BJ, Schlosser K, Schoors
S, Dos Santos C, Perez-Iratxeta C, Lavoie JR, Zhang H, Long L,
Flockton AR, et al: Identification of MicroRNA-124 as a major
regulator of enhanced endothelial cell glycolysis in pulmonary
arterial hypertension via PTBP1 (Polypyrimidine Tract Binding
Protein) and Pyruvate Kinase M2. Circulation. 136:2451–2467. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Belanger K, Nutter CA, Li J, Yu P and
Kuyumcu-Martinez MN: A developmentally regulated spliced variant of
PTBP1 is upregulated in type 1 diabetic hearts. Biochem Biophys Res
Commun. 509:384–389. 2019. View Article : Google Scholar :
|
|
48
|
Li AY, Yang Q and Yang K: MiR-133a
mediates the hypoxia-induced apoptosis by inhibiting TAGLN2
expression in cardiac myocytes. Mol Cell Biochem. 400:173–181.
2015. View Article : Google Scholar
|