Most S100 proteins are involved not only in cell
proliferation, differentiation, motility and apoptosis, but also in
physiological processes such as the assembly-disassembly state of
cytoskeletal components, phagocytosis, expression and activity of
transcription factors, redox balance, Ca2+ release from
Ca2+ stores, ion channel activity, protein degradation
and immune cell responses, which function in a
Ca2+-dependent manner (16,47). Certain S100 proteins are
constitutively secreted as extracellular signals by specific types
of cell, including passive release from damaged or dying cells or
immune cells during inflammatory events (16,48,49). For example, during acute or
chronic local inflammation, myeloid cells actively secrete
S100A8/A9 (49). Moreover,
several extracellular S100 proteins serve as damage-associated
molecular patterns and activate a series of membrane receptors,
including pattern recognition and Toll-like receptor-4, G-protein
coupled receptors, as well as receptor for advanced glycation end
products (RAGE) (16,21,48-50). Under normal and pathological
conditions, extracellular S100 proteins affect the activity of
numerous types of cell, including neurons, astrocytes,
cardiomyocytes, microglia, adipocytes, epithelial and smooth muscle
cells and skeletal muscle fibroblasts, which suggests that S100
serves an important role in inflammation (16,21,32,49,51-55). In addition, S100 protein has been
associated with fibrosis in multiple organs, such as the kidney
(56), lung (57), skin (58) and liver (59) and may serve as a biomarker of
fibrosis. It has also been reported that S100 protein expression is
elevated in tumors, including such as breast (60), lung (61), colorectal (62) and pancreatic cancer (63), as well as HCC (28). Therefore, S100 proteins serve a
key role in tumorigenesis and cancer progression and can also be
used as potential markers and therapeutic targets for various types
of tumors, such as osteosarcoma and lung, colorectal and breast
cancer (64-68).
As aforementioned, the S100 protein family is
involved in numerous physiological processes. Previous studies have
reported that S100 proteins serve an important role in the onset
and progression of disease, including fibrosis, inflammatory
disease and tumor (21,56-58,60-63). Similarly, S100 protein exerts a
key role in liver disease (Table
II), especially liver fibrosis and HCC, and may be a biomarker
and therapeutic target for these diseases (28,59,69). Thus, the S100 protein family may
facilitate the diagnosis and treatment of liver disease in the
future.
There are >100 million patients with liver
fibrosis worldwide; this disease seriously affects patient health
(70). Liver fibrosis involves
abnormal proliferation of connective tissue in the liver and is
caused by various factors, including viral and autoimmune
hepatitis, NAFLD and ALD (71).
The pathological mechanism of liver fibrosis is complex and is
primarily driven by inflammation and immune regulation mechanisms
(72,73). Previous studies identified the
activation of hepatic stellate cells (HSCs) and excessive
deposition of extracellular matrix (ECM) components as the two main
processes leading to the development of liver fibrosis (72,74). In liver fibrosis, fibroblasts are
primarily derived from activated (a)HSCs (75). The physiological role of
quiescent (q)HSCs is storage of vitamin A in the liver (76). When external factors (such as
viral infection, alcohol or drugs) cause liver damage, qHSC are
activated by inflammatory mediators and differentiate into
myofibroblasts (75).
Subsequently, ECM proteins and MMPs secreted by aHSC begin to
remodel tissue in the liver (77,78). Activated fibroblasts are the
primary source of ECM following liver injury. Thus, activation and
proliferation of myofibroblasts causes liver fibrosis (75,77). The onset and progression of liver
fibrosis are associated with activation and proliferation of HSCs;
therefore, a key target for the treatment of liver fibrosis is the
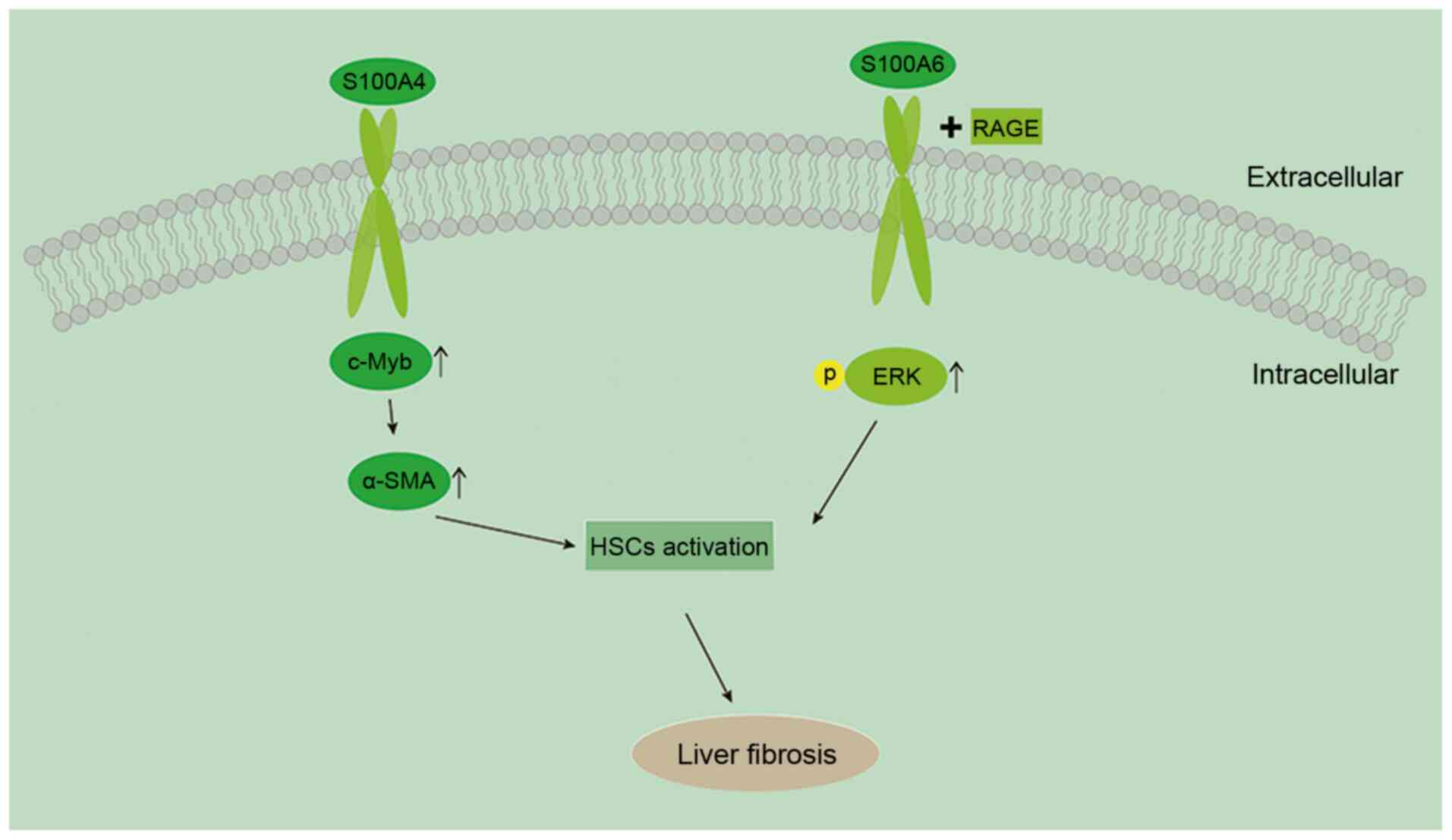
inactivation of HSCs and apoptosis (79,80). Chen et al (59) reported that S100A4 is upregulated
in liver tissue during the progression of liver fibrosis and that
the degree of liver fibrosis decreases after blocking S100A4
expression in vivo. They also found that S100A4 promotes HSC
activation via upregulation of c-Myb (Fig. 1). Chen et al (59) analyzed S100A4 levels in patients
with cirrhosis and identified that serum S100A4 levels were
significantly elevated. The same results were reported in a study
by Louka and Ramzy (69), in
which expression levels of S100A4 in liver tissue were positively
correlated with the degree of liver fibrosis.
In addition to inactivation of HSCs, another
important step to reverse fibrosis is degradation of the ECM
(81). MMPs comprise a series of
enzymes with different substrate affinities for matrix components
and are the most important effector for the degradation of ECM
(81). The interstitial
collagenase MMP-13 is a highly specific protease derived from
stellate cells (82). MMP-13
serves a key role in liver fibrosis by mediating the initial
inflammatory response in the liver and accelerating the formation
of cholestatic liver fibrosis (83). Previous studies have shown that
S100A4 is directly involved in the transcription of the MMP-13 gene
(84) and in human chondrocytes,
extracellular S100A4 protein directly increases expression levels
of MMP-13 by interacting with RAGE (85). Similarly, in liver fibrosis,
MMP-13 expression levels are positively correlated with
upregulation of S100A4 (69).
Thus, it should be further investigated whether S100A4 also binds
to RAGE in liver fibrosis, thereby promoting the expression of
MMP-13.
Similar to S100A4, S100A6 facilitates liver fibrosis
by promoting HSC proliferation, primarily by binding to RAGE and
inducing ERK activation (86).
S100B also serves a role in liver disease; previous studies have
reported that S100B expression is decreased in the early stages of
liver fibrosis and chronic liver disease and its expression levels
during the course of chronic liver disease do not significantly
change (87,88).
Viral hepatitis is a major global public health
problem affecting hundreds of millions of individuals and is
associated with significant morbidity and mortality, with an
estimated 257 million people worldwide living with HBV and 71
million with HCV in 2017 (89).
Global mortality from chronic HBV and HCV infection is rising, with
>1.4 million deaths/year (90). The liver stores vitamin A
(retinol) and produces large amounts of all-trans retinoic acid
(RA) (91). Previous studies
have revealed that RA serves an important role in liver
regeneration, fibrosis and tumor formation (92,93). Moreover, it has been shown that
RA inhibits dendritic cell function via an S100A4-mediated
mechanism, leading to downregulation of T-cell responses and
ultimately decreasing hepatitis-induced viral liver injury
(91). A recent study by Yan
et al (94) compared
liver biopsy results and serum S100A4 levels in patients with
chronic hepatitis B (CHB; n=175); serum S100A4 levels were higher
in CHB cases with significant fibrosis compared with those in CHB
cases without fibrosis. Furthermore, S100A9 expression is increased
during HBV infection and can be used as a marker to distinguish the
severity of liver necrotizing inflammation (95). In addition, S100A12 reflects the
level of oxidative stress and inflammation in HBV-associated acute
and chronic liver failure and its elevated expression may be an
important marker of poor prognosis (96).
NAFLD is the most common chronic liver disease
worldwide. A meta-analysis of studies from 1989 to 2015 reported
that the global prevalence of NAFLD was ~25% (15), ranging from 13% in Africa to 42%
in Southeast Asia (15,97). Its histological features include
non-alcoholic steatohepatitis (NASH) and simple steatosis and it is
characterized by fat accumulation, swelling of hepatocytes and the
development of inflammation and/or fibrosis (98). Furthermore, NAFLD may ultimately
progress to liver cirrhosis and HCC (99). Recently, Zhang et al
(100) established a mouse
model of NAFLD and found that S100A4 expression was increased in
NAFLD, whereas decreased inflammation and liver fibrosis were
observed in S100A4 knockout mice. In a NAFLD rat model, serum
S100A9 levels are positively correlated with the degree of hepatic
steatosis, lobular inflammation and NAFLD activity score,
indicating that these may be a potential biomarker of liver and
metabolic progress (101).
Zhang et al (102)
established a tree shrew model of NAFLD and identified that
overexpression of S100A11 promoted FOXO1-mediated autophagy and
adipogenesis via the S100A11/histone deacetylase 6/FOXO1 axis,
thereby promoting hepatic steatosis and providing a potential
therapeutic target for NAFLD. However, further experiments are
required to identify the role of autophagy and lipogenesis in the
pathogenesis of NAFLD (103).
Another study reported that S100A11 promotes liver inflammation and
fibrosis in a mouse model of NAFLD and that upregulation of S100A11
may be a key feature in the transition from steatosis to
NASH/fibrosis in mice (104).
However, current studies on the role of the S100 family in NAFLD
primarily use cellular and animal models and further analysis of
clinical samples is required.
ALD often occurs in individuals who drink a
significant amount alcohol over a long period of time. There were
1,256,900 deaths in 2016 due to cirrhosis and chronic liver disease
(105). Among those, 334,900
(27%) were attributed to alcohol (105). The initial stage of ALD is
alcoholic fatty liver (AFL), which is caused by alcohol and its
metabolites (106). The primary
effect of AFL on the liver includes synthesis and breakdown of
fatty acids, which leads to hepatitis and fibrosis, and ultimately
cirrhosis and HCC, in a subset of drinkers (those with viral
hepatitis, diabetes or a history of smoking) (106-108). Studies have shown an increased
risk of alcoholic hepatitis and cirrhosis among individuals
drinking >40 g/day (109-111). A previous study reported that
ethanol-fed mice exhibit higher levels of S100A4 in the liver,
whereas S100A4-knockout mice have decreased liver inflammation, and
that S100A4 promotes early alcoholic hepatitis primarily by
activating the STAT3 pathway (112). At the same time, S100A4
inhibits lipid accumulation in chronic alcohol-induced fatty liver
(112). To be best of our
knowledge, no studies have analyzed clinical samples of ALD.
As one of the most common types of malignancy
worldwide, recent statistics have shown that liver cancer is the
sixth most commonly diagnosed cancer and is the fourth leading
cause of cancer-associated mortality (113). There were 905,677 new cases of
liver cancer and 830,180 deaths in 2020, which accounted for 4.7%
of new cancer cases and 8.3% of deaths, respectively (114). The most common type of primary
liver cancer is HCC, which accounts for 80-90% of cases and usually
presents following chronic liver disease (113). Currently, the most effective
treatments for HCC are surgery, liver transplantation, chemotherapy
and targeted therapy, but the overall survival rate remains
unsatisfactory (115). The
highest 5-year survival rate in the world was 27.9% in Taiwan,
China (116). Furthermore, for
patients with recurrence or distant metastases, prognosis is
particularly poor (117).
Therefore, the treatment of HCC requires further research and
investigation.
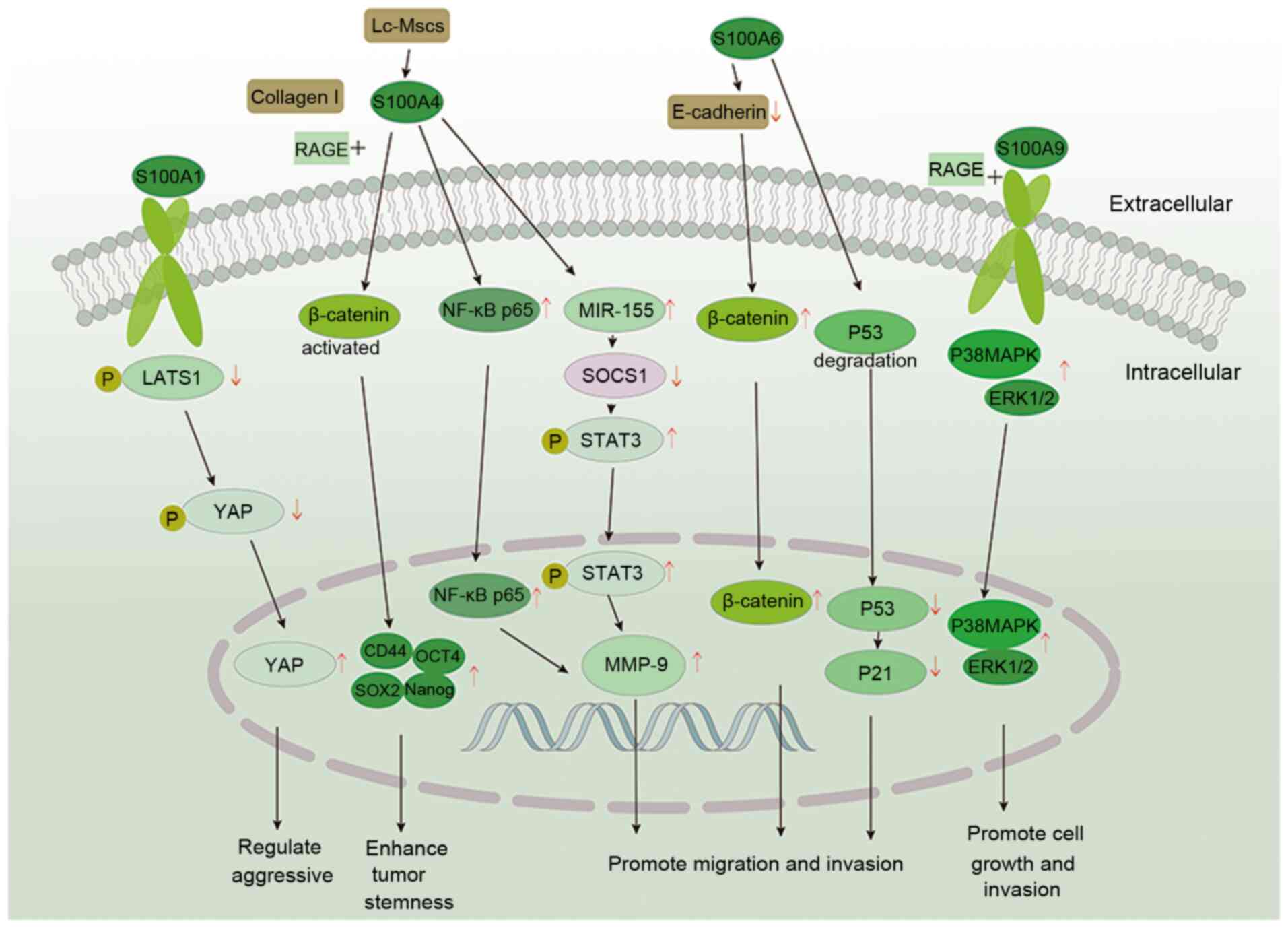
S100A4 also serves a key role in the occurrence of
HCC and is associated with the development, invasion and recurrence
of HCC (28,30). A previous study analyzed HCC
tissue (n=72) and revealed that S100A4 expression was significantly
increased and negatively correlated with overall survival, which
was associated with differentiation, invasion and recurrence of HCC
(30). Similarly, Zhang et
al (120) analyzed invasive
(n=20) and non-invasive HCC (n=20) tissue; S100A4 was highly
expressed in invasive HCC tissue and was positively correlated with
the invasiveness of HCC. Moreover, it was suggested its underlying
mechanism may involve activation and translocation of NF-κB p65
into the nucleus, thereby upregulating MMP-9 to promote cell
migration and invasion. Zhai et al (121) reported that upregulation of
S100A4 expression is associated with the aggressive and malignant
phenotype of HCC, as determined by analyzing HCC tissue (n=113). A
recent study showed that tumor size in HBV-HCC is associated with
S100A4 expression; elevated expression of S100A4 promotes HCC cell
proliferation and is associated with prognosis of patients with HCC
(122).
A 2002 study reported that S100A6 expression is
upregulated in 10% (n=20) of patients with HCC and its expression
is significantly lower compared with that in cholangiocarcinoma
(135). Hua et al
(136) showed that the
difference in S100A6 expression between HCC (n=51) and normal liver
tissue (n=10) was >10-fold and S100A6 expression was upregulated
in 31.4% of HCC samples. The different results of these two studies
may be due to different sample sources and sizes, thus additional
clinical samples are required for further investigation. At the
cellular level, S100A6 is highly expressed in HepG2 cells (137). S100A6 is highly expressed in
36.2% (n=47) of HCC tissue samples (138). Following silencing of S100A6 in
HepG2 cells, cell proliferation is inhibited; following restoration
S100A6 expression, the proliferation and motility of HepG2.2.15
cells was restored (138). The
proposed mechanism is that S100A6 expression results in
downregulation of E-cadherin on the plasma membrane and promotes
nuclear accumulation of β-catenin in cells, which ultimately leads
to proliferation and invasion of liver cancer cells (138). A recent study also reported
that the expression levels of S100A6 are higher in HCC (n=6)
compared with adjacent liver tissue and that overexpression of
S100A6 in HepG2 cells promotes p53 ubiquitin-dependent proteasomal
degradation, which regulates expression of p21 and ultimately
promotes proliferation and migration of HCC cells (139).
S100A9 was demonstrated to be upregulated in HCC as
early as 2000; it is also associated with poorly differentiated HCC
(140). Studies have reported
that S100A9 is upregulated in mouse HCC models and liver cancer
cells (141,142). Wu et al (142) confirmed that S100A9 is highly
expressed in HepG2 cells (primarily in the cytoplasm) and promotes
proliferation and invasion of liver cancer cells by upregulating
ERK1/2 and p38, thereby activating the MAPK pathway. These
functions of S100A9 were primarily generated via interaction with
RAGE (143). In a study of
S100A9 and HBV-associated HCC, S100A9 served a key role in
HBV-encoded X protein-induced HCC growth and metastasis, while TNM
stage and liver metastasis status of HBV-associated HCC were
associated with serum S100A9 expression (144). Moreover, a recent study showed
that S100A9 secretion is upregulated by tumor-associated
macrophages, which subsequently enhances stem cell-like properties
of liver cancer cells to promote tumor development (145).
In a previous study, S100A10 was observed to be
highly expressed in liver cancer cell lines (146). The same results were reported
in a recent study by Zhao et al (147). However, these studies were
limited to in vitro experiments, and further in vivo
experiments and analysis of clinical samples are required. EGFR
belongs to the family of growth factor receptor tyrosine kinases
and serves an important role in the survival, proliferation and
motility of tumor cells (148).
The type III EGFR deletion mutant (EGFRvIII) is the most common
EGFR mutant (149). A previous
study demonstrated that S100A11 expression is increased in 68.6% of
HCC tissue samples (n=51) and promotes HCC cell invasion and
migration primarily via the EGFRvIII/-STAT3 pathway (150). Recent studies have also shown
that S100A11 is secreted by cancer cells and is a marker of liver
cell dedifferentiation and also promotes cell proliferation and
migration (104,151). Furthermore, increased
expression of S100A11 is associated with poor prognosis in
high-grade HCC cases (104). By
analyzing HCC tissue (n=130), Cai et al (152) reported that S100A12 is
expressed only in the cytoplasm of stromal cells. In addition, high
expression of S100A12 in patients with HCC is an independent
prognostic factor for overall and progression-free survival
(152).
The treatment of liver disease is an area of ongoing
research. When progressing to end-stage liver disease, patient
survival rates remain low, although current treatments, such as
surgical techniques and therapeutic regimens, have improved on
previous approaches (153,154). The research and clinical
treatment of liver disease is limited by the lack of sensitive
markers of liver fibrosis, which also limits the development of
anti-fibrotic drugs (155).
Previous studies have suggested that S100A4 may be a potential
marker and therapeutic target for liver fibrosis (59,69). A recent study by Yan et al
(94) demonstrated that the
combination of serum S100A4 and liver stiffness assay improved the
accuracy of diagnosis of severe fibrosis in hepatitis B. Therefore,
serum S100A4 levels may be used as a marker of liver fibrosis in
patients with CHB. S100A9 may also be a potential marker of NAFLD
and metabolic progression (101). Among other S100 proteins,
S100A11 may provide a potential therapeutic target for NAFLD
(102) and the upregulation of
S100A11 may be amarker of the transition from steatosis to
NASH/fibrosis (104).
The treatment of HCC is a hot topic of research.
Treatment outcomes remain poor when progressing to end-stage HCC.
In particular, the prognosis remains poor for patients with
recurrence or distant metastasis (117). Among the S100 protein family,
S100A1, S100A3, S100A4, S100A6, S100A9, S100A10, S100A11 and
S100A12 are upregulated in HCC and may be potential therapeutic
targets and/or markers for HCC (30,118-121,138,141,147,150,152). Among these, high expression of
S100A1 can be used as a predictive marker for poor prognosis of HCC
(118). Furthermore, S100A4 can
be used as a therapeutic target and a useful indicator of tumor
aggressiveness and prognosis (30). In addition, S100A6 can be used as
an important marker for HCC (139), while S100A9 can be used as a
marker of HCC metastasis (144). Huang et al (156) revealed that assessment of
urinary S100A9 and granulocyte protein levels may be helpful in the
diagnosis of early HCC. Moreover, S100A9 may be a potential target
for the treatment of HCC (145). In a recent retrospective study,
Meng et al (157)
examined patients with HCC (n=379) who underwent radical resection
and found that elevated serum S100A9 levels were associated with
poor prognosis, suggesting that S100A9 may be a prognostic
indicator following HCC resection. In HepG2 cells, S100A10 inhibits
proliferation by upregulating miR-590-5p (146). At the cellular level, LINC00174
is be associated with the development of HCC and its downstream
genes included S100A10 and miR-320 (147). Thus, S100A10 may be a
therapeutic target and biomarker for HCC. Similarly, S100A11 and
S100A12 may be potential targets and biomarkers for immunotherapy
for HCC (104,150,152).
In the study of S100 protein as a therapeutic
target, sodium cantharidinate inhibits expression of S100A3 in
HepG2 cells and suppresses cell viability (119). Zhang et al (158) reported that decreased tumor
incidence and growth following anti-miR-21 treatment in a mouse
model may be associated with decreased fibrosis and high
consumption of S100A4. Moreover, decreased expression of S100A4
following anti-miR-21 treatment suggests that anti-miR-21 may be
effective in tumor-targeted therapy, but further studies are
required to determine whether anti-miR-21 treatment is effective as
an adjuvant therapy to drugs that kill tumor cells. A study by Jiao
et al (159) using a
mouse model of HCC treated with ganciclovir injection, reported a
decrease in incidence of HCC and tumor size in model mice compared
with controls, but this change was not statistically significant.
Therefore, S100A4 as a target for treatment of HCC requires further
investigation. At present, there are only therapeutic approaches
targeting S100A3 and S100A4 (158,159) and additional experiments are
needed to target S100 protein for the treatment of HCC.
In numerous types of liver disease, especially
liver fibrosis and HCC, S100 protein is highly upregulated and
promotes the development and progression of liver disease. Both
S100A4 and S100A6 promote the development and progression of liver
fibrosis by activating HSC and may be potential therapeutic targets
and markers for liver fibrosis (59,86). S100A1, S 100A3, S100A4, S100A6
S100A9, S100A10, S100A11, and S100A12 are all upregulated during
HCC and may be potential targets and/or markers of HCC (30, 118-121,138,141,147,150,152). However, the mechanisms of
action remain unknown and further research is required. In mouse
experiments, inhibition of S100A4 expression decreases
tumorigenesis and development. Moreover, inhibition of S100A10 at
the cellular level inhibits proliferation. In conclusion, S100
protein is a promising target and potential marker for liver
disease treatment in future. However, drugs targeting S100 protein
need to be examined further in animal experiments and clinical
studies.
Not applicable.
SY and XY made substantial contributions to the
conception and design of the study. JA, HJ, GW, HW and BT were
involved in revising the manuscript critically for important
intellectual content. Data authentication is not applicable. All
authors read and approved the final manuscript.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
Not applicable.
|
1
|
Wakim KG: Physiology of the liver. Am J
Med. 16:256–271. 1954. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lee S, Mardinoglu A, Zhang C, Lee D and
Nielsen J: Dysregulated signaling hubs of liver lipid metabolism
reveal hepatocellular carcinoma pathogenesis. Nucleic Acids Res.
44:5529–5539. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Han HS, Kang G, Kim JS, Choi BH and Koo
SH: Regulation of glucose metabolism from a liver-centric
perspective. Exp Mol Med. 48:e2182016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wang Y, Song L, Liu M, Ge R, Zhou Q, Liu
W, Li R, Qie J, Zhen B, Wang Y, et al: A proteomics landscape of
circadian clock in mouse liver. Nat Commun. 9:15532018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Evarts RP, Hu Z, Fujio K, Marsden ER and
Thorgeirsson SS: Activation of hepatic stem cell compartment in the
rat: Role of transforming growth factor alpha, hepatocyte growth
factor, and acidic fibroblast growth factor in early proliferation.
Cell Growth Differ. 4:555–561. 1993.PubMed/NCBI
|
|
6
|
Kang JH, Toita R and Murata M: Liver
cell-targeted delivery of therapeutic molecules. Crit Rev
Biotechnol. 36:132–143. 2016. View Article : Google Scholar
|
|
7
|
Gao B: Hepatoprotective and
anti-inflammatory cytokines in alcoholic liver disease. J
Gastroenterol Hepatol. 27(Suppl 2): S89–S93. 2012. View Article : Google Scholar
|
|
8
|
Asrani SK, Devarbhavi H, Eaton J and
Kamath PS: Burden of liver diseases in the world. J Hepatol.
70:151–171. 2019. View Article : Google Scholar
|
|
9
|
GBD 2017 Disease and Injury Incidence and
Prevalence Collaborators: Global, regional, and national incidence,
prevalence, and years lived with disability for 354 diseases and
injuries for 195 countries and territories, 1990-2017: A systematic
analysis for the Global Burden of Disease Study 2017. Lancet.
392:1789–1858. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Paik JM, Golabi P, Younossi Y, Mishra A
and Younossi ZM: Changes in the Global Burden of Chronic Liver
Diseases From 2012 to 2017: The Growing Impact of NAFLD.
Hepatology. 72:1605–1616. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang FS, Fan JG, Zhang Z, Gao B and Wang
HY: The global burden of liver disease: The major impact of China.
Hepatology. 60:2099–2108. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cooke GS, Andrieux-Meyer I, Applegate TL,
Atun R, Burry JR, Cheinquer H, Dusheiko G, Feld JJ, Gore C,
Griswold MG, et al: Accelerating the elimination of viral
hepatitis: A Lancet Gastroenterology & Hepatology Commission.
Lancet Gastroenterol Hepatol. 4:135–184. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Xiao J, Wang F, Wong NK, He J, Zhang R,
Sun R, Xu Y, Liu Y, Li W, Koike K, et al: Global liver disease
burdens and research trends: Analysis from a Chinese perspective. J
Hepatol. 71:212–221. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Avila MA, Dufour JF, Gerbes AL, Zoulim F,
Bataller R, Burra P, Cortez-Pinto H, Gao B, Gilmore I, Mathurin P,
et al: Recent advances in alcohol-related liver disease (ALD):
Summary of a Gut round table meeting. Gut. 69:764–780. 2020.
View Article : Google Scholar
|
|
15
|
Younossi ZM, Koenig AB, Abdelatif D, Fazel
Y, Henry L and Wymer M: Global epidemiology of nonalcoholic fatty
liver disease-Meta-analytic assessment of prevalence, incidence,
and outcomes. Hepatology. 64:73–84. 2016. View Article : Google Scholar
|
|
16
|
Donato R, Cannon BR, Sorci G, Riuzzi F,
Hsu K, Weber DJ and Geczy CL: Functions of S100 proteins. Curr Mol
Med. 13:24–57. 2013. View Article : Google Scholar :
|
|
17
|
Chan JK, Roth J, Oppenheim JJ, Tracey KJ,
Vogl T, Feldmann M, Horwood N and Nanchahal J: Alarmins: Awaiting a
clinical response. J Clin Invest. 122:2711–2719. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kraus C, Rohde D, Weidenhammer C, Qiu G,
Pleger ST, Voelkers M, Boerries M, Remppis A, Katus HA and Most P:
S100A1 in cardiovascular health and disease: Closing the gap
between basic science and clinical therapy. J Mol Cell Cardiol.
47:445–455. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bresnick AR, Weber DJ and Zimmer DB: S100
proteins in cancer. Nat Rev Cancer. 15:96–109. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Cristóvão JS and Gomes CM: S100 proteins
in Alzheimer's disease. Front Neurosci. 13:4632019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Austermann J, Spiekermann C and Roth J:
S100 proteins in rheumatic diseases. Nat Rev Rheumatol. 14:528–541.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Foell D, Kucharzik T, Kraft M, Vogl T,
Sorg C, Domschke W and Roth J: Neutrophil derived human S100A12
(EN-RAGE) is strongly expressed during chronic active inflammatory
bowel disease. Gut. 52:847–853. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yang Z, Tao T, Raftery MJ, Youssef P, Di
Girolamo N and Geczy CL: Proinflammatory properties of the human
S100 protein S100A12. J Leukoc Biol. 69:986–994. 2001.PubMed/NCBI
|
|
24
|
Turnier JL, Fall N, Thornton S, Witte D,
Bennett MR, Appenzeller S, Klein-Gitelman MS, Grom AA and Brunner
HI: Urine S100 proteins as potential biomarkers of lupus nephritis
activity. Arthritis Res Ther. 19:2422017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ren ZG, Zhao JD, Gu K, Wang J and Jiang
GL: Hepatic proliferation after partial liver irradiation in
Sprague-Dawley rats. Mol Biol Rep. 39:3829–3836. 2012. View Article : Google Scholar
|
|
26
|
Dixon LJ, Barnes M, Tang H, Pritchard MT
and Nagy LE: Kupffer cells in the liver. Compr Physiol. 3:785–797.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chen L, Hu X, Wu H, Jia Y, Liu J, Mu X, Wu
H and Zhao Y: Over-expression of S100B protein as a serum marker of
brain metastasis in non-small cell lung cancer and its prognostic
value. Pathol Res Pract. 215:427–432. 2019. View Article : Google Scholar
|
|
28
|
Yan XL, Jia YL, Chen L, Zeng Q, Zhou JN,
Fu CJ, Chen HX, Yuan HF, Li ZW, Shi L, et al: Hepatocellular
carcinoma-associated mesenchymal stem cells promote hepatocarcinoma
progression: Role of the S100A4-miR155-SOCS1-MMP9 axis. Hepatology.
57:2274–2286. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Maletzki C, Bodammer P, Breitrück A and
Kerkhoff C: S100 proteins as diagnostic and prognostic markers in
colorectal and hepatocellular carcinoma. Hepat Mon.
12:e72402012.PubMed/NCBI
|
|
30
|
Liu Z, Liu H, Pan H, Du Q and Liang J:
Clinicopathological significance of S100A4 expression in human
hepatocellular carcinoma. J Int Med Res. 41:457–462. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Moore BW: A soluble protein characteristic
of the nervous system. Biochem Biophys Res Commun. 19:739–744.
1965. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Donato R: S100: A multigenic family of
calcium-modulated proteins of the EF-hand type with intracellular
and extracellular functional roles. Int J Biochem Cell Biol.
33:637–668. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Marenholz I, Heizmann CW and Fritz G: S100
proteins in mouse and man: From evolution to function and pathology
(including an update of the nomenclature). Biochem Biophys Res
Commun. 322:1111–1122. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Chow KH, Park HJ, George J, Yamamoto K,
Gallup AD, Graber JH, Chen Y, Jiang W, Steindler DA, Neilson EG, et
al: S100A4 is a biomarker and regulator of glioma stem cells that
is critical for mesenchymal transition in glioblastoma. Cancer Res.
77:5360–5373. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Dahlmann M, Kobelt D, Walther W, Mudduluru
G and Stein U: S100A4 in cancer metastasis: Wnt signaling-driven
interventions for metastasis restriction. Cancers (Basel).
8:592016. View Article : Google Scholar
|
|
36
|
Donato R: Intracellular and extracellular
roles of S100 proteins. Microsc Res Tech. 60:540–551. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Österreicher CH, Penz-Österreicher M,
Grivennikov SI, Guma M, Koltsova EK, Datz C, Sasik R, Hardiman G,
Karin M and Brenner DA: Fibroblast-specific protein 1 identifies an
inflammatory subpopulation of macrophages in the liver. Proc Natl
Acad Sci USA. 108:308–313. 2011. View Article : Google Scholar :
|
|
38
|
Zhang J, Chen L, Liu X, Kammertoens T,
Blankenstein T and Qin Z: Fibroblast-specific protein
1/S100A4-positive cells prevent carcinoma through collagen
production and encapsulation of carcinogens. Cancer Res.
73:2770–2781. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhang J, Chen L, Xiao M, Wang C and Qin Z:
FSP1+ fibroblasts promote skin carcinogenesis by
maintaining MCP-1-mediated macrophage infiltration and chronic
inflammation. Am J Pathol. 178:382–390. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Kuźnicki J, Kordowska J, Puzianowska M and
Woźniewicz BM: Calcyclin as a marker of human epithelial cells and
fibroblasts. Exp Cell Res. 200:425–430. 1992. View Article : Google Scholar
|
|
41
|
Markowitz J and Carson WE III: Review of
S100A9 biology and its role in cancer. Biochim Biophys Acta.
1835:100–109. 2013.
|
|
42
|
Saiki Y and Horii A: Multiple functions of
S100A10, an important cancer promoter. Pathol Int. 69:629–636.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
He H, Li J, Weng S, Li M and Yu Y:
S100A11: Diverse function and pathology corresponding to different
target proteins. Cell Biochem Biophys. 55:117–126. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Guignard F, Mauel J and Markert M:
Identification and characterization of a novel human neutrophil
protein related to the S100 family. Biochem J. 309:395–401. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Bagheri V: S100A12: Friend or foe in
pulmonary tuberculosis? Cytokine. 92:80–82. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Donato R, Sorci G, Riuzzi F, Arcuri C,
Bianchi R, Brozzi F, Tubaro C and Giambanco I: S100B's double life:
Intracellular regulator and extracellular signal. Biochim Biophys
Acta. 1793:1008–1022. 2009. View Article : Google Scholar
|
|
47
|
Santamaria-Kisiel L, Rintala-Dempsey AC
and Shaw GS: Calcium-dependent and -independent interactions of the
S100 protein family. Biochem J. 396:201–214. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Goyette J and Geczy CL:
Inflammation-associated S100 proteins: New mechanisms that regulate
function. Amino Acids. 41:821–842. 2011. View Article : Google Scholar
|
|
49
|
Pruenster M, Vogl T, Roth J and Sperandio
M: S100A8/A9: From basic science to clinical application. Pharmacol
Ther. 167:120–131. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Lim SY, Raftery MJ and Geczy CL: Oxidative
modifications of DAMPs suppress inflammation: The case for S100A8
and S100A9. Antioxid Redox Signal. 15:2235–2248. 2011. View Article : Google Scholar
|
|
51
|
Averill MM, Kerkhoff C and Bornfeldt KE:
S100A8 and S100A9 in cardiovascular biology and disease.
Arterioscler Thromb Vasc Biol. 32:223–229. 2012. View Article : Google Scholar
|
|
52
|
Gross SR, Sin CG, Barraclough R and
Rudland PS: Joining S100 proteins and migration: For better or for
worse, in sickness and in health. Cell Mol Life Sci. 71:1551–1579.
2014. View Article : Google Scholar
|
|
53
|
Donato R, Sorci G and Giambanco I: S100A6
protein: Functional roles. Cell Mol Life Sci. 74:2749–2760. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Riuzzi F, Sorci G, Arcuri C, Giambanco I,
Bellezza I, Minelli A and Donato R: Cellular and molecular
mechanisms of sarcopenia: The S100B perspective. J Cachexia
Sarcopenia Muscle. 9:1255–1268. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Wang S, Song R, Wang Z, Jing Z, Wang S and
Ma J: S100A8/A9 in inflammation. Front Immunol. 9:12982018.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Sun H, Zhao A, Li M, Dong H, Sun Y, Zhang
X, Zhu Q, Bukhari A, Cao C, Su D, et al: Interaction of calcium
binding protein S100A16 with myosin-9 promotes cytoskeleton
reorganization in renal tubulointerstitial fibrosis. Cell Death
Dis. 11:1462020. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Akiyama N, Hozumi H, Isayama T, Okada J,
Sugiura K, Yasui H, Suzuki Y, Kono M, Karayama M, Furuhashi K, et
al: Clinical significance of serum S100 calcium-binding protein A4
in idiopathic pulmonary fibrosis. Respirology. 25:743–749. 2020.
View Article : Google Scholar
|
|
58
|
Zhong A, Xu W, Zhao J, Xie P, Jia S, Sun
J, Galiano RD, Mustoe TA and Hong SJ: S100A8 and S100A9 are induced
by decreased hydration in the epidermis and promote fibroblast
activation and fibrosis in the dermis. Am J Pathol. 186:109–122.
2016. View Article : Google Scholar
|
|
59
|
Chen L, Li J, Zhang J, Dai C, Liu X, Wang
J, Gao Z, Guo H, Wang R, Lu S, et al: S100A4 promotes liver
fibrosis via activation of hepatic stellate cells. J Hepatol.
62:156–164. 2015. View Article : Google Scholar
|
|
60
|
Cancemi P, Buttacavoli M, Di Cara G,
Albanese NN, Bivona S, Pucci-Minafra I and Feo S: A multiomics
analysis of S100 protein family in breast cancer. Oncotarget.
9:29064–29081. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Liu Y, Cui J, Tang YL, Huang L, Zhou CY
and Xu JX: Prognostic roles of mRNA expression of S100 in
non-small-cell lung cancer. Biomed Res Int.
2018:98158062018.PubMed/NCBI
|
|
62
|
Moravkova P, Kohoutova D, Rejchrt S,
Cyrany J and Bures J: Role of S100 proteins in colorectal
carcinogenesis. Gastroenterol Res Pract. 2016:26327032016.
View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Fang D, Zhang C, Xu P, Liu Y, Mo X, Sun Q,
Abdelatty A, Hu C, Xu H, Zhou G, et al: S100A16 promotes metastasis
and progression of pancreatic cancer through FGF19-mediated AKT and
ERK1/2 pathways. Cell Biol Toxicol. Jan 2–2021.Epub ahead of print.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Liu Y, Luo G and He D: Clinical importance
of S100A9 in osteosarcoma development and as a diagnostic marker
and therapeutic target. Bioengineered. 10:133–141. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Wang T, Huo X, Chong Z, Khan H, Liu R and
Wang T: A review of S100 protein family in lung cancer. Clin Chim
Acta. 476:54–59. 2018. View Article : Google Scholar
|
|
66
|
Destek S and Gul VO: S100A4 may be a good
prognostic marker and a therapeutic target for colon cancer. J
Oncol. 2018:18287912018. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Sun X, Wang T, Zhang C, Ning K, Guan ZR,
Chen SX, Hong TT and Hua D: S100A16 is a prognostic marker for
colorectal cancer. J Surg Oncol. 117:275–283. 2018. View Article : Google Scholar
|
|
68
|
Yuan W, Goldstein LD, Durinck S, Chen YJ,
Nguyen TT, Kljavin NM, Sokol ES, Stawiski EW, Haley B, Ziai J, et
al: S100a4 upregulation in Pik3caH1047R;Trp53R270H;MMTV-C re-driven
mammary tumors promotes metastasis. Breast Cancer Res. 21:1522019.
View Article : Google Scholar
|
|
69
|
Louka ML and Ramzy MM: Involvement of
fibroblast-specific protein 1 (S100A4) and matrix
metalloproteinase-13 (MMP-13) in CCl4-induced reversible liver
fibrosis. Gene. 579:29–33. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Zadorozhna M, Di Gioia S, Conese M and
Mangieri D: Neovascularization is a key feature of liver fibrosis
progression: Anti-angiogenesis as an innovative way of liver
fibrosis treatment. Mol Biol Rep. 47:2279–2288. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Hernandez-Gea V and Friedman SL:
Pathogenesis of liver fibrosis. Annu Rev Pathol. 6:425–456. 2011.
View Article : Google Scholar
|
|
72
|
Pellicoro A, Ramachandran P, Iredale JP
and Fallowfield JA: Liver fibrosis and repair: Immune regulation of
wound healing in a solid organ. Nat Rev Immunol. 14:181–194. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Song LJ, Yin XR, Mu SS, Li JH, Gao H,
Zhang Y, Dong PP, Mei CJ and Hua ZC: The differential and dynamic
progression of hepatic inflammation and immune responses during
liver fibrosis induced by Schistosoma japonicum or carbon
tetrachloride in mice. Front Immunol. 11:5705242020. View Article : Google Scholar :
|
|
74
|
Parola M and Pinzani M: Liver fibrosis:
Pathophysiology, pathogenetic targets and clinical issues. Mol
Aspects Med. 65:37–55. 2019. View Article : Google Scholar
|
|
75
|
Zhang CY, Yuan WG, He P, Lei JH and Wang
CX: Liver fibrosis and hepatic stellate cells: Etiology,
pathological hallmarks and therapeutic targets. World J
Gastroenterol. 22:10512–10522. 2016. View Article : Google Scholar
|
|
76
|
Blomhoff R, Rasmussen M, Nilsson A, Norum
KR, Berg T, Blaner WS, Kato M, Mertz JR, Goodman DS, Eriksson U, et
al: Hepatic retinol metabolism. Distribution of retinoids, enzymes,
and binding proteins in isolated rat liver cells. J Biol Chem.
260:13560–13565. 1985. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Li D, He L, Guo H, Chen H and Shan H:
Targeting activated hepatic stellate cells (aHSCs) for liver
fibrosis imaging. EJNMMI Res. 5:712015. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Puche JE, Saiman Y and Friedman SL:
Hepatic stellate cells and liver fibrosis. Compr Physiol.
3:1473–1492. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Henderson NC and Iredale JP: Liver
fibrosis: Cellular mechanisms of progression and resolution. Clin
Sci (Lond). 112:265–280. 2007. View Article : Google Scholar
|
|
80
|
Kisseleva T, Cong M, Paik Y, Scholten D,
Jiang C, Benner C, Iwaisako K, Moore-Morris T, Scott B, Tsukamoto
H, et al: Myofibroblasts revert to an inactive phenotype during
regression of liver fibrosis. Proc Natl Acad Sci USA.
109:9448–9453. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Iredale JP, Thompson A and Henderson NC:
Extracellular matrix degradation in liver fibrosis: Biochemistry
and regulation. Biochim Biophys Acta. 1832:876–883. 2013.
View Article : Google Scholar
|
|
82
|
Schaefer B, Rivas-Estilla AM, Meraz-Cruz
N, Reyes-Romero MA, Hernández-Nazara ZH, Domínguez-Rosales JA,
Schuppan D, Greenwel P and Rojkind M: Reciprocal modulation of
matrix metalloproteinase-13 and type I collagen genes in rat
hepatic stellate cells. Am J Pathol. 162:1771–1780. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Uchinami H, Seki E, Brenner DA and
D'Armiento J: Loss of MMP 13 attenuates murine hepatic injury and
fibrosis during cholestasis. Hepatology. 44:420–429. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Miranda KJ, Loeser RF and Yammani RR:
Sumoylation and nuclear translocation of S100A4 regulate
IL-1beta-mediated production of matrix metalloproteinase-13. J Biol
Chem. 285:31517–31524. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Yammani RR, Carlson CS, Bresnick AR and
Loeser RF: Increase in production of matrix metalloproteinase 13 by
human articular chondrocytes due to stimulation with S100A4: Role
of the receptor for advanced glycation end products. Arthritis
Rheum. 54:2901–2911. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Xia P, He H, Kristine MS, Guan W, Gao J,
Wang Z, Hu J, Han L, Li J, Han W and Yu Y: Therapeutic effects of
recombinant human S100A6 and soluble receptor for advanced
glycation end products(sRAGE) on CCl(4)-induced liver fibrosis in
mice. Eur J Pharmacol. 833:86–93. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Baik SJ, Kim TH, Yoo K, Moon IH, Choi JY,
Chung KW and Song DE: Decreased S100B expression in chronic liver
diseases. Korean J Intern Med. 32:269–276. 2017. View Article : Google Scholar :
|
|
88
|
Park JW, Kim MJ, Kim SE, Kim HJ, Jeon YC,
Shin HY, Park SJ, Jang MK, Kim DJ, Park CK and Choi EK: Increased
expression of S100B and RAGE in a mouse model of bile duct
ligation-induced liver fibrosis. J Korean Med Sci. 36:e902021.
View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Lanini S, Ustianowski A, Pisapia R, Zumla
A and Ippolito G: Viral hepatitis: Etiology, epidemiology,
transmission, diagnostics, treatment, and prevention. Infect Dis
Clin North Am. 33:1045–1062. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Thomas DL: Global elimination of chronic
hepatitis. N Engl J Med. 380:2041–2050. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Jie Z, Liang Y, Yi P, Tang H, Soong L,
Cong Y, Zhang K and Sun J: Retinoic acid regulates immune responses
by promoting IL-22 and modulating S100 proteins in viral hepatitis.
J Immunol. 198:3448–3460. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Radaeva S, Wang L, Radaev S, Jeong WI,
Park O and Gao B: Retinoic acid signaling sensitizes hepatic
stellate cells to NK cell killing via upregulation of NK cell
activating ligand RAE1. Am J Physiol Gastrointest Liver Physiol.
293:G809–G816. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Lee YS and Jeong WI: Retinoic acids and
hepatic stellate cells in liver disease. J Gastroenterol Hepatol.
27(Suppl 2): S75–S79. 2012. View Article : Google Scholar
|
|
94
|
Yan LB, Zhang QB, Zhu X, He M and Tang H:
Serum S100 calcium binding protein A4 improves the diagnostic
accuracy of transient elastography for assessing liver fibrosis in
hepatitis B. Clin Res Hepatol Gastroenterol. 42:64–71. 2018.
View Article : Google Scholar
|
|
95
|
Wu R, Zhang Y, Xiang Y, Tang Y, Cui F, Cao
J, Zhou L, You Y and Duan L: Association between serum S100A9
levels and liver necroinflammation in chronic hepatitis B. J Transl
Med. 16:832018. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Cai J, Han T, Nie C, Jia X, Liu Y, Zhu Z
and Gao Y: Biomarkers of oxidation stress, inflammation, necrosis
and apoptosis are associated with hepatitis B-related
acute-on-chronic liver failure. Clin Res Hepatol Gastroenterol.
40:41–50. 2016. View Article : Google Scholar
|
|
97
|
Li J, Zou B, Yeo YH, Feng Y, Xie X, Lee
DH, Fujii H, Wu Y, Kam LY, Ji F, et al: Prevalence, incidence, and
outcome of non-alcoholic fatty liver disease in Asia, 1999-2019: A
systematic review and meta-analysis. Lancet Gastroenterol Hepatol.
4:389–398. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Sayiner M, Koenig A, Henry L and Younossi
ZM: Epidemiology of nonalcoholic fatty liver disease and
nonalcoholic steatohepatitis in the united states and the rest of
the world. Clin Liver Dis. 20:205–214. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Afonso MB, Rodrigues PM, Simão AL and
Castro RE: Circulating microRNAs as potential biomarkers in
non-alcoholic fatty liver disease and hepatocellular carcinoma. J
Clin Med. 5:302016. View Article : Google Scholar :
|
|
100
|
Zhang YH, Ma Q, Ding P, Li J, Chen LL, Ao
KJ and Tian YY: S100A4 gene is crucial for
methionine-choline-deficient diet-induced non-alcoholic fatty liver
disease in mice. Yonsei Med J. 59:1064–1071. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Liu X, Wang Y, Ming Y, Song Y, Zhang J,
Chen X, Zeng M and Mao Y: S100A9: A potential biomarker for the
progression of non-alcoholic fatty liver disease and the diagnosis
of non-alcoholic steatohepatitis. PLoS One. 10:e01273522015.
View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Zhang L, Zhang Z, Li C, Zhu T, Gao J, Zhou
H, Zheng Y, Chang Q, Wang M, Wu J, et al: S100A11 promotes liver
steatosis via FOXO1-mediated autophagy and lipogenesis. Cell Mol
Gastroenterol Hepatol. 11:697–724. 2021. View Article : Google Scholar :
|
|
103
|
Ni HM, Chao X and Ding WX: S100A11
overexpression promotes fatty liver diseases via increased
autophagy? Cell Mol Gastroenterol Hepatol. 11:885–886. 2021.
View Article : Google Scholar :
|
|
104
|
Sobolewski C, Abegg D, Berthou F, Dolicka
D, Calo N, Sempoux C, Fournier M, Maeder C, Ay AS, Clavien PA, et
al: S100A11/ANXA2 belongs to a tumour suppressor/oncogene network
deregulated early with steatosis and involved in inflammation and
hepatocellular carcinoma development. Gut. 69:1841–1854. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
105
|
GBD 2016 Causes of Death Collaborators:
Global, regional, and national age-sex specific mortality for 264
causes of death, 1980-2016: A systematic analysis for the Global
Burden of Disease Study 2016. Lancet. 390:1151–1210. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Purohit V, Gao B and Song BJ: Molecular
mechanisms of alcoholic fatty liver. Alcohol Clin Exp Res.
33:191–205. 2009. View Article : Google Scholar :
|
|
107
|
Anstee QM, Daly AK and Day CP: Genetics of
alcoholic and nonalcoholic fatty liver disease. Semin Liver Dis.
31:128–146. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Kwon HJ, Won YS, Park O, Chang B, Duryee
MJ, Thiele GE, Matsumoto A, Singh S, Abdelmegeed MA, Song BJ, et
al: Aldehyde dehydrogenase 2 deficiency ameliorates alcoholic fatty
liver but worsens liver inflammation and fibrosis in mice.
Hepatology. 60:146–157. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Rehm J, Taylor B, Mohapatra S, Irving H,
Baliunas D, Patra J and Roerecke M: Alcohol as a risk factor for
liver cirrhosis: A systematic review and meta-analysis. Drug
Alcohol Rev. 29:437–445. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Roerecke M, Vafaei A, Hasan OSM, Chrystoja
BR, Cruz M, Lee R, Neuman MG and Rehm J: Alcohol consumption and
risk of liver cirrhosis: A systematic review and meta-analysis. Am
J Gastroenterol. 114:1574–1586. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Seitz HK, Bataller R, Cortez-Pinto H, Gao
B, Gual A, Lackner C, Mathurin P, Mueller S, Szabo G and Tsukamoto
H: Alcoholic liver disease. Nat Rev Dis Primers. 4:162018.
View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Yuan Q, Hou S, Zhai J, Tian T, Wu Y, Wu Z,
He J, Chen Z and Zhang J: S100A4 promotes inflammation but
suppresses lipid accumulation via the STAT3 pathway in chronic
ethanol-induced fatty liver. J Mol Med (Berl). 97:1399–1412. 2019.
View Article : Google Scholar
|
|
113
|
Llovet JM, Kelley RK, Villanueva A, Singal
AG, Pikarsky E, Roayaie S, Lencioni R, Koike K, Zucman-Rossi J and
Finn RS: Hepatocellular carcinoma. Nat Rev Dis Primers. 7:62021.
View Article : Google Scholar : PubMed/NCBI
|
|
114
|
International Agency for Research on
Cancer: Liver: GLOBOCAN. 2020, https://gco.iarc.fr/today/data/factsheets/cancers/11-Liver-fact-sheet.pdf.
|
|
115
|
Chen Z, Xie H, Hu M, Huang T, Hu Y, Sang N
and Zhao Y: Recent progress in treatment of hepatocellular
carcinoma. Am J Cancer Res. 10:2993–3036. 2020.PubMed/NCBI
|
|
116
|
Allemani C, Matsuda T, Di Carlo V,
Harewood R, Matz M, Nikšić M, Bonaventure A, Valkov M, Johnson CJ,
Estève J, et al: Global surveillance of trends in cancer survival
2000-14 (CONCORD-3): Analysis of individual records for 37 513 025
patients diagnosed with one of 18 cancers from 322 population-based
registries in 71 countries. Lancet. 391:1023–1075. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Goyal L, Muzumdar MD and Zhu AX: Targeting
the HGF/c-MET pathway in hepatocellular carcinoma. Clin Cancer Res.
19:2310–2318. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Guo Q, Wang J, Cao Z, Tang Y, Feng C and
Huang F: Interaction of S100A1 with LATS1 promotes cell growth
through regulation of the Hippo pathway in hepatocellular
carcinoma. Int J Oncol. 53:592–602. 2018.PubMed/NCBI
|
|
119
|
Tao R, Wang ZF, Qiu W, He YF, Yan WQ, Sun
WY and Li HJ: Role of S100A3 in human hepatocellular carcinoma and
the anticancer effect of sodium cantharidinate. Exp Ther Med.
13:2812–2818. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Zhang J, Zhang DL, Jiao XL and Dong Q:
S100A4 regulates migration and invasion in hepatocellular carcinoma
HepG2 cells via NF-κB-dependent MMP-9 signal. Eur Rev Med Pharmacol
Sci. 17:2372–2382. 2013.PubMed/NCBI
|
|
121
|
Zhai X, Zhu H, Wang W, Zhang S, Zhang Y
and Mao G: Abnormal expression of EMT-related proteins, S100A4,
vimentin and E-cadherin, is correlated with clinicopathological
features and prognosis in HCC. Med Oncol. 31:9702014. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Zhu K, Huang W, Wang W, Liao L, Li S, Yang
S, Xu J, Li L, Meng M, Xie Y, et al: Up-regulation of S100A4
expression by HBx protein promotes proliferation of hepatocellular
carcinoma cells and its correlation with clinical survival. Gene.
749:1446792020. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Hanahan D and Coussens LM: Accessories to
the crime: Functions of cells recruited to the tumor
microenvironment. Cancer Cell. 21:309–322. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Malanchi I, Santamaria-Martínez A, Susanto
E, Peng H, Lehr HA, Delaloye JF and Huelsken J: Interactions
between cancer stem cells and their niche govern metastatic
colonization. Nature. 481:85–89. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Tsai KS, Yang SH, Lei YP, Tsai CC, Chen
HW, Hsu CY, Chen LL, Wang HW, Miller SA, Chiou SH, et al:
Mesenchymal stem cells promote formation of colorectal tumors in
mice. Gastroenterology. 141:1046–1056. 2011. View Article : Google Scholar
|
|
126
|
Karnoub AE, Dash AB, Vo AP, Sullivan A,
Brooks MW, Bell GW, Richardson AL, Polyak K, Tubo R and Weinberg
RA: Mesenchymal stem cells within tumour stroma promote breast
cancer metastasis. Nature. 449:557–563. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Huang S and He X: The role of microRNAs in
liver cancer progression. Br J Cancer. 104:235–240. 2011.
View Article : Google Scholar :
|
|
128
|
Gallardo M, Kemmerling U, Aguayo F, Bleak
TC, Muñoz JP and Calaf GM: Curcumin rescues breast cells from
epithelial-mesenchymal transition and invasion induced by
anti-miR-34a. Int J Oncol. 56:480–493. 2020.
|
|
129
|
Datta J, Islam M, Dutta S, Roy S, Pan Q
and Teknos TN: Suberoylanilide hydroxamic acid inhibits growth of
head and neck cancer cell lines by reactivation of tumor suppressor
microRNAs. Oral Oncol. 56:32–39. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Wang B, Majumder S, Nuovo G, Kutay H,
Volinia S, Patel T, Schmittgen TD, Croce C, Ghoshal K and Jacob ST:
Role of microRNA-155 at early stages of hepatocarcinogenesis
induced by choline-deficient and amino acid-defined diet in C57BL/6
mice. Hepatology. 50:1152–1161. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Xie Q, Chen X, Lu F, Zhang T, Hao M, Wang
Y, Zhao J, McCrae MA and Zhuang H: Aberrant expression of microRNA
155 may accelerate cell proliferation by targeting sex-determining
region Y box 6 in hepatocellular carcinoma. Cancer. 118:2431–2442.
2012. View Article : Google Scholar
|
|
132
|
Chen G, Wang D, Zhao X, Cao J, Zhao Y,
Wang F, Bai J, Luo D and Li L: miR-155-5p modulates malignant
behaviors of hepatocellular carcinoma by directly targeting CTHRC1
and indirectly regulating GSK-3β-involved Wnt/β-catenin signaling.
Cancer Cell Int. 17:1182017. View Article : Google Scholar
|
|
133
|
Reya T, Morrison SJ, Clarke MF and
Weissman IL: Stem cells, cancer, and cancer stem cells. Nature.
414:105–111. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Li Y, Wang J, Song K, Liu S, Zhang H, Wang
F, Ni C, Zhai W, Liang J, Qin Z and Zhang J: S100A4 promotes
hepatocellular carcinogenesis by intensifying fibrosis-associated
cancer cell stemness. Oncoimmunology. 9:17253552020. View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Kim J, Kim J, Yoon S, Joo J, Lee Y, Lee K,
Chung J and Choe I: S100A6 protein as a marker for differential
diagnosis of cholangiocarcinoma from hepatocellular carcinoma.
Hepatol Res. 23:2742002. View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Hua Z, Chen J, Sun B, Zhao G, Zhang Y,
Fong Y, Jia Z and Yao L: Specific expression of osteopontin and
S100A6 in hepatocellular carcinoma. Surgery. 149:783–791. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Tong A, Gou L, Lau QC, Chen B, Zhao X, Li
J, Tang H, Chen L, Tang M, Huang C and Wei YQ: Proteomic profiling
identifies aberrant epigenetic modifications induced by hepatitis B
virus X protein. J Proteome Res. 8:1037–1046. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
138
|
Li Z, Tang M, Ling B, Liu S, Zheng Y, Nie
C, Yuan Z, Zhou L, Guo G, Tong A and Wei Y: Increased expression of
S100A6 promotes cell proliferation and migration in human
hepatocellular carcinoma. J Mol Med (Berl). 92:291–303. 2014.
View Article : Google Scholar
|
|
139
|
Song D, Xu B, Shi D, Li S and Cai Y:
S100A6 promotes proliferation and migration of HepG2 cells via
increased ubiquitin-dependent degradation of p53. Open Med (Wars).
15:317–326. 2020. View Article : Google Scholar
|
|
140
|
Arai K, Yamada T and Nozawa R:
Immunohistochemical investigation of migration inhibitory
factor-related protein (MRP)-14 expression in hepatocellular
carcinoma. Med Oncol. 17:183–188. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Németh J, Stein I, Haag D, Riehl A,
Longerich T, Horwitz E, Breuhahn K, Gebhardt C, Schirmacher P, Hahn
M, et al: S100A8 and S100A9 are novel nuclear factor kappa B target
genes during malignant progression of murine and human liver
carcinogenesis. Hepatology. 50:1251–1262. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
142
|
Wu R, Duan L, Ye L, Wang H, Yang X, Zhang
Y, Chen X, Zhang Y, Weng Y, Luo J, et al: S100A9 promotes the
proliferation and invasion of HepG2 hepatocellular carcinoma cells
via the activation of the MAPK signaling pathway. Int J Oncol.
42:1001–1010. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
143
|
Wu R, Duan L, Cui F, Cao J, Xiang Y, Tang
Y and Zhou L: S100A9 promotes human hepatocellular carcinoma cell
growth and invasion through RAGE-mediated ERK1/2 and p38 MAPK
pathways. Exp Cell Res. 334:228–238. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
144
|
Duan L, Wu R, Zhang X, Wang D, You Y,
Zhang Y, Zhou L and Chen W: HBx-induced S100A9 in NF-κB dependent
manner promotes growth and metastasis of hepatocellular carcinoma
cells. Cell Death Dis. 9:6292018. View Article : Google Scholar
|
|
145
|
Wei R, Zhu WW, Yu GY, Wang X, Gao C, Zhou
X, Lin ZF, Shao WQ, Wang SH, Lu M and Qin LX: S100 calcium-binding
protein A9 from tumor-associated macrophage enhances cancer stem
cell-like properties of hepatocellular carcinoma. Int J Cancer.
148:1233–1244. 2021. View Article : Google Scholar
|
|
146
|
Shan X, Miao Y, Fan R, Qian H, Chen P, Liu
H, Yan X, Li J and Zhou F: MiR-590-5P inhibits growth of HepG2
cells via decrease of S100A10 expression and Inhibition of the Wnt
pathway. Int J Mol Sci. 14:8556–8569. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
147
|
Zhao JT, Chi BJ, Sun Y, Chi NN, Zhang XM,
Sun JB, Chen Y and Xia Y: LINC00174 is an oncogenic lncRNA of
hepatocellular carcinoma and regulates miR-320/S100A10 axis. Cell
Biochem Funct. 38:859–869. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
148
|
Takeuchi K and Ito F: EGF receptor in
relation to tumor development: Molecular basis of responsiveness of
cancer cells to EGFR-targeting tyrosine kinase inhibitors. FEBS J.
277:316–326. 2010. View Article : Google Scholar
|
|
149
|
Wheeler SE, Suzuki S, Thomas SM, Sen M,
Leeman-Neill RJ, Chiosea SI, Kuan CT, Bigner DD, Gooding WE, Lai SY
and Grandis JR: Epidermal growth factor receptor variant III
mediates head and neck cancer cell invasion via STAT3 activation.
Oncogene. 29:5135–5145. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
150
|
Luo X, Xie H, Long X, Zhou M, Xu Z, Shi B,
Jiang H and Li Z: EGFRvIII mediates hepatocellular carcinoma cell
invasion by promoting S100 calcium binding protein A11 expression.
PLoS One. 8:e833322013. View Article : Google Scholar :
|
|
151
|
Mitsui Y, Tomonobu N, Watanabe M,
Kinoshita R, Sumardika IW, Youyi C, Murata H, Yamamoto KI, Sadahira
T, Rodrigo AGH, et al: Upregulation of mobility in pancreatic
cancer cells by secreted S100A11 through activation of surrounding
fibroblasts. Oncol Res. 27:945–956. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
152
|
Cai H, Ye BG, Ao JY, Zhu XD, Zhang YY,
Chai ZT, Wang CH and Sun HC: High expression of S100A12 on
intratumoral stroma cells indicates poor prognosis following
surgical resection of hepatocellular carcinoma. Oncol Lett.
16:5398–5404. 2018.PubMed/NCBI
|
|
153
|
Shen H, Wu H, Sun F, Qi J and Zhu Q: A
novel four-gene of iron metabolism-related and methylated for
prognosis prediction of hepatocellular carcinoma. Bioengineered.
12:240–251. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
154
|
Sun XJ, Wang MC, Zhang FH and Kong X: An
integrated analysis of genome-wide DNA methylation and gene
expression data in hepatocellular carcinoma. FEBS Open Bio.
8:1093–1103. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
155
|
Schuppan D and Pinzani M: Anti-fibrotic
therapy: Lost in translation? J Hepatol. 56(Suppl 1): S66–74. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
156
|
Huang CH, Kuo CJ, Liang SS, Chi SW, Hsi E,
Chen CC, Lee KT and Chiou SH: Onco-proteogenomics identifies
urinary S100A9 and GRN as potential combinatorial biomarkers for
early diagnosis of hepatocellular carcinoma. BBA Clin. 3:205–213.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
157
|
Meng J, Gu F, Fang H and Qu B: Elevated
serum S100A9 indicated poor prognosis in hepatocellular carcinoma
after curative resection. J Cancer. 10:408–415. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
158
|
Zhang J, Jiao J, Cermelli S, Muir K, Jung
KH, Zou R, Rashid A, Gagea M, Zabludoff S, Kalluri R and Beretta L:
miR-21 inhibition reduces liver fibrosis and prevents tumor
development by inducing apoptosis of CD24+ progenitor
cells. Cancer Res. 75:1859–1867. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
159
|
Jiao J, González Á, Stevenson HL, Gagea M,
Sugimoto H, Kalluri R and Beretta L: Depletion of
S100A4+ stromal cells does not prevent HCC development
but reduces the stem cell-like phenotype of the tumors. Exp Mol
Med. 50:e4222018. View Article : Google Scholar
|