Introduction
E74-like ETS transcription factor 5 (ELF5), an ETS
transcription factor, has been shown to be expressed in lung
epithelial cells and to function as a crucial regulator of lung
development (1,2). ELF5 regulates the specification and
differentiation of epithelial cells in the fetal lung (1). ELF5 expression was also found to
exhibit a dynamic expression pattern in the developing mouse lung,
where its expression was restricted to distal epithelium during
early development (1,2). By the end of the gestation period,
ELF5 expression is enriched in proximal epithelial cells, although
its expression is lost in the distal epithelium. Overexpression of
ELF5 in the embryonic lung has been shown to disrupt branching
morphogenesis, leading to a block in alveolar and airway
differentiation (1).
An increasing body of evidence has revealed that
ELF5 fulfills a tumor-suppressive role in prostate (3,4),
bladder (5) and ovarian (6) cancer. It was found that ELF5 was
lowly expressed in ovarian cancer tissues and highly expressed in
adjacent tissues, indicating that the low expression of ELF5 may be
closely associated with the occurrence of ovarian cancer (7). Both tumor-promoting and
tumor-suppressive roles for ELF5 have been reported in breast
cancer, and these differential functions may be linked to the
subtype of the disease. ELF5 was reported to function as a tumor
suppressor in basal and mesenchymal triple-negative breast cancer
by preventing interferon-γ signaling-driven immunosuppressive
alterations in the tumor microenvironment (8); nevertheless, another study
demonstrated that an increased expression of ELF5 could drive
metastasis and progression of estrogen receptor-positive
(ER+) breast cancer, rendering it resistant to endocrine
therapy (9).
ELF5 is a key transcriptional effector in the lung
(1,2); however, to the best of the authors'
knowledge, the role of ELF5 in lung cancer has yet to be fully
elucidated. Therefore, the aim of the present study was to
investigate the role of ELF5 in lung cancer. It is shown that the
expression of ELF5 in lung adenocarcinoma was significantly higher
compared with that in corresponding adjacent normal tissues.
Furthermore, an association was identified between ELF5 and lung
adenocarcinoma growth both in vitro and in vivo, and
ELF5 could promote the proliferation of lung adenocarcinoma cells.
Mechanistically, adenomatous polyposis coli 2 (APC2) was found to
be directly transcriptionally regulated by ELF5 during the
regulation of lung cancer cells. Taken together, these findings
provide new insights into the functions of ELF5, and its role in
lung adenocarcinoma.
Materials and methods
Tissue microarray analysis
A human lung adenocarcinoma tissue microarray (cat.
no. HLugA150CS03) that included 72 formalin-fixed,
paraffin-embedded lung adenocarcinoma tissues and their
corresponding adjacent lung tissues was randomly selected from the
National Engineering Center for Biochip at Shanghai (Shanghai Outdo
Biotech Co., Ltd.). The clinical characteristics of patients,
including age, sex, histological grade and tumor-node-metastasis
(TNM) stage, were obtained from the medical records of the
patients, with certain missing clinical data excluded. Ethics
approval (approval no. 81402373) was granted by the Human Research
Ethics Committee of Taizhou Hospital of Zhejiang Province (Taizhou,
China), and all patients or their next of kin provided their
informed consent prior to the study. The present study was
conducted in accordance with the principles and guidelines of The
Declaration of Helsinki.
Immunohistochemical (IHC) analysis
Tissues were fixed in 4% paraformaldehyde at room
temperature for 24 h or 10% formalin, embedded in paraffin, and
3-µm sections were cut for IHC analysis. Antigen retrieval
was performed by boiling the tissue slices in sodium citrate buffer
at 100°C for 5 min. The activity of endogenous peroxide was blocked
with 3% hydrogen peroxide for 10 min at room temperature, and 1%
goat serum (cat. no. AR0009; Wuhan Boster Biological Technology,
Ltd.) was applied to the sections at room temperature for 1 h as a
blocking reagent to reduce non-specific binding. The ELF5 (1:100;
cat. no. NHA6603; Novogene Co., Ltd.) and Ki-67 (1:200; cat. no.
9129S; Cell Signaling Technology, Inc.) antibodies were used as
primary antibodies at 4°C overnight, and subsequently the secondary
antibody [goat anti-rabbit IgG (H+L)-HRP; cat. no. RM3002; Beijing
Ray Antibody Biotech] was incubated with the tissue slices at room
temperature for 1 h and stained with 3,3-diaminobenzidine (DAB) at
room temperature for 2 min. After washing in phosphate-buffered
saline (PBS) solution, the tissue slices were counterstained with
conventional hematoxylin solution at room temperature for 2
min.
Each sample was evaluated and scored independently
by two pathologists who were blinded to clinical, pathological and
molecular data at the time of analysis. A total of two high-power
visual fields were observed on each slice, and the IHC score was
evaluated using a semi-quantitative method. Briefly, the scoring
method was as follows: <1% positively stained cells was scored
as 0; 1-30% positively stained cells was scored as 1; 31-60%
positively stained cells was scored as 2; and >60% positively
stained cells was scored as 3. Positive staining intensity was
graded as follows: Colorless was scored as 0 (negative); light
yellow was scored as 1 (weak); brown-yellow was scored as 2
(moderate); and brown was scored as 3 (strong). The two scores were
multiplied to generate the IHC score (in the range 0-9). Positive
expression of ELF5 was defined as the tissue having an IHC score
≥5. The images were acquired using an Olympus SZX16
stereomicroscope (Olympus Corporation) and analyzed using ZEN
Imaging Software (version Zen 2011 Service Pack 1; Carl Zeiss
AG).
Cell culture
The human lung adenocarcinoma cell line A549
(American Type Culture Collection), the murine Lewis lung carcinoma
(LLC) (Procell Life Science & Technology Co., Ltd.) cell line
and the human embryonic kidney cell line 293 (American Type Culture
Collection) were cultured in DMEM with 10% Gibco® fetal
bovine serum (Thermo Fisher Scientific, Inc.) and 1%
Penicillin-Streptomycin (cat. no. 15140122; Gibco™; Thermo Fisher
Scientific, Inc.). All cells were cultured in a 37°C incubator with
5% CO2 and 95% humidity. The cell lines were not
authenticated after purchase, but routinely tested negative for
mycoplasma contamination.
Plasmid construction and
transfection
The open reading frame of ELF5 was amplified from
293 cells (American Type Culture Collection) cDNA and cloned into
the pcDNA3.1 vector (Thermo Fisher Scientific, Inc.). The sequences
were confirmed by Sanger sequencing. The recombinant plasmid and
vector plasmid were transfected into A549 cells using
Invitrogen® Lipofectamine 3000™ transfection reagent
(Thermo Fisher Scientific, Inc.) at 37°C for 6 h, following the
manufacturer's protocol. The concentration of nucleic acid used was
500 ng/ml. At 48 h after transfection, the cells were collected for
subsequent experiments.
Lentivirus infection
The 3rd generation of lentiviral packaging system
was used to produce a lentivirus carrying the ELF5 shRNA. The
lentivirus carrying the ELF5 shRNA was purchased from GeneCopoeia,
Inc. The ELF5 small hairpin RNA (shRNA) (Table SI) was subcloned via a
BamHI-EcoRI restriction digest into a psi-LVRU6GP
vector (GeneCopoeia, Inc.). The psi-LVRU6GP-scramble (GeneCopoeia,
Inc.) was used as a control. The Calcium Phosphate Cell
Transfection Kit (Beyotime Institute of Biotechnology) was used to
co-transfect 5 µg of recombined lentiviral vectors with 3.75
µg of psPAX2 and 1.25 µg of pMD2 VSV-G packaging
vectors in 293T cells (American Type Culture Collection) at 37°C
for 4 h. The culture supernatants were collected after 48 h after
transfection. Following centrifugation to remove cell debris, the
supernatant was filtered through 0.45-µm polyethersulfone
low protein-binding filters. The virus stock was aliquoted and kept
at -80°C until use. A549 and LLC cells were transduced with the
lentivirus containing the ELF5 shRNA at a multiplicity of infection
(MOI) of 6. The nucleotide sequence is GCC CTG AGA TAC TAC TAT AAA
(Table SI). Briefly,
2×105 cells were seeded in each well of a 12-well plate
and incubated at 37°C in an atmosphere of 5% CO2
overnight. Subsequently, the cells were infected with either
lentivirus containing the ELF5 shRNA or vector virus, respectively.
After 24 h, the cell culture medium was replaced with fresh medium.
Fresh medium containing puromycin was added, and the medium was
replaced with fresh puromycin-containing media every 2 days.
Western blot analysis
Cells were lysed in buffer containing 1 mM
phenylmethylsulfonylfluoride (PMSF), 1X phosphatase inhibitor and
1X protease inhibitor cocktail. Following centrifugation at 12,000
× g for 10 min at 4°C, the protein concentration was determined
using a BCA Protein Assay Kit (Thermo Fisher Scientific, Inc.), and
total proteins (the mass of protein loaded per lane, ≥20 µg)
were further analyzed using 10% SDS-PAGE. The samples were then
transferred to a polyvinylidene difluoride membrane and 5% BSA was
applied to the sections at room temperature for 1 h as a blocking
reagent to reduce non-specific binding. The samples were then
probed with antibodies against β-actin (1:4,000; cat. no. RM2001;
Beijing Ray Antibody Biotech), ELF5 (1:500; cat. no. NHA1145;
Novogene Co, Ltd.), proliferating cell nuclear antigen (PCNA)
(1:1,500; cat. no. 13110; Cell Signaling Technology, Inc.), APC2
(1:500; cat. no. ab233753; Abcam) and cyclin D1 (1:1,000; cat. no.
55506; Cell Signaling Technology, Inc.). Following the incubation
with primary antibodies at room temperature for 1 h, the membranes
were washed with TBS/0.05% Tween-20 and incubated with
HRP-conjugated secondary antibodies (1:3,000; cat. nos. RM3001 and
RM3002; Beijing Ray Antibody Biotech) at room temperature for 1 h.
Proteins were detected using enhanced chemiluminescence substrates
(PerkinElmer, Inc.). The results were quantified using ImageJ
software (version 1.52A; National Institutes of Health).
Cell Counting Kit-8 (CCK-8) assay
Cell proliferation was measured using CCK-8 assay.
Briefly, 2,000 cells were plated into each well of a 96-well plate
and incubated at 37°C in an atmosphere containing 5%
CO2. Subsequently, 10 µl CCK-8 solution (Vazyme
Biotech Co., Ltd.) was added to each well. The cells were then
incubated for 2 h at 37°C, and the absorbance was measured using a
microplate reader at 450 nm. Three independent experiments were
performed. The data were plotted in order to derive the cell
proliferation curves.
Real-time cell proliferation assay
The proliferative ability of the cells was monitored
using the xCELLigence R Real-Time Cell Analyzer S16 (RTCA S16)
(ACEA Bioscience, Inc.). This platform is able to measure cellular
growth status in real time. Briefly, 2,000 cells were plated into
the special plate (E-Plate 16) per well. After a 30-min incubation,
E-Plate 16 and the xCELLigence RTCA S16 system were connected for
the purposes of scanning the plate. Cell growth was monitored
continuously and recorded as a cell index. The whole device was
placed in an incubator at 37°C containing 5% CO2 for 104
h. The cell index was derived from the change in electrical
impedance as the living cells interacted with the biocompatible
microelectrode surface in the microplate well, which provided an
effective means of measuring the cell number. The cell index was
read automatically, and the recorded curves are shown as the cell
index ± SEM.
Animal studies
C57BL/6, five-week-old male mice (n=6, weight,
20±1.5 g) were purchased from The Animal Center of Southern Medical
University (Guangzhou, China). All mice were housed in specific
pathogen-free conditions in an environment with regulated
temperature (22±1°C) and humidity (40-70%) and exposure to a
constant 12-h light-dark cycle in the animal facility. A total of
1×106 LLC cells suspended in 100 µl saline were
injected subcutaneously (and intraperitoneally) into the lateral
thighs of the mice (n=6), and this operation was performed under
anesthesia [60 mg/kg sodium pentobarbital (body weight)]. The cells
were implanted on the right side for the control group, whereas for
the ELF5-shRNA group, they were implanted on the left side. The
size of the tumors was evaluated for the first time nine days after
injection using calipers, according to the following formula: Tumor
volume=(length x width2)/2. The health and behavior of
the animals were monitored every 2 days. No mice succumbed and
there were no abnormal signs of humane endpoints over the course of
the experiment. At 24 days after injection of the cells, the mice
were sacrificed via cervical dislocation, and all tumors were
identified to be <2,000 cm3 in size. Following
confirmation that the experimental animals had no heartbeat or had
ceased breathing, the tumors were isolated and weighed. The humane
endpoints for the experiment were designated as follows: A marked
reduction in food or water intake, labored breathing, an inability
to stand and no response to external stimuli; however, no abnormal
signs that were indicative of the humane endpoints of the
experiment were observed in any of the mice during these
experiments. All mouse experiments lasted one month and included
acclimatization to the feeding environment, tumor implantation and
growth, which were performed in compliance with the Institutional
Animal Care and Use Committee guidelines of Southern Medical
University, Guangzhou, China [approval no. SYXK (Guangdong)
2016-0167].
Promoter activity assay
Transfections and dual luciferase reporter assays
were performed as previously described (10). Briefly, human genomic DNA was used
as a template to amplify the mice APC2 fragments covering ~2 kb of
the 5'-flanking sequence by PCR. Subsequently, the PCR products
were inserted into the pGL3.0 basic vector (Promega Corporation),
and their efficient insertion was confirmed by sequencing. To
detect the potential binding sites, different constructs of mutated
seed sequences for ELF5-binding sites in the promoter regions of
the APC2 were cloned. For the luciferase reporter assay, 293T cells
were cultured in 24-well plates and each well was transfected with
0.5 µg firefly luciferase reporter plasmid, 0.05 µg
pRL-CMV plasmid (Promega Corporation) and 0.5 µg control or
Flag-ELF5 construct using Invitrogen™ Lipofectamine 3000 (Thermo
Fisher Scientific, Inc.). Luciferase activities were measured at 36
h following transfection using luciferase assay kits (Promega
Corporation).
RNA sequencing (RNA-seq) and data
processing
Total amounts and integrity of RNA which was
extracted from cells using the TRIzol Lysis Reagent (cat. no.
10296028; Invitrogen; Thermo Fisher Scientific, Inc.) according to
the user guidelines, were assessed using the RNA Nano 6000 Assay
kit (cat. no. 5067-1511; Agilent Technologies, Inc.) of the
Bioanalyzer 2100 system (Agilent Technologies, Inc.). Briefly, mRNA
was purified from total RNA using poly-T oligo-attached magnetic
beads (cat. no. S1419S; New England BioLabs, Inc.). Second strand
cDNA synthesis was subsequently performed using DNA Polymerase I
and dNTP (cat. nos. M0210L and N0447L, respectively; New England
BioLabs, Inc.). PCR amplification was then performed. The PCR
product was purified using AMPure XP beads (cat. no. A63882;
Beckman Coulter, Inc.), and the library was finally obtained.
Following the construction of the library, reverse
transcription-quantitative (RT-q) PCR was used to accurately
quantify the effective concentration of the library (the effective
concentration of the library was >2 nM) to ensure the quality of
the library. After the library was qualified, it was sequenced
using the Illumina NovaSeq 6000 (cat. no. 20012850; Illumina,
Inc.). To generate the end reading of 150 bp. The basic principle
of sequencing was to synthesize and sequence concurrently
(sequencing by synthesis). Differential expression analysis of two
conditions was performed using the edgeR package (3.22.5;
https://bioconductor.org/packages/release/bioc/html/edgeR.html).
RT-qPCR analysis
Total RNA from cell lines was isolated using Life
Technologies® TRIzol® reagent (Thermo Fisher
Scientific, Inc.), according to the manufacturer's instructions.
The reverse-transcription of RNA (2.0 µg) to cDNA was
performed using the PrimeScript RT reagent kit (Takara
Biotechnology Co., Ltd.) following the manufacturer's instructions.
For the qPCR analysis, SYBR Green qPCR master mix (Takara
Biotechnology Co., Ltd.) was used, following the protocol supplied
by the manufacturer. The RT-qPCR thermocycling conditions were as
follows: Initial denaturation at 95°C for 30 sec, followed by 40
cycles at 95°C for 5 sec and 60°C for 20 sec. The 2−ΔΔCq
method (11) was used for the
semi-quantification of the target genes, with GAPDH as the internal
reference compound. The sequences of the primers used for qPCR are
shown in Table SII.
Statistical analysis
Statistical analyses were performed using SPSS 20.0
software (IBM Corp.). All experiments were repeated at least three
times and quantitative data were expressed as the mean ± standard
error.Unpaired Student's t-test was used to compare means between
groups. Chi-squared test was used to analyze the association
between ELF5 expression and the clinicopathological parameters.
P<0.05 was considered to indicate a statistically significant
difference.
Results
ELF5 expression in lung adenocarcinoma is
significantly higher compared with that in adjacent normal
tissues
To identify the role of ELF5 in lung adenocarcinoma,
the association between ELF5 expression and clinicopathological
features in patients with lung adenocarcinoma based on the results
from the tissue microarray (TMA) analysis was first retrospectively
assessed (Fig. 1A and Table SII). Since ELF5 is a nuclear
transcription factor, the evaluation of ELF5 expression was based
exclusively on distinct nuclear staining intensity, through
examining its staining rate in the nucleus (Fig. 1B). The results indicated that the
ELF5 expression in lung adenocarcinoma was significantly higher
compared with that in corresponding adjacent lung tissues. The
association between the expression of ELF5 and the pathological
characteristics of patients with lung adenocarcinoma was
subsequently analyzed using the chi-square test. The results
obtained revealed that the high expression of ELF5 was positively
associated with stage classification and lymph node metastasis, but
not with other pathological characteristics (Table SIII). Taken together, these
results suggested that a strong association exists between ELF5 and
lung adenocarcinoma growth.
ELF5 promotes the proliferation of lung
adenocarcinoma cells
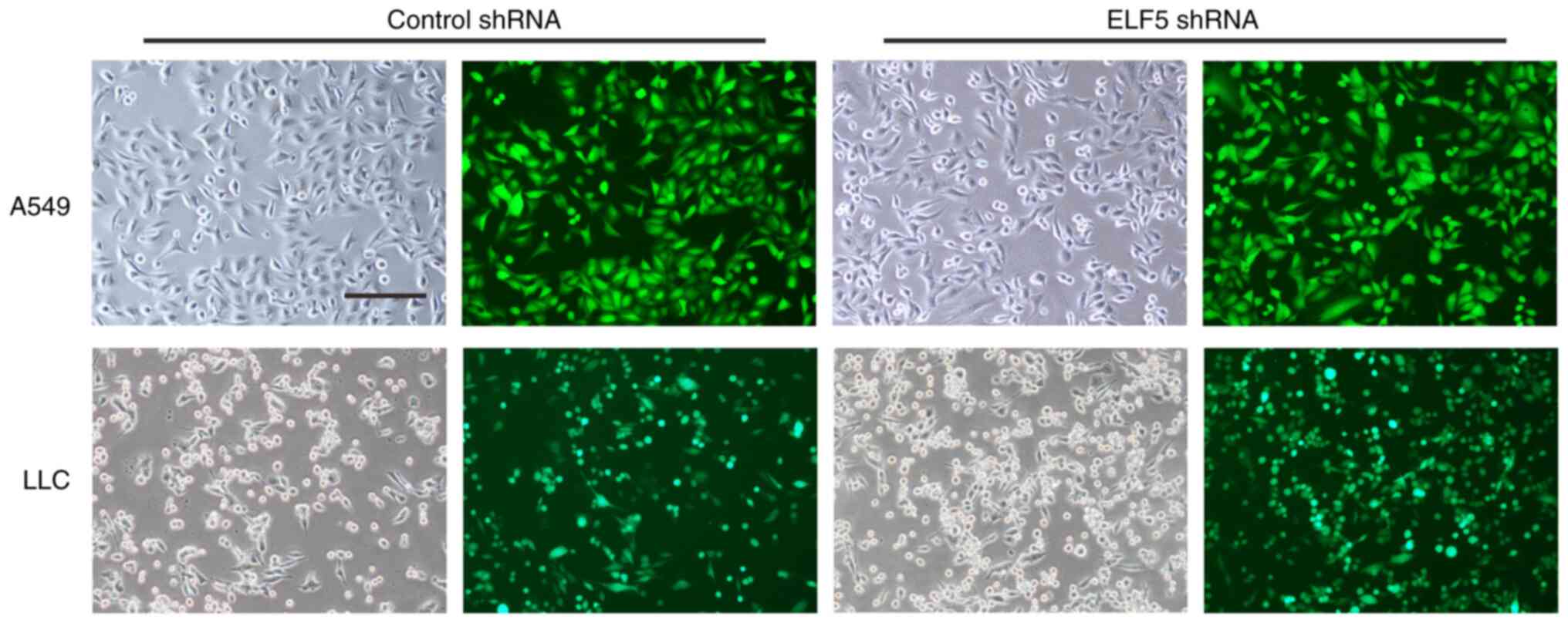
Subsequently, A549 cells were engineered to stably
express the shRNA of ELF5 by infection of lentivirus with GFP
fluorescence (Fig. 2). In
addition, to identify the role of ELF5 in lung adenocarcinoma
growth, lung adenocarcinoma A549 cells were transfected with pcDNA
3.1-ELF5 or the vector plasmid. Western blot analysis was employed
to confirm the overexpression of ELF5 in A549 cells following
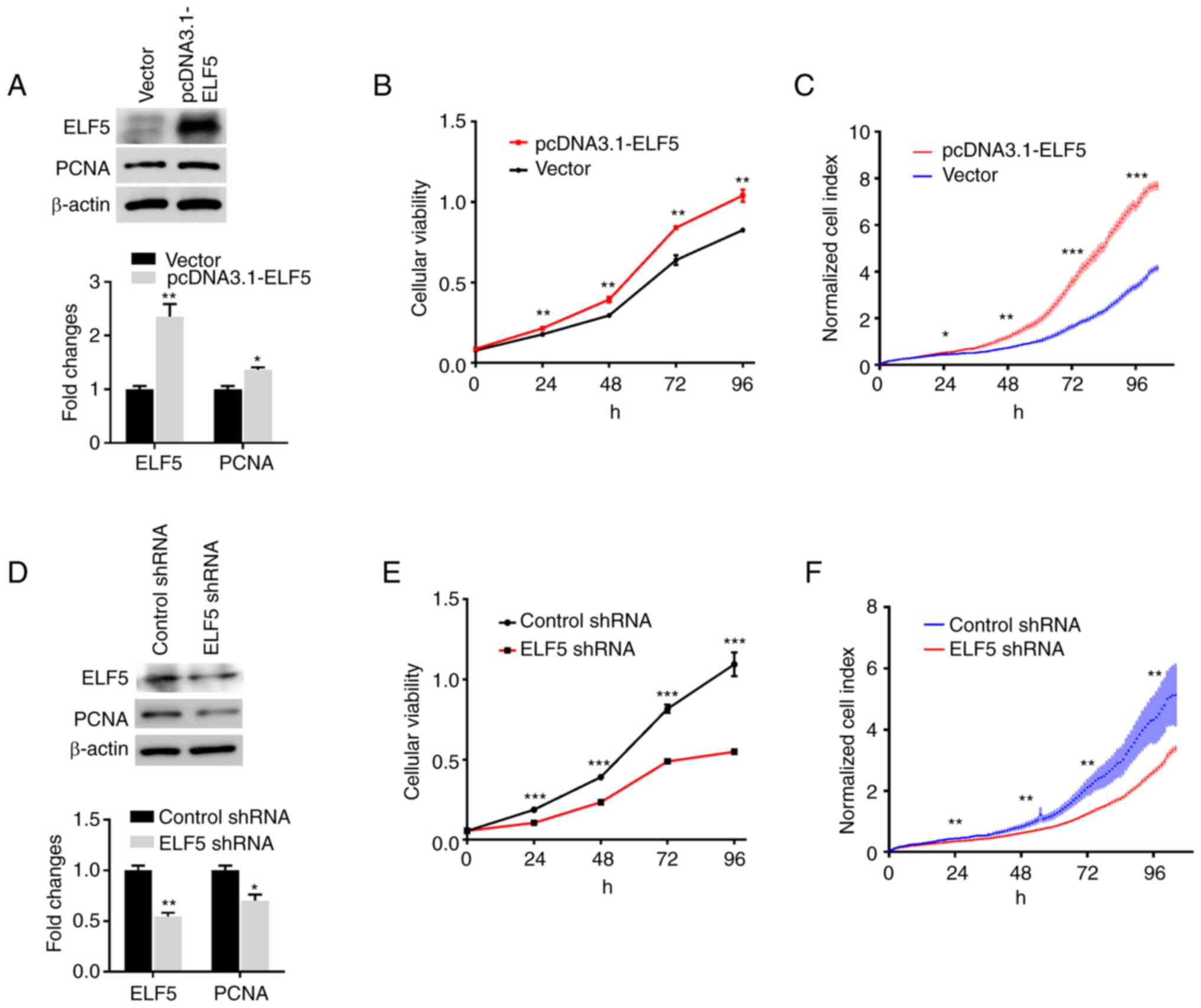
transfection with pcDNA 3.1-ELF5 (Fig. 3A). Furthermore, the overexpression
of ELF5 increased the expression of PCNA (Fig. 3A), a marker of cell proliferation
that is commonly used to assess the growth fraction of a cell
population (12,13). As shown in Fig. 3B and C, the overexpression of ELF5
led to an increase in the proliferation of A549 cells.
The results of western blot analysis verified a
significant reduction of ELF5 in A549 cells following lentivirus
infection (Fig. 3D). Furthermore,
the low expression of ELF5 led to a decrease in the expression of
PCNA. The proliferation of A549 cells was measured using CCK-8
assay (Fig. 3E) and the RTCA S16
system (Fig. 3F). The results
showed that knocking down ELF5 significantly blocked the growth of
A549 cells.
Knocked down expression of ELF5 decreases
the proliferation of lung adenocarcinoma in vitro and in vivo
To further investigate the role of ELF5 in lung
adenocarcinoma, mouse LLC cells were engineered to stably express
the ELF5 shRNA via infection of lentivirus with green fluorescent
protein (GFP) fluorescence (Fig.
2). Western blot analysis was used to confirm that a
significant reduction in ELF5 expression in LLC cells occurred
following lentivirus infection (Fig.
4A). Furthermore, the knocked down expression of ELF5 caused a
decrease in the expression of PCNA. The proliferation of LLC cells
was subsequently measured by CCK-8 assay (Fig. 4B) and the RTCA S16 system
(Fig. 4C). The results obtained
revealed that the reduction of ELF5 led to a decrease in the
proliferation of LLC cells, findings that were consistent with
those obtained in A549 cells.
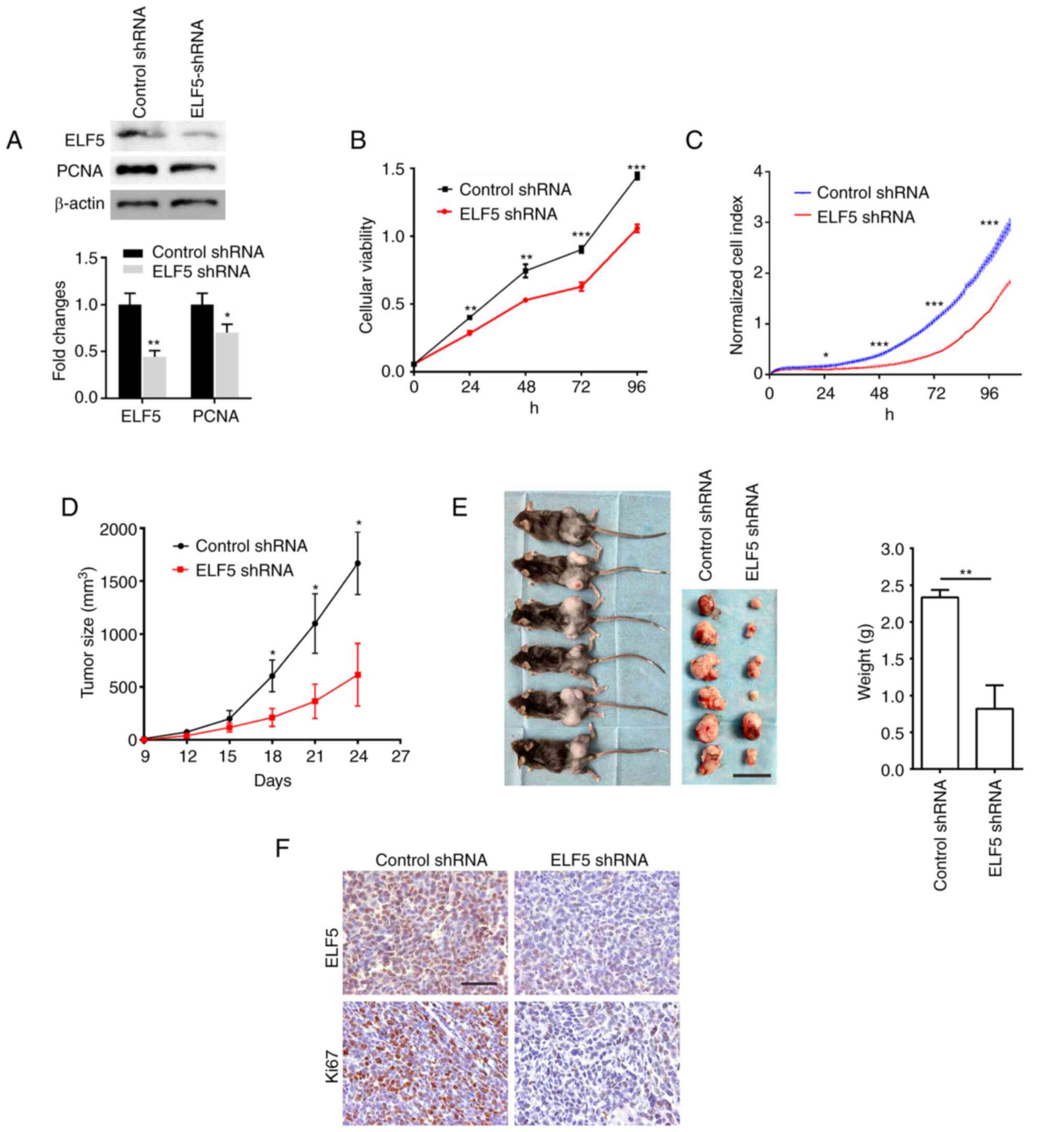 | Figure 4Knockdown of the expression of ELF5
decreases the proliferation of lung adenocarcinoma in vitro
and in vivo. Mouse LLC cells were engineered to stably
express the shRNA of ELF5 through infection of lentivirus with GFP
fluorescence. (A) The expression of ELF5 and PCNA was detected
using western blot analysis in LLC cells, with β-actin serving as a
loading control. The proliferation of LLC cells was assessed using
(B) Cell Counting Kit-8 assay and (C) the Real-Time Cell Analyzer
S16 system. To construct the animal tumor model, 1×106
LLC cells were subcutaneously injected into the lateral thigh of
mice (n=6). (D) The size of the tumor was evaluated for the first
time nine days after injection, and every 3 days thereafter, using
calipers, according to the formula: Tumor volume=(length x
width2)/2. (E) After 24 days, the tumor tissue was
isolated from the mice and weighed (scale bar, 20 mm). (F) The
expression levels of ELF5 and Ki-67 in the tumor tissue were
detected by immunohistochemical staining analysis (scale bars
represent 100 µm). The data were analyzed using unpaired
t-test (*P<0.05, **P<0.01 and
***P<0.001). ELF5, E74-like ETS transcription factor
5; LLC, Lewis lung cancer; PCNA, proliferating cell nuclear
antigen. |
Subsequently, an animal tumor model was constructed
via the subcutaneous injection of LLC cells. The results obtained
revealed that the knocked down expression of ELF5 led to a marked
decrease in tumor growth in vivo (Fig. 4D and E). Subsequent IHC analysis
of the tumor tissue showed that ELF5 knockdown led to a marked
decrease in ELF5 levels in the tumor tissue (Fig. 4F). Moreover, the knocked down
expression of ELF5 caused a decrease in the expression level of
Ki-67, a commonly used marker of cell proliferation. Taken
together, these results suggested that ELF5 is essential for lung
adenocarcinoma cell growth.
APC2 acts as the direct target of ELF5 in
the regulation of lung cancer cells
To further explore the underlying mechanism of ELF5
in lung cancer cells, RNA-Seq was performed following transfection
of vector control or pcDNA3.1-ELF5 plasmid in LLC cells. A total of
323 DEGs, including 136 downregulated DEGs and 187 upregulated
DEGs, were revealed in LLC cells following ELF5 overexpression
(log2 |fold change|>1; P<0.05) (Fig. S1). A total of 6 DEGs were further
confirmed using RT-qPCR analysis in LLC cells (Fig. 5A). Among these, APC2 was found to
be involved in tumorigenesis, and was strongly suppressed by ELF5
overexpression.
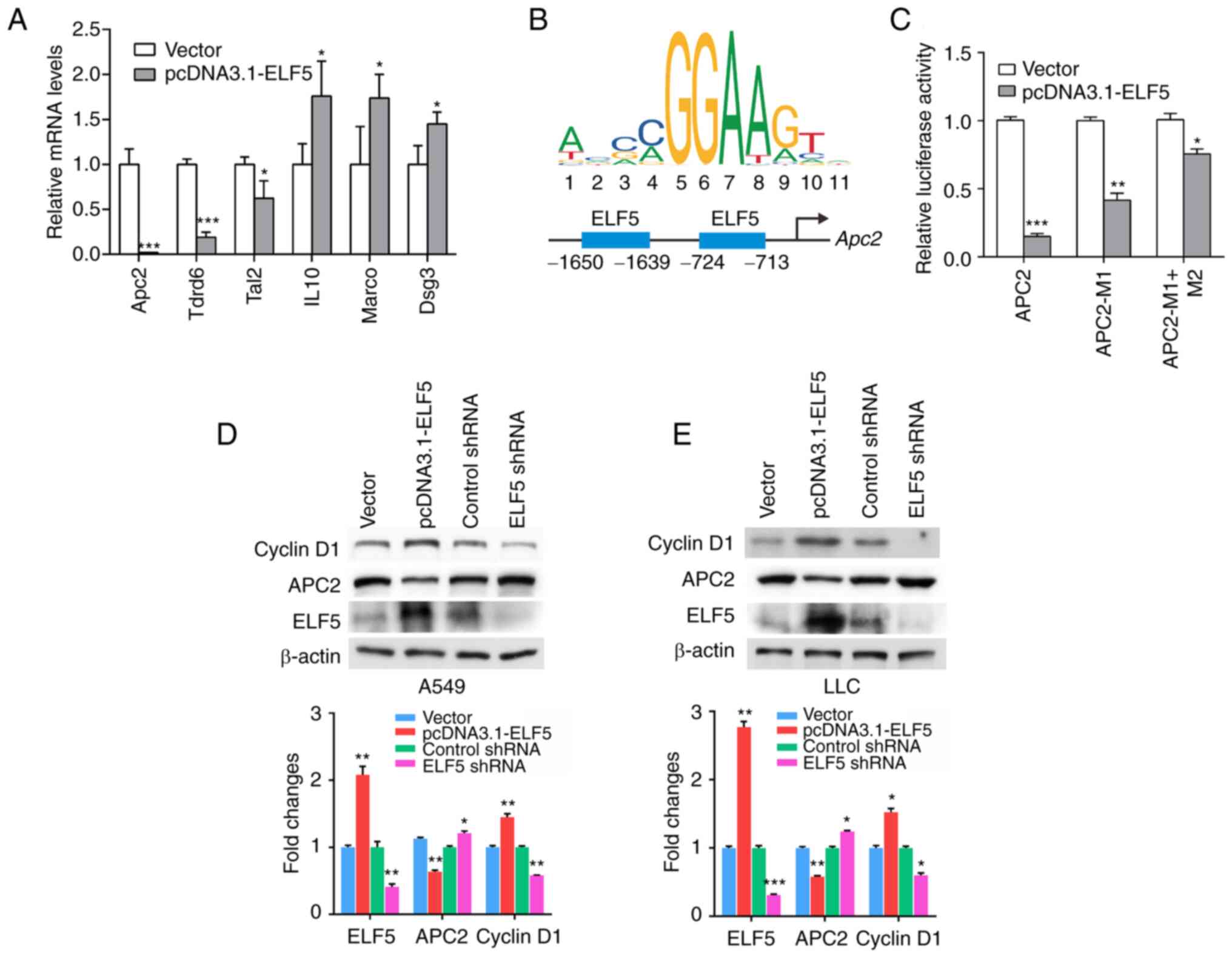 | Figure 5APC2 acts as the direct target of
ELF5 in the regulation of lung cancer cells. (A) Reverse
transcription-quantitative PCR analysis was performed, and the mRNA
levels of 6 genes (Apc2, Tdrd6, Tal2, Il10, Marco and Dsg3) in LLC
cells transfected with vector control or pcDNA3.1-ELF5 plasmid are
shown (n=3; *P<0.05 and ***P<0.001).
(B) ELF5 binding sites in the Apc2 promoter were predicted using
JASPAR, and the binding sites are shown schematically in the
diagrams. (C) Relative luciferase activity was detected in 293T
cells that were co-transfected with pcDNA3.1-ELF5 plasmid or vector
control and the wild-type Apc2 promoter or the mutated constructs
(n=3; *P<0.05, **P<0.01 and
***P<0.001). Western blot analysis was performed, and
the protein levels of cyclin D1, APC2 and ELF5 in (D) A549 cells
and (E) LLC cells transfected with plasmid or infected with
lentivirus containing the ELF5 shRNA are shown.
*P<0.05, **P<0.01 and
***P<0.001. ELF5, E74-like ETS transcription factor
5; APC2, adenomatous polyposis coli 2. |
In order to investigate the directly acting
mechanism through which ELF5 promotes the proliferation of lung
adenocarcinoma cells, JASPAR 2022 (14) was employed to predict APC2 as one
of the downstream target genes of ELF5 (Fig. 5B). Subsequently, luciferase
reporter assays were performed to validate that ELF5 could directly
target the promoter of APC2. Through the transfection of wild-type
(WT) or mutant (M1 or M2; i.e., mutations of the ELF5 binding
sites) APC2 promoter reporter constructs into control or
ELF5-overexpressed cells, ELF5 overexpression was revealed to
significantly suppress the luciferase activity of the WT APC2
promoter reporter constructs compared to the vector, whereas the M1
or M1 + M2 constructs resulted in an increase in luciferase
activity compared with that in the wild-type (Fig. 5C). These findings clearly
demonstrated that ELF5 supresses the transcription of APC2 through
the ELF5-binding site motifs. Western blot analysis revealed that
overexpression of ELF5 led to a marked decrease in APC2 expression
in lung adenocarcinoma cells, whereas the silencing of ELF5
resulted in an increase in APC2 expression (Fig. 5D and E). Furthermore, the results
obtained demonstrated that ELF5 could increase cyclin D1
expression, and silencing of ELF5 caused a decrease in cyclin D1
expression in lung adenocarcinoma cells (Fig. 5D and E).
Discussion
Lung cancer is one of the leading causes of
cancer-associated mortality globally (15,16). Non-small cell lung cancer (NSCLC)
is the prominent type of lung cancer, which accounts for >80% of
all cases of lung cancer. Despite some advances that have been made
in terms of early detection of the tumor, and recent improvements
in treatment, lung cancer remains a significant cause of both the
mortality and morbidity of patients with cancer. Therefore, there
is an urgent need to elucidate the molecular mechanisms underlying
lung cancer progression, and to develop efficient therapies against
lung cancer.
ELF5 was initially found to be expressed
predominantly in epithelial cells, and it was shown to function as
a crucial regulator of mammary gland development and milk
production (17,18). During the course of normal breast
development, ELF5 drives a cell fate decision of the mammary
progenitor cells, causing them to establish the estrogen
receptor-negative (ER−) cell lineage responsible for
alveolar development and milk production (18). ELF5 can not only induce mammary
gland tumor cell transformation from the luminal subtypes to
basal-like subtypes, but it can also inhibit the cell phenotype,
effectively leading to an increase in estrogen sensitivity in
breast cancer (19). Both
tumor-promoting and tumor-suppressive roles for ELF5 have been
reported in breast cancer, which may be linked to the subtype of
the disease. Mutations in ELF5 are relatively rare in human cancers
(20). Missense mutations occur
in melanoma, mesothelioma, uterine, stomach and colorectal cancers,
which are predominantly non-recurring missense mutations of unknown
function. Amplification of ELF5 occurs in cancers of the upper
gastrointestinal tract (esophageal and stomach), ovary, head and
neck, and breast in 2-6% of cases, whereas occasional deletions of
ELF5 occur in prostate, sarcoma, bladder, and lung cancers, as well
as in acute myeloid leukemia and gliomas (20).
Although previous studies have assessed ELF5 in a
variety of different cancer types (3-7),
to the best of our knowledge there have been no studies published
on determining the relevance of ELF5 in lung cancer tissues or its
clinicopathological significance. In the present study, it was
possible to determine that the expression of ELF5 in lung
adenocarcinoma was significantly higher compared with its
expression in corresponding adjacent normal tissues, thereby
suggesting that ELF5 may exert an important role in the development
of lung adenocarcinoma. Due to the limited number of cases in the
present study, however, especially the number of lung
adenocarcinoma patients with metastasis, the real prognostic
significance of ELF5 remains to be elucidated in larger cohorts in
lung adenocarcinoma.
In vitro experiments indicated that
overexpression of ELF5 led to an increase in the proliferation of
lung adenocarcinoma cells. On the other hand, a reduction in ELF5
expression led to a decrease in the proliferation of lung
adenocarcinoma cells, showing that ELF5 was required for lung
adenocarcinoma cell proliferation. Subsequent in vivo
experiments showed that the low expression of ELF5 caused a marked
decrease in tumor growth, a finding that was consistent with the
experimental results identified in vitro. Therefore, these
experiments revealed that ELF5 fulfills an essential role in the
proliferation of lung adenocarcinoma cells.
APC2 has been shown to be closely associated with
multiple aspects of lung development and lung tumor progression
(21). The loss of APC2 has been
observed previously in lung cancer (22), where it acts as a critical
negative regulator of the Wnt signaling pathway (23,24). Downregulation of APC2 leads to the
stabilization of active β-catenin, thereby increasing the activity
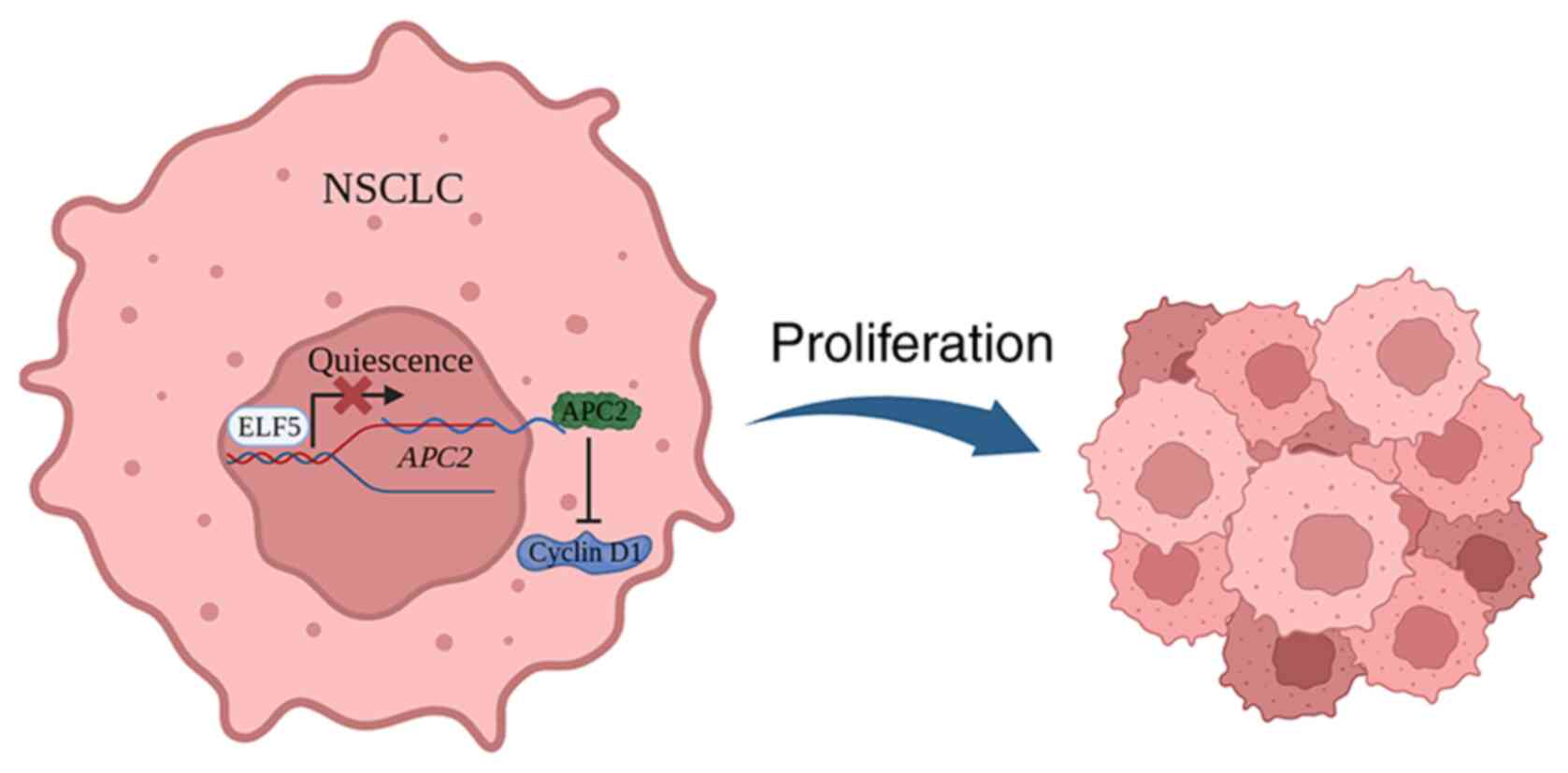
of the Wnt pathway to promote lung cancer progression (23-26). In the present study, it was shown
that ELF5 could promote lung cancer cell proliferation through
inhibiting APC2. The mechanistic experiments revealed that ELF5
caused an increase in cyclin D1 expression, which is a critical
downstream target of Wnt pathway (27,28), confirming that ELF5 could promote
lung cancer cell proliferation through inhibiting APC2 and
activating the Wnt pathway (Fig.
6).
Grainyhead-like 2 (GRHL2) plays multiple roles in
lung morphogenesis that are essential for respiratory function
(29-31). Loss of GRHL2 function in the lung
epithelium of mice was shown to result in their death within hours
of birth due to respiratory distress. Gene transcription profiling
of GRHL2-deficient lung epithelial cells revealed a significant
downregulation of ELF5. GRHL2 has been shown to control normal lung
morphogenesis through tightly regulating the activity of distal tip
progenitor cells through the direct regulation of ELF5 (30). GRHL2 has been implicated in tumor
development in lung cancer, breast cancer, oral cancer and various
gastric cancers (32,33). However, the function of GRHL2
appears to be complex and controversial in the context of cancer,
varying with cancer type (34).
GRHL2 was shown to be implicated as an oncogene in the etiology and
progression of NSCLC (32). A
higher level of GRHL2 expression in NSCLC is a predictor of poor
prognosis. GRHL2 fulfills a regulatory role, participating in both
cell proliferation and metastasis. GRHL2 was also shown to promote
cell growth and colony formation, simultaneously suppressing the
cell migration of NSCLC cells. Since ELF5 is regulated by GRHL2,
GRHL2 may regulate the proliferation of lung cancer cells by
regulating ELF5, although this aspect requires further study.
In conclusion, the findings of the present study
have demonstrated that ELF5 exerts an essential role in the
proliferation of lung adenocarcinoma cells. Furthermore, ELF5 was
shown to promote lung cancer cell proliferation through inhibiting
APC2. Taken together, the present study has shown that a higher
expression level of ELF5 in lung adenocarcinoma may have potential
as a therapeutic target option for the treatment of human lung
adenocarcinoma.
Supplementary Data
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request. Data used for RNA-seq analysis in this study have been
deposited into the Sequence Read Archive at the National Center for
Biotechnology Information database under accession code
PRJNA936676.
Authors' contributions
JW, GQ and ZJ contributed to the conceptualization,
investigation, methodology, validation, and writing of the original
draft. ZL, RZ, HD and ZX contributed to the formal analysis,
visualization of the study and reviewing and editing. WC and QS
contributed to the conceptualization, funding acquisition,
resources, as well as the supervision of the study. All of the
authors confirm the authenticity of all the raw data. All authors
read and approved the final manuscript.
Ethics approval and consent to
participate
The studies involving human tissue samples were
approved (approval no. 81402373) by the Human Research Ethics
Committee of Taizhou Hospital of Zhejiang Province (Taizhou,
China), and all patients or their next of kin provided their
informed consent prior to the study. The present study was
conducted in accordance with the principles and guidelines of The
Declaration of Helsinki. Animal experiments were conducted in
accordance with the guidelines of the Institutional Animal Care and
Use Committee of Southern Medical University, Guangzhou, China
[approval no. SYXK (Guangdong) 2016-0167].
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interest.
Abbreviations:
|
ELF5
|
E74-like ETS transcription factor
5
|
|
LLC
|
Lewis lung carcinoma
|
|
CCK-8
|
Cell Counting Kit-8
|
|
TMA
|
tissue microarray
|
|
PCNA
|
proliferating cell nuclear antigen
|
|
RTCA
|
real-time cell analyzer
|
|
NSCLC
|
non-small cell lung cancer
|
|
APC2
|
adenomatous polyposis coli 2
|
Acknowledgments
Not applicable.
Funding
The present study was supported by the National Natural Science
Foundation of China (grant no. 82202930), Tongling Science and
Technology Project (grant no. 20200203037), the National Key
Research and Development Program of China (grant no.
2019YFC0117301) and the Natural Science Foundation of Guangdong
Province (grant no. 2019A1515011168).
References
|
1
|
Metzger DE, Stahlman MT and Shannon JM:
Misexpression of ELF5 disrupts lung branching and inhibits
epithelial differentiation. Dev Biol. 320:149–160. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Metzger DE, Xu Y and Shannon JM: Elf5 is
an epithelium-specific, fibroblast growth factor-sensitive
transcription factor in the embryonic lung. Dev Dyn. 236:1175–1192.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yao B, Zhao J, Li Y, Li H, Hu Z, Pan P,
Zhang Y, Du E, Liu R and Xu Y: Elf5 inhibits TGF-β-driven
epithelial-mesenchymal transition in prostate cancer by repressing
SMAD3 activation. Prostate. 75:872–882. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Li K, Guo Y, Yang X, Zhang Z, Zhang C and
Xu Y: ELF5-mediated AR activation regulates prostate cancer
progression. Sci Rep. 7:427592017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wu B, Cao X, Liang X, Zhang X, Zhang W,
Sun G and Wang D: Epigenetic regulation of Elf5 is associated with
epithelial-mesenchymal transition in urothelial cancer. PLoS One.
10:e01175102015. View Article : Google Scholar :
|
|
6
|
Yan H, Qiu L, Xie X, Yang H, Liu Y, Lin X
and Huang H: ELF5 in epithelial ovarian carcinoma tissues and
biological behavior in ovarian carcinoma cells. Oncol Rep.
37:1412–1418. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hu Y, Yan Y, Xu Y, Yang H, Fang L, Liu Y,
Li X, Li Q and Yan H: Expression and clinical significance of WWOX,
Elf5, Snail1 and EMT related factors in epithelial ovarian cancer.
Oncol Lett. 19:1281–1290. 2020.
|
|
8
|
Singh S, Kumar S, Srivastava RK, Nandi A,
Thacker G, Muralil H, Kim S, Baldeon M, Tobias J, Blanco MA, et al:
Loss of ELF5-FBXW7 stabilizes IFNGR1 to promote the growth and
metastasis of triple-negative breast cancer through interferon-γ
signalling. Nat Cell Biol. 22:591–602. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Piggin CL, Roden DL, Law AMK, Molloy MP,
Krisp C, Swarbrick A, Naylor MJ, Kalyuga M, Kaplan W, Oakes SR, et
al: ELF5 modulates the estrogen receptor cistrome in breast cancer.
PLoS Genet. 16:e10085312020. View Article : Google Scholar :
|
|
10
|
Yao F, Wang X, Cui ZK, Lan H, Ai X, Song
Q, Chen Z, Yang J, Wu B and Bai X: ETS2 promotes
epithelial-to-mesenchymal transition in renal fibrosis by targeting
JUNB transcription. Lab Invest. 100:438–453. 2020. View Article : Google Scholar
|
|
11
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
12
|
Takasaki Y, Deng JS and Tan EM: A nuclear
antigen associated with cell proliferation and blast
transformation. J Exp Med. 154:1899–1909. 1981. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Juríková M, Danihel Ľ, Polák Š and Varga
I: Ki67, PCNA, and MCM proteins: Markers of proliferation in the
diagnosis of breast cancer. Acta Histochem. 118:544–552. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Castro-Mondragon JA, Riudavets-Puig R,
Rauluseviciute I, Lemma RB, Turchi L, Blanc-Mathieu R, Lucas J,
Boddie P, Khan A, Manosalva Pérez N, et al: JASPAR 2022: The 9th
release of the open-access database of transcription factor binding
profiles. Nucleic Acids Res. 50(D1): D165–D173. 2022. View Article : Google Scholar :
|
|
15
|
Barta JA, Powell CA and Wisnivesky JP:
Global epidemiology of lung cancer. Ann Glob Health. 85:82019.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhou J, Chehab R, Tkalcevic J, Naylor MJ,
Harris J, Wilson TJ, Tsao S, Tellis I, Zavarsek S, Xu D, et al:
Elf5 is essential for early embryogenesis and mammary gland
development during pregnancy and lactation. EMBO J. 24:635–644.
2005. View Article : Google Scholar :
|
|
18
|
Oakes SR, Naylor MJ, Asselin-Labat ML,
Blazek KD, Gardiner-Garden M, Hilton HN, Kazlauskas M, Pritchard
MA, Chodosh LA, Pfeffer PL, et al: The Ets transcription factor
Elf5 specifies mammary alveolar cell fate. Gene Dev. 22:581–586.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kalyuga M, Gallego-Ortega D, Lee HJ, Roden
DL, Cowley MJ, Caldon CE, Stone A, Allerdice SL, Valdes-Mora F,
Launchbury R, et al: ELF5 suppresses estrogen sensitivity and
underpins the acquisition of antiestrogen resistance in luminal
breast cancer. PLoS Biol. 10:e10014612012. View Article : Google Scholar
|
|
20
|
Luk IY, Reehorst CM and Mariadason JM:
ELF3, ELF5, EHF and SPDEF transcription factors in tissue
homeostasis and cancer. Molecules. 23:21912018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Bonner AE, Lemon WJ, Devereux TR, Lubet RA
and You M: Molecular profiling of mouse lung tumors: Association
with tumor progression, lung development, and human lung
adenocarcinomas. Oncogene. 23:1166–1176. 2004. View Article : Google Scholar
|
|
22
|
Miura K, Bowman ED, Simon R, Peng AC,
Robles AI, Jones RT, Katagiri T, He P, Mizukami H, Charboneau L, et
al: Laser capture microdissection and microarray expression
analysis of lung adenocarcinoma reveals tobacco smoking- and
prognosis-related molecular profiles. Cancer Res. 62:3244–3250.
2002.PubMed/NCBI
|
|
23
|
Zhang K, Wang J, Yang L, Yuan YC, Tong TR,
Wu J, Yun X, Bonner M, Pangeni R, Liu Z, et al: Targeting histone
methyltransferase G9a inhibits growth and Wnt signaling pathway by
epigenetically regulating HP1α and APC2 gene expression in
non-small cell lung cancer. Mol Cancer. 17:1532018. View Article : Google Scholar
|
|
24
|
Croy HE, Fuller CN, Giannotti J, Robinson
P, Foley AVA, Yamulla RJ, Cosgriff S, Greaves BD, von Kleeck RA, An
HH, et al: The Poly(ADP-ribose) polymerase enzyme tankyrase
antagonizes activity of the β-catenin destruction complex through
ADP-ribosylation of axin and APC2. J Biol Chem. 291:12747–12760.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Dai B, Kong DL, Tian J, Liu TW, Zhou H and
Wang ZF: microRNA-1205 promotes cell growth by targeting APC2 in
lung adenocarcinoma. Eur Rev Med Pharmacol Sci. 23:1125–1133.
2019.PubMed/NCBI
|
|
26
|
Dong Y, Wu B, Wang X, Lu F, Li Q and Zhao
Q: High miR-3648 expression and low APC2 expression are associated
with shorter survival and tumor progression in NSCLC. Histol
Histopathol. 37:355–364. 2022.
|
|
27
|
Ge YX, Wang CH, Hu FY, Pan LX, Min J, Niu
KY, Zhang L, Li J and Xu T: New advances of TMEM88 in cancer
initiation and progression, with special emphasis on Wnt signaling
pathway. J Cell Physiol. 233:79–87. 2018. View Article : Google Scholar
|
|
28
|
Rimerman RA, Gellert-Randleman A and Diehl
JA: Wnt1 and MEK1 cooperate to promote cyclin D1 accumulation and
cellular transformation. J Biol Chem. 275:14736–14742. 2000.
View Article : Google Scholar
|
|
29
|
Varma S, Cao Y, Tagne JB, Lakshminarayanan
M, Li J, Friedman TB, Morell RJ, Warburton D, Kotton DN and Ramirez
MI: The transcription factors Grainyhead-like 2 and NK2-homeobox 1
form a regulatory loop that coordinates lung epithelial cell
morphogenesis and differentiation. J Biol Chem. 287:37282–37295.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kersbergen A, Best SA, Dworkin S, Ah-Cann
C, de Vries ME, Asselin-Labat ML, Ritchie ME, Jane SM and
Sutherland KD: Lung morphogenesis is orchestrated through
Grainyhead-like 2 (Grhl2) transcriptional programs. Dev Biol.
443:1–9. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Gao X, Bali AS, Randell SH and Hogan BLM:
GRHL2 coordinates regeneration of a polarized mucociliary
epithelium from basal stem cells. J Cell Biol. 211:669–682. 2015.
View Article : Google Scholar :
|
|
32
|
Pan X, Zhang R, Xie C, Gan M, Yao S, Yao
YB, Jin J, Han T, Huang Y, Gong Y, et al: GRHL2 suppresses tumor
metastasis via regulation of transcriptional activity of RhoG in
non-small cell lung cancer. Am J Transl Res. 9:4217–4226.
2017.PubMed/NCBI
|
|
33
|
Werner S, Frey S, Riethdorf S, Schulze C,
Alawi M, Kling L, Vafaizadeh V, Sauter G, Terracciano L, Schumacher
U, et al: Dual roles of the transcription factor grainyhead-like 2
(GRHL2) in breast cancer. J Biol Chem. 288:22993–23008. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
He J, Feng C, Zhu H, Wu S, Jin P and Xu T:
Grainyhead-like 2 as a double-edged sword in development and
cancer. Am J Transl Res. 12:310–331. 2020.PubMed/NCBI
|