Introduction
Since its spread at the beginning of 2020, the
coronavirus disease 2019 (COVID-19) pandemic represents one of the
major health problems, causing radical changes in the social
behavior of the affected population. Consequently, research efforts
have been made to characterize the severe acute respiratory
syndrome coronavirus 2 (SARS-CoV-2) sequences and develop novel
therapeutic options (1-3). Despite the approval, testing, and
worldwide distribution of anti-SARS-CoV-2 vaccines, the COVID-19
pandemic still represents one of the most important challenges in
developing specific antiviral agents targeting the SARS-CoV-2 life
cycle with high efficiency (4-9).
Despite the majority of individuals show moderate symptoms, certain
patients develop severe disease, which is generally associated with
clinical and laboratory signs of inflammation (10-13). From a molecular point of view,
SARS-CoV-2 entry within the cells is mediated by the interaction
between the viral surface spike protein and the host cells
angiotensin-converting enzyme receptor (14-16). Following its entry into the
respiratory epithelial cells, SARS-CoV-2 causes an immune response
with inflammatory cytokine production, followed by infiltration of
macrophages and neutrophils into the lung tissue, which results in
the cytokine storm (17-20). SARS-CoV-2 activates T lymphocytes,
which in turn secrete proinflammatory cytokines, including
granulocyte-macrophage colony-stimulating factor and IL-6 (21,22). The cytokine storm in COVID-19 was
proposed to occur due to SARS-CoV-2-mediated activation of
transcription factors, such as NF-κB and STAT3, which in turn
regulate expression of genes involved in inflammation, including
vascular endothelial growth factor, monocyte chemoattractant
protein 1, IL-8 and IL-6 (23-26).
Among the large variety of pharmaceutical
strategies, an increasing number of studies have focused on
repurposed drugs and bioactive molecules from natural sources. The
isothiocyanate sulforaphane (SFN) is one of the most abundant
bioactive components of Brassicaceae plants (for example,
broccoli) (27,28). SFN is derived from the hydrolysis
of its biogenic precursor glucoraphanin, which is mediated by
myrosinase. Myrosinases are present not only in plants but also in
gastrointestinal microflora; for this reason, they can be
administered directly in their active forms or as glucoraphanin
(29). As already and extensively
reported in previous studies, SFN exhibits a wide range of
biological effects including anticancer (30), antioxidant (31), antimicrobial (32), neuroprotective (33), cardioprotective (34), and anti-inflammatory (35) activities. As demonstrated by
several studies, the anti-inflammatory activity of SFN is mediated
by NF-κB inhibition (36-38). Following its translocation to the
nucleus, NF-κB is able to induce the expression of proinflammatory
cytokines (39) including, but
not limited to IL-6 (40,41), IL-8 (42) and IL-1β (39). It is interesting to note that the
results from several phase I and II clinical trials investigating
the safety and tolerability of SFN are currently available
(43-46).
Recently, it was reported that SFN inhibited the
expression of IL-6 and IL-8 genes induced by the treatment of IB3-1
bronchial cells with a recombinant spike protein of SARS-CoV-2
(47). The data reported were in
full agreement with the results published by Ordonez et al
(48) in which SFN was able to
inhibit the in vitro replication of six SARS-CoV-2 strains,
including delta and omicron. In the present study, the ability of
SFN to inhibit SARS-CoV-2 replication in SARS-CoV-2 infected Calu-3
cells was investigated along with its effects on the expression
levels of NF-κB induced pro-inflammatory genes.
Materials and methods
Cellular models
Experiments were conducted in epithelial respiratory
model: Calu-3 cells (cat. no. HTB-55; American Type Culture
Collection; cell passage: 36 when they were purchased), isolated
from 25-year-old Caucasian man with lung adenocarcinoma (49,50). Cells were cultured in a humidified
atmosphere of 5% CO2 in DMEM/F12 medium (Gibco; Thermo
Fisher Scientific, Inc.) supplemented with 10% fetal bovine serum
(Biowest), 100 units/ml penicillin, 100 µg/ml streptomycin
(Lonza Group, Ltd.), and 1% NEEA (100X) (Non-Essential Amino Acids
Solution; Gibco; Thermo Fisher Scientific, Inc.). The number of
cells to be seeded was determined using a Z2 Coulter Counter
(Beckman Coulter, Inc.).
Chemical compounds
Stock solutions of SFN (D,L-Sulforaphane; cat. no.
574215-25MG; MilliporeSigma) were prepared at final concentration
of 150 mM in DMSO (cat. no. D8418; MilliporeSigma). Aliquots of
stock solution were prepared and stored at -20°C (protected from
light). Each stock solution of SFN was diluted 1:10 in DMSO just
before cell treatment (51,52). Control cell populations cultured
in the presence of only DMSO were accordingly employed. The
concentration of DMSO in the control population was always
identical to that used for the SFN treatment [maximum DMSO
concentration used 0.1% (v/v)].
Infection of Calu-3 cells with
SARS-CoV-2
SARS-CoV-2 manipulation was performed in the BSL-3
laboratory following the biosafety requirements. SARS-CoV-2 was
isolated from a nasopharyngeal swab retrieved from a patient with
COVID-19 (Caucasian man of Italian origin, genome sequences
available at GenBank (SARS-CoV-2-UNIBS-AP66:ERR4145453, https://trace.ncbi.nlm.nih.gov/Traces/?view=run_browser&acc=ERR4145453&display=metadata).
This SARS-CoV-2 isolate clustered in the B1 clade which includes
most of the Italian sequences, together with sequences derived from
other European countries and USA. The susceptibility of Calu-3
cells to SARS-CoV-2 infection was assayed by infecting single type
cell with a MOI of 0.1 for 2 h at 37°C (~2×105
infectious virus particles per well, containing 106
cells). A total of 24 and 48 h after infection, the
infected/treated cells were collected. Viral load within Calu-3
cells was detected 24 and 48 h post-infection (hpi) by reverse
transcription-droplet-digital (RT-dd) PCR using SARS-CoV-2 RUO qPCR
Primer & Probe kit, according to the manufacturer's
instructions (Integrated DNA Technologies, Inc.) which detects two
sequences in SARS-CoV-2 N region (N1 and N2) and uses the human
RNAse P as a normalizer. Number of copies of SARS-CoV-2 are in the
order of 109 copies/µg of isolated RNA.
Cellular RNA extraction
Calu-3 SARS-CoV-2-infected cells were lysed in TRI
Reagent® (cat. no. T9424; MilliporeSigma) according to
the manufacturer's instructions. Obtained RNA pellets were washed
with 1 ml of 75% ethanol and centrifuged at 12,000 × g for 5 min at
4°C. Finally, the pellets were suspended in RNAse-free water and
checked for RNA integrity on 1% agarose gel, employing FluoroVue
Nucleic Acid Stain as intercalating agent and following the
manufacturer's instructions (SMOBIO Technology, Inc.; cat. no.
NS1000).
Viral RNA isolation and analysis
RNA was extracted from clarified cell culture
supernatants 24 and 48 hpi using PureLink Viral RNA/DNA Mini kit
(cat. no. 1228050; Thermo Fisher Scientific, Inc.) according to the
manufacturer's instructions. SARS-CoV-2 quantification was
performed using the PowerUp SYBR Green Master Mix (Thermo Fisher
Scientific, Inc.) targeting the S gene using the following primers:
RBD-q forward, 5′-CAATGGTTTAACAGGCACAGG-3′ and reverse,
5′-CTCAAGTGTCTGTGGATCACG-3′ (Integrated DNA Technologies, Inc.)
(53). The standard curve was
obtained by determination of copy numbers derived from serial
dilutions (102-108 copies) of the
corresponding gene block (Integrated DNA Technologies, Inc.).
Viral load determination in Calu-3
infected cells
Viral load within Calu-3 cells was detected 24 and
48 hpi by RT-dd PCR using SARS-CoV-2 RUO qPCR Primer & Probe
kit (Integrated DNA Technologies, Inc.) which detects two sequences
in SARS-CoV-2 N region and in the human RNAse P as normalizer
(sequences are reported in Table
I). Number of copies of SARS-CoV-2 are in the order of
109 copies/µg of isolated RNA.
 | Table IComplete list of employed TaqMan PCR
assays. |
Table I
Complete list of employed TaqMan PCR
assays.
| Target | Assay ID | Sequence |
|---|
| IL-1β |
Hs.PT.58.1518186 | Purchased from
Integrated DNA Technologies, Inc. |
| IL-6 |
Hs.PT.58.40226675 | Purchased from
Integrated DNA Technologies, Inc. |
| IL-8 |
Hs.PT.58.38869678.g | Purchased from
Integrated DNA Technologies, Inc. |
| NF-kB p65 |
Hs.PT.58.22880470 | Purchased from
Integrated DNA Technologies, Inc. |
| NF-kB p50 |
Hs.PT.58.38905484 | Purchased from
Integrated DNA Technologies, Inc. |
| β-Actin | Home-made
assay | Forward Primer:
ACGATGGAGGGGAAGACG |
| Reverse Primer:
ACAGAGCCTCGCCTTTG |
| Probe:
CCTTGCACATGCCGGAGC |
| RPL13A | Home-made
assay | Forward Primer:
GGCAATTTCTACAGAAACAAGTTG |
| Reverse Primer:
GTTTTGTGGGGCAGCATACC |
| Probe:
CGCACGGTCCGCCAGAAGAT |
| GAPDH | Home-made
assay | Forward Primer:
ACATCGCTCAGACACCATG |
| Reverse Primer:
TGTAGTTGAGGTCAATGAAGGG |
| Probe:
AAGGTCGGAGTCAACGGATTTGGTC |
| N1-SARS-CoV-2 | SARS-CoV-2 Research
Use | Forward Primer:
GACCCCAAAATCAGCGAAAT |
| Protein | Only Primer and
Probe Sets | Reverse Primer:
TCTGGTTACTGCCAGTTGAATCTG |
| Probe:
ACCCCGCATTACGTTTGGTGGACC |
| N2-SARS-CoV-2 | SARS-CoV-2 Research
Use | Forward Primer:
TTACAAACATTGGCCGCAAA |
| Protein | Only Primer and
Probe Sets | Reverse Primer:
GCGCGACATTCCGAAGAA |
| Probe:
CAATTTGCCCCCAGCGCTTCA |
| RNAse P | SARS-CoV-2 Research
Use | Forward Primer:
AGATTTGGACCTGCGAGCG |
| Only Primer and
Probe Sets | Reverse Primer:
GAGCGGCTGTCTCCACAAGT |
| Probe:
TTCTGACCTGAAGGCTCTGCGCG |
Reverse transcription-quantitative (RT-q)
PCR analysis
For the synthesis of cDNA, a combination of both
random hexamers and oligo dT (TaqMan Reverse Transcription
Reagents; Thermo Fisher Scientific, Inc.) and 500 ng of total RNA
were used. RT-qPCR assay was carried out using gene-specific double
fluorescently labelled probes in a CFX96 Touch Real-Time PCR
Detection System (Bio-Rad Laboratories, Inc.). Relative expression
was calculated using the comparative cycle threshold method
(2−ΔΔCq method) (54)
and the endogenous control human β-actin was used as normalizer.
Sequences of the employed TaqMan Real Time PCR assay are reported
in Table I and more detailed
information about the assays are available at: https://eu.idtdna.com/site/order/qpcr/predesignedassay.
Western blotting
For NF-κB (p105/p50 and p65) protein quantification,
20 micrograms of total protein extract (quantified by the BCA
method employing Pierce BCA Protein Assay Kit; cat. no. 23225;
Thermo Fisher Scientific, Inc.) were denatured for 5 min at 98°C
and loaded on SDS polyacrylamide (8%) gel in Tris-glycine buffer
(25 mM Tris, 192 mM glycine, 0.1% SDS). The electro-transfer to
0.2-µm nitrocellulose membrane was performed overnight at
360 mA and 4°C in electro-transfer with CAPS buffer (25 mM Tris,
192 mM glycine, CAPS 10 mM, 10% methanol). Obtained membranes were
stained in Ponceau S solution (MilliporeSigma) to verify protein
transfer and incubated in 25 ml of blocking buffer for 1 h at room
temperature. After three washes in TBST 1X (0.1% Tween-20),
membranes were incubated overnight at 4°C in primary antibody
(complete list of employed antibodies and catalogue number are
reported in Table II). The
following day, membranes were washed in TBST 1X and incubated for 1
h at room temperature, with an appropriate horseradish
peroxidase-conjugated secondary antibody (anti-rabbit IgG
HRP-conjugated; 1:2,000; cat no. 7074P3; Cell Signaling Technology,
Inc.). The primary antibody against β-actin (cat. no. 4970S; Cell
Signaling Technology, Inc.) was used as normalization control
(1:1,000 dilution). Nitrocellulose membrane was incubated with 5 ml
LumiGLO® detection working solution (cat. no. 7003; Cell
Signalling Technology, Inc.) and exposed to x-ray film (Hyperfilm™;
cat. no. 28906836; Cytiva). The original uncropped western blotting
gels and the relative representative Ponceau S staining of the
membranes are shown in Figs. S1,
S2, S4 and S5 (uncropped gels) and Figs. S3 and S6 (Ponceau staining). Images of the
blots were acquired and analyzed using Bio-Rad Image Lab Software
v.6.1 (Bio-Rad Laboratories, Inc.).
 | Table IIComplete list of employed antibodies
in western blot analysis. |
Table II
Complete list of employed antibodies
in western blot analysis.
| Antibody name | Cat. no. | Clonality | Supplier |
|---|
| NF-kB p65 Ab | GTX102090 | Rabbit
polyclonal | GeneTex, Inc. |
| NF-kB p105/p50
Ab | GTX133711 | Rabbit
polyclonal | GeneTex, Inc. |
| β-actin Ab | 4970S | Rabbit
polyclonal | Cell Signaling
Technology, Inc. |
IL-6 protein quantification by ELISA
Cell supernatants collected from Calu-3 cells
infected or not infected with SARS-CoV-2 were collected 24 and 48 h
after the infection. IL-6 released into cell culture supernatants
was measured by ELISA (cat. no. ab46027; Abcam) following the
manufacturer's protocol. Samples were subsequently analyzed on a
Sunrise microplate reader (Tecan Group, Ltd.).
Computational studies
All the computational methodologies were carried out
on a 32 Core AMD Ryzen 9 3905X, 3.5 GHz Linux Workstation (O.S.
Ubuntu 20.04) equipped with GPU (Nvidia Quadro RTX 4000, 8 GB). The
3D structure of NF-κB was obtained from the Protein Data Bank (PDB
code: 1NFK), and the structure of SFN was prepared starting from
its SMILES code with SeeSAR 12.1.0 software [SeeSAR version 12.1.0;
BioSolveIT GmbH, 2022, www.biosolveit.de/SeeSAR]. The docking simulation was
performed with SeeSAR 12.1.0 software considering only the protein
chain A. Binding sites were identified through the 'find unoccupied
binding pockets' option in SeeSAR. A total of 50 independent poses
were generated with the default parameters.
Molecular dynamics (MD) simulations were conducted
with the Gromacs 2021.1 software (55,56) under the Martini 2 CG force field.
CG parametrization of NF-κB and DNA were obtained with martinize2
tool [http://cgmartini.nl/index.php/tools2/proteins-and-bilayers/204-martinize],
setting the martini22 as force field and the activating the
'elastic' option for both the protein and the DNA. Protein chains
were not merged. CG parametrization of SFN and n-nonane were
obtained through the automartini tool (57,58). All the CG systems were subjected
to: in vacuo energy minimization for at most 500 steps;
solvation with CG-water (W) containing the 10% of CG-antifreeze
water (WF); neutralization with the appropriate number of
CG-chlorine ions (CL-); energy minimization for at most 10,000
steps; NVT equilibration with position restraints for 10 nanosec
and dt=0.01 ps; NVT equilibration with position restraints for 10
nanosec and dt=0.02 ps; NPT equilibration with position restraints
for 50 nanosec and dt=0.02 ps. Production MDs were run for 100
nanosec with dt=0.02 ps at T=300°K. Barostat, thermostat and the
other MD parameters were set according to the general indications
reported for the Martini CG force field. Distances and RMSD were
obtained using the 'gmx distance' and 'gmx rms' tools in Gromacs
(57,58).
Statistical analysis
All the data were normally distributed and presented
as the mean ± S.D. Comparison of NF-κB (p105/p50 and p65), IL-1β,
IL-6 and IL-8 expression levels between SARS-Cov-2-infected Calu-3
cells and SFN-treated cells was performed using paired Student's
t-test. Comparison among intracellular production of SARS-CoV-2
genomes under different treatment conditions, measured after 24 and
48 hpi, was performed using a two-way analysis of variance (ANOVA),
followed by Bonferroni's post-hoc tests. For statistical analyses,
the STATISTICA version 7.1 software (StatSoft, Inc.) was employed.
P<0.05 was considered to indicate a statistically significant
difference.
Results
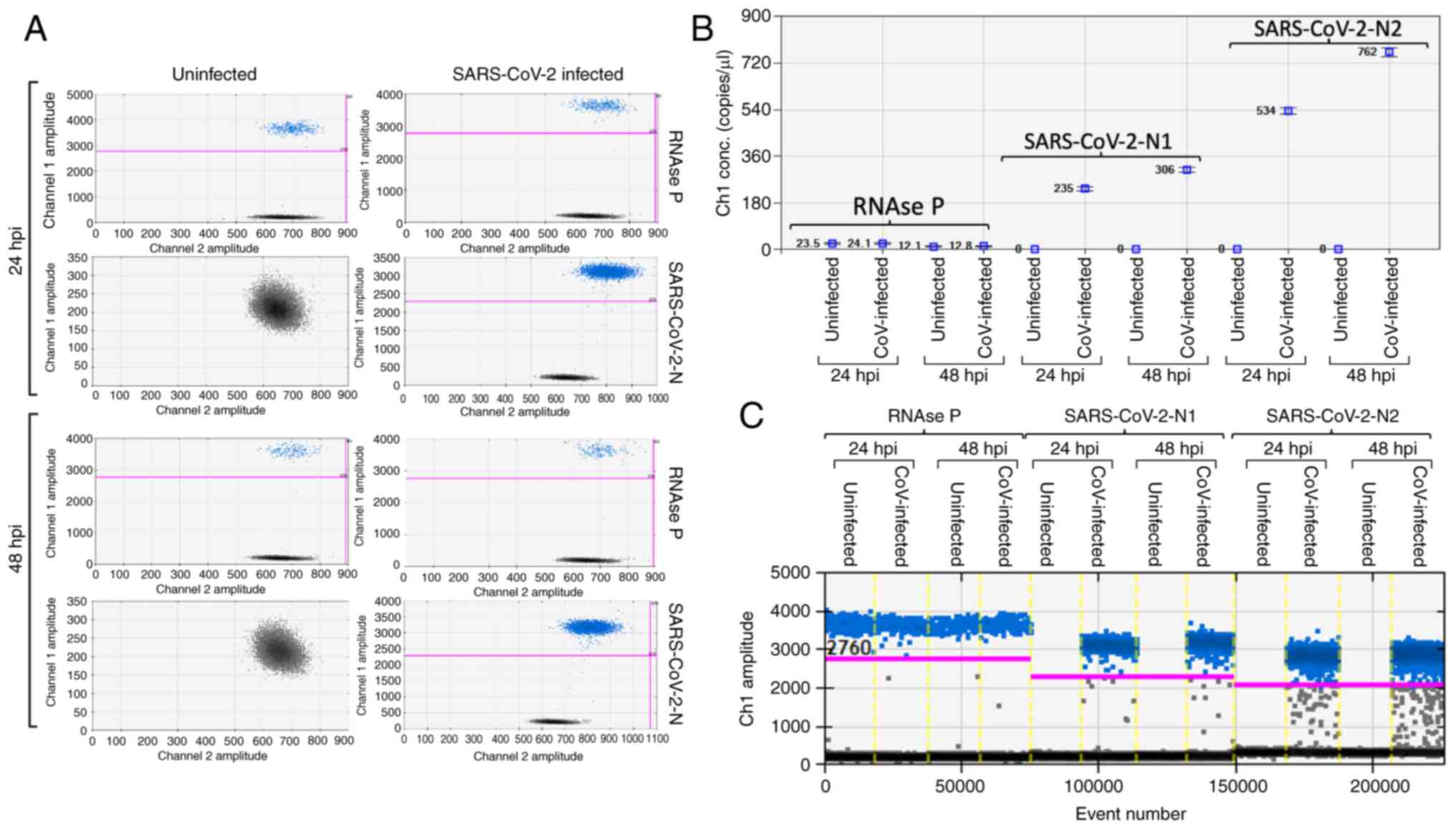
Infection of Calu-3 cells with SARS-CoV-2
is associated with upregulation of NF-κB and NF-κB dependent gene
expression levels
The susceptibility of Calu-3 cells to SARS-CoV-2
infection was assayed by infecting a single type cell with a MOI of
0.1 for 2 h at 37°C (~2×105 infectious viral particles
per well, containing 106 cells). Following infection,
the cells were cultured for 24 and 48 h, and the infected cells
were collected following washing in order to remove the cell-free
viral particles. For viral RNA detection, RNA extraction was
performed 24 and 48 hpi as aforementioned and the viral load within
Calu-3 cells was detected at 24 and 48 hpi by RT-ddPCR using
SARS-CoV-2 RUO qPCR Primer & Probe kit (Integrated DNA
Technologies, Inc.). This method can detect two sequences in the
SARS-CoV-2 nucleocapsid (N) protein region (N1 and N2) and uses the
human RNAse P as a normalizer. The number of copies of the
SARS-CoV-2 were in the order of 109 copies/µg of
isolated RNA. The data obtained fully supported the conclusion that
efficient intracellular replication of SARS-CoV-2 was fully
achieved within 24 h (Fig.
1A-C).
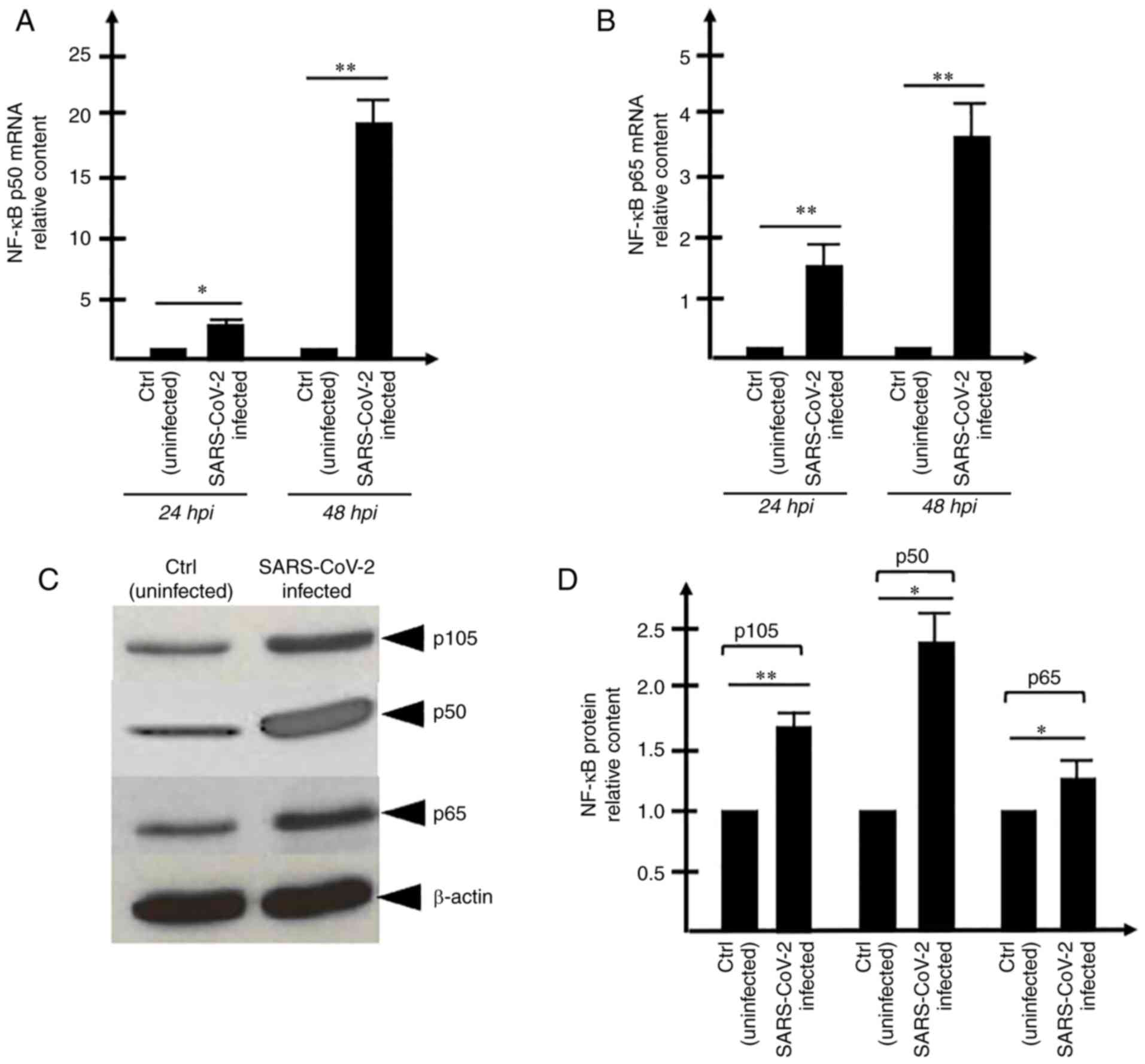
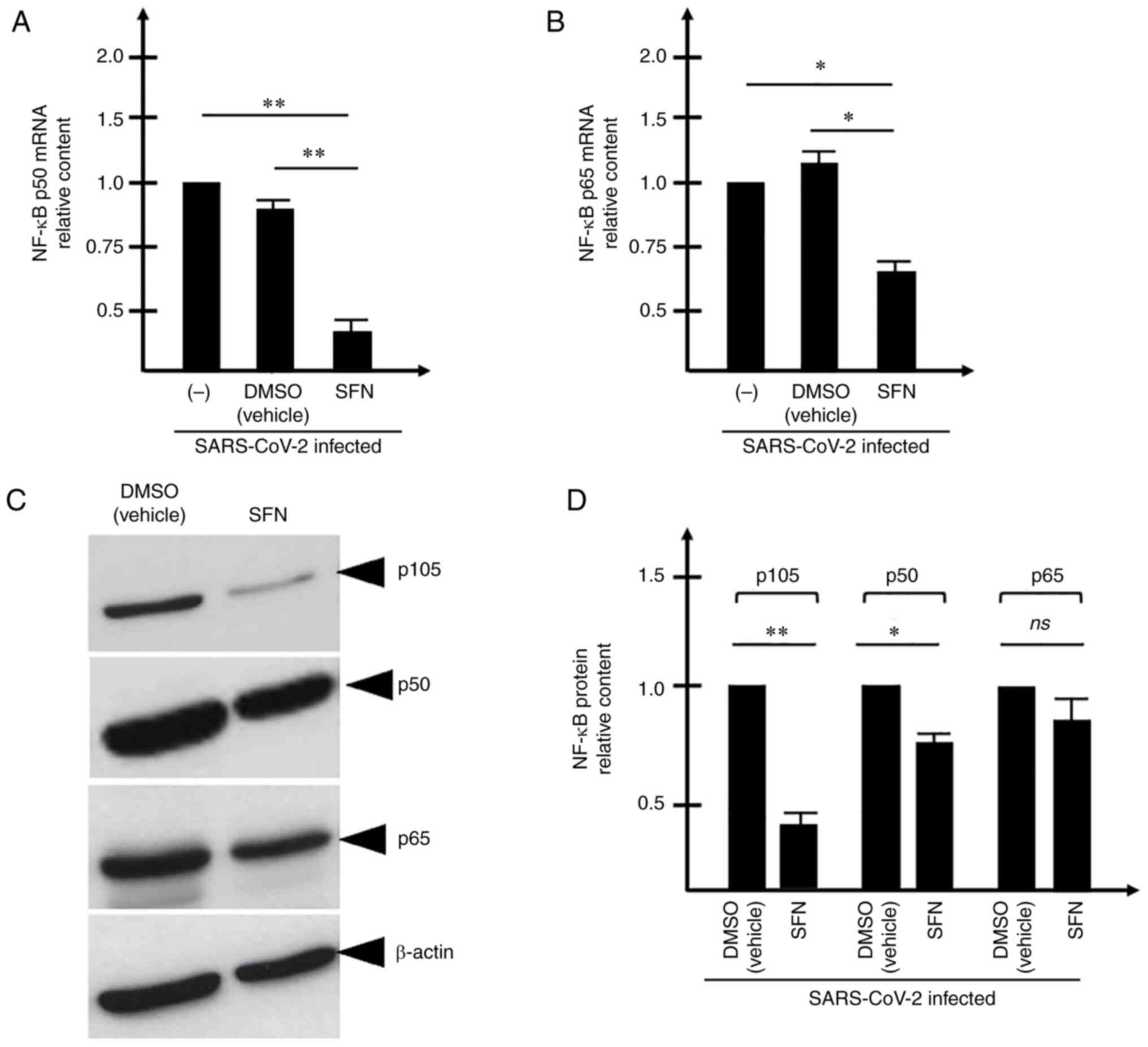
The concept that increase in the expression levels
of the pro-inflammatory transcription factor NF-κB occurs and is
detectable following 24 and 48 h of SARS-CoV-2 infection was
supported by the present findings (Fig. 2). This was particularly evident
when the analysis was performed on NF-κB p50 mRNA (Fig. 2A) and NF-κBp65 mRNA (Fig. 2B) using RNA isolated from 48-h
infected cells. A trend similar to these RT-qPCR data was obtained
by performing the western blot experiment (Fig. 2C and D), indicating that the
relative content of p105, p50 and p65 NF-κB proteins is increased
in SARS-CoV-2 infected cells. The differences in the increased
levels found following RT-qPCR and western blotting were
expectable, as the sensitivities of these two experimental
approaches are different and western blotting, unlike RT-qPCR, is
not quantitative. Since the increased content of the NF-κB mRNAs
(Fig. 2A and B) and proteins
(Fig. 2C and D) was assessed in
SARS-infected Calu-3 cells, the possible effects of SARS-CoV-2
infection were examined on the expression levels of NF-κB regulated
genes. The first effects noted on the changes of the transcription
factors are expected to involve the contents of the transcripts on
the regulated genes. Therefore, the present study focused on
examining the expression levels of IL-1β, IL6 and IL-8 mRNAs, which
encode for proteins that belong to the so-called 'COVID-19 cytokine
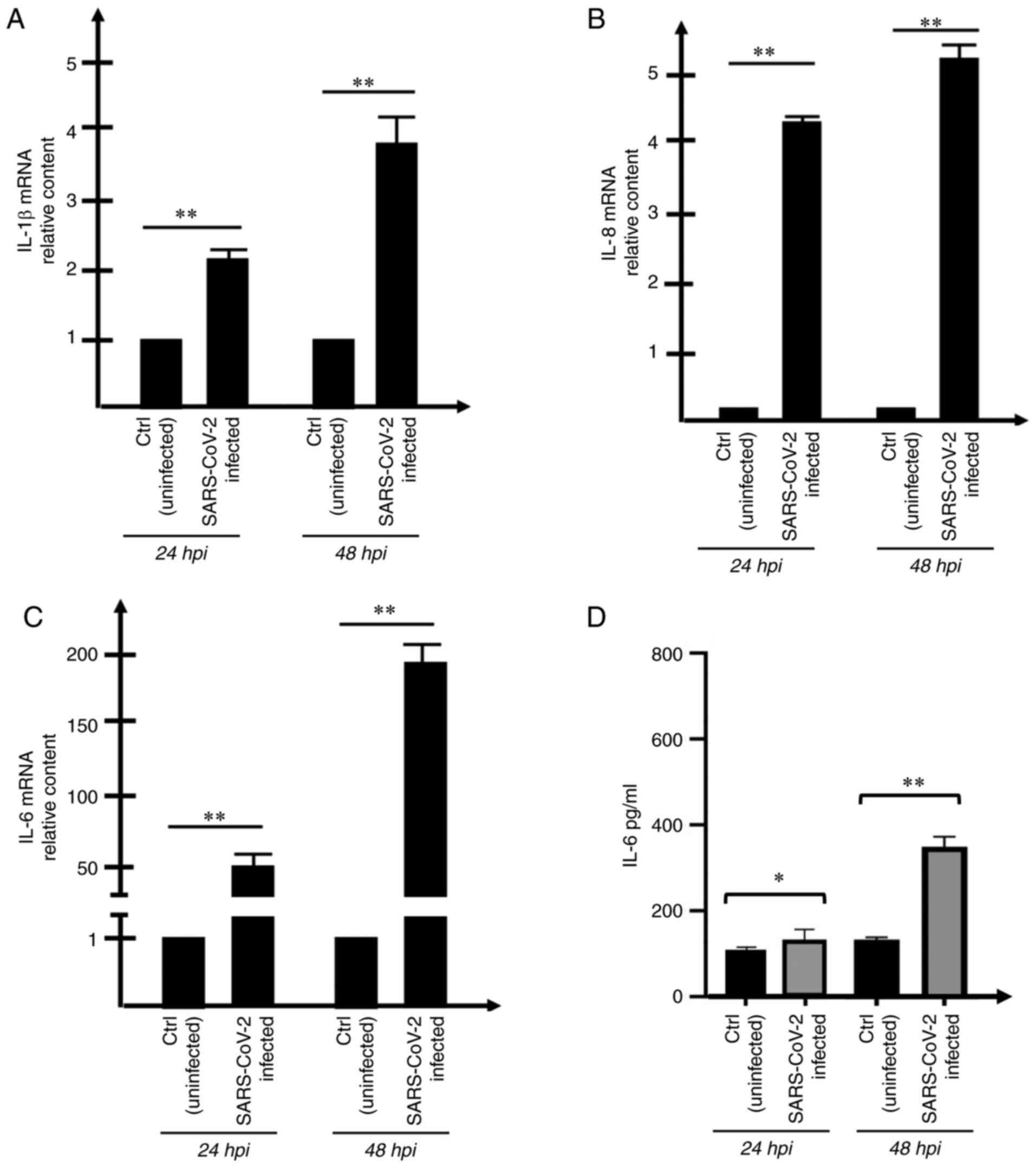
storm' (Fig. 3A-D).
The increase in the extracellular release of the
protein encoded by the most upregulated gene (IL-6; Fig. 3C) was confirmed by ELISA following
quantification of the released IL-6 protein in the supernatants of
the SARS-CoV-2-infected cell cultures (Fig. 3D). As expected, the increased
release of IL-6 was found, in particular, in Calu-3 cultures
exposed for 48 h to SARS-CoV-2. Focus was particularly addressed on
IL-6 because the release of this protein has a central role in
COVID-19 cytokine storm (13,14). IL-1β, that together with IL-6
plays a major role in the cytokine storm (17) was not considered, since it is
released at very low level in Calu3 cells (data not shown).
Binding of SFN to NF-kB: a bioinformatic
analysis
One of the possible mechanisms of action of SFN was
the inhibition of the activity of certain transcription factors. In
this regard, it is well established that SFN induces nuclear factor
erythroid 2-related factor. Among possible transcription factors
involved in the SARS-CoV-2 life cycle and in the activation of the
'cytokine storm', NF-κB is of relevance, since inhibitors of NF-κB
and STAT-3 were found to be particularly effective against
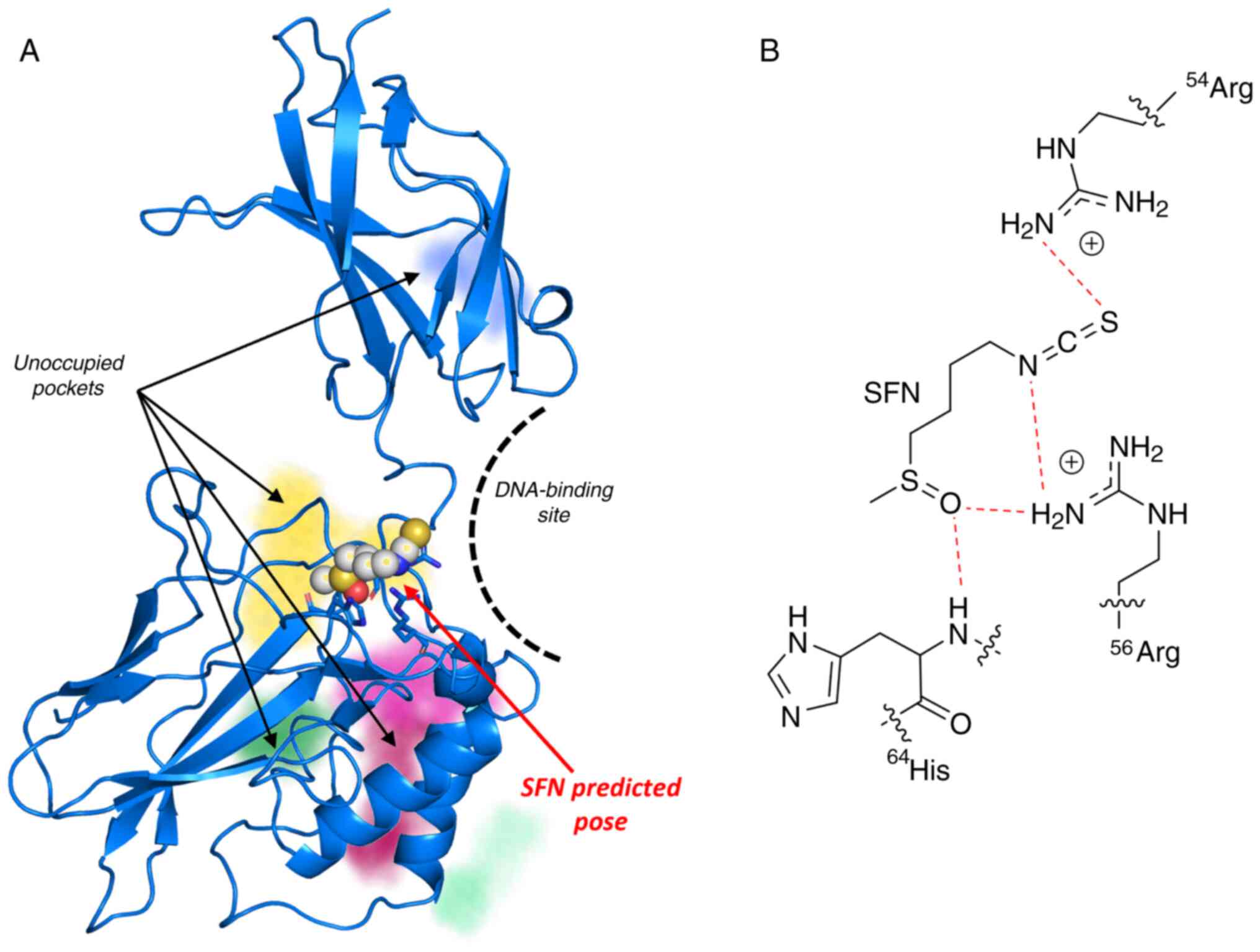
SARS-CoV-2. As a first approach, it was assessed by
docking-experiments whether SFN can bind to NF-kB. The docking
studies were conducted using the software SeeSAR, starting from the
crystallographic complex of NF-κB with DNA (PDB-id: 1NFK). The p60
monomer structure was extracted from the complex and the potential
binding sites were automatically determined through the 'find
unoccupied binding pockets' option in SeeSAR (Fig. 4A). SFN was subsequently docked
against all the binding sites, generating 50 poses. A total of 48
poses were generated in the same region, suggesting that the most
probable binding site was surrounded by Arg54, Arg56 and His64
(Fig. 4B). The superimposition of
the binding pose of SFN with the full 1NFK structure indicated that
the compound-protein interaction occurred in the DNA binding
region, which impaired the interaction with the nucleic acid. These
results are in consistency with the data reported in a previous
study demonstrating that SFN inhibits NF-κB binding to DNA and
NF-kB-dependent luciferase activity when luc-reporter plasmids were
used under the control of NF-κB regulatory promoter regions
(52).
To further explore the mechanism of the inhibitory
action of SFN on NF-kB, MD studies were conducted. Due to the large
dimensions of the system under consideration, the Martini
coarse-grained (CG) force field was used. Indeed, CG-MD allows the
conduct of long simulations at a reasonable computational cost,
while retaining the majority of the chemical information. Indeed,
the Martini force field has been extensively used to investigate
protein-protein as well as ligand-protein interactions, yielding
results in great accordance with all-atoms simulations (59-61). In the present study, the Martini 2
force field was used along with the elastic network approach in
order to retain the secondary structures of NF-κB and that of the
DNA molecule (62).
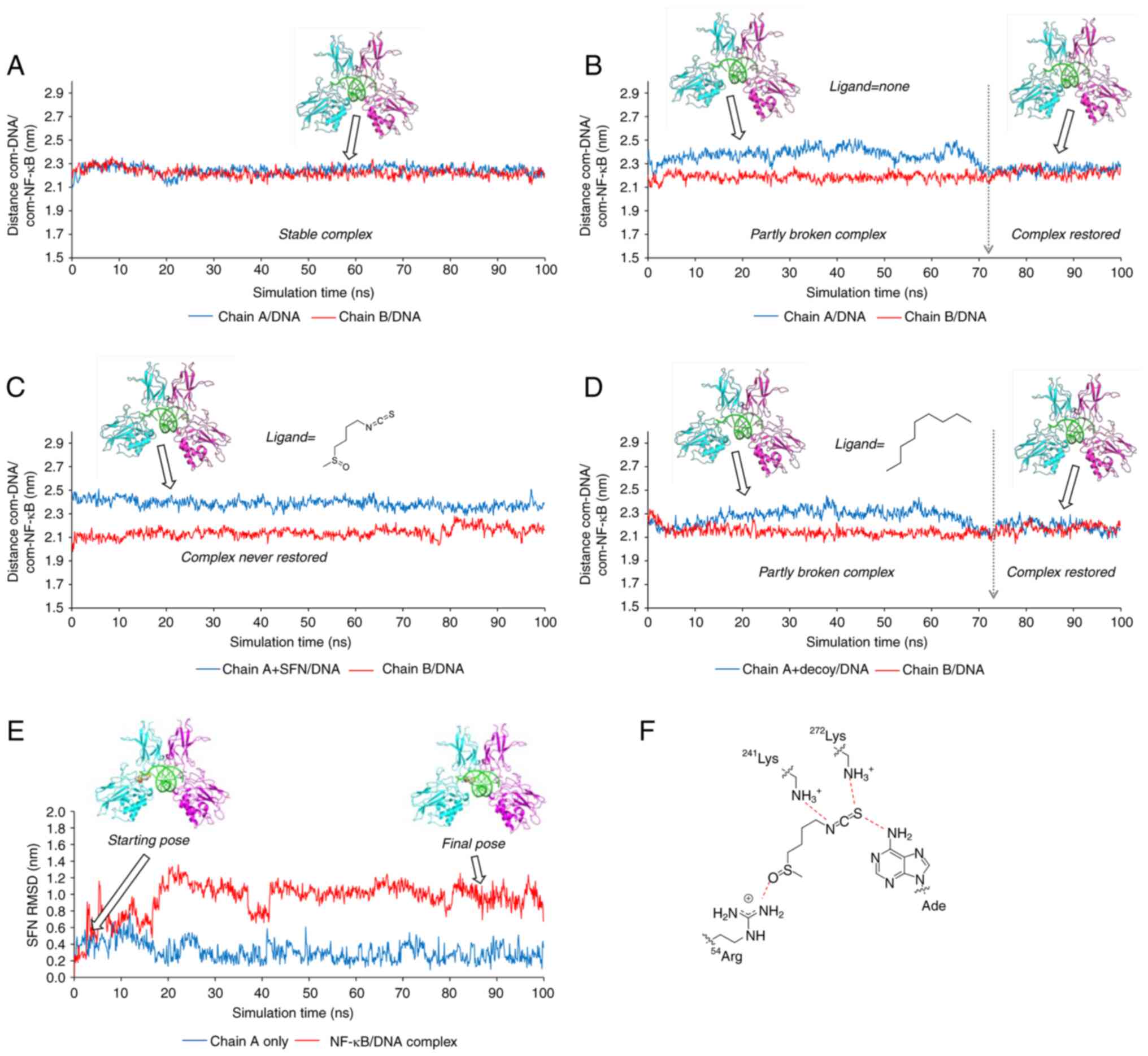
A preliminary CG-MD simulation was run using the
crystallographic complex NF-kB/DNA, measuring the distance between
the centers of the mass (com) of the DNA molecule and each chain of
the NF-κB protein. The system resulted in a highly stable
formulation during the entire simulation time (100 nanosec;
Fig. 5A), with the distances
com-DNA/com-chain A and com-DNA/com-chain B both fluctuating around
the same value (2.23±0.04 nm). Since the molecular docking studies
suggested that SFN could bind to the chain A of NF-κB impairing the
interaction with the DNA, the chain A was slightly shifted from the
nucleic acid, while maintaining the contact with chain B. In this
way a putative structure was prepared where the SFN could be
accommodated in the complex NF-kB/DNA without any clashes with the
DNA. When a CG-MD simulation was run without the SFN, the original
NF-kB/DNA complex was restored within ~72 nanosec (Fig. 5B) as demonstrated by the variation
of the values of com-DNA/com-chain A (t=0-72 nanosec: 2.38±0.06 nm;
t=72-100 nanosec: 2.25±0.03 nm). Conversely, in the presence of
SFN, the original complex was not restored during the 100 nanosec
of the simulation and the distance com-DNA/com-chain A remained
always closed to the displaced value (2.39±0.04 nm), further
suggesting that SFN could impair the correct binding of the
transcription factor with the DNA (Fig. 5C). To exclude that the MD
simulation was biased by the simplification introduced by the CG
approach, the SFN was replaced with a 'decoy' ligand (namely the
n-nonane) characterized by the same number of CG particles
as that of the SFN but with different features (i.e., the
absence of any H-bond acceptor capability and consequently the
absence of the binding ability with the protein). As revealed in
Fig. 5D, the presence of the
nonane (readily displaced from the protein) had no effect on the
time required to restore the complex (~72 nanosec; Fig. 5D with Fig. 5B).
The inspection of the ligand root mean square
deviation along the trajectory revealed that the SFN changed its
position and interactions during the simulation (red curve in
Fig. 5E). This displacement was
not observed when the CG-MD was run considering only SFN and
chain-A (blue curve in Fig. 5E).
Accordingly, the following conclusions could be postulated that: i)
SFN was stably bound to the NF-κB monomer; ii) a ternary
NF-kB/SFN/DNA complex was formed; iii) SFN interacted with both the
protein and the nucleic acid modifying its binding mode (Fig. 5F) and impairing the full
interaction between the NF-κB and DNA molecules, which finally
stabilized the inactive complex.
SFN inhibits SARS-CoV-2 intracellular
replication and viral release from infected cells
In order to determine the effects of SFN on
SARS-CoV-2 infection, Calu-3 cells were infected with SARS-CoV-2
and the effects of SFN were determined on the extracellular release
of SARS-CoV-2 genomes after 24 and 48 hpi and on the intracellular
production of SARS-CoV-2 genomes. The results obtained in
Calu-3-infected, SFN-treated cells were compared with
Calu-3-infected cells cultured in the presence of DMSO (the vehicle
of SFN), as DMSO affects both extracellular release and
intracellular production of SARS-CoV-2 genomes. As demonstrated in
Fig. 6A-C, treatment of the
SARS-CoV-2-infected Calu-3 cells with SFN was associated with
inhibition of SARS-CoV-2 replication. As expected, the
extracellular release of SARS-CoV-2 genomes was significantly
higher 48 hpi (Fig. 6A).
Moreover, SFN was able to inhibit the extracellular release of
SARS-CoV-2 genomes both at 24 and 48 hpi.
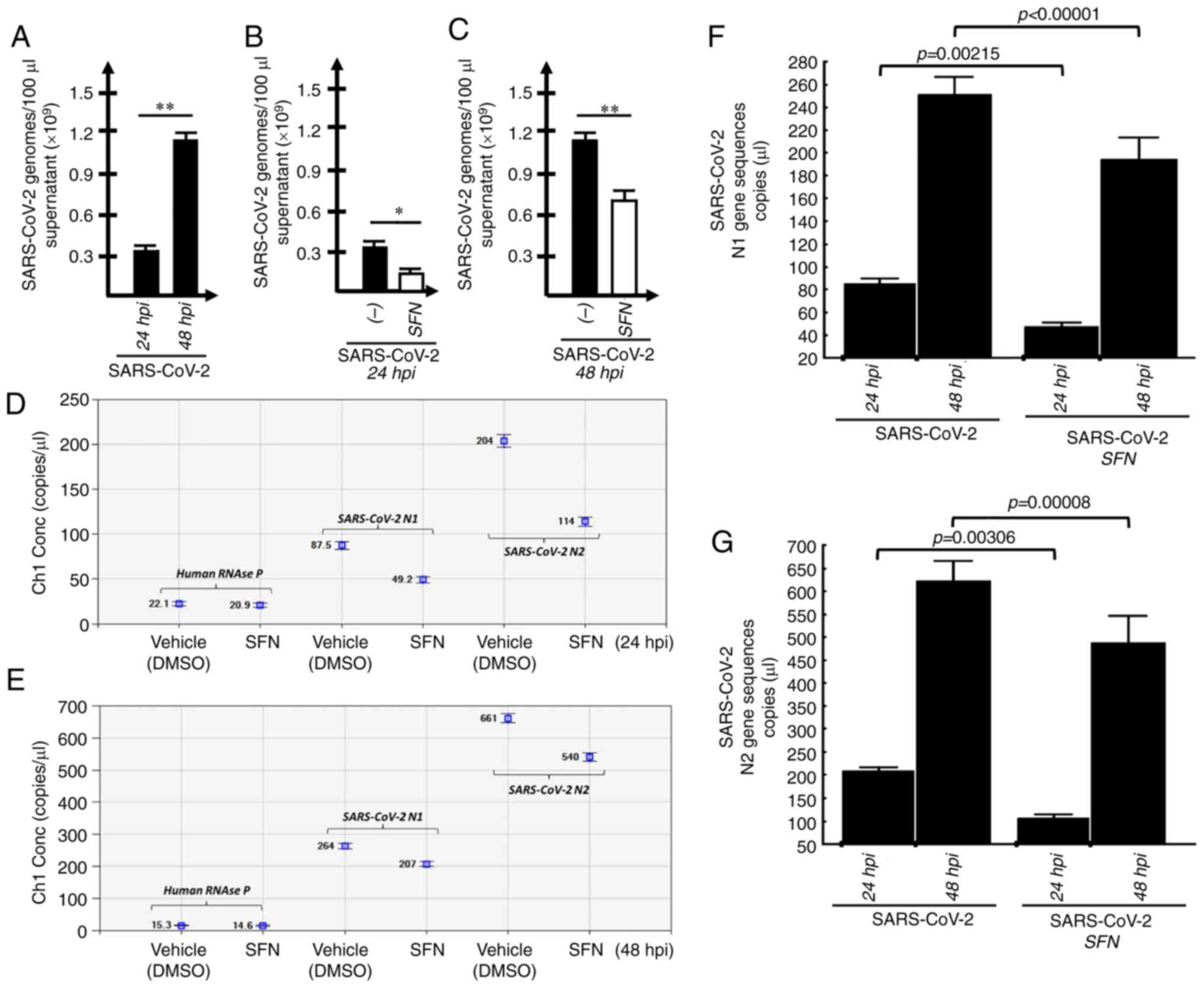 | Figure 6Quantification of SARS-CoV-2 genomes
in Calu-3 SARS-Cov-2-infected and SFN-treated cells. (A-C) The
amount of SARS-CoV-2-released genomes was determined by absolute
reverse transcription-quantitative PCR and compared in (A) Calu-3
infected cells analyzed 24 and 48 hpi, or in Calu-3 infected cells
treated with SFN or with the SFN vehicle DMSO and analyzed (B) 24
and (C) 48 hpi (n=3) *P<0.05, significant;
**P<0.01, highly significant. (D and E)
Representative reverse transcription-digital-droplet dPCR 2D plots
quantifying intracellular SARS-CoV-2 genomic N1 and N2 sequences
(as indicated); samples were isolated at (D) 24 and (E) 48 hpi time
points. (F and G) Two-way ANOVA, followed by post hoc Bonferroni
test (see also Fig. S7)
performed on samples from SARS-CoV-2-infected Calu-3 cells in the
absence or in the presence of SFN, as indicated. Cells were
harvested at 24 and 48 hpi. The results represent the mean ± S.D.
(n=6). The analysis of the data shown in panels F and G is further
detailed in Fig. S7. For
SARS-CoV-2 genome quantification, the N1 (F) and N2 (G) sequences
have been considered. The P-values reported in panels F and G
(two-way ANOVA, followed by post hoc Bonferroni test) were obtained
comparing untreated infected cells (SARS-CoV-2) vs. SFN-treated
infected cells (SARS-CoV-2 + SFN). Results represent the mean ±
S.D. SARS-CoV-2, severe acute respiratory syndrome coronavirus 2;
hpi, h post-infection; SFN, sulforaphane. |
This latter conclusion was fully supported by the
results obtained studying the intracellular SARS-CoV-2 genome
copies detected by RT-ddPCR (representative examples are shown in
Fig. 6D and E). In order to
quantify the intracellular SARS-CoV-2 content, two assays, which
were both able to amplify nucleocapsid protein mRNA, yet in
different regions, were employed. The human RNAse P sequence was
used as a template loading control. The results obtained
demonstrated a decrease of intracellular SARS-CoV-2 genomes
following SFN treatment of SARS-CoV-2 infected cells studied 24 and
48 hpi (Fig. 6F and G).
Two-way ANOVA, shown in Fig. S7 and summarized in (Fig. 6F and G), demonstrated a strong
significance (P<0.0001) of both main effects (24 vs. 48 hpi and
different treatments) for both N1 (Fig. S7A) and N2 (Fig. S7B) gene sequences. Specifically,
post-hoc comparisons revealed that the difference found when
analysing samples at 24 and 48 hpi was highly significant
(P<0.0001 for both N1 and N2 gene sequences, Fig. S7) considering time, as expected,
as a major parameter affecting the total accumulation of
intracellular SARS-CoV-2 sequences. In addition, the differences
between SFN-treated SARS-CoV-2-infected cells and control
DMSO-cultured SARS-CoV-2-infected cells were also found highly
significant both at 24 and 48 hpi, considering both N1 (Fig. 6F) and N2 (Fig. 6G) gene sequences.
Collectively, the data shown in Fig. 6 demonstrated that SFN inhibited
SARS-CoV-2 intracellular replication (Fig. 6F and G) as well as the viral
release in the extracellular environment (Fig. 6B and C). Despite the fact that the
inhibitory effects were slightly different when the two RT-PCR
methods (absolute RT-qPCR and RT-ddPCR) were compared, the results
obtained were consistent with the hypothesis stating that SFN
inhibits SARS-CoV-2 life cycle.
Expression of NF-κB is downregulated in
SARS-CoV-2 infected, SFN-treated Calu-3 cells
Since it has been reported that the SARS-CoV
N-protein binds to the NF-κB promoter, thereby stimulating the
NF-κB transcription (63),
further investigations were conducted to determine whether lower
expression of the N-protein gene (Fig. 6F and G) was associated with lower
expression of NF-kB. Therefore, the content of NF-κB p50 and p65
mRNA was initially analyzed in SARS-CoV-2-infected, SFN-treated
Calu-3 cells (Fig. 7A and B).
The results demonstrated that the contents of NF-κB
p50 and p65 mRNA were significantly lower when samples from
SARS-CoV-2-infected, SFN-treated Calu-3 cells were compared with
samples of SARS-CoV-2-infected cells. Western blotting confirmed
this trend, as indicated in Fig. 7C
and D.
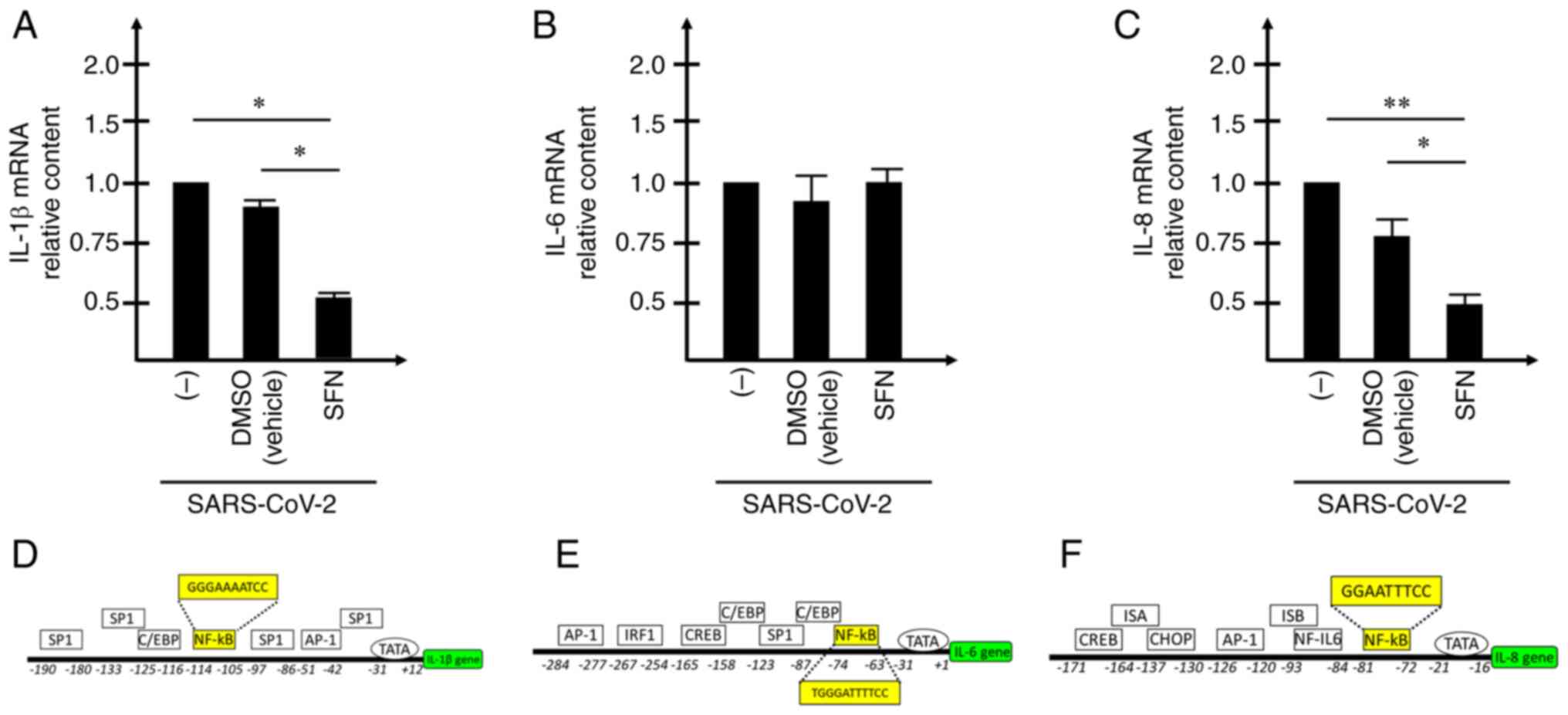
Expression of NF-κBregulated
pro-inflammatory genes in SARS-CoV-2-infected, SFN-treated Calu-3
cells
Considering the inhibitory effects on NF-κB
(Fig. 7A and B), subsequent
studies aimed to determined whether the expression levels of
pro-inflammatory genes were modified following treatment of
SARS-CoV-2-infected Calu-3 cells with SFN. To this end, the
expression levels of the mRNAs encoding IL-1β, IL-6 and IL-8 were
assessed by RT-qPCR in SARS-CoV-2 infected, SFN-treated Calu-3
cells (Fig. 8). The results
obtained demonstrated that the expression levels of IL-1β (Fig. 8A) and IL-8 (Fig. 8C) were significantly reduced when
samples from SARS-CoV-2-infected, SFN-treated Calu-3 cells were
compared with samples of SARS-CoV-2-infected cells. It is notable
that all these regulatory proteins belong to the 'COVID-19 cytokine
storm'. Regarding the expression of IL-6, non-significant changes
were detected (Fig. 8B); however,
this may be caused by the high activation of this gene following
SARS-CoV-2 infection (Fig.
3).
Discussion
In the present study, the effects of SFN were
characterized on the bronchial epithelial cell line Calu-3 infected
by SARS-CoV-2 with regard to the replication of the viral genome
and the possible inhibition of gene encoding pro-inflammatory
proteins. The present study is related to previously published
observations indicating that SFN was able to inhibit the expression
levels of IL-6 and IL-8 genes induced by the treatment of IB3-1
bronchial cells with a recombinant spike protein of SARS-CoV-2
(46).
In the current study, the Calu-3 cellular system was
characterized with regard to the SARS-CoV-2-induced increase in the
expression levels of NF-κB and NF-κB-regulated pro-inflammatory
genes. A highly significant increase in the expression levels of
these genes has been demonstrated to be associated with SARS-CoV-2
infection of Calu-3 cells. SFN-mediated inhibition of SARS-CoV-2
replication was analyzed by RT-ddPCR and by RT-qPCR quantification
of the release of SARS-CoV-2 genomes by the infected cells. It was
found that SFN could inhibit SARS-CoV-2 replication by analyzing
either the intracellular amount of N-protein sequences or the
release of the virus; this led to the subsequent analysis of the
SARS-CoV-2 genomes present in the supernatants of SFN-treated,
SARS-CoV-2-infected Calu-3 cells. With the regard to its effects on
the pro-inflammatory genes, SFN was able to inhibit upregulation of
NF-κB expression in SARS-CoV-2-infected cells. It is worth noting
that molecular docking studies suggested a direct interaction of
SFN to the DNA binding region of NF-κB. Further MD simulations
provided additional information on the potential mechanism of
action of SFN at the molecular level, suggesting that this compound
could impair the formation of the effective protein/DNA complex
(Video S1). The results obtained
from the quantification of mRNAs encoding IL-1β, IL-6 and IL-8,
indicated that SFN exerted an inhibitory effect on IL-1β and IL-8
mRNA levels. The assessment of the SFN effects on IL-6 should be
further analyzed, as the downregulation of this pro-inflammatory
gene was not detectable under these experimental conditions,
possibly due to the very high levels of SARS-CoV-2-mediated
induction (>180 fold, when infected cells were compared with
uninfected cells).
The data obtained in the present study are in strong
agreement with other studies indicating inhibitory effects of SFN
on SARS-CoV-2 life cycle (48)
and the possible use of this compound in the management of patients
with COVID-19 patients, as recently suggested in different studies
(64-67).
Based on the evidence, it is important to note that
the management of the COVID-19 pandemic requires antiviral agents
targeting SARS-CoV-2 life cycle with high efficiency despite the
approval, testing, and worldwide distribution of anti-SARS-CoV-2
vaccines (4-8).
The present study contains certain limitations, the
most relevant being that only a low number of pro-inflammatory
genes were studied in SARS-CoV-2-infected cells. Therefore, the
present study should be considered a proof-of-principle study
indicating that SFN may be a double-acting agent (a SARS-CoV-2
replication inhibitor and an anti-inflammatory compound).
The analysis of other pro-inflammatory genes
involved in the COVID-19 'cytokine storm' should be conducted in
order to determine whether the SFN-mediated inhibitory effects
noted on the expression levels of IL-1β and IL-8 can be generalized
to all the other genes involved in the COVID-19 associated
hyper-inflammatory state. This experimental plan will also clarify
the relationship between the SFN-mediated effects on genes involved
in the COVID-19 'cytokine storm' and the corresponding alterations
noted in the regulation of the NF-κB pathway.
Supplementary Data
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
JG, AF, GM, VG and RG curated data. JG, GM, CS, VG,
RG and AF performed formal analysis. RG acquired funding. JG, CP,
MZ, GM and VG conducted investigation. JG, CP, CS, VG, RR and GM
developed methodology. RG, AF, RR, CS, and GM supervised the study.
JG, RG, AF, GM, VG and RR wrote the original draft. JG, GM, CP, MZ,
VG, RR, CS, RG and AF wrote, reviewed and edited the manuscript.
All authors have read and approved the final version of the
manuscript. JG, AF and RG confirm the authenticity of all the raw
data.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Abbreviations:
|
SFN
|
sulforaphane
|
|
COVID-19
|
coronavirus disease 2019
|
|
SARS-CoV-2
|
severe acute respiratory syndrome
coronavirus 2
|
|
IL
|
interleukin
|
|
RT-qPCR
|
reverse transcription-quantitative
polymerase-chain reaction
|
|
NF-kB
|
nuclear factor-kappa B
|
Acknowledgments
Not applicable.
Funding
The present study was supported by the MUR-FISR COVID-miRNAPNA
Project (grant no. FISR2020IP_04128), the Interuniversity
Consortium for Biotechnologies in Italy (grant no.
CIB-Unife-2020-1), the CARIPARO Foundation (grant no.
MARZ_CARIVARI20_01 C94I20002500007) and the FIRC-AIRC 'Michele e
Carlo Ardizzone' fellowship (grant no. 25528).
References
|
1
|
Walker PGT, Whittaker C, Watson OJ,
Baguelin M, Winskill P, Hamlet A, Djafaara BA, Cucunubá Z, Olivera
Mesa D, Green W, et al: The impact of COVID-19 and strategies for
mitigation and suppression in low- and middle-income countries.
Science. 369:413–422. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Khan S, Siddique R, Ali A, Bai Q, Li Z, Li
H, Shereen MA, Xue M and Nabi G: The spread of novel coronavirus
has created an alarming situation worldwide. J Infect Public
Health. 13:469–471. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Killerby ME, Biggs HM, Midgley CM, Gerber
SI and Watson JT: Middle east respiratory syndrome coronavirus
transmission. Emerg Infect Dis. 26:191–198. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Watson OJ, Barnsley G, Toor J, Hogan AB,
Winskill P and Ghani AC: Global impact of the first year of
COVID-19 vaccination: A mathematical modelling study. Lancet Infect
Dis. 22:1293–1302. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Tulimilli SV, Dallavalasa S, Basavaraju
CG, Kumar Rao V, Chikkahonnaiah P, Madhunapantula SV and Veeranna
RP: Variants of severe acute respiratory syndrome coronavirus 2
(SARS-CoV-2) and vaccine effectiveness. Vaccines (Basel).
10:17512022. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Villamagna AH, Gore SJ, Lewis JS and
Doggett JS: The need for antiviral drugs for pandemic coronaviruses
from a global health perspective. Front Med (Lausanne).
7:5965872020. View Article : Google Scholar
|
|
7
|
Takashita E, Kinoshita N, Yamayoshi S,
Sakai-Tagawa Y, Fujisaki S, Ito M, Iwatsuki-Horimoto K, Halfmann P,
Watanabe S, Maeda K, et al: Efficacy of antiviral agents against
the SARS-CoV-2 omicron subvariant BA.2. N Engl J Med.
386:1475–1477. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kumari M, Lu RM, Li MC, Huang JL, Hsu FF,
Ko SH, Ke FY, Su SC, Liang KH, Yuan JP, et al: A critical overview
of current progress for COVID-19: Development of vaccines,
antiviral drugs, and therapeutic antibodies. J Biomed Sci.
29:682022. View Article : Google Scholar
|
|
9
|
Zhu C, Lee JY, Woo JZ, Xu L, Nguyenla X,
Yamashiro LH, Ji F, Biering SB, Van Dis E, Gonzalez F, et al: An
intranasal ASO therapeutic targeting SARS-CoV-2. Nat Commun.
13:45032022. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Mondal S, Quintili AL, Karamchandani K and
Bose S: Thromboembolic disease in COVID-19 patients: A brief
narrative review. J Intensive Care. 8:702020. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zong X, Gu Y, Yu H, Li Z and Wang Y:
Thrombocytopenia is associated with COVID-19 severity and outcome:
An updated meta-analysis of 5637 patients with multiple outcomes.
Lab Med. 52:10–15. 2021. View Article : Google Scholar
|
|
12
|
Smilowitz NR, Kunichoff D, Garshick M,
Shah B, Pillinger M, Hochman JS and Berger JS: C-reactive protein
and clinical outcomes in patients with COVID-19. Eur Heart J.
42:2270–2279. 2021. View Article : Google Scholar
|
|
13
|
Santa Cruz A, Mendes-Frias A, Oliveira AI,
Dias L, Matos AR, Carvalho A, Capela C, Pedrosa J, Castro AG and
Silvestre R: Interleukin-6 is a biomarker for the development of
fatal severe acute respiratory syndrome coronavirus 2 pneumonia.
Front Immunol. 12:6134222021. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hu B, Huang S and Yin L: The cytokine
storm and COVID-19. J Med Virol. 93:250–256. 2021. View Article : Google Scholar
|
|
15
|
Iwasaki M, Saito J, Zhao H, Sakamoto A,
Hirota K and Ma D: Inflammation triggered by SARS-CoV-2 and ACE2
augment drives multiple organ failure of severe COVID-19: Molecular
mechanisms and implications. Inflammation. 44:13–34. 2021.
View Article : Google Scholar
|
|
16
|
Yang J, Petitjean SJL, Koehler M, Zhang Q,
Dumitru AC, Chen W, Derclaye S, Vincent SP, Soumillion P and
Alsteens D: Molecular interaction and inhibition of SARS-CoV-2
binding to the ACE2 receptor. Nat Commun. 11:45412020. View Article : Google Scholar :
|
|
17
|
Song P, Li W, Xie J, Hou Y and You C:
Cytokine storm induced by SARS-CoV-2. Clin Chim Acta. 509:280–287.
2020. View Article : Google Scholar :
|
|
18
|
Hazeldine J and Lord JM: Neutrophils and
COVID-19: Active participants and rational therapeutic targets.
Front Immunol. 12:6801342021. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Veenith T, Martin H, Le Breuilly M,
Whitehouse T, Gao-Smith F, Duggal N, Lord JM, Mian R, Sarphie D and
Moss P: High generation of reactive oxygen species from neutrophils
in patients with severe COVID-19. Sci Rep. 12:104842022. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Sefik E, Qu R, Junqueira C, Kaffe E, Mirza
H, Zhao J, Brewer JR, Han A, Steach HR, Israelow B, et al:
Inflammasome activation in infected macrophages drives COVID-19
pathology. Nature. 606:585–593. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Moss P: The T cell immune response against
SARS-CoV-2. Nat Immunol. 23:186–193. 2022. View Article : Google Scholar
|
|
22
|
Mohammed RN, Tamjidifar R, Rahman HS,
Adili A, Ghoreishizadeh S, Saeedi H, Thangavelu L, Shomali N,
Aslaminabad R, Marofi F, et al: A comprehensive review about immune
responses and exhaustion during coronavirus disease (COVID-19).
Cell Commun Signal. 20:792022. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Majidpoor J and Mortezaee K: Interleukin-6
in SARS-CoV-2 induced disease: Interactions and therapeutic
applications. Biomed Pharmacother. 145:1124192022. View Article : Google Scholar
|
|
24
|
Zizzo G, Tamburello A, Castelnovo L, Laria
A, Mumoli N, Faggioli PM, Stefani I and Mazzone A: Immunotherapy of
COVID-19: Inside and beyond IL-6 signalling. Front Immunol.
13:7953152022. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Khan S, Shafiei MS, Longoria C, Schoggins
JW, Savani RC and Zaki H: SARS-CoV-2 spike protein induces
inflammation via TLR2-dependent activation of the NF-κB pathway.
Elife. 10:e685632021. View Article : Google Scholar
|
|
26
|
Matsuyama T, Kubli SP, Yoshinaga SK,
Pfeffer K and Mak TW: An aberrant STAT pathway is central to
COVID-19. Cell Death Differ. 27:3209–3225. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kaiser AE, Baniasadi M, Giansiracusa D,
Giansiracusa M, Garcia M, Fryda Z, Wong TL and Bishayee A:
Sulforaphane: A broccoli bioactive phytocompound with cancer
preventive potential. Cancers (Basel). 13:47962021. View Article : Google Scholar
|
|
28
|
Barba FJ, Nikmaram N, Roohinejad S, Khelfa
A, Zhu Z and Koubaa M: Bioavailability of glucosinolates and their
breakdown products: Impact of processing. Front Nutr. 3:242016.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Narbad A and Rossiter JT: Gut
glucosinolate metabolism and isothiocyanate production. Mol Nutr
Food Res. 62:e17009912018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lenzi M, Fimognari C and Hrelia P:
Sulforaphane as a promising molecule for fighting cancer. Cancer
Treat Res. 159:207–223. 2014. View Article : Google Scholar
|
|
31
|
Kubo E, Chhunchha B, Singh P, Sasaki H and
Singh DP: Sulforaphane reactivates cellular antioxidant defense by
inducing Nrf2/ARE/Prdx6 activity during aging and oxidative stress.
Sci Rep. 7:141302017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Haristoy X, Angioi-Duprez K, Duprez A and
Lozniewski A: Efficacy of sulforaphane in eradicating Helicobacter
pylori in human gastric xenografts implanted in nude mice.
Antimicrob Agents Chemother. 47:3982–3984. 2003. View Article : Google Scholar
|
|
33
|
Schepici G, Bramanti P and Mazzon E:
Efficacy of sulforaphane in neurodegenerative diseases. Int J Mol
Sci. 21:86372020. View Article : Google Scholar :
|
|
34
|
Li YP, Wang SL, Liu B, Tang L, Kuang RR,
Wang XB, Zhao C, Song XD, Cao XM, Wu X, et al: Sulforaphane
prevents rat cardiomyocytes from hypoxia/reoxygenation injury in
vitro via activating SIRT1 and subsequently inhibiting ER stress.
Acta Pharmacol Sin. 37:344–353. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Qi T, Xu F, Yan X, Li S and Li H:
Sulforaphane exerts anti-inflammatory effects against
lipopolysaccharide-induced acute lung injury in mice through the
Nrf2/ARE pathway. Int J Mol Med. 37:182–188. 2016. View Article : Google Scholar
|
|
36
|
Folkard DL, Marlow G, Mithen RF and
Ferguson LR: Effect of sulforaphane on NOD2 via NF-κB: Implications
for Crohn's disease. J Inflamm (Lond). 12:62015. View Article : Google Scholar
|
|
37
|
Negi G, Kumar A and Sharma SS: Nrf2 and
NF-κB modulation by sulforaphane counteracts multiple
manifestations of diabetic neuropathy in rats and high
glucose-induced changes. Curr Neurovasc Res. 8:294–304. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Xu C, Shen G, Chen C, Gélinas C and Kong
ANT: Suppression of NF-kappaB and NF-kappaB-regulated gene
expression by sulforaphane and PEITC through IkappaBalpha, IKK
pathway in human prostate cancer PC-3 cells. Oncogene.
24:4486–4495. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Liu T, Zhang L, Joo D and Sun SC: NF-κB
signaling in inflammation. Signal Transduct Target Ther.
2:170232017. View Article : Google Scholar
|
|
40
|
Libermann TA and Baltimore D: Activation
of interleukin-6 gene expression through the NF-kappa B
transcription factor. Mol Cell Biol. 10:2327–2334. 1990.PubMed/NCBI
|
|
41
|
McFarland BC, Hong SW, Rajbhandari R,
Twitty GB Jr, Gray GK, Yu H, Benveniste EN and Nozell SE:
NF-κB-induced IL-6 ensures STAT3 activation and tumor
aggressiveness in glioblastoma. PLoS One. 8:e787282013. View Article : Google Scholar
|
|
42
|
Elliott CL, Allport VC, Loudon JA, Wu GD
and Bennett PR: Nuclear factor-kappa B is essential for
up-regulation of interleukin-8 expression in human amnion and
cervical epithelial cells. Mol Hum Reprod. 7:787–790. 2001.
View Article : Google Scholar
|
|
43
|
Fahey JW and Kensler TW: The challenges of
designing and implementing clinical trials with broccoli sprouts…
and turning evidence into public health action. Front Nutr.
8:6487882021. View Article : Google Scholar
|
|
44
|
Yagishita Y, Fahey JW, Dinkova-Kostova AT
and Kensler TW: Broccoli or sulforaphane: Is it the source or dose
that matters? Molecules. 24:35932019. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Yagishita Y, Gatbonton-Schwager TN,
McCallum ML and Kensler TW: Current landscape of NRF2 biomarkers in
clinical trials. Antioxidants (Basel). 9:7162020. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Zimmerman AW, Singh K, Connors SL, Liu H,
Panjwani AA, Lee LC, Diggins E, Foley A, Melnyk S, Singh IN, et al:
Randomized controlled trial of sulforaphane and metabolite
discovery in children with autism spectrum disorder. Mol Autism.
12:382021. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Gasparello J, D'Aversa E, Papi C, Gambari
L, Grigolo B, Borgatti M, Finotti A and Gambari R: Sulforaphane
inhibits the expression of interleukin-6 and interleukin-8 induced
in bronchial epithelial IB3-1 cells by exposure to the SARS-CoV-2
spike protein. Phytomedicine. 87:1535832021. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Ordonez AA, Bullen CK, Villabona-Rueda AF,
Thompson EA, Turner ML, Merino VF, Yan Y, Kim J, Davis SL, Komm O,
et al: Sulforaphane exhibits antiviral activity against pandemic
SARS-CoV-2 and seasonal HCoV-OC43 coronaviruses in vitro and in
mice. Commun Biol. 5:2422022. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Bortolotti D, Gentili V, Rizzo S, Schiuma
G, Beltrami S, Strazzabosco G, Fernandez M, Caccuri F, Caruso A and
Rizzo R: TLR3 and TLR7 RNA sensor activation during SARS-CoV-2
infection. Microorganisms. 9:18202021. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Papi C, Gasparello J, Zurlo M, Manicardi
A, Corradini R, Cabrini G, Gambari R and Finotti A: Combined
treatment of bronchial epithelial Calu-3 cells with peptide nucleic
acids targeting miR-145-5p and miR-101-3p: Synergistic enhancement
of the expression of the cystic fibrosis transmembrane conductance
regulator (CFTR) gene. Int J Mol Sci. 23:93482022. View Article : Google Scholar
|
|
51
|
Gasparello J, Papi C, Zurlo M, Gambari L,
Rozzi A, Manicardi A, Corradini R, Gambari R and Finotti A:
Treatment of human glioblastoma U251 cells with sulforaphane and a
peptide nucleic acid (PNA) targeting miR-15b-5p: Synergistic
effects on induction of apoptosis. Molecules. 27:12992022.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Heiss E, Herhaus C, Klimo K, Bartsch H and
Gerhäuser C: Nuclear factor kappa B is a molecular target for
sulforaphane-mediated anti-inflammatory mechanisms. J Biol Chem.
276:32008–32015. 2001. View Article : Google Scholar
|
|
53
|
Caruso A, Caccuri F, Bugatti A, Zani A,
Vanoni M, Bonfanti P, Cazzaniga ME, Perno CF, Messa C and
Alberghina L: Methotrexate inhibits SARS-CoV-2 virus replication
'in vitro'. J Med Virol. 93:1780–1785. 2021. View Article : Google Scholar
|
|
54
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
55
|
Van Der Spoel D, Lindahl E, Hess B,
Groenhof G, Mark AE and Berendsen HJC: GROMACS: Fast, flexible, and
free. Comput Chem. 26:1701–1718. 2005. View Article : Google Scholar
|
|
56
|
Pronk S, Páll S, Schulz R, Larsson P,
Bjelkmar P, Apostolov R, Shirts MR, Smith JC, Kasson PM, van der
Spoel D, et al: GROMACS 4.5: A high-throughput and highly parallel
open source molecular simulation toolkit. Bioinformatics.
29:845–854. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Marrink SJ, Risselada HJ, Yefimov S,
Tieleman DP and de Vries AH: The MARTINI force field: Coarse
grained model for biomolecular simulations. J Phys Chem B.
111:7812–7824. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Bereau T and Kremer K: Automated
parametrization of the coarse-grained Martini force field for small
organic molecules. J Chem Theory Comput. 11:2783–2791. 2015.
View Article : Google Scholar
|
|
59
|
Lamprakis C, Andreadelis I, Manchester J,
Velez-Vega C, Duca JS and Cournia Z: Evaluating the efficiency of
the Martini force field to study protein dimerization in aqueous
and membrane environments. J Chem Theory Comput. 17:3088–3102.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Souza PCT, Thallmair S, Conflitti P,
Ramírez-Palacios C, Alessandri R, Raniolo S, Limongelli V and
Marrink SJ: Protein-ligand binding with the coarse-grained Martini
model. Nat Commun. 11:37142020. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Honorato RV, Roel-Touris J and Bonvin
AMJJ: MARTINI-based protein-DNA coarse-grained HADDOCKing. Front
Mol Biosci. 6:1022019. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Periole X, Cavalli M, Marrink SJ and
Ceruso MA: Combining an elastic network with a coarse-grained
molecular force field: Structure, dynamics, and intermolecular
recognition. J Chem Theory Comput. 5:2531–2543. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Zhang X, Wu K, Wang D, Yue X, Song D, Zhu
Y and Wu J: Nucleocapsid protein of SARS-CoV activates
interleukin-6 expression through cellular transcription factor
NF-kappaB. Virology. 365:324–335. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
du Preez HN, Aldous C, Kruger HG and
Johnson L: N-Acetylcysteine and other sulfur-donors as a
preventative and adjunct therapy for COVID-19. Adv Pharmacol Pharm
Sci. 2022:45554902022.PubMed/NCBI
|
|
65
|
Zinovkin RA and Grebenchikov OA:
Transcription factor Nrf2 as a potential therapeutic target for
prevention of cytokine storm in COVID-19 patients. Biochemistry
(Mosc). 85:833–837. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Cuadrado A, Pajares M, Benito C,
Jiménez-Villegas J, Escoll M, Fernández-Ginés R, Garcia Yagüe AJ,
Lastra D, Manda G, Rojo AI and Dinkova-Kostova AT: Can activation
of NRF2 Be a strategy against COVID-19? Trends Pharmacol Sci.
41:598–610. 2020. View Article : Google Scholar :
|
|
67
|
Kow CS, Ramachandram DS and Hasan SS: Use
of sulforaphane in COVID-19: Clinical trials are needed. Mol
Immunol. 145:78–79. 2022. View Article : Google Scholar :
|