Ovarian cancer (OC) is the most lethal tumor of the
female genital system, and caused an estimated 13,270 deaths in the
USA in 2023 (1). The poor
prognosis of OC is largely attributed to the insidious onset of the
disease leading to a delayed diagnosis. Furthermore, the
pathogenesis of OC has not been well characterized. The known risk
factors of OC include genetic, hormonal, reproductive and
lifestyle-related factors (2).
At present, surgery and chemotherapy are the standard treatment
modalities for OC (3).
Chemotherapy is the first-choice treatment for advanced and
recurrent OC, which includes conventional platinum-based drugs and
paclitaxel, targeted anti-angiogenic drugs, poly (ADP-ribose)
polymerase inhibitors and immunotherapy (4). Despite the efficacy of these drugs,
the 5-year survival rate of patients with OC remains low, which is
50% in USA (1). Therefore, the
identification of novel therapeutic targets for OC is
imperative.
Circular RNAs (circRNAs) are highly conserved and
stable structures that may serve as novel diagnostic and prognostic
biomarkers, as well as chemotherapeutic targets, in the context of
tumors (5). The proposed concept
of competing endogenous RNA (ceRNA) illustrates a new mechanism of
post-transcriptional gene regulation. The phenomenon wherein
non-coding RNA (ncRNA) sequester microRNAs (miRNAs) and remove the
inhibition of their targets was initially observed in both plant
and animal cells (6,7). This phenomenon is the core
mechanism of ceRNAs. circRNAs can act as ceRNAs (8), and certain studies have identified
numerous circRNAs as potential prognostic biomarkers and
therapeutic targets for OC (9,10). The present review aims to discuss
current knowledge on the molecular mechanisms by which circRNAs,
through miRNA sponging, influence the pathobiology of OC.
Furthermore, the present review highlights the clinical
applicability of circRNAs as predictive markers and therapeutic
targets in OC.
ncRNAs refer to the RNA molecules that do not encode
proteins transcribed from the genome (11). circRNAs belong to the class of
regulatory ncRNAs and have both the commonality of ncRNAs and a
unique structure and function. circRNAs were first found in viroids
by Sanger et al (12)
over 40 years ago and were shown to have a covalently closed loop
structure, initially thought to be the aberrant product of
mis-splicing (13). The loop
structure formed by the reversed splicing of circRNA results in the
absence of a 5' cap or 3' poly A tail (14). This confers high stability to the
molecular structure of circRNA. circRNAs can exist and accumulate
in specific cells for long periods due to their ability to avoid
digestion by RNA enzymes (15).
Most circRNAs are located in the cytoplasm. circRNAs are
significantly enriched in the brain, human platelets, during
epithelial-mesenchymal transition (EMT) and during the
differentiation of hematopoietic progenitor cells into lymphoid and
myeloid cells (16).
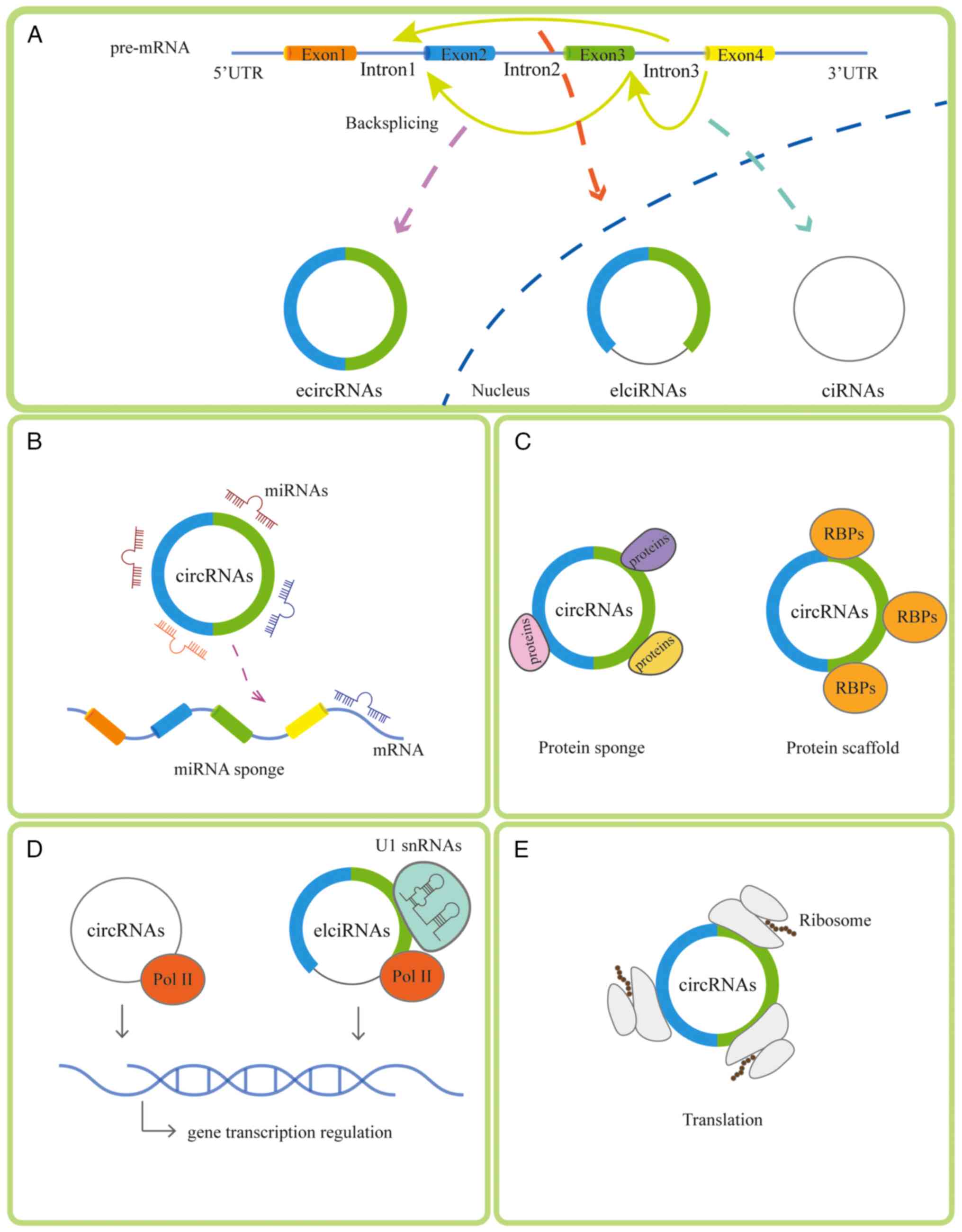
circRNAs can be categorized based on the composition
of exons and introns as follows: Exonic circRNA (ecircRNA),
intronic circRNA and exon-intron circRNA (17). ecircRNAs are reported to function
in the circRNA sponging of miRNA (18). There are four main mechanisms of
circRNAs: i) Sponging miRNA (19); ii) binding to diverse RNA binding
proteins (20) or acting as a
protein sponge; iii) regulating transcription (21); and iv) participating in the
translation progress (22). The
generation and mechanism of circRNAs is presented in Fig. 1. The ability of circRNAs to act
as ceRNAs has been verified in a number of diseases such as
neurological diseases, cardiovascular diseases and tumors, as well
as in the context of the immune response (such as the antivirus and
antitumor response) (23-26).
Functionally, circRNAs have been observed to exert oncogenic or
tumor-suppressive effects. At the cellular level, they are
instrumental in modulating processes such as proliferation,
migration, invasion, apoptosis and chemoresistance (27). In vivo studies,
particularly those utilizing animal models, have demonstrated that
circRNAs influence tumor growth and metastasis (28,29). Moreover, akin to other ncRNAs
such as miRNAs (30,31), circRNAs are emerging as vital
prognostic and diagnostic biomarkers for a multitude of diseases
such as neurologic diseases (32), diabetic complications (33) and tumors (34).
The ceRNA hypothesis has been proposed to describe
the mechanism by which ncRNAs that harbor miRNA response elements
(MREs) can sequester miRNAs from other targets that share the same
MREs, thereby regulating their expression (35). Specifically, ncRNAs competitively
bind to miRNAs to reduce the inhibition of mRNA and regulate the
corresponding gene expression. Long ncRNAs (lncRNAs), mRNAs,
pseudogenic RNAs and circRNAs can act as ceRNAs, among which
circRNAs have the strongest binding ability to miRNA, and therefore
may be the main type of ncRNAs participating in observed oncogenic
effects.
The resulting effect of an interaction between
circRNAs and miRNAs depends on the number of binding sites among
them (36). The higher the
number of competitive binding sites, the greater the potential of a
circRNA to act as a ceRNA. ciRS-7 is the most well-known ceRNA, has
>70 conserved miR-7 binding sites and has high levels of stable
expression in the brain (37).
Generally, circRNAs that act as ceRNAs negatively regulate miRNA.
This implies that when the expression of circRNAs, which act as
ceRNAs, increases, the expression of miRNA is reduced (38) and vice versa. The presence of
miRNA binding sites on a circRNA does not necessarily imply that it
will inhibit miRNA, as whether a circRNA negatively or positively
regulates miRNA is related to the stoichiometric relationship
between the MREs in the potential sponge and the target mRNA
(39).
A number of studies have demonstrated the role of
circRNAs in respiratory system (40), digestive system (41,42) and female reproductive system
tumors (43,44). circRNAs can be upregulated or
downregulated in tumors, which regulates mRNA expression in tumor
cells by regulating miRNA. ceRNAs of malignant tumors competitively
bind to miRNAs in the MREs of the 3' untranslated region (UTR)
(35). 3'UTR shortening caused
by the reduction of MREs alters the ability of ceRNAs to compete
for miRNAs and function as ceRNAs. In a study by Sang et al
(45), hsa_circ_0025202 was
found to act as a ceRNA in breast cancer by sponging miR-182-5p,
further regulating the expression and activity of FOXO3a. Moreover,
functional studies demonstrated that hsa_circ_0025202 suppresses
tumorigenesis and improves sensitivity to tamoxifen via the
miR-182-5p/FOXO3a axis. In a study by Wang et al (46), has-circRNA-002178 was found to
enhance programmed death-ligand 1 expression in lung cancer cells
by sponging miR-34, which induced T cell failure. In addition,
hsa_circRNA_104348 may act as a ceRNA to promote the progression of
hepatocellular carcinoma by targeting the miR-187-3p/rhotekin2 axis
and activating the Wnt/β-catenin pathway (47). In conclusion, circRNAs play
important roles as ceRNAs in tumors and influence the biological
activity of tumor cells.
Cell growth is a critical factor in the
proliferation of tumors and sustained growth is one of the key
attributes of malignant tumors. Proliferation of normal cells in
the human body is closely regulated; however, tumor cells evade
this regulation and achieve sustained proliferation through four
mechanisms: i) Autocrine growth signals; ii) stimulation of normal
cells to secrete growth signals; iii) increase in receptor
expression and therefore amplification of growth signals; and iv)
altered receptor structure, resulting in receptor activation
(48). The studies described
below demonstrate that circRNAs can act as ceRNAs, sponging miRNAs
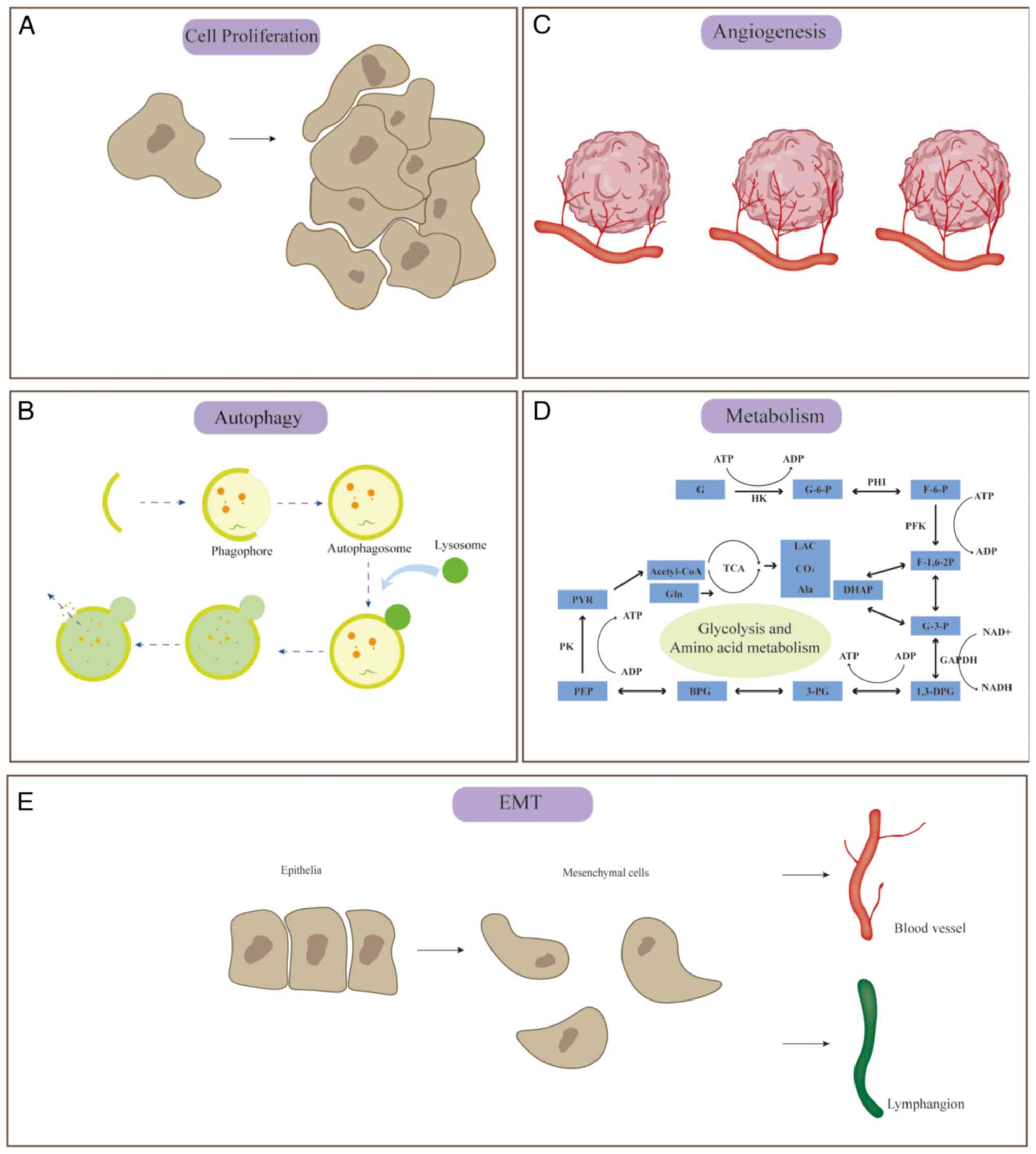
as well as targeting mRNAs to affect cell proliferation (Fig. 2A).
The cell cycle, which spans from the conclusion of
one cell division to the completion of the next, is a critical
phase during which circRNAs can exert influence. circ-BNC2 can
inhibit the transition of OC cells from the G0/G1 phase into the
G2/M phase through the miR-223-3p/F-box/WD repeat-containing
protein 7 axis (49). The
circUBAP2/miR-382-5p/pre-mRNA-processing-splicing factor 8 axis is
another regulatory pathway inducing G0/G1 phase arrest (50). Additionally, circPVT1 promotes OC
cell viability through the miR-149-5p/forkhead box protein M1
(FOXM1) pathway (51).
Furthermore, circ_0013958, found to be upregulated in OC, targets
plexin-B2 (PLXNB2) via miR-637 sponging, promoting OC proliferation
(52). PLXNB2 mediates
intracellular RNA processing and contributes to cell proliferation
and survival (53).
Autophagy is an evolutionarily conserved,
self-degrading, normal cellular metabolic process. Cells can
degrade harmful intracellular components to ensure normal cell
growth and operation via autophagy. As such, autophagy is a
mechanism with a dual role in both the apoptosis and survival of
cells. Autophagic levels can be assessed in studies by detecting
proteins such as autophagy-related gene (ATG), LC3, p62 and Beclin1
(54). circRNAs regulate
autophagy-related proteins to mediate autophagy (55). The studies demonstrating that
circRNAs can act as ceRNAs, sponging miRNAs as well as targeting
mRNAs, to affect cellular autophagy are described below (Fig. 2B).
CircRAB11FIP1, which is upregulated in OC, has been
implicated in targeting ATG7 and ATG14 by sponging miR-129, and
therefore engaging in the autophagic process (56). Meanwhile, circRAB11FIP1 also
directly binds to desmocollin 1 to mediate autophagy. It has been
demonstrated that circMUC16 is upregulated in OC tissues compared
with normal ovary tissues, which inhibits the autophagy flux of OC
(57). The assessment of
autophagic flux in the aforementioned study was based on the
expression of Beclin1, a key autophagy-related factor active in the
initial stages of autophagy. circRNF144B has been shown to sponge
miR-342-3p, thereby regulating F-box and leucine-rich repeat
protein 11 (FBXL11) and influencing the ubiquitination and
subsequent degradation of Beclin1 (58). The autophagy of OC can be
inhibited through the circRNF144B/miR-342-3p/FBXL11 axis. In
summary, although all three aforementioned circRNAs are upregulated
in OC, they exhibit divergent effects on autophagy; while circMUC16
and circRNF144B suppress autophagy, circRAB11FIP1 enhances ATG
expression and promotes the autophagic pathway.
Angiogenesis is a physiological process wherein
capillary or post-capillary veins form following
neovascularization. circRNAs are notably abundant in
angiogenesis-related diseases, which indicates an association
between circRNA and angiogenesis. The studies described below
demonstrate that circRNAs can act as ceRNAs, sponging miRNAs as
well as targeting mRNAs, to affect angiogenesis (Fig. 2C).
Tumor angiogenesis is largely dependent on vascular
endothelial growth factor A (VEGFA)-driven responses, which largely
contribute to dysregulation of vascular function (59). circATRNL1 targets Smad4 by
sponging miR-378, which mediates the AKT signaling pathway and
inhibits cell proliferation, migration and invasion, and is
involved in anti-angiogenesis in OC (60). Angiogenesis is affected by the
restoration of Smad, which downregulates the expression of VEGF and
upregulates the expression of thrombospondin-1 to inhibit
angiogenesis (61). In addition,
circASH2L silencing was found to inhibit the invasion and growth of
OC cells and to inhibit the angiogenesis and lymphangiogenesis of
transplanted tumors via the miR-665/VEGFA axis (62). Furthermore, circ_0061140 is
elevated in OC tissues and cells, and promotes proliferation,
migration, invasion and angiogenesis through the miR-761/leucine
zipper-EF-hand containing transmembrane protein 1 axis (63).
EMT is a highly complex phenotypic transition during
embryonic development that drives tissue formation and is important
for tumor metastasis (64). EMT
is also a cause of tumor progression and poor prognosis in patients
with cancer. A study has suggested that circRNAs are associated
with EMT transcription factors (such as snail, vimentin and twist)
and EMT-related signaling pathways (such as the TGF-β/Smad and Wnt
signaling pathways) (65). The
studies described below demonstrate that circRNAs can act as
ceRNAs, sponging miRNAs as well as targeting mRNAs, to affect EMT
(Fig. 2E).
EMT can be assessed via the detection of EMT-related
proteins such as E-cadherin, N-cadherin and Vimentin (65). circPLEKHM3 mediates OC cell
migration and EMT by sponging miR-9 and targeting BRCA1/DNAJ
homolog subfamily B member 6 (DNAJB6)/Krüppel-like factor 4 (KLF4)
(66). BRCA1, DNAJB6 and KLF4
contribute to the metastasis of tumors. GATA binding protein 3
mediates BRCA1 to suppress EMT, inhibiting the metastasis of breast
cancer (67). DNAJB6, a member
of HSP40 family, inhibits the EMT of tumors (68). KLF4 activates the expression of
epithelial genes, playing a notable role in EMT (69). circCELSR1 targets
bromodomain-containing protein 4 (BRD4) by sponging miR-598 to
promote the EMT of OC cells (70). BRD4 also inhibits EMT in renal
cell carcinoma (71). In
summary, circRNAs mediate the EMT of OC cells by targeting
EMT-related mRNA. circFGFR3 (72), circPTK2 (73), ciRS-7 (74) and hsa_circ_0061140 (75) all regulate downstream genes to
promote EMT in OC (76-79).
Tumor metabolism mainly includes glucose, lipid and
amino acid metabolism. Tumor cells mainly produce nutrients through
aerobic glycolysis to maintain the basal cellular requirements for
upregulated proliferation. The 'Warburg effect' is a process in
which tumor cells convert glucose to lactate in the presence of
oxygen (80). It is considered
that circRNAs are associated with lipid metabolism in hepatocytes,
adipocytes and macrophages (81). Glutamine is an essential source
of energy for cell survival, and tumor cells will overtake
glutamine to maintain abnormal cell growth. A previous study has
indicated that ncRNAs can regulate tumor metabolism and thus
participate in the biological functions of tumors (82). The studies described below
demonstrate that circRNAs can act as ceRNAs, sponging miRNAs as
well as targeting mRNAs, to affect tumor metabolism (Fig. 2D).
circRNAs have a significant impact on glycolysis in
OC. For example, circITCH has been shown to attenuate cell
proliferation, invasion and glycolysis in OC through the
miR-106a/E-cadherin axis (83).
Additionally, has_circ_0002711 facilitates aerobic glycolysis via
the miR-1244/Rho-associated protein kinase 1 pathway in OC
(84). circ_0025033 targets LSM4
through the sponging of miR-184 to promote glycolytic metabolism in
OC (85). Furthermore, LSM4 is
closely associated with cell cycle, cell replication, focal
adhesion and multiple metabolism-related pathways, including fatty
acid metabolism, in hepatocellular carcinoma (86). Moreover, silencing circ_0023033
enhances glutamine metabolism in OC (87). However, the role of circRNA in
the regulation of lipid metabolism within tumors remains to be
thoroughly investigated. The role of circRNAs as ceRNAs in OC is
shown in Fig. 2.
Due to the highly stable and specific expression of
circRNAs in OC, certain circRNAs may serve as useful diagnostic and
prognostic biomarkers. A study demonstrated that circ-ABCB10 may
help distinguish OC tissue from adjacent tissue, affirming its
value as a diagnostic biomarker [area under the curve (AUC)=0.766;
95% confidence interval (CI), 0.690-0.842] (88). circBNC2 has also demonstrated
value in distinguishing OC tissue from benign ovarian cysts
[receiver operating characteristic (ROC) AUC=0.879; 95% CI,
0.822-0.937; sensitivity, 96.4%; specificity, 80.7%] (89). Plasma circN4BP2L2 expression
significantly differentiated OC tissue from benign ovarian tumor
tissue (ROC AUC=0.82; sensitivity, 72%; specificity, 87%) and from
normal ovarian tissue (ROC AUC=0.90; sensitivity, 77%; specificity,
88%) (90). In addition, high
expression of circ-ABCB10 was associated with significantly
decreased overall survival (OS) time in patients with OC [hazard
ratio (HR)=2.994; P<0.05] (88), and high expression of circ-ITCH
was associated with a significantly longer OS in patients with OC
(HR=0.207; 95% CI, 0.066-0.065; P<0.05) (83). A recent study has also
demonstrated specific expression of circRNAs in the blood (91). Therefore, this non-invasive test
(just withdrawing blood) may provide portability in detecting
circRNAs as tumor markers in the future, meaning that it would not
be necessary to obtain pathological tissue through invasive methods
to detect circRNA.
The aforementioned studies indicate that circRNAs
play an important role in the progression of OC. Therefore,
circRNAs may also act as molecular targets for targeted therapy, in
which the target circRNA is overexpressed or knocked down to affect
tumor progression or drug resistance, thus impacting patient
treatment outcomes. Upregulation of circEXOC6B tends to imply an
earlier TNM stage, less lymph node metastasis and a higher survival
rate, and is associated with decreased paclitaxel resistance in OC
(92). Conversely, circ_0061140
is known to enhance paclitaxel resistance by sponging miR-136 and
targeting chromobox 2, showing significant upregulation of
circ_0061140 in paclitaxel-resistant OC tissues and cells (93). circ_0025033 targets FOXM1 by
sponging miR-532-3p, and circ_0025033 is significantly upregulated
in paclitaxel-resistant cells of OC (94). Furthermore, downregulation of
circ_0025033 in the exosomes of paclitaxel-resistant cells inhibits
the malignant effects of OC cells. circTNPO3 and circCELSR1 have
also been implicated in paclitaxel resistance, sponging miR-1299
and miR-1252, respectively, to target NIMA related kinase 2 and
FOXR2 in OC, respectively (16,95). Resistance to cisplatin (DDP),
another cornerstone in OC chemotherapy, has also been associated
with circRNAs. Downregulation of circ_0067934 was shown to decrease
DDP resistance in OC (96),
while overexpression of circFoxp1 was associated with increased DDP
drug resistance (97). In
addition, circ-LPAR3 promotes DDP resistance by sponging miR-634
and targeting pyruvate dehydrogenase kinase 1 in OC (98). The aforementioned studies have
therefore demonstrated that circRNAs may be used as therapeutic
targets in OC, to affect drug resistance such as for the two
classic chemotherapy drugs, paclitaxel and DDP. Beyond OC, circRNAs
synthesized in vitro have shown promise in gastric cancer
treatment strategies by sponging miRNAs to target genes (99). This insight verifies the
possibility of circRNAs, functioning as ceRNAs, as targeted
therapeutic markers in the management of OC.
As aforementioned, circRNAs mediate biological
processes, including proliferation, migration and invasion,in
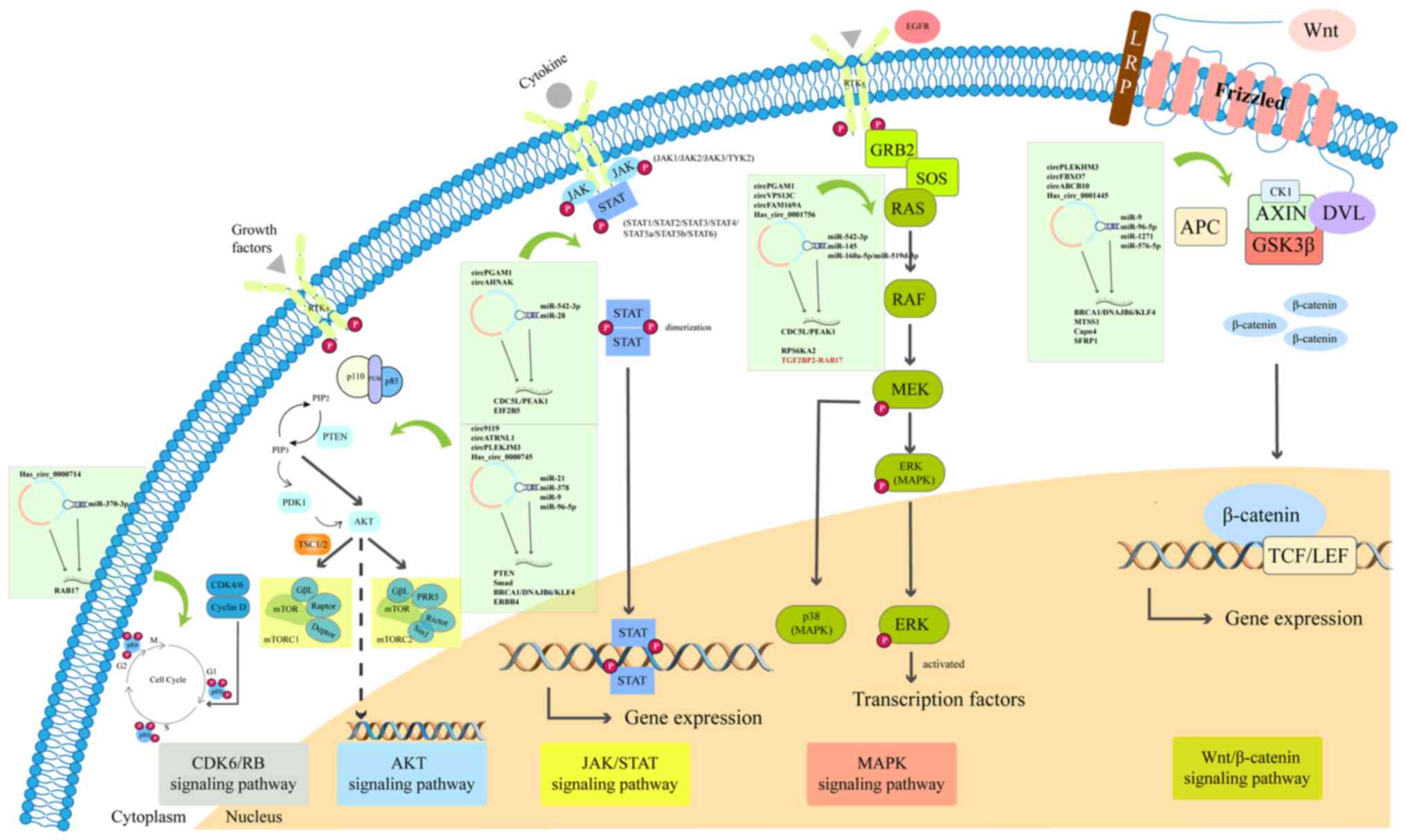
various diseases through regulating signaling pathways. The
signaling pathways mainly include Notch, janus kinase (JAK)/STAT,
Wnt/β-catenin, TGF-β/Smad and AMP-activated protein kinase
signaling pathways (100). Wang
et al (101)
demonstrated that differential circRNAs in the serum of 20 patients
with OC were enriched in Fcγ R-mediated phagocytosis, VEGF
signaling, transcriptional misregulation in cancer, chemokine
signaling, ErbB signaling and TNF signaling pathways, according to
a Kyoto Encyclopedia of Genes and Genomes analysis. Recent studies
have demonstrated that circRNAs mainly participate in the
Wnt/β-catenin, AKT, MAPK, JAK/STAT and CDK/retinoblastoma protein
signaling pathways in OC (Table
I). The specific signaling pathways involving circRNAs are
presented in Fig. 3.
In recent years, the role of circRNAs in tumors has
become a research hotspot. In OC, circRNAs are mainly studied
extensively as ceRNAs. circRNAs affect the proliferation, invasion,
migration and apoptosis of OC cells through sponging miRNA and
subsequently modulating the target genes, thus changing the
clinical course of solid tumors. circRNAs can also regulate OC
progression through mediating autophagy, angiogenesis, the cell
cycle, EMT and metabolism. Autophagy, a process with dichotomous
roles in tumor promotion and suppression, requires further
investigation to elucidate its dual regulatory mechanisms within
the OC context. Currently, research regarding circRNA-mediated
metabolism mainly focuses on glycometabolism and amino acid
metabolism, and lipid/nuclear acid metabolism is rarely involved.
circRNAs mainly participate in the Wnt/β-catenin, AKT and MAPK
signaling pathways. Moreover, circRNAs may serve as valuable
predictive markers and therapeutic targets for OC. In addition,
recent studies have not only focused on the downstream mechanisms
of circRNAs, but also on the upstream mechanisms. One study found
that circRNAs also play a role in the tumor microenvironment, which
is involved in the immunotherapy of tumors (102). circUHRF1, secreted by
hepatocellular hepatoma cells in exosomes, is involved in
immunosuppression by inducing natural killer cell dysfunction.
Moreover, overexpression of circUHRF1 was found to decrease the
effect of anti-programmed cell death 1 (PD-1) drug therapy, while
the targeting of circUHRF1 restored the sensitivity of anti-PD1
therapy (102). However, this
research is still in the nascent phase and requires further
exploration. In view of the research described in the present
review, circRNAs have a certain prospect as biomarkers and
therapeutic targets not only in OC but also in other tumors and
diseases.
Not applicable.
NX, QW and WLY contributed to the study conception
and design, material preparation, literature collection and
conclusion. The first draft of the manuscript was written by YML
and WLY, and all authors commented on subsequent versions of the
manuscript. All authors read and approved the final version of the
manuscript. Data authentication is not applicable.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
Not applicable.
This work was supported by the National Natural Science
Foundation of Liaoning Province (2019ZD0790).
|
1
|
Siegel RL, Miller KD, Wagle NS and Jemal
A: Cancer statistics, 2023. CA Cancer J Clin. 73:17–48. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
La Vecchia C: Ovarian cancer: Epidemiology
and risk factors. Eur J Cancer Prev. 26:55–62. 2017. View Article : Google Scholar
|
|
3
|
Kuroki L and Guntupalli SR: Treatment of
epithelial ovarian cancer. BMJ. 371:m37732020. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Armstrong DK, Alvarez RD, Backes FJ,
Bakkum-Gamez JN, Barroilhet L, Behbakht K, Berchuck A, Chen LM,
Chitiyo VC, Cristea M, et al: NCCN guidelines® insights:
Ovarian cancer, version 3.2022. J Natl Compr Canc Netw. 20:972–980.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Meng S, Zhou H, Feng Z, Xu Z, Tang Y, Li P
and Wu M: Circrna: Functions and properties of a novel potential
biomarker for cancer. Mol Cancer. 16:942017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Franco-Zorrilla JM, Valli A, Todesco M,
Mateos I, Puga MI, Rubio-Somoza I, Leyva A, Weigel D, García JA and
Paz-Ares J: Target mimicry provides a new mechanism for regulation
of microRNA activity. Nat Genet. 39:1033–1037. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ebert MS, Neilson JR and Sharp PA:
MicroRNA sponges: Competitive inhibitors of small RNAs in mammalian
cells. Nat Methods. 4:721–726. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tang X, Ren H, Guo M, Qian J, Yang Y and
Gu C: Review on circular RNAs and new insights into their roles in
cancer. Comput Struct Biotechnol J. 19:910–928. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ding J, Wang Q, Guo N, Wang H, Chen H, Ni
G and Li P: CircRNA circ_0072995 promotes the progression of
epithelial ovarian cancer by modulating miR-147a/CDK6 axis. Aging
(Albany NY). 12:17209–17223. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Guan X, Zong ZH, Liu Y, Chen S, Wang LL
and Zhao Y: circPUM1 promotes tumorigenesis and progression of
ovarian cancer by sponging miR-615-5p and miR-6753-5p. Mol Ther
Nucleic Acids. 18:882–892. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mattick JS and Makunin IV: Non-coding RNA.
Hum Mol Genet. 15:R17–R29. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sanger HL, Klotz G, Riesner D, Gross HJ
and Kleinschmidt AK: Viroids are single-stranded covalently closed
circular RNA molecules existing as highly base-paired rod-like
structures. Proc Natl Acad Sci USA. 73:3852–3856. 1976. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Cocquerelle C, Mascrez B, Hétuin D and
Bailleul B: Mis-splicing yields circular RNA molecules. FASEB J.
7:155–160. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Jeck WR and Sharpless NE: Detecting and
characterizing circular RNAs. Nat Biotechnol. 32:453–461. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Suzuki H and Tsukahara T: A view of
pre-mRNA splicing from RNase R resistant RNAs. Int J Mol Sci.
15:9331–9342. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhang S, Cheng J, Quan C, Wen H, Feng Z,
Hu Q, Zhu J, Huang Y and Wu X: circCELSR1 (hsa_circ_0063809)
contributes to paclitaxel resistance of ovarian cancer cells by
regulating FOXR2 expression via miR-1252. Mol Ther Nucleic Acids.
19:718–730. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang F, Nazarali AJ and Ji S: Circular
RNAs as potential biomarkers for cancer diagnosis and therapy. Am J
Cancer Res. 6:1167–1176. 2016.PubMed/NCBI
|
|
18
|
Yang X, Mei J, Wang H, Gu D, Ding J and
Liu C: The emerging roles of circular RNAs in ovarian cancer.
Cancer Cell Int. 20:2652020. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hansen TB, Wiklund ED, Bramsen JB,
Villadsen SB, Statham AL, Clark SJ and Kjems J: miRNA-dependent
gene silencing involving Ago2-mediated cleavage of a circular
antisense RNA. EMBO J. 30:4414–4422. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Huang A, Zheng H, Wu Z, Chen M and Huang
Y: Circular RNA-protein interactions: Functions, mechanisms, and
identification. Theranostics. 10:3503–3517. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Qu S, Yang X, Li X, Wang J, Gao Y, Shang
R, Sun W, Dou K and Li H: Circular RNA: A new star of noncoding
RNAs. Cancer Lett. 365:141–148. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Panda AC, Grammatikakis I, Munk R, Gorospe
M and Abdelmohsen K: Emerging roles and context of circular RNAs.
Wiley Interdiscip Rev RNA. 8: View Article : Google Scholar : 2017.
|
|
23
|
Su Q and Lv X: Revealing new landscape of
cardiovascular disease through circular RNA-miRNA-mRNA axis.
Genomics. 112:1680–1685. 2020. View Article : Google Scholar
|
|
24
|
Su L, Li R, Zhang Z, Liu J, Du J and Wei
H: Identification of altered exosomal microRNAs and mRNAs in
Alzheimer's disease. Ageing Res Rev. 73:1014972022. View Article : Google Scholar
|
|
25
|
Zhang J, Luo Q, Li X, Guo J, Zhu Q, Lu X,
Wei L, Xiang Z, Peng M, Ou C and Zou Y: Novel role of
immune-related non-coding RNAs as potential biomarkers regulating
tumour immunoresponse via MICA/NKG2D pathway. Biomark Res.
11:862023. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lu S, Zhu N, Guo W, Wang X, Li K, Yan J,
Jiang C, Han S, Xiang H, Wu X, et al: RNA-Seq revealed a circular
RNA-microRNA-mRNA regulatory network in hantaan virus infection.
Front Cell Infect Microbiol. 10:972020. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yang X, Ye T, Liu H, Lv P, Duan C, Wu X,
Jiang K, Lu H, Xia D, Peng E, et al: Expression profiles,
biological functions and clinical significance of circRNAs in
bladder cancer. Mol Cancer. 20:42021. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Najafi S: Circular RNAs as emerging
players in cervical cancer tumorigenesis; A review to roles and
biomarker potentials. Int J Biol Macromol. 206:939–953. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Najafi S: The emerging roles and potential
applications of circular RNAs in ovarian cancer: A comprehensive
review. J Cancer Res Clin Oncol. 149:2211–2234. 2023. View Article : Google Scholar
|
|
30
|
Fattahi M, Shahrabi S, Saadatpour F,
Rezaee D, Beyglu Z, Delavari S, Amrolahi A, Ahmadi S,
Bagheri-Mohammadi S, Noori E, et al: microRNA-382 as a tumor
suppressor? Roles in tumorigenesis and clinical significance. Int J
Biol Macromol. 250:1258632023. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Pordel S, Khorrami M, Saadatpour F, Rezaee
D, Cho WC, Jahani S, Aghaei-Zarch SM, Hashemi E and Najafi S: The
role of microRNA-185 in the pathogenesis of human diseases: A focus
on cancer. Pathol Res Pract. 249:1547292023. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Najafi S, Aghaei Zarch SM, Majidpoor J,
Pordel S, Aghamiri S, Fatih Rasul M, Asemani Y, Vakili O, Mohammadi
V, Movahedpour A and Arghiani N: Recent Insights into the roles of
circular rnas in human brain development and neurologic diseases.
Int J Biol Macromol. 225:1038–1048. 2023. View Article : Google Scholar
|
|
33
|
Xu YX, Pu SD, Li X, Yu ZW, Zhang YT, Tong
XW, Shan YY and Gao XY: Exosomal ncRNAs: Novel therapeutic target
and biomarker for diabetic complications. Pharmacol Res.
178:1061352022. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wang Y, Liu J, Ma J, Sun T, Zhou Q, Wang
W, Wang G, Wu P, Wang H, Jiang L, et al: Exosomal circRNAs:
Biogenesis, effect and application in human diseases. Mol Cancer.
18:1162019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Karreth FA and Pandolfi PP: ceRNA
cross-talk in cancer: When ce-bling rivalries go awry. Cancer
Discov. 3:1113–1121. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Salmena L, Poliseno L, Tay Y, Kats L and
Pandolfi PP: A ceRNA hypothesis: the Rosetta Stone of a hidden RNA
language? Cell. 146:353–358. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Hansen TB, Jensen TI, Clausen BH, Bramsen
JB, Finsen B, Damgaard CK and Kjems J: Natural RNA circles function
as efficient microRNA sponges. Nature. 495:384–388. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Lan C, Peng H, Hutvagner G and Li J:
Construction of competing endogenous RNA networks from paired
RNA-seq data sets by pointwise mutual information. BMC Genomics.
20(Suppl 9): S9432019. View Article : Google Scholar
|
|
39
|
Kristensen LS, Andersen MS, Stagsted LVW,
Ebbesen KK, Hansen TB and Kjems J: The biogenesis, biology and
characterization of circular RNAs. Nat Rev Genet. 20:675–691. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Peng Z, Fang S, Jiang M, Zhao X, Zhou C
and Gong Z: Circular RNAs: Regulatory functions in respiratory
tract cancers. Clin Chim Acta. 510:264–271. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Li R, Jiang J, Shi H, Qian H, Zhang X and
Xu W: CircRNA: A rising star in gastric cancer. Cell Mol Life Sci.
77:1661–1680. 2020. View Article : Google Scholar
|
|
42
|
Rong Z, Xu J, Shi S, Tan Z, Meng Q, Hua J,
Liu J, Zhang B, Wang W, Yu X and Liang C: Circular RNA in
pancreatic cancer: A novel avenue for the roles of diagnosis and
treatment. Theranostics. 11:2755–2769. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Chaichian S, Shafabakhsh R, Mirhashemi SM,
Moazzami B and Asemi Z: Circular RNAs: A novel biomarker for
cervical cancer. J Cell Physiol. 235:718–274. 2020. View Article : Google Scholar
|
|
44
|
Razavi ZS, Tajiknia V, Majidi S, Ghandali
M, Mirzaei HR, Rahimian N, Hamblin MR and Mirzaei H: Gynecologic
cancers and non-coding RNAs: Epigenetic regulators with emerging
roles. Crit Rev Oncol Hematol. 157:1031922021. View Article : Google Scholar
|
|
45
|
Sang Y, Chen B, Song X, Li Y, Liang Y, Han
D, Zhang N, Zhang H, Liu Y, Chen T, et al: circRNA_0025202
regulates tamoxifen sensitivity and tumor progression via
regulating the miR-182-5p/FOXO3a axis in breast cancer. Mol Ther.
27:1638–1652. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Wang J, Zhao X, Wang Y, Ren F, Sun D, Yan
Y, Kong X, Bu J, Liu M and Xu S: circRNA-002178 act as a ceRNA to
promote PDL1/PD1 expression in lung adenocarcinoma. Cell Death Dis.
11:322020. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Huang G, Liang M, Liu H, Huang J, Li P,
Wang C, Zhang Y, Lin Y and Jiang X: CircRNA hsa_circRNA_104348
promotes hepatocellular carcinoma progression through modulating
miR-187-3p/RTKN2 axis and activating Wnt/β-catenin pathway. Cell
Death Dis. 11:10652020. View Article : Google Scholar
|
|
48
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Liu T, Yuan L and Zou X: Circular RNA
circ-BNC2 (hsa_ circ_0008732) inhibits the progression of ovarian
cancer through microRNA-223-3p/FBXW7 axis. J Ovarian Res.
15:952022. View Article : Google Scholar
|
|
50
|
Xu Q, Deng B, Li M, Chen Y and Zhuan L:
circRNA-UBAP2 promotes the proliferation and inhibits apoptosis of
ovarian cancer though miR-382-5p/PRPF8 axis. J Ovarian Res.
13:812020. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Li M, Chi C, Zhou L, Chen Y and Tang X:
Circular PVT1 regulates cell proliferation and invasion via
miR-149-5p/FOXM1 axis in ovarian cancer. J Cancer. 12:611–621.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Liang Y, Meng K and Qiu R: Circular RNA
Circ_0013958 functions as a tumor promoter in ovarian cancer by
regulating miR-637/PLXNB2 axis. Front Genet. 12:6444512021.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Yu W, Goncalves KA, Li S, Kishikawa H, Sun
G, Yang H, Vanli N, Wu Y, Jiang Y, Hu MG, et al: Plexin-B2 mediates
physiologic and pathologic functions of angiogenin. Cell.
171:849–864.e25. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Li X, He S and Ma B: Autophagy and
autophagy-related proteins in cancer. Mol Cancer. 19:122020.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Wang Y, Mo Y, Peng M, Zhang S, Gong Z, Yan
Q, Tang Y, He Y, Liao Q, Li X, et al: The influence of circular
RNAs on autophagy and disease progression. Autophagy. 18:240–253.
2022. View Article : Google Scholar :
|
|
56
|
Zhang Z, Zhu H and Hu J: CircRAB11FIP1
promoted autophagy flux of ovarian cancer through DSC1 and miR-129.
Cell Death Dis. 12:2192021. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Gan X, Zhu H, Jiang X, Obiegbusi SC, Yong
M, Long X and Hu J: CircMUC16 promotes autophagy of epithelial
ovarian cancer via interaction with ATG13 and miR-199a. Mol Cancer.
19:452020. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Song W, Zeng Z, Zhang Y, Li H, Cheng H,
Wang J and Wu F: CircRNF144B/miR-342-3p/FBXL11 axis reduced
autophagy and promoted the progression of ovarian cancer by
increasing the ubiquitination of Beclin-1. Cell Death Dis.
13:8572022. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Claesson-Welsh L and Welsh M: Vegfa and
tumour angiogenesis. J Intern Med. 273:114–127. 2013. View Article : Google Scholar
|
|
60
|
Wang J, Li Y, Zhou JH, Shen FR, Shi X and
Chen YG: CircATRNL1 activates Smad4 signaling to inhibit
angiogenesis and ovarian cancer metastasis via miR-378. Mol Oncol.
15:1217–1233. 2021. View Article : Google Scholar :
|
|
61
|
Schwarte-Waldhoff I and Schmiegel W: Smad4
transcriptional pathways and angiogenesis. Int J Gastrointest
Cancer. 31:47–59. 2002. View Article : Google Scholar
|
|
62
|
Chen J, Li X, Yang L, Li M, Zhang Y and
Zhang J: CircASH2L promotes ovarian cancer tumorigenesis,
angiogenesis, and lymphangiogenesis by regulating the miR-665/VEGFA
axis as a competing endogenous RNA. Front Cell Dev Biol.
8:5955852020. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Ma L, Liu W and Li M: Circ_0061140
contributes to ovarian cancer progression by targeting
miR-761/LETM1 signaling. Biochem Genet. 61:628–650. 2023.
View Article : Google Scholar
|
|
64
|
Pastushenko I and Blanpain C: EMT
transition states during tumor progression and metastasis. Trends
Cell Biol. 29:212–226. 2019. View Article : Google Scholar
|
|
65
|
Shang BQ, Li ML, Quan HY, Hou PF, Li ZW,
Chu SF, Zheng JN and Bai J: Functional roles of circular RNAs
during epithelial-to-mesenchymal transition. Mol Cancer.
18:1382019. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Zhang L, Zhou Q, Qiu Q, Hou L, Wu M, Li J,
Li X, Lu B, Cheng X, Liu P, et al: CircPLEKHM3 acts as a tumor
suppressor through regulation of the miR-9/BRCA1/DNAJB6/KLF4/AKT1
axis in ovarian cancer. Mol Cancer. 18:1442019. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Bai F, Zhang LH, Liu X, Wang C, Zheng C,
Sun J, Li M, Zhu WG and Pei XH: GATA3 functions downstream of BRCA1
to suppress EMT in breast cancer. Theranostics. 11:8218–8233. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Menezes ME, Mitra A, Shevde LA and Samant
RS: DNAJB6 governs a novel regulatory loop determining
Wnt/β-catenin signalling activity. Biochem J. 444:573–580. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Tiwari N, Meyer-Schaller N, Arnold P,
Antoniadis H, Pachkov M, van Nimwegen E and Christofori G: Klf4 is
a transcriptional regulator of genes critical for EMT, including
Jnk1 (Mapk8). PLoS One. 8:e573292013. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Zeng XY, Yuan J, Wang C, Zeng D, Yong JH,
Jiang XY, Lan H and Xiao SS: circCELSR1 facilitates ovarian cancer
proliferation and metastasis by sponging miR-598 to activate BRD4
signals. Mol Med. 26:702020. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Li X, Lin S, Mo Z, Jiang J, Tang H, Wu C
and Song J: CircRNA_100395 inhibits cell proliferation and
metastasis in ovarian cancer via regulating
miR-1228/p53/epithelial-mesenchymal transition (EMT) axis. J
Cancer. 11:599–609. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Zhou J, Dong ZN, Qiu BQ, Hu M, Liang XQ,
Dai X, Hong D and Sun YF: CircRNA FGFR3 induces
epithelial-mesenchymal transition of ovarian cancer by regulating
miR-29a-3p/E2F1 axis. Aging (Albany NY). 12:14080–14091. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Wu SG, Zhou P, Chen JX, Lei J, Hua L, Dong
Y, Hu M, Lian CL, Yang LC and Zhou J: circ-PTK2 (hsa_circ_0008305)
regulates the pathogenic processes of ovarian cancer via miR-639
and FOXC1 regulatory cascade. Cancer Cell Int. 21:2772021.
View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Zhang F, Xu Y, Ye W, Jiang J and Wu C:
Circular RNA S-7 promotes ovarian cancer EMT via sponging miR-641
to up-regulate ZEB1 and MDM2. Biosci Rep. 40:BSR202008252020.
View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Chen Q, Zhang J, He Y and Wang Y:
hsa_circ_0061140 knockdown reverses FOXM1-mediated cell growth and
metastasis in ovarian cancer through miR-370 sponge activity. Mol
Ther Nucleic Acids. 13:55–63. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Wang T, Chen X, Qiao W, Kong L, Sun D and
Li Z: Transcription factor E2F1 promotes EMT by regulating ZEB2 in
small cell lung cancer. BMC Cancer. 17:7192017. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Li Y and Chen X: miR-4792 inhibits
epithelial-mesenchymal transition and invasion in nasopharyngeal
carcinoma by targeting FOXC1. Biochem Biophys Res Commun.
468:863–869. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Caramel J, Ligier M and Puisieux A:
Pleiotropic roles for ZEB1 in cancer. Cancer Res. 78:30–35. 2018.
View Article : Google Scholar
|
|
79
|
Katoh M, Igarashi M, Fukuda H, Nakagama H
and Katoh M: Cancer genetics and genomics of human FOX family
genes. Cancer Lett. 328:198–206. 2013. View Article : Google Scholar
|
|
80
|
Wang Y and Patti GJ: The Warburg effect: A
signature of mitochondrial overload. Trends Cell Boil.
33:1014–1020. 2023. View Article : Google Scholar
|
|
81
|
Yu G, Yang Z, Peng T and Lv Y: Circular
RNAs: Rising stars in lipid metabolism and lipid disorders. J Cell
Physiol. 236:4797–4806. 2021. View Article : Google Scholar
|
|
82
|
Yu T, Wang Y, Fan Y, Fang N, Wang T, Xu T
and Shu Y: CircRNAs in cancer metabolism: A review. J Hematol
Oncol. 12:902019. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Lin C, Xu X, Yang Q, Liang L and Qiao S:
Circular RNA ITCH suppresses proliferation, invasion, and
glycolysis of ovarian cancer cells by up-regulating CDH1 via
sponging miR-106a. Cancer Cell Int. 20:3362020. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Xie W, Liu LU, He C, Zhao M, Ni R, Zhang Z
and Shui C: Circ_0002711 knockdown suppresses cell growth and
aerobic glycolysis by modulating miR-1244/ROCK1 axis in ovarian
cancer. J Biosci. 46:212021. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Hou W and Zhang Y: Circ_0025033 promotes
the progression of ovarian cancer by activating the expression of
LSM4 via targeting miR-184. Pathol Res Pract. 217:1532752021.
View Article : Google Scholar
|
|
86
|
Chen L, Lin YH, Liu GQ, Huang JE, Wei W,
Yang ZH, Hu YM, Xie JH and Yu HZ: Clinical significance and
potential role of LSM4 overexpression in hepatocellular carcinoma:
An integrated analysis based on multiple databases. Front Genet.
12:8049162022. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Ma H, Qu S, Zhai Y and Yang X:
circ_0025033 promotes ovarian cancer development via regulating the
hsa_miR-370-3p/SLC1A5 axis. Cell Mol Biol Lett. 27:942022.
View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Chen Y, Ye X, Xia X and Lin X: Circular
RNA ABCB10 correlates with advanced clinicopathological features
and unfavorable survival, and promotes cell proliferation while
reduces cell apoptosis in epithelial ovarian cancer. Cancer
Biomark. 26:151–161. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Hu Y, Zhu Y, Zhang W, Lang J and Ning L:
Utility of plasma circBNC2 as a diagnostic biomarker in epithelial
ovarian cancer. Onco Targets Ther. 12:9715–9723. 2019. View Article : Google Scholar
|
|
90
|
Ning L, Lang J and Wu L: Plasma
circN4BP2L2 is a promising novel diagnostic biomarker for
epithelial ovarian cancer. BMC Cancer. 22:62022. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Wang S, Zhang K, Tan S, Xin J, Yuan Q, Xu
H, Xu X, Liang Q, Christiani DC, Wang M, et al: Circular RNAs in
body fluids as cancer biomarkers: the new frontier of liquid
biopsies. Mol Cancer. 20:132021. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Zheng Y, Li Z, Yang S, Wang Y and Luan Z:
CircEXOC6B suppresses the proliferation and motility and sensitizes
ovarian cancer cells to paclitaxel through miR-376c-3p/FOXO3 axis.
Cancer Biother Radiopharm. 37:802–814. 2022.
|
|
93
|
Zhu J, Luo JE, Chen Y and Wu Q:
Circ_0061140 knockdown inhibits tumorigenesis and improves PTX
sensitivity by regulating miR-136/CBX2 axis in ovarian cancer. J
Ovarian Res. 14:1362021. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Huang H, Yan L, Zhong J, Hong L, Zhang N
and Luo X: Circ_0025033 deficiency suppresses paclitaxel resistance
and malignant development of paclitaxel-resistant ovarian cancer
cells by modulating the miR-532-3p/FOXM1 network. Immunopharmacol
Immunotoxicol. 44:275–286. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Xia B, Zhao Z, Wu Y, Wang Y, Zhao Y and
Wang J: Circular RNA circTNPO3 regulates paclitaxel resistance of
ovarian cancer cells by miR-1299/NEK2 signaling pathway. Mol Ther
Nucleic Acids. 21:780–791. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Yuan D, Guo T, Qian H, Ge H, Zhao Y, Huang
A, Wang X, Cao X, Zhu D, He C and Yu H: Icariside II suppresses the
tumorigenesis and development of ovarian cancer by regulating
miR-144-3p/IGF2R axis. Drug Dev Res. 83:1383–1393. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Luo Y and Gui R: Circulating exosomal
circFoxp1 confers cisplatin resistance in epithelial ovarian cancer
cells. J Gynecol Oncol. 31:e752020. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Liu X, Yin Z, Wu Y, Zhan Q, Huang H and
Fan J: Circular RNA lysophosphatidic acid receptor 3 (circ-LPAR3)
enhances the cisplatin resistance of ovarian cancer. Bioengineered.
13:3739–3750. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Cheng Y, Ban R, Liu W, Wang H, Li S, Yue
Z, Zhu G, Zhuan Y and Wang C: MiRNA-409-3p enhances
cisplatin-sensitivity of ovarian cancer cells by blocking the
autophagy mediated by Fip200. Oncol Res. Jan 2–2018.Epub ahead of
print. View Article : Google Scholar
|
|
100
|
Ghafouri-Fard S, Khoshbakht T, Bahranian
A, Taheri M and Hallajnejad M: CircMTO1: A circular RNA with roles
in the carcinogenesis. Biomed Pharmacother. 142:1120252021.
View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Wang J, Wu A, Yang B, Zhu X, Teng Y and Ai
Z: Profiling and bioinformatics analyses reveal differential
circular RNA expression in ovarian cancer. Gene. 724:1441502020.
View Article : Google Scholar
|
|
102
|
Zhang PF, Gao C, Huang XY, Lu JC, Guo XJ,
Shi GM, Cai JB and Ke AW: Cancer cell-derived exosomal circUHRF1
induces natural killer cell exhaustion and may cause resistance to
anti-PD1 therapy in hepatocellular carcinoma. Mol Cancer.
19:1102020. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Gong J, Xu X, Zhang X and Zhou Y: Circular
RNA-9119 suppresses in ovarian cancer cell viability via targeting
the microRNA-21-5p-PTEN-Akt pathway. Aging (Albany NY).
12:14314–14328. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Guo M, Li S, Zhao X, Yuan Y, Zhang B and
Guan Y: Knockdown of circular RNA Hsa_circ_0000714 can regulate
RAB17 by sponging miR-370-3p to reduce paclitaxel resistance of
ovarian cancer through CDK6/RB pathway. Onco Targets Ther.
13:13211–13224. 2020. View Article : Google Scholar :
|
|
105
|
Ji J, Li C, Wang J, Wang L, Huang H, Li Y
and Fang J: Hsa_circ_0001756 promotes ovarian cancer progression
through regulating IGF2BP2-mediated RAB5A expression and the
EGFR/MAPK signaling pathway. Cell Cycle. 21:685–696. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Zhang C, Li Y, Zhao W, Liu G and Yang Q:
Circ-PGAM1 promotes malignant progression of epithelial ovarian
cancer through regulation of the miR-542-3p/CDC5L/PEAK1 pathway.
Cancer Med. 9:3500–3521. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
He SL, Zhao X and Yi SJ: CircAHNAK
upregulates EIF2B5 expression to inhibit the progression of ovarian
cancer by modulating the JAK2/STAT3 signaling pathway.
Carcinogenesis. 43:941–955. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Lu H, Zheng G, Gao X, Chen C, Zhou M and
Zhang L: Propofol suppresses cell viability, cell cycle progression
and motility and induces cell apoptosis of ovarian cancer cells
through suppressing MEK/ERK signaling via targeting
circVPS13C/miR-145 axis. J Ovarian Res. 14:302021. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Fu Z, Ding C, Gong W and Lu C: ncRNAs
mediated RPS6KA2 inhibits ovarian cancer proliferation via p38/MAPK
signaling pathway. Front Oncol. 13:10283012023. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Wang S, Li Z, Zhu G, Hong L, Hu C, Wang K,
Cui K and Hao C: RNA-binding protein IGF2BP2 enhances circ_0000745
abundancy and promotes aggressiveness and stemness of ovarian
cancer cells via the microRNA-3187-3p/ERBB4/PI3K/AKT axis. J
Ovarian Res. 14:1542021. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Wu M, Qiu Q, Zhou Q, Li J, Yang J, Zheng
C, Luo A, Li X, Zhang H, Cheng X, et al: circFBXO7/miR-96-5p/MTSS1
axis is an important regulator in the Wnt signaling pathway in
ovarian cancer. Mol Cancer. 21:1372022. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Lin X, Chen Y, Ye X and Xia X: Circular
RNA ABCB10 promotes cell proliferation and invasion, but inhibits
apoptosis via regulating the microRNA-1271-mediated
Capn4/Wnt/β-catenin signaling pathway in epithelial ovarian cancer.
Mol Med Rep. 23:3872021. View Article : Google Scholar
|
|
113
|
Wu Y, Zhou J, Li Y, Shi X, Shen F, Chen M,
Chen Y and Wang J: Hsa_circ_0001445 works as a cancer suppressor
via miR-576-5p/SFRP1 axis regulation in ovarian cancer. Cancer Med.
12:5736–5750. 2023. View Article : Google Scholar
|