Introduction
Osteosarcoma is a highly aggressive primary bone
malignancy characterized by the production of osteoid and bone
tissue, predominantly affecting the metaphyseal regions of long
bones, particularly the distal femur, proximal tibia and proximal
humerus (1). It generally arises
in adolescents and young adults, peaking between the ages of 10 and
20 years, during periods of rapid skeletal growth (1). Epidemiologically, osteosarcoma has
an annual incidence of ~3-4 cases per million individuals in the
general population, with a notable male predominance (~1.5:1)
(2). Geographic variations in
incidence have been observed, with certain studies indicating
higher rates in African populations and lower frequencies in Asian
cohorts (2). Recent advancements
in genomic studies have identified possible genetic risk factors,
including mutations in the tumor protein 53 (TP53) gene linked to
Li-Fraumeni syndrome (3,4). Furthermore, previous exposure to
radiation and certain chemotherapeutic agents have been implicated
in the etiology of osteosarcoma. Understanding these
epidemiological patterns and genetic predispositions is crucial for
developing targeted screening and prevention strategies in at-risk
populations (5).
The current standard of care for osteosarcoma
primarily involves a multidisciplinary approach, combining
neoadjuvant chemotherapy, surgical resection and adjuvant
chemotherapy (6). High-dose
multi-agent chemotherapy, typically including doxorubicin,
cisplatin and methotrexate, aims to shrink tumors before surgery
and eliminate micrometastatic disease postoperatively (7). However, despite advances in
treatment, outcomes remain suboptimal, with a 5-year survival rate
of ~60-70% for localized disease and significantly lower for
metastatic cases. Limitations in treatment efficacy arise from
tumor heterogeneity, development of chemoresistance and late
diagnosis (8). Furthermore, the
potential for severe adverse effects from aggressive chemotherapy
regimens poses significant challenges to patient quality of life
(9). Emerging strategies that
incorporate multi-omics analyses are being explored to identify
novel therapeutic targets and improve individualized treatment
plans.
Multi-omics is an integrative approach that combines
various layers of biological information, such as genomics,
transcriptomics, proteomics and metabolomics, to gain comprehensive
insights into cancer biology (10). This concept facilitates the
understanding of the complex molecular interplay driving
tumorigenesis, progression and response to therapy (11). By simultaneously analyzing the
genetic profiles (including gene mutations and epigenetic changes),
gene (mRNA) expression, protein expression, other RNA expression
and metabolic profiles of tumors, researchers can identify novel
biomarkers and therapeutic targets that may be overlooked by
traditional methods such as isolated genomic, transcriptomic or
proteomic analyses (12). Recent
studies have highlighted the potential of multi-omics to refine
patient stratification and personalize treatment strategies,
leading to improved outcomes in cancers like osteosarcoma (4,13). The advent of advanced
technologies and data analytics further enhances the capacity to
integrate these diverse data types, positioning multi-omics as a
cornerstone of precision medicine in oncology (14).
The purpose of this review is to synthesize current
advances in multi-omics technologies and their application in
osteosarcoma research, highlighting how these integrative
approaches can enhance the understanding of tumor biology, improve
prognostic stratification and inform the development of
personalized treatment strategies for achieving better patient
outcomes.
The complexity of osteosarcoma
Genetic landscape of osteosarcoma
The genetic landscape of osteosarcoma is
characterized by multiple recurrent mutations and dysregulated
pathways that contribute to its pathogenesis (15). Key mutations commonly observed in
osteosarcoma include alterations in the TP53, retinoblastoma
protein 1 (RB1) and mouse double minute 2 (MDM2) genes (Fig. 1) (16-18). TP53, a well-established tumor
suppressor gene, is frequently mutated in numerous cancers,
including osteosarcoma, leading to the loss of its normal function
in cell cycle regulation and apoptosis (19). Mutations in RB1, another critical
tumor suppressor, disrupt the Rb-E2F pathway, promoting
uncontrolled cellular proliferation and contributing to tumor
growth (20). Meanwhile, MDM2
overexpression results in the degradation of p53, further
exacerbating the dysregulation of apoptosis and cell survival
(21).
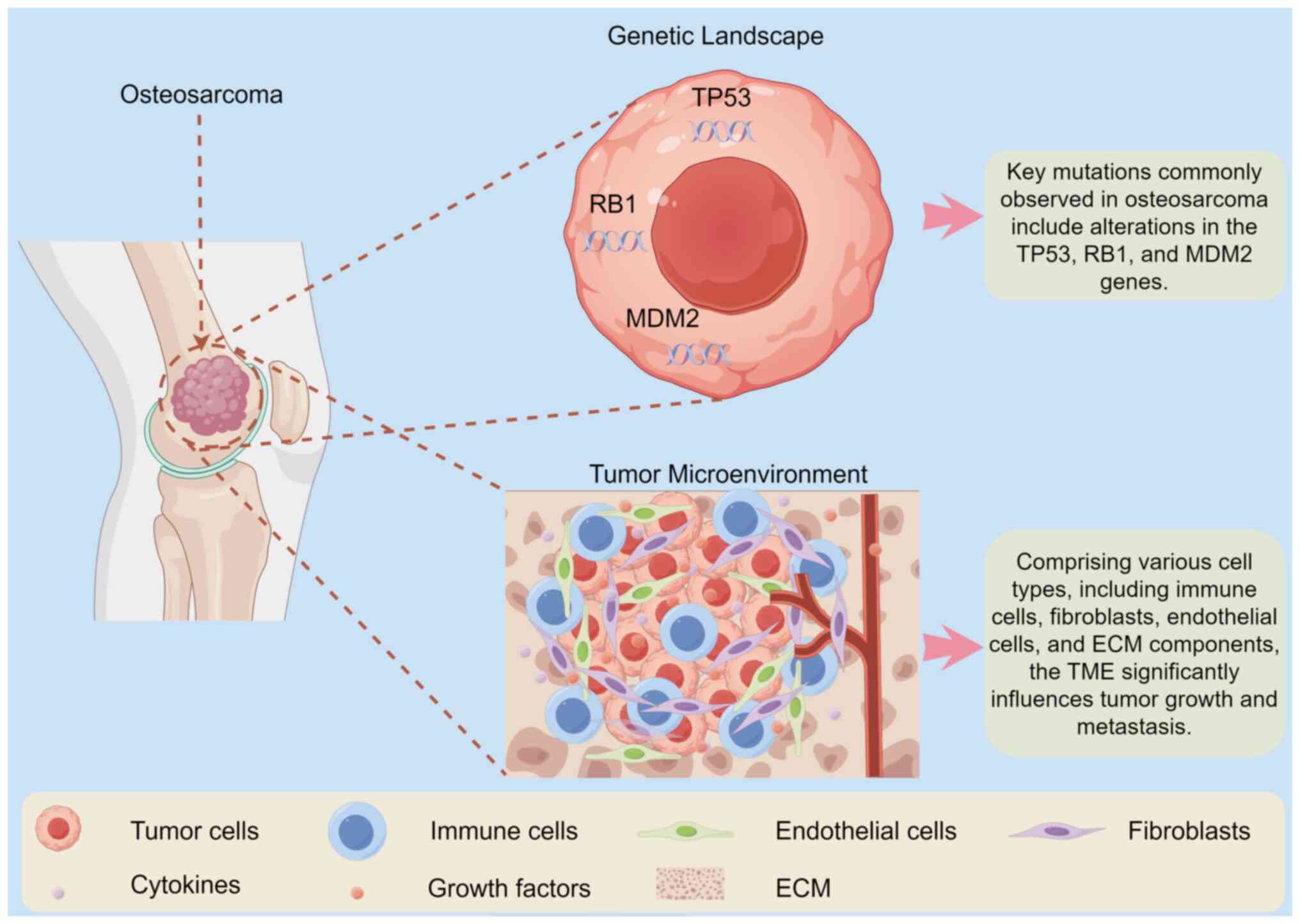 | Figure 1Schematic illustrating the
characteristics of the genetic landscape and the TME of
osteosarcoma. Osteosarcoma is characterized by a complex genetic
landscape and TME. The figure shows key genetic alterations,
including mutations in TP53, RB1 and MDM2, which contribute to
osteosarcoma pathogenesis. In addition, it depicts the TME
comprising immune cells, fibroblasts, endothelial cells and ECM
components that influence tumor growth and metastasis. The
interplay between genetic mutations and microenvironmental factors
creates a conducive niche for tumor progression and therapeutic
resistance. The figure was generated using Figdraw (https://www.figdraw.com). RB1, retinoblastoma protein
1; MDM2, mouse double minute 2; ECM, extracellular matrix; TME,
tumor microenvironment; ECM, extracellular matrix. |
These genetic alterations not only inform tumor
behavior but also influence treatment response. For example,
osteosarcoma tumors with TP53 mutations may exhibit resistance to
conventional chemotherapy agents, necessitating alternative
therapeutic approaches (22).
Furthermore, specific molecular subtypes identified through genomic
profiling can be targeted with novel therapeutic agents,
emphasizing the importance of personalizing treatment based on the
genetic characteristics of the tumor (23).
Role of the tumor microenvironment
(TME)
The TME plays a crucial role in shaping the behavior
and progression of osteosarcoma (24). Comprising various cell types,
including immune cells, fibroblasts, endothelial cells and
extracellular matrix (ECM) components, the TME significantly
influences tumor growth and metastasis (Fig. 1) (25). Interactions between osteosarcoma
cells and immune cells, such as macrophages and T-cells, can
modulate the immune response, either promoting tumor progression
through immune evasion or enhancing anti-tumor activity (26).
Certain soluble factors within the TME can also
affect the behavior of osteosarcoma cells (27). For instance, the release of
cytokines and growth factors from stromal cells can promote
angiogenesis and tumor-supportive inflammation, creating a
favorable niche for tumor growth (28). Additionally, components of the
ECM, such as collagen and fibronectin, can interact with integrins
on the surface of osteosarcoma cells, triggering signaling pathways
that enhance proliferation and migration (29). Understanding the complex
interplay between osteosarcoma cells and their microenvironment
holds potential for therapeutic intervention. Strategies aimed at
modulating the TME, such as targeted therapies that disrupt
tumor-stroma interactions or immune checkpoint inhibitors, are
being explored in ongoing clinical trials (30).
Tumor heterogeneity
Tumor heterogeneity is a hallmark feature of
osteosarcoma, characterized by variations in the cellular
composition, genetic makeup and functional properties of the tumor
(31). Histopathological
analyses often reveal a mixture of osteoblastic, chondroblastic and
fibroblastic cell types within osteosarcoma, reflecting the diverse
lineage origins and differentiation states of the tumor (31).
This heterogeneity has profound implications for
treatment resistance, as subpopulations of cells may exhibit
differential responses to therapy. For instance, a subset of cancer
stem-like cells has been identified in osteosarcoma that is thought
to be responsible for tumor recurrence and metastasis due to their
enhanced survival capabilities and resistance to conventional
therapies (32). The presence of
these resistant subpopulations can lead to incomplete responses to
treatment, ultimately resulting in unfavorable prognostic outcomes
(33).
Furthermore, the dynamic nature of tumor evolution
complicates the understanding of treatment resistance (34). As tumors are exposed to
therapeutic pressures, they may undergo clonal selection, leading
to the emergence of resistant variants that contribute to disease
treatment resistance progression (35). Integrating multi-omics approaches
to profile the genetic, transcriptomic and proteomic landscapes of
osteosarcoma will provide insights into the mechanisms of
heterogeneity and resistance, paving the way for the development of
targeted therapeutic strategies tailored to specific tumor
characteristics (36).
The complexity of osteosarcoma is a multifaceted
challenge that necessitates a deeper understanding of its genetic
landscape, the role of the TME and the heterogeneity within the
tumor itself (37). Advances in
multi-omics technologies hold promise for elucidating the
underlying mechanisms driving osteosarcoma biology and can aid in
the development of innovative therapeutic strategies that enhance
patient outcomes (38). As we
move towards precision medicine, a thorough comprehension of these
complexities will be indispensable in addressing the limitations of
current treatment paradigms.
Overview of multi-omics approaches
Genomics
Recent advances in whole-genome sequencing (WGS)
have significantly enhanced our understanding of the genomic
landscape of osteosarcoma (39).
WGS allows for the identification of genetic alterations,
comprehensively analyzing both coding and non-coding regions of the
genome (40). Studies have
identified critical driver mutations in key genes such as TP53
(16), RB1 (41) and MDM2 (42), which play pivotal roles in tumor
development and progression (Table
I).
 | Table IGenomics findings in osteosarcoma
research. |
Table I
Genomics findings in osteosarcoma
research.
| Author/s, year | Potential
targets | Samples | Major findings | (Refs.) |
|---|
| Mirabello et
al, 2015 | TP53 | 765 osteosarcoma
cases | The genetic
susceptibility of osteosarcoma with early onset differs from that
of osteosarcoma with late onset, characterized by a high frequency
of exon mutations in the TP53 gene. | (16) |
| Li et al,
2022 | RB1 | Mouse model | RB1 functions as a
tumor suppressor in osteosarcoma by regulating glucose metabolism
through the inhibition of YAP, with its loss leading to increased
expression of Glut1 and enhanced tumor progression. | (41) |
| Mokánszki et
al, 2020 | RB1 | A patient with
retinoblastoma | RB1 functions as a
tumor suppressor in osteosarcoma by regulating cell cycle
progression and preventing tumorigenesis, with mutations leading to
tumor development in patients with retinoblastoma. | (17) |
| Feng et al,
2020 | MDM2 | 596 patients with
osteosarcoma and 1,696 healthy controls | MDM2 acts as an
oncogene in osteosarcoma by inhibiting the tumor suppressor
function of p53, leading to enhanced tumor progression and poor
patient outcomes. | (42) |
| He et al,
2018 | MDM2 | 22 patients with
low-grade osteosarcoma | MDM2 functions as
an oncogene in low-grade osteosarcoma by promoting cellular
proliferation through amplification, thereby contributing to
tumorigenesis. | (43) |
| Limbach et
al, 2020 | MDM2 | 4 cases of primary
osteosarcoma | MDM2 acts as an
oncogene in craniofacial osteosarcoma by facilitating tumor
differentiation and progression through gene amplification, aiding
in distinguishing malignant tumors from benign lesions. | (44) |
| Kaur et al,
2022 | MDM2 | 101 tissue samples
diagnosed as osteosarcoma, ossifying fibroma, fibrous dysplasia or
fibrous hyperplasia | MDM2 functions as
an oncogene in osteosarcoma by promoting cell proliferation through
gene amplification, aiding in differentiation from benign
lesions. | (45) |
| Ren et al,
2020 | CLEC3A | Tissue samples from
patients with osteosarcoma from Gene Expression Omnibus dataset
GSE99671 | CLEC3A functions as
an oncogene in osteosarcoma by promoting cell proliferation and
reducing chemosensitivity through the AKT1/mTOR/HIF1α signaling
pathway. | (46) |
| Wang et al,
2019 | MAT1 | Osteosarcoma tissue
samples with and without lung metastasis | MAT1 promotes lung
metastasis in osteosarcoma by enhancing cell proliferation,
migration and invasion through increased AKT1 expression,
correlating with poor patient prognosis. | (47) |
| Huang et al,
2023 | FGF2 | Human osteosarcoma
tissue sections | FGF2 promotes
osteosarcoma metastasis by enhancing cell migration and invasion
through the activation of FGFR signaling and upregulation of ICAM-1
expression. | (49) |
| Kim et al,
2022 | FGFR1 | Osteosarcoma
tissues | FGFR1 facilitates
radiation resistance in osteosarcoma by promoting cell survival and
polyploidy through nuclear localization and activation of specific
signaling pathways. | (50) |
| Makise et
al, 2018 | H3K27me3 | 19 cases of
extraskeletal osteosarcoma | H3K27me3 functions
as a transcriptional repressor in osteosarcoma, with its deficiency
correlating with aggressive disease and poor patient outcomes. | (52) |
| Saba et al,
2019 | FN1-FGFR1 | A single case of
chondroblastoma-like osteosarcoma | The FN1-FGFR1 gene
fusion promotes oncogenic signaling in chondroblastoma-like
osteosarcoma, potentially contributing to its distinct genetic
profile and malignant behaviors. | (53) |
Mutations in key tumor suppressor
genes
Mutations in the TP53 gene are frequently observed
in osteosarcoma, which are critical for tumor progression (22). These mutations lead to the
inactivation of its tumor suppressor function, compromising the
cellular mechanisms of apoptosis and DNA repair (22). A comprehensive study has
established that germline TP53 mutations significantly increase
susceptibility to osteosarcoma and correlate with poorer survival
outcomes (16). The RB1 is
another crucial tumor suppressor associated with osteosarcoma.
Deletions or mutations in RB1, frequently observed in conjunction
with TP53 mutations, enhance cell cycle progression and contribute
to oncogenesis (41). It is
noteworthy that the co-mutation of TP53 and RB1 is often linked to
aggressive disease phenotypes and metastasis (17).
Amplification of oncogenes
Amplification of the MDM2 gene, which encodes a
protein that inhibits TP53, is present in ~10-15% of osteosarcoma
cases. Increased MDM2 expression facilitates the bypass of
TP53-mediated apoptosis, aiding in tumor growth and resistance to
therapies (42-44). Its role in the differentiation of
osteosarcoma from benign lesions highlights its diagnostic
significance (45). The PI3K/AKT
signaling pathway is frequently activated in osteosarcoma. For
instance, a study on C-type lectin domain family 3 member A
demonstrated that its suppression led to reduced cell proliferation
through the AKT serine/threonine kinase 1 (AKT1)/mammalian target
of rapamycin (mTOR) axis (46).
Furthermore, ménage à trios 1 has been shown to facilitate lung
metastasis via upregulation of AKT1 expression, underscoring the
pathway's role in metastatic spread (47).
Alterations in growth factor
receptors
The fibroblast growth factor receptor (FGFR)
signaling pathway has gained attention due to its involvement in
osteosarcoma progression (48).
The FGFR signaling pathway, particularly FGFR1, plays a critical
role in the progression of osteosarcoma. Overexpression of FGF2 and
FGFRs has been linked to increased cell migration, invasion and
metastasis, notably through intercellular adhesion molecule-1
expression (49). Furthermore,
nuclear FGFR1 contributes to radiation resistance by inducing cell
survival mechanisms, such as G-2 checkpoint adaptation and histone
modifications. Targeting FGFR signaling may not only hinder tumor
progression but also enhance the efficacy of radiation therapy in
osteosarcoma, providing a promising therapeutic strategy (50).
Epigenetic modifications
Aberrant epigenetic modifications have been
identified in osteosarcoma that may contribute to its
aggressiveness (51). For
instance, alterations in histone markers, such as H3K27me3, have
been noted in various osteosarcoma subtypes, indicating
heterogeneous disease behavior (52). Such findings emphasize the need
for integrating epigenetic profiling into the genomic landscape of
osteosarcoma. Recent genomic profiling has identified fusion genes,
such as fibronectin 1-FGFR1, which present new therapeutic targets
(53). Such fusions indicate a
need for targeted sequencing in osteosarcoma patients, particularly
those with refractory disease, as they may provide insights into
novel treatment strategies.
The integration of genomic findings into clinical
practice presents an opportunity for personalized medicine in
osteosarcoma. Genetic profiling can direct the selection of
targeted therapies, optimize chemotherapeutic regimens and enhance
the monitoring of disease progression. For instance, the detection
of MDM2 overexpression may prompt the use of specific MDM2
inhibitors, tailoring treatment to the genetic landscape of the
tumor (43,45).
The totality of genomic research on osteosarcoma
illustrates a landscape of intricate genetic alterations that drive
tumorigenesis (54). The
mutations in tumor suppressor genes like TP53 and RB1, the
amplification of oncogenes such as MDM2 and AKT1, and disturbances
to growth factor signaling pathways underscore the heterogeneous
nature of this malignancy (16,41,42,47). Advancements in understanding
these changes are crucial not only in elucidating osteosarcoma
biology but also in providing a framework for developing targeted
therapies and precision medicine approaches. Future research should
focus on synthesizing these genomic insights with clinical outcomes
to enhance treatment efficacy and patient survival rates.
Transcriptomics
The application of transcriptomics in the study of
osteosarcoma has unveiled significant insights into the molecular
mechanisms underlying this aggressive bone tumor (55). By analyzing various RNA types,
particularly messenger RNA, researchers have identified crucial
genes and pathways involved in osteosarcoma tumorigenesis,
metastasis and response to therapy (Table II). This summary highlights key
findings related to specific gene features, emphasizing their
implications for precision medicine in osteosarcoma management.
 | Table IITranscriptomics findings in
osteosarcoma research. |
Table II
Transcriptomics findings in
osteosarcoma research.
| Author/s, year | Potential
targets | Samples | Major findings | (Refs.) |
|---|
| Das et al,
2021 | TP53 | 26 samples of
osteosarcoma from dogs | TP53 functions as a
critical tumor suppressor in osteosarcoma by regulating cell cycle
progression and apoptosis, with its mutations linked to poorer
outcomes. | (57) |
| Luo et al,
2023 | TP53 | Several
osteosarcoma cell lines | TP53 functions as a
tumor suppressor in osteosarcoma and its R156P mutation promotes
tumorigenicity by inhibiting rigidity sensing, facilitating anoikis
resistance. | (58) |
| Feng et al,
2020 | Myc | A tissue microarray
constructed from 70 patient samples | Myc functions as an
oncogene in osteosarcoma, promoting tumor growth and metastasis,
with its overexpression linked to poor patient prognosis. | (59) |
| Akkawi et
al, 2024 | Myc | A traceable
osteosarcoma mouse model | Myc acts as an
oncogenic driver in osteosarcoma, promoting tumorigenesis through
upregulation of its target genes, particularly in the context of
WWOX and TP53 deletion. | (60) |
| Ma et al,
2018 | GATA3 | GATA3 expression in
osteosarcoma cells and tissues | GATA3 functions as
a tumor suppressor in osteosarcoma by inhibiting cell
proliferation, migration and invasion through the transcriptional
regulation of the EMT-associated factor slug. | (61) |
| Wu et al,
2019 | VEGF | Osteosarcoma
samples from a cohort of 53 patients | VEGF promotes
angiogenesis in osteosarcoma, and its high expression is associated
with aggressive tumor behavior and poorer patient prognosis. | (62) |
| Gu et al,
2019 | VEGF | Two human
osteosarcoma cell lines | VEGF facilitates
angiogenesis in osteosarcoma, and its inhibition can significantly
reduce tumor proliferation and invasion. | (63) |
| Quan et al,
2023 | VEGF | 18 patients with
osteosarcoma | VEGF promotes
osteosarcoma progression by enhancing cell proliferation,
migration, invasion and angiogenesis, while its isoform VEGF exerts
antitumor effects and is downregulated in the disease. | (64) |
| Luo et al,
2022 | HIF-1α | Human osteosarcoma
tissues and cell lines | HIF-1α promotes
osteosarcoma development by inducing the expression of miR-18b-5p,
which inhibits the tumor suppressor PHF2, thereby enhancing cell
proliferation and metastasis. | (65) |
| Zeng et al,
2023 | HIF-1α | Human osteosarcoma
cell lines | HIF-1α contributes
to osteosarcoma progression by regulating the expression of key
angiogenic and metastatic factors, including VEGF, through the mTOR
signaling pathway. | (66) |
| Zhang et al,
2013 | CXCR4 | Human osteosarcoma
cells | CXCR4 facilitates
osteosarcoma growth and pulmonary metastasis by mediating the
interaction between tumor cells and MSCs, enhancing VEGF secretion
and promoting tumorigenic processes. | (68) |
| Li et al,
2024 | CXCR4 |
Doxorubicin-resistant osteosarcoma cell
lines | CXCR4 contributes
to osteosarcoma doxorubicin resistance by regulating the CARM1-YAP
signaling axis, which controls aerobic glycolysis and affects tumor
cell survival and metabolism. | (69) |
| Gong et al,
2020 | CXCR4 | Pathological
samples of 73 patients with osteosarcoma | CXCR4 facilitates
the invasion and metastasis of osteosarcoma by correlating with
MMP-2 expression, serving as a significant prognostic indicator for
patient outcomes. | (70) |
| Liu et al,
2020 | MMP9 | Osteosarcoma cell
lines | MMP-9 facilitates
osteosarcoma progression and metastasis by mediating cell migration
and invasion pathways regulated by MCP-1 signaling. | (71) |
Oncogenes and tumor suppressor genes
Mutations in the TP53 gene are prevalent in numerous
cancers, including osteosarcoma (56). Research indicates that TP53
missense mutations are associated with longer survival in a canine
model of osteosarcoma, suggesting a potential survival advantage
linked to specific mutations (57). Furthermore, the impairment of
rigid sensing due to mutant TP53 gain-of-function mutations has
been noted, implicating TP53 in the modulation of cellular
mechanics and tumor progression (58). The myelocytomatosis viral
oncogene homolog (Myc) oncogene is pivotal in osteosarcoma
pathogenesis. A study demonstrated that Myc not only acts as a poor
prognostic biomarker but also serves as a potential therapeutic
target, highlighting its critical role in the proliferation of
osteosarcoma cells (59).
Furthermore, the promotion of osteosarcoma development through WW
domain containing oxidoreductase-mediated upregulation of Myc adds
another layer of complexity to Myc's role in this disease (60). The downregulation of GATA binding
protein 3 has been linked to the epithelial-to-mesenchymal
transition (EMT) and migration of osteosarcoma cells. Its
regulatory effects on Slug indicate its significance in managing
metastatic potential in osteosarcoma (61).
Pathways and molecular mechanisms
Vascular endothelial growth factor (VEGF)/VEGF
receptors: VEGF is essential for angiogenesis and has been shown to
be associated with poor prognosis in patients with osteosarcoma
(62). The dual inhibition of
VEGF and survivin has exhibited proliferation inhibition and
induced apoptosis in osteosarcoma cells (63), thereby presenting a promising
therapeutic strategy. In addition, the splicing factor YBX1
promotes osteosarcoma progression by upregulating VEGF and
downregulating its anti-angiogenic isoform VEGF (64). Hypoxia-inducible factor 1α
(HIF-1α) mediates several processes involved in osteosarcoma,
including cellular response to hypoxia and metabolism. Of note,
HIF-1α-mediated augmentation of certain microRNAs facilitates
proliferation and metastasis, reflecting its role in EMT pathways
(65). Furthermore, the
interplay between HIF-1α and mTOR signaling pathways is critical,
as demonstrated by the synergistic anti-tumor activity of
ginsenoside Rg3 alongside doxorubicin (66).
Cell migration and invasion
The C-X-C motif chemokine receptor 4 (CXCR4) is
crucial for osteosarcoma cell migration and metastasis (67). Studies have shown that CXCR4
interacts with mesenchymal stem cells to promote tumor growth and
pulmonary metastasis, primarily through VEGF signaling (68). Additionally, the
CXCR4-coactivator associated arginine methyltransferase 1
(CARM1)-Yes-associated protein 1 (YAP) axis has been implicated in
overcoming doxorubicin resistance in osteosarcoma by suppressing
aerobic glycolysis, underscoring potential therapeutic pathways
targeting these interactions (69). Expression of matrix
metalloproteinase (MMP)-2 and MMP-9 is associated with poor
prognosis and higher metastatic potential in osteosarcoma (70). Furthermore, the macrophage
recruitment factor monocyte chemoattractant protein-1 enhances
cancer cell migration through the c-Raf/MAPK/AP-1 pathway, further
indicating the role of MMPs in the metastatic cascade (71).
Therapeutic implications and precision
medicine
The integration of transcriptomic data in
understanding the biology of osteosarcoma heralds a shift towards
precision medicine approaches (72). Identifying specific gene
expression patterns and their associated pathways allows clinicians
to tailor treatment strategies based on individual molecular
profiles, potentially improving outcomes. For example, targeting
pathways involving TP53, HIF-1α and Myc provides a framework for
developing targeted therapies that address the unique
characteristics of each patient's tumor (59,73,74). Furthermore, the combination of
transcriptomic and other omics data (e.g., proteomics,
metabolomics) is expected to revolutionize the understanding and
management of osteosarcoma. It can enable the identification of
novel biomarkers for early diagnosis, prognosis and the development
of innovative therapeutic strategies (75).
The application of transcriptomics in osteosarcoma
research has significantly contributed to unravelling the
complexities of this malignancy. Key findings regarding oncogenes,
tumor suppressor genes, signaling pathways and non-coding RNAs
provide a rich resource for future therapeutic strategies (76). The path toward precision medicine
in osteosarcoma is being paved by these insights, which have the
potential to enhance patient outcomes and transform clinical
practices in oncology.
Proteomics
Recent advances in proteomics have illuminated the
complexity of its pathology, leading to insights into specific
proteins that drive cell proliferation, survival, invasion and
metastasis (77). In the chapter
below, key findings from proteomic studies in osteosarcoma are
being discussed, focusing on significant protein classes such as
cyclins, signaling proteins, apoptosis-related proteins,
invasion/metastasis proteins, metabolic proteins, transcription
factors and other targeted proteins (Table III).
 | Table IIIProteomics findings in osteosarcoma
research. |
Table III
Proteomics findings in osteosarcoma
research.
| Author/s, year | Potential
targets | Samples | Major findings | (Refs.) |
|---|
| Chang et al,
2024 | CML | Human osteosarcoma
tissues | CML promotes
osteosarcoma progression by enhancing cell migration, invasion and
stemness through the activation of the RAGE receptor and downstream
ERK/NF-κB signaling pathways. | (79) |
| Jiang et al,
2019 | CXCR4 | Osteosarcoma
tissues and murine models | CXCR4 facilitates
immune evasion in osteosarcoma by promoting the migration of MDSCs,
which inhibit cytotoxic T-cell expansion and reduce their apoptosis
through SDF-1 signaling. | (80) |
| Liao et al,
2021 | CXCR4 | Osteosarcoma cell
lines | CXCR4 facilitates
osteosarcoma chemoresistance by regulating autophagy processes,
with its inhibition enhancing doxorubicin-induced apoptosis and
sensitizing osteosarcoma cells to chemotherapy. | (81) |
| Zhang et al,
2024 | MATN4 | Osteosarcoma
tissues and cell lines | MATN4 promotes
osteosarcoma progression by enhancing cell proliferation, migration
and invasion under hypoxic conditions, regulated by HIF-1α. | (82) |
| Gao et al,
2019 | SIRT6 | Osteosarcoma
tissues and cell lines | SIRT6 functions as
a tumor suppressor in osteosarcoma by inhibiting cell proliferation
and invasion through the downregulation of N-cadherin. | (84) |
| Masuelli et
al, 2020 | Bcl-2 | Human and murine
osteosarcoma cells | Bcl-2 promotes
osteosarcoma cell survival and resistance to therapy by inhibiting
apoptosis, making it a critical target for therapeutic
interventions. | (85) |
| Jiang et al,
2021 | LDHA | Osteosarcoma
specimens | LDHA promotes
osteosarcoma metastasis by enhancing glycolysis and facilitating
tumor cell migration, regulated by the demethylation activity of
KDM6B. | (87) |
| Mei et al,
2024 | NAT10 | Human osteosarcoma
tissues | NAT10 promotes
osteosarcoma progression by regulating m(6)A modification and glycolysis through
the stabilization of mRNA for key glycolytic enzymes, mediated by
YTHDC1. | (88) |
| Xia et al,
2023 | TRIM26 | Osteosarcoma
tissues and cell lines | TRIM26 inhibits
osteosarcoma progression by promoting the degradation of RACK1,
leading to the inactivation of MEK/ERK signaling and suppression of
EMT. | (89) |
Role of TME-related proteins
The TME plays a crucial role in osteosarcoma
progression and metastasis, largely driven by signaling pathways
mediated by proteins such as RAGE, HIF-1α and CXCR4 (78). Chang et al (79) elucidate that
Nε-(1-carboxymethyl)-L-lysine activates RAGE signaling, driving
osteosarcoma cell metastasis via ERK/NFκB axis. This suggests a
significant link between advanced glycation end products and
sarcomagenesis. Overexpression of CXCR4 has been tightly correlated
with disease progression and poor prognosis in osteosarcoma.
Specifically, CXCR4 signaling has been shown to facilitate
myeloid-derived suppressor cell accumulation, which can blunt
responses to immunotherapy (80). Furthermore, CXCR4 blockade has
been proven to sensitize osteosarcoma cells to doxorubicin through
autophagic cell death induction and inhibition of the PI3K-Akt-mTOR
pathway (81). HIF-1α, a key
regulator in the tumor response to hypoxic conditions, enhances
osteosarcoma proliferation and metastasis through various proteins,
such as matrilin 4 (MATN4). HIF-1α mediates the expression of
MATN4, promoting progression (82), indicating the dual role of
hypoxic signaling in metabolic reprogramming and tumor
aggressiveness.
Metastasis-related proteins
Metastasis is a complex biological process that
involves multiple signaling pathways, with several proteins acting
as key facilitators. MMPs, particularly MMP-2 and MMP-9, are
crucial for ECM degradation, contributing to metastasis (83). Overexpression of mucin (MUC)15
has been associated with enhanced osteosarcoma cell proliferation
and invasiveness via the Wnt/β-catenin signaling pathway and
regulated through MMP-2 and MMP-9 (74). A study indicated that sirtuin 6
negatively regulates proliferation and invasion in osteosarcoma by
targeting N-cadherin (84),
showcasing how proteins involved in EMT are leveraged by
osteosarcoma cells to facilitate metastatic spread.
Apoptosis and survival pathways
Proteomic studies have illuminated various pathways
by which osteosarcoma cells evade programmed cell death, revealing
potential targets for apoptosis-inducing therapies (77). Proteins such as survivin and
Livin have been highlighted as important mediators of apoptosis
resistance in osteosarcoma. MUC15 was shown to promote cell
proliferation through the Livin protein, linking the Wnt/β-catenin
pathway with survival (74). The
B-cell lymphoma-2 (Bcl-2) inhibitor AT-101 has demonstrated
efficacy in inhibiting osteosarcoma growth in preclinical models
(85). Further exploration of
how survival pathways modulated by Bcl-2 family members interact
with proteomics will enhance our understanding of therapeutic
resistance.
Metabolic regulation of osteosarcoma
The metabolic profile of osteosarcoma cells is
essential for their proliferation and survival. Critical proteins
involved in glycolysis and oxidative phosphorylation are frequent
subjects of investigation (86).
A study indicated that lysine-specific demethylase 6B-mediated
histone demethylation of lactate dehydrogenase A (LDHA) promotes
lung metastasis in osteosarcoma. This highlights the role of LDHA
in shaping the metabolic landscape (87). Furthermore, N-acetyltransferase
10-mediated ac4C acetylation of LDHA was shown to upregulate
glycolytic metabolism in osteosarcoma (88). Aerobic glycolysis: The
CXCR4-CARM1-YAP axis has been found to be critical in regulating
glucose metabolism, linking aerobic glycolysis to chemoresistance
(69). Targeting these metabolic
pathways may thus provide another avenue for intervention.
Potential therapeutic targets
Overall, the insights gleaned from proteomics
research reveal numerous potential therapeutic targets for
osteosarcoma. Inhibition of motif-containing family genes 26
(TRIM26) offers a potential therapeutic strategy by destabilizing
RACK1 and inactivating MEK/ERK signaling, demonstrating its role in
osteosarcoma progression (89).
Evidence suggests that Zyxin and TRIM22 restrict proliferation and
metastasis via diverse signaling pathways, providing promising
avenues for therapeutic explorations (90,91). The splicing factor Y-box binding
protein 1 (YBX1) has been identified as a promoter of osteosarcoma
progression through the upregulation of VEGF (64). Intervening in YBX1 pathways may
effectively inhibit angiogenesis and tumor growth.
The ongoing proteomics research into osteosarcoma
extends our understanding of tumor biology, highlighting crucial
proteins that underlie molecular pathways involved in tumor
progression, metastasis and survival (92). These proteins offer promising
targets for new therapeutic strategies, directing future research
toward enhancing targeted therapies and personalized medicine
approaches for osteosarcoma. As proteomic data are being harnessed,
integrating these insights with other omics layers will be pivotal
in advancing precision medicine for osteosarcoma management.
Metabolomics
The exploration of metabolomic changes in
osteosarcoma has opened up several metabolic pathways that are
responsible for the disease's progression and therapeutic
resistance (93). The summary
provided in the chapter below delineates findings from recent
studies, emphasizing various metabolic pathways related to
osteosarcoma, with particular attention paid to lipid and glucose
metabolism, amino acid metabolism and the role of TME interactions
(Table IV).
 | Table IVMetabolomics findings in osteosarcoma
research. |
Table IV
Metabolomics findings in osteosarcoma
research.
| Author/s, year | Metabolites | Samples | Major findings | (Refs.) |
|---|
| Hu et al,
2022 | Sphingolipid | Data from The
Cancer Genome Atlas | Sphingolipid
metabolism promotes osteosarcoma metastasis and worsens prognosis
by activating notch signaling and angiogenesis pathways. | (95) |
| Cai et al,
2024 | Lipid | Osteosarcoma
tissues | Lipid metabolism
promotes osteosarcoma proliferation and tumor growth by regulating
lipogenic enzymes and the Akt/mTOR pathway, with RPARP-AS1 acting
as a critical oncogenic lncRNA. | (96) |
| Bispo et al,
2023 | Lipid | Osteosarcoma
cells | Lipid metabolism
influences osteosarcoma cell responses to chemotherapy by affecting
membrane fluidity and triglyceride levels, with cisplatin enhancing
apoptosis while Pd2Spermine primarily modulates membrane
properties. | (97) |
| Fritsche-Guenther
et al, 2020 | Fatty acid
oxidation | A female
osteosarcoma cell line | Fatty acid
oxidation supports osteosarcoma metastasis by enhancing energy
metabolism and metabolic flux in metastatic cells compared to
benign counterparts. | (98) |
| Shen et al,
2023 | Glycolysis | An osteosarcoma
model | Glycolysis
facilitates osteosarcoma progression by promoting a hypoxia-induced
feedback loop involving circRNA Hsa_circ_0000566 and HIF-1α,
enhancing the Warburg effect under hypoxic conditions. | (99) |
| Li et al,
2021 | Glucose | miR-329-3p
expression in osteosarcoma tissue | Glucose metabolism
increases cisplatin resistance in osteosarcoma by enhancing LDHA
activity, which is negatively regulated by miR-329-3p. | (100) |
| Wang et al,
2023 | Glucose | Osteosarcoma
cells | Glucose metabolism
promotes osteosarcoma growth and cancer stem-like properties by
facilitating aerobic glycolysis, which solasonine inhibits in an
ALDOA-dependent manner. | (101) |
| Ren et al,
2024 | Amino acids | A novel
three-dimensional model of osteosarcoma cell lines | Amino acids,
particularly glutamine, enhance osteosarcoma drug resistance by
promoting metabolic changes in a three-dimensional
microenvironment, which can be targeted to overcome
resistance. | (102) |
| Wang et al,
2024 | Glutamine | Single-cell
transcriptomic data | Glutamine
metabolism enhances the resistance of osteosarcoma to chemotherapy
by activating the YAP1 pathway, which serves as a bypass mechanism
against drug effects. | (103) |
| Lin et al,
2022 | Branched-chain
amino acids | Osteosarcoma
samples and cell lines | BCAAs promote
osteosarcoma progression by activating the mTOR signaling pathway
in the absence of ANGPTL4, which negatively regulates BCAA
catabolism. | (104) |
| Li et al,
2024 | Copper | Osteosarcoma
samples of patients | Copper metabolism,
particularly through cuproptosis-related sphingolipid metabolism,
regulates osteosarcoma progression and drug resistance by
influencing immune cell infiltration and signaling pathways that
govern cell migration and apoptosis. | (107) |
| Lin et al,
2023 | Copper | Datasets GSE21257
including 50 osteosarcoma patients | Copper metabolism
influences osteosarcoma progression and drug sensitivity by
establishing a prognostic signature through CMRGs that correlates
with immune activity and patient outcomes. | (108) |
Lipid metabolism
Lipid metabolism has emerged as a critical player in
osteosarcoma pathophysiology. Research indicates that deregulated
lipid homeostasis is linked to cancer progression and metastasis in
osteosarcoma (94). For
instance, Hu et al (95)
demonstrated that sphingolipid metabolism is intricately associated
with osteosarcoma metastasis and patient prognosis. Furthermore,
studies suggest that long non-coding RNAs, such as RPARP-antisense
RNA 1, can regulate lipid metabolism, thus promoting osteosarcoma
progression (96). Additionally,
Bispo et al (97) have
shown that novel metal-based drugs can impact lipid metabolism in
MG-63 osteosarcoma cells, presenting a potential therapeutic
avenue. Notably, fatty acid oxidation and the associated signaling
pathways are essential for the survival and growth of osteosarcoma
cells. Research by Fritsche-Guenther et al (99) highlighted the shifting nutrient
dependencies of osteosarcoma cells, revealing that certain
metabolic adaptations facilitate their aggressive traits. The
interplay between lipid droplets and the osteosarcoma
microenvironment is thus critical for understanding the tumor's
metabolism.
Glucose metabolism
The Warburg effect, characterized by increased
glycolysis in the presence of oxygen, is prevalent among
osteosarcoma cells. Shen et al (99) noted that circular RNAs, such as
Hsa_circ_0000566, promote glycolysis and contribute to osteosarcoma
progression by feedback regulation on HIF-1α. Similarly, Li et
al (100) indicated that
aberrations in glucose metabolism, specifically through the
upregulation of LDHA by miR-329-3p, sensitize osteosarcoma cells to
cisplatin treatment, underscoring the relevance of metabolic
pathways for therapeutic responses. Furthermore, elevated glucose
levels have been correlated with enhanced stemness and metastatic
potential in osteosarcoma. Wang et al (101) demonstrated that modulating
glucose metabolism affects the cancer stem cells' properties and
their tumorigenicity. Targeting these metabolic pathways provides
opportunities to enhance the efficacy of existing therapies.
Amino acid metabolism
Amino acid metabolism, particularly glutamine
metabolism, has gained attention in cancer research due to its
association with tumor growth and survival. Ren et al
(102) showed that altered
glutamine usage in osteosarcoma elevates drug resistance by
sustaining metabolic demands under stress conditions. In addition,
research by Wang et al (103) highlighted the role of
YAP1-mediated glutamine metabolism in osteosarcoma, indicating that
disrupting this pathway could enhance therapeutic lethality. The
branched-chain amino acids (BCAAs) are also implicated in
osteosarcoma progression. Lin et al (104) found that angiopoietin like 4
regulates BCAA metabolism, presenting a potential target for
disrupting metabolic communications in osteosarcoma. The metabolic
flexibility conferred by amino acid utilization aids osteosarcoma
cells in adapting to various environmental stressors.
Interactions with the TME
The TME significantly influences the metabolic
landscape of osteosarcoma. A study has shown that metabolic gene
expression correlates with immune microenvironment alterations,
impacting clinical outcomes (105). Wu et al (106) explored the metabolic interplay
between osteosarcoma cells and the immune microenvironment,
suggesting that a thorough understanding of these interactions
could lead to novel immunotherapy approaches. Moreover, copper
metabolism, highlighted in recent work, affects both the immune
landscape and the viability of osteosarcoma cells (107). Lin et al (108) emphasized the need to assess the
copper-metabolism-related genes for further insights into
osteosarcoma prognosis and treatment responses.
As the understanding of the metabolic alterations in
osteosarcoma deepens, it brings forth exciting prospects for
targeted metabolic interventions. The integration of multi-omics
approaches is essential for elucidating the complex metabolic
networks underlying osteosarcoma, potentially leading to
personalized therapeutic strategies (93). Future research should continue to
explore the interactions among different metabolic pathways and
their implications in the TME, providing a foundation for
innovative treatment modalities based on metabolic
reprogramming.
Integrative multi-omics approaches
Combining genomic, transcriptomic,
proteomic and metabolomic data
The integration of multi-omics data provides unique
challenges and opportunities. Techniques such as machine learning,
bioinformatics and systems biology are becoming essential for
managing the vast datasets generated by each omics layer (10). Machine learning methods,
particularly those based on supervised and unsupervised algorithms,
can be employed to identify patterns and correlations among
genomic, transcriptomic, proteomic and metabolomic profiles. These
methods facilitate the extraction of meaningful insights that a
single omics approach may overlook (10). Recent advancements in
computational frameworks, such as weighted gene co-expression
network analysis and pathway enrichment tools, have further enabled
the identification of cross-omics interactions, enhancing the
interpretability of complex datasets (109).
One illustrative case study is the combined omics
analysis conducted by Lin et al (110), which investigated the molecular
landscape of osteosarcoma. The study utilized RNA-seq data to
identify differentially expressed genes, while proteomic profiling
revealed novel proteins associated with disease progression. By
integrating these findings with metabolomic data, they were able to
identify specific metabolic pathways, such as glycolysis and
lactate metabolism, that were altered in osteosarcoma. This
integrative approach provided a more holistic understanding of the
TME and its metabolic adaptation, highlighting the critical
interplay between different molecular layers.
A recent multi-omics study employed single-cell RNA
sequencing (scRNA-seq) and spatial transcriptomics to dissect
intratumoral heterogeneity in advanced osteosarcoma (111). The study identified distinct
cellular subpopulations enriched in chemotherapy-resistant cells,
characterized by upregulated glycolysis and PI3K/AKT pathway
activity. Proteomic validation further confirmed the overexpression
of glycolytic enzymes (e.g., LDHA) and AKT1 in these
subpopulations, providing a mechanistic link between metabolic
reprogramming and therapeutic resistance. This study exemplifies
how integrative approaches can uncover actionable targets, such as
combined inhibition of AKT and glycolysis, to overcome
resistance.
Another compelling example involves a study by Jia
et al (112), which
integrated genomic, proteomic and metabolomic data to explore
cuproptosis-related pathways in osteosarcoma. The authors
identified a copper metabolism signature correlated with immune
suppression and poor prognosis. Functional experiments demonstrated
that copper chelators synergized with immune checkpoint inhibitors
to enhance T-cell infiltration and reduce tumor growth in
preclinical models. This translational application underscores the
potential of multi-omics in guiding combination therapies tailored
to metabolic-immune crosstalk.
Furthermore, Truong et al (113) mapped the differentiation
trajectories of osteosarcoma cells using scRNA-seq and chromatin
accessibility assays. They revealed that epigenetic remodeling of
SOX9-driven chondrogenic pathways promotes metastatic niche
formation. Complementary metabolomic profiling highlighted elevated
glutamine utilization in metastatic cells, suggesting therapeutic
vulnerability to glutaminase inhibitors. This study not only
clarified the role of SOX9 in osteosarcoma progression but also
demonstrated how multi-omics can bridge molecular mechanisms to
therapeutic strategies.
Impact on understanding tumor biology and
treatment options
Integrative multi-omics approaches significantly
enhance our understanding of tumor biology, yielding crucial
insights that can inform treatment decisions (114). For instance, the identification
of specific genetic and proteomic alterations linked to signaling
pathways, such as the mTOR and PI3K/AKT pathways, allows for the
development of targeted therapies aimed at these pathways (115). Wang et al (103) combined CRISPR-Cas9 screens with
proteomic profiling to identify YAP1-mediated glutamine addiction
in chemotherapy-resistant osteosarcoma. They demonstrated that
disrupting the YAP1-glutamine axis using the small-molecule
inhibitor CA3 restored cisplatin sensitivity in vivo,
highlighting the translational value of pathway-centric
multi-omics.
Furthermore, studies leveraging integrative
approaches can identify patient stratification markers,
facilitating personalized treatment regimens (116). By categorizing patients based
on distinct molecular signatures, clinicians can tailor therapies
that specifically target the identified vulnerabilities of each
tumor. For instance, patients exhibiting specific metabolic
dysregulations could be treated with agents designed to exploit
those metabolic weaknesses, potentially enhancing therapeutic
efficacy (117).
Furthermore, the integrative analysis of multi-omics
data can assist in predicting treatment responses and resistance
mechanisms in osteosarcoma (107). Understanding the interactions
between genetic alterations, changes in gene expression and
alterations in protein levels can provide insights into why certain
patients may not respond to conventional therapies (118). For instance, the presence of
specific mutations alongside corresponding protein changes can
indicate an adaptive resistance mechanism, signaling the need for
alternative treatment options (118).
The integrative analysis of multi-omics data can
also assist in predicting treatment responses and resistance
mechanisms in osteosarcoma (104). For example, Audinot et
al (119) utilized
circulating tumor DNA sequencing and longitudinal metabolomics to
monitor osteosarcoma evolution during chemotherapy. They identified
recurrent mutations in TP53 and RB1 alongside increased kynurenine
pathway activity as hallmarks of acquired resistance. Targeting
these pathways with poly(ADP-ribose) polymerase inhibitors and
indoleamine 2,3-dioxygenase 1 blockers in patient-derived
xenografts (PDXs) significantly delayed relapse, illustrating the
power of dynamic multi-omics monitoring.
In summary, integrative multi-omics approaches not
only provide a comprehensive view of the complex biological
landscape of osteosarcoma but also foster innovation in treatment
strategies. The ability to couple genomic, transcriptomic,
proteomic and metabolomic data through advanced computational
techniques can lead to the discovery of novel biomarkers and
therapeutic targets, ultimately guiding the implementation of
precision medicine in osteosarcoma management.
Translational applications of multi-omics in
osteosarcoma
The integration of multi-omics approaches
encompassing genomics, transcriptomics, proteomics and metabolomics
holds great promise in enhancing the diagnosis and management of
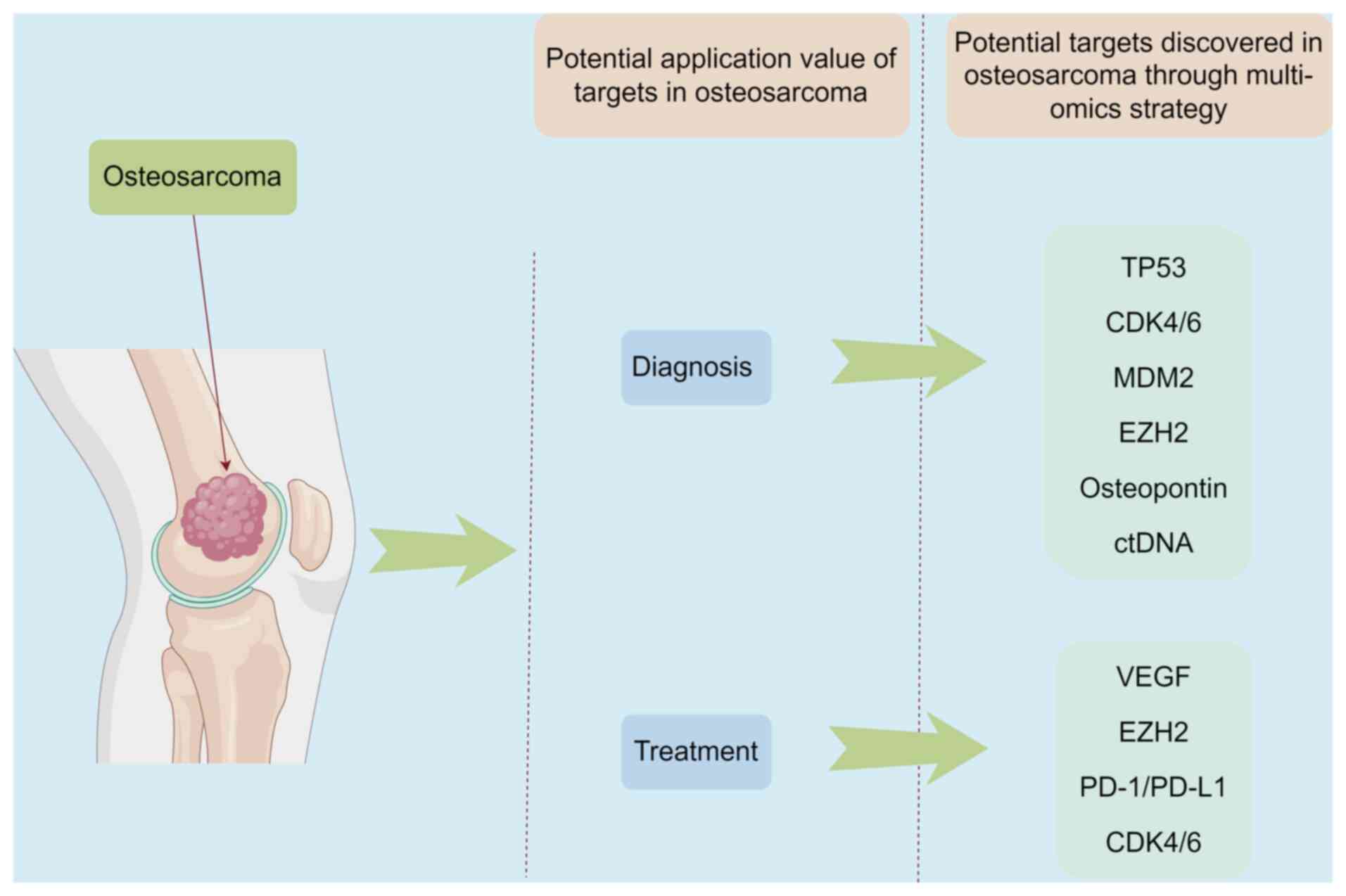
osteosarcoma (Table V) (4). The targets identified through
multi-omics strategies show significant potential in the diagnosis
and treatment of osteosarcoma, providing direction for targeted
therapy and combination therapy for this condition (Fig. 2).
 | Table VThe clinical value of multi-omics
findings in osteosarcoma. |
Table V
The clinical value of multi-omics
findings in osteosarcoma.
| Author/s, year | Biomarker | Clinical
application | Expected
effect | (Refs.) |
|---|
| Chen et al,
2016 | TP53 | Diagnosis | TP53 mutations
serve as a significant prognostic marker indicating poor 2-year
overall survival in patients with osteosarcoma. | (121) |
| Ru et al,
2015 | TP53 | Diagnosis | TP53 genetic
mutations serve as significant indicators of osteosarcoma risk and
prognosis in the Chinese population. | (122) |
| Jeon et al,
2015 | CDK4 and MDM2 | Diagnosis | CDK4 immunostaining
may serve as a valuable diagnostic adjunct for low-grade central
osteosarcoma, although its negative results do not exclude the
diagnosis. | (123) |
| Iwata et al,
2021 | CDK4 | Diagnosis | CDK4 amplification
and overexpression in osteosarcoma tumors serve as predictive
biomarkers for resistance to conventional chemotherapy. | (124) |
| Schubert et
al, 2022 | CDK4/6 | Diagnosis | CDK4/6 genomic
aberrations in osteosarcoma indicate potential as therapeutic
targets, though their correlation with treatment efficacy requires
further evaluation. | (125) |
| Sun et al,
2016 | EZH2 | Diagnosis and
treatment | EZH2 overexpression
serves as a potential prognostic marker for aggressive behavior and
poor outcomes in osteosarcoma, highlighting its role as a promising
therapeutic target. | (126) |
| Nazarizadeh et
al, 2021 | Osteopontin | Diagnosis | Osteopontin shows
promise as a sensitive and specific biomarker for diagnosing bone
cancers and predicting their clinicopathological features. | (128) |
| Barris et
al, 2018 | ctDNA | Diagnosis | ctDNA analysis
offers a promising non-invasive approach for detecting somatic
alterations in osteosarcoma, which may aid in patient management
and treatment monitoring. | (129) |
| Audinot et
al, 2024 | ctDNA | Diagnosis | ctDNA
quantification enhances the prognostic stratification of
osteosarcoma patients, serving as a significant independent
prognostic factor alongside traditional clinical parameters. | (119) |
| Zhao et al,
2015 | VEGF | Treatment | VEGF serves as a
crucial target in osteosarcoma therapy, as its expression is
directly regulated by miR-410, which, when overexpressed, inhibits
tumor growth and promotes apoptosis in osteosarcoma cells. | (131) |
| Tawbi et al,
2017 | VEGF | Treatment | VEGF-targeted
therapy holds potential in bone sarcoma treatment, where it may
enhance response rates and overall survival, particularly when
combined with immunotherapy approaches like pembrolizumab. | (132) |
| Mickymaray et
al, 2021 | PI3K/AKT/mTOR
pathway | Treatment | The PI3K/AKT/mTOR
pathway is a promising target in osteosarcoma therapy, as its
downregulation by Rhaponticin effectively induces cytotoxic effects
and apoptosis in osteosarcoma cells. | (134) |
| Lu et al,
2020 | EZH2 | Treatment | EZH2 represents a
valuable therapeutic target in osteosarcoma treatment, as its
regulation by miR-449a is linked to inhibited tumor progression and
improved patient prognosis. | (135) |
| Chen et al,
2023 | EZH2 | Treatment | EZH2 is a key
target in osteosarcoma therapy, as its downregulation by capsaicin
significantly inhibits cancer stemness and metastatic potential,
potentially improving patient outcomes. | (136) |
| Liu et al,
2019 | PD-1/PD-L1
axis | Treatment | Targeting the
PD-1/PD-L1 axis enhances the effectiveness of cisplatin
chemotherapy in osteosarcoma, improving tumor control and promoting
apoptosis in cancer cells. | (137) |
| Yoshida et
al, 2019 | PD-L1 | Treatment | PD-L1 expression in
osteosarcoma correlates with early metastatic formation and may
serve as a predictive marker for the effectiveness of anti-PD-1
therapy, highlighting its potential in targeted treatment
strategies. | (138) |
| Davis et al,
2020 | PD-1/PD-L1 | Treatment | Targeting PD-1 with
nivolumab demonstrates a favorable safety profile in children and
young adults, indicating its potential as a therapeutic option for
osteosarcoma and other pediatric malignancies, particularly in
combination therapies. | (139) |
| Zhou et al,
2018 | CDK4 | Treatment | CDK4 represents a
promising therapeutic target in osteosarcoma, as its inhibition
significantly suppresses tumor growth and migratory potential, and
induces apoptosis in cancer cells. | (140) |
| Oshiro et
al, 2021 | CDK4/6 | Treatment | The combination of
CDK4/6 and mTOR inhibitors shows promising therapeutic potential
for treating doxorubicin-resistant osteosarcoma by significantly
reducing tumor volume and inducing tumor necrosis. | (141) |
Mutations in TP53 have been extensively studied as
potential biomarkers. Various studies indicate that TP53 mutations
are associated with aggressive disease and poor outcomes in
osteosarcoma (120). For
instance, a meta-analysis identified TP53 mutations as significant
prognostic factors, suggesting their potential as biomarkers for
patient stratification (121).
Specific polymorphisms in TP53 have also been linked to
susceptibility to osteosarcoma among certain populations, providing
insights into genetic predisposition (122). MDM2, an antagonist of TP53, has
emerged as another potential biomarker. Its overexpression has been
implicated in various malignancies, including osteosarcoma.
Research demonstrates that MDM2 overexpression can be detected via
immunohistochemistry and correlates with lower survival rates,
making it a valuable adjunct in the diagnosis and prognostic
assessment of patients with osteosarcoma (123). Cyclin-dependent kinases (CDKs),
particularly CDK4, have garnered attention as biomarkers in
osteosarcoma. Overexpression of CDK4 is associated with resistance
to conventional chemotherapy and correlates with aggressive disease
features (124). Furthermore,
CDK4/6 inhibitors are being explored as potential therapeutic
options, thus reinforcing the clinical relevance of CDK4 as a
biomarker in osteosarcoma (125).
Enhancer of zeste homologue 2 (EZH2), a histone
methyltransferase, has been implicated in osteosarcoma progression
and poor prognosis. High levels of EZH2 expression have been
associated with advanced disease stages and metastasis,
establishing its role as a prognostic marker (126). Furthermore, EZH2 inhibitors are
being evaluated in clinical trials, showing promise in targeted
therapy approaches for osteosarcoma (127). Osteopontin has been identified
as a significant serum biomarker for osteosarcoma. Elevated levels
of osteopontin are associated with poorer survival outcomes, making
it a potential marker for disease monitoring and therapy response
(128). Its measurement in
biofluids may provide real-time insights into the disease state.
The use of circulating tumor DNA (ctDNA) represents a breakthrough
in the non-invasive monitoring of osteosarcoma. ctDNA analysis
enables the detection of tumor-specific mutations and informs about
minimal residual disease and the risk of recurrence (129). Recent studies have validated
the clinical utility of ctDNA quantification in predicting
outcomes, indicating its integration into routine management
protocols (119).
Recent advancements in metabolomics have identified
various metabolites that exhibit significant alterations in
patients with osteosarcoma compared to healthy controls. Specific
metabolic signatures associated with tumor presence and progression
may serve as novel diagnostic biomarkers, although research in this
area is still in its infancy (130). Multi-omics technologies have
significantly expanded the understanding of osteosarcoma,
identifying various biomarkers with diagnostic and prognostic
potential. Integrating these biomarkers into clinical workflows may
facilitate earlier diagnosis, enhance personalized treatment
strategies and ultimately improve patient outcomes. Future research
should focus on validating these biomarkers in larger cohorts and
elucidating their mechanisms of action, propelling the field toward
precision medicine in osteosarcoma management.
VEGF has emerged as a pivotal biomarker in
osteosarcoma due to its role in tumor angiogenesis. Studies have
demonstrated that inhibition of VEGF not only reduces tumor growth
but also enhances the sensitivity of osteosarcoma cells to
therapeutic agents. For instance, silencing VEGF through microRNAs,
such as miR-410, has been shown to suppress proliferation and
invasion of osteosarcoma cells (131). Furthermore, the application of
anti-VEGF therapies, such as bevacizumab, in combination with other
agents may provide a synergistic effect, as evidenced in recent
clinical trials (132,133).
The PI3K/AKT/mTOR pathway is frequently dysregulated
in osteosarcoma, making it a suitable target for intervention.
Various studies have explored this pathway's role in cell survival
and proliferation. For instance, the natural compound rhaponticin
was shown to inhibit osteosarcoma via suppression of the
PI3K-Akt-mTOR signaling axis (134). Furthermore, selective
inhibition of EZH2 has been demonstrated to effectively regulate
the PI3K/AKT signaling pathway, impacting cancer cell behavior
(135,136).
The blockade of immune checkpoints, particularly
programmed death-1 (PD-1)/programmed death-ligand 1 (PD-L1), has
been explored in osteosarcoma and demonstrates significant
therapeutic potential. Research indicates that combining PD-1/PD-L1
inhibitors with traditional chemotherapeutics can enhance
anti-tumor efficacy by overcoming immune escape mechanisms
(137,138). Furthermore, the combination of
immune checkpoint blockade and T-cell therapies has shown promising
results in clinical settings (139).
The CDK pathway, specifically CDK4/6, has gained
attention as a therapeutic target due to its involvement in cell
cycle regulation in osteosarcoma (140). Targeting CDK4/6 has been shown
to enhance the effects of chemotherapy and may help in the
management of drug-resistant osteosarcoma variants (125,141).
In conclusion, multi-omics approaches have
significantly advanced the understanding of the molecular
underpinnings of osteosarcoma. By translating these findings into
clinical applications, it may be possible to better harness
targeted therapies, ultimately improving patient outcomes in this
challenging malignancy. Continuous research and clinical trials
will be paramount to validate these strategies and ensure their
efficacy in the broader patient population.
Future directions
One of the most pressing needs in osteosarcoma
research is the establishment of collaborative research networks
that promote data sharing among institutions worldwide (142). The heterogeneity of
osteosarcoma, combined with the complexity of multi-omics data,
necessitates a collaborative approach to facilitate large-scale
studies and the development of robust databases that integrate
genomic, transcriptomic, proteomic and metabolomic information
(13).
Current efforts, such as the Pediatric Cancer Genome
Project, provide a framework for collaborative research but need to
be expanded to include international participants and diverse
patient populations (143).
Furthermore, data sharing platforms, such as the European
Genome-phenome Archive (https://ega-archive.org) and cBioPortal (https://www.cbioportal.org), play a crucial role in
providing researchers access to valuable datasets, fostering
collaborations that can lead to meaningful discoveries in
osteosarcoma research (144).
By encouraging multi-institutional collaborations,
it may be possible to facilitate the standardization of omics data
collection protocols, ensuring consistency and reproducibility
among disparate studies. As multi-omics research evolves, a shared
commitment to open science will enhance the overall quality of
research findings and enable faster translation into clinical
applications. The future of multi-omics in osteosarcoma research is
closely tied to the rapid advancements in emerging technologies,
particularly single-cell omics (113). Single-cell sequencing
technologies allow for the analysis of individual cells within a
tumor, providing unprecedented insights into tumor heterogeneity,
cell lineage and microenvironment interactions that traditional
bulk analysis cannot achieve (145).
These technologies hold the potential to uncover
specific cell populations that drive tumor growth and metastasis,
which can lead to the identification of novel biomarkers and
therapeutic targets (146).
Recent studies employing scRNA-seq in osteosarcoma have revealed
distinct cellular subpopulations that contribute to the TME's
immunosuppressive nature, potentially guiding the development of
more effective immunotherapy strategies (111,147,148). Furthermore, integrating
single-cell proteomics and metabolomics with transcriptomics can
help create a multi-layered understanding of the molecular dynamics
within tumors (111). Future
initiatives should focus on standardizing these methodologies and
developing comprehensive platforms that seamlessly integrate
heterogeneous single-cell data for further analysis.
Longitudinal studies that utilize patient-derived
samples are critical for advancing the current understanding of
osteosarcoma's progression and treatment responses. Monitoring
tumors over time through the collection of biopsies, blood samples
for ctDNA and other biofluids can provide valuable insights into
the temporal dynamics of genomic and phenotypic alterations
(149). Such studies can
enhance our understanding of how tumors evolve under specific
therapies, allowing for the identification of resistance mechanisms
and the development of adaptive treatment approaches. For instance,
a prospective trial analyzing ctDNA in patients with osteosarcoma
demonstrated that detecting mutations associated with chemotherapy
resistance provided critical insights for treatment modifications
(119).
Furthermore, incorporating PDXs and organoids
derived from osteosarcoma tumors into research initiatives allows
for a more accurate representation of tumor biology in the
laboratory (150,151). These models can be utilized for
drug screening and validation of treatment strategies before
clinical implementation, ultimately promoting personalized therapy.
To successfully implement these initiatives, securing funding and
collaboration from stakeholders, including academic institutions,
pharmaceutical companies and patient advocacy groups, will be
vital. Engaging patients in the research process, ensuring their
informed consent and privacy are respected, will further promote
patient-centric research endeavors that align with modern ethical
standards.
The future directions and initiatives in
multi-omics research for osteosarcoma promise to enhance the
current understanding and management of this complex disease. By
fostering collaborative research and data sharing, harnessing the
potential of emerging technologies such as single-cell omics, and
implementing longitudinal studies using patient-derived samples, it
may be possible to pave the way for precision medicine that caters
to the individual needs of patients with osteosarcoma. As these
initiatives evolve, they will play a vital role in overcoming the
challenges currently faced by osteosarcoma research and ultimately
improve patient outcomes.
Conclusion
In conclusion, the integration of multi-omics
approaches holds transformative potential for osteosarcoma research
and treatment, allowing for a comprehensive understanding of tumor
heterogeneity and the development of targeted therapies. By
elucidating the intricate molecular networks underlying
osteosarcoma, it may be possible to enhance early diagnosis and
personalize treatment strategies, ultimately improving patient
outcomes. It is imperative for clinicians and researchers to
collaborate and advocate for the incorporation of multi-omics into
clinical practice to advance precision medicine and offer hope to
patients with osteosarcoma.
Availability of data and materials
Not applicable.
Authors' contributions
XC, BT, YW and XK made substantial contributions to
the conception and design of the article. XC and XK were involved
in the acquisition, analysis and interpretation of data. XC, BT,
YW, JZ and XK wrote the manuscript. Data authentication is not
applicable. All authors have read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Abbreviations:
|
TME
|
tumor microenvironment
|
|
ECM
|
extracellular matrix
|
|
EMT
|
epithelial-mesenchymal transition
|
|
VEGF
|
vascular endothelial growth
factor
|
|
HIF-1α
|
hypoxia-inducible factor 1α
|
|
CXCR4
|
C-X-C chemokine receptor type 4
|
|
MMPs
|
matrix metalloproteinases
|
|
LDHA
|
lactate dehydrogenase A
|
|
YAP1
|
Yes-associated protein 1
|
|
mTOR
|
mammalian target of rapamycin
|
|
FGFR
|
fibroblast growth factor receptor
|
|
FN1
|
fibronectin 1
|
|
ICAM-1
|
intercellular adhesion molecule-1
|
|
BCAA
|
branched-chain amino acid
|
|
AKT1
|
AKT serine/threonine kinase 1
|
|
CLEC3A
|
C-type lectin domain family 3 member
A
|
|
MAT1
|
ménage à trios 1
|
|
PD-1/PD-L1
|
programmed death-1/programmed
death-ligand 1
|
|
CDK
|
cyclin-dependent kinase
|
|
EZH2
|
enhancer of zeste homologue 2
|
|
ctDNA
|
circulating tumor DNA
|
|
CMRG
|
copper metabolism-related gene
|
Acknowledgments
Not applicable.
Funding
No funding was received.
References
|
1
|
Ritter J and Bielack SS: Osteosarcoma. Ann
Oncol. 21(Suppl 7): vii320–vii325. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Belayneh R, Fourman MS, Bhogal S and Weiss
KR: Update on osteosarcoma. Curr Oncol Rep. 23:712021. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Czarnecka AM, Synoradzki K, Firlej W,
Bartnik E, Sobczuk P, Fiedorowicz M, Grieb P and Rutkowski P:
Molecular biology of osteosarcoma. Cancers (Basel). 12:21302020.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Jiang Y, Wang J, Sun M, Zuo D, Wang H,
Shen J, Jiang W, Mu H, Ma X, Yin F, et al: Multi-omics analysis
identifies osteosarcoma subtypes with distinct prognosis indicating
stratified treatment. Nat Commun. 13:72072022. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Mirabello L, Troisi RJ and Savage SA:
Osteosarcoma incidence and survival rates from 1973 to 2004: Data
from the surveillance, epidemiology, and end results program.
Cancer. 115:1531–1543. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Meltzer PS and Helman LJ: New horizons in
the treatment of osteosarcoma. N Engl J Med. 385:2066–2076. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Garcia-Ortega DY, Cabrera-Nieto SA,
Caro-Sánchez HS and Cruz-Ramos M: An overview of resistance to
chemotherapy in osteosarcoma and future perspectives. Cancer Drug
Resist. 5:762–793. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Panez-Toro I, Muñoz-García J,
Vargas-Franco JW, Renodon-Cornière A, Heymann MF, Lézot F and
Heymann D: Advances in osteosarcoma. Curr Osteoporos Rep.
21:330–343. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Peled Y, Levin D, Manisterski M, Kollander
N, Shukrun R and Elhasid R: Weight loss and response to
chemotherapy in pediatric patients with osteosarcoma. Eur J Clin
Nutr. 78:541–543. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
He X, Liu X, Zuo F, Shi H and Jing J:
Artificial intelligence-based multi-omics analysis fuels cancer
precision medicine. Semin Cancer Biol. 88:187–200. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang L, Qu J, Harari O, Boddey JA, Wang Z
and Linna-Kuosmanen S: The impact of multi-omics in medicine. Cell
Rep Med. 5:1017422024. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mohammadi-Shemirani P, Sood T and Paré G:
From omics to multi-omics technologies: The discovery of novel
causal mediators. Curr Atheroscler Rep. 25:55–65. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tang S, Roberts RD, Cheng L and Li L:
Osteosarcoma multi-omics landscape and subtypes. Cancers (Basel).
15:49702023. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Donisi C, Pretta A, Pusceddu V, Ziranu P,
Lai E, Puzzoni M, Mariani S, Massa E, Madeddu C and Scartozzi M:
Immunotherapy and cancer: The multi-omics perspective. Int J Mol
Sci. 25:35632024. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Urban W, Krzystańska D, Piekarz M, Nazar J
and Jankowska A: Osteosarcoma's genetic landscape painted by genes'
mutations. Acta Biochim Pol. 70:671–678. 2023.PubMed/NCBI
|
|
16
|
Mirabello L, Yeager M, Mai PL,
Gastier-Foster JM, Gorlick R, Khanna C, Patiño-Garcia A,
Sierrasesúmaga L, Lecanda F, Andrulis IL, et al: Germline TP53
variants and susceptibility to osteosarcoma. J Natl Cancer Inst.
107:djv1012015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Mokánszki A, Chang Chien YC, Mótyán JA,
Juhász P, Bádon ES, Madar L, Szegedi I, Kiss C and Méhes G: Novel
RB1 and MET gene mutations in a case with bilateral retinoblastoma
followed by multiple metastatic osteosarcoma. Diagnostics (Basel).
11:282020. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Righi A, Gambarotti M, Benini S, Gamberi
G, Cocchi S, Picci P and Bertoni F: MDM2 and CDK4 expression in
periosteal osteosarcoma. Hum Pathol. 46:549–553. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Monti P, Menichini P, Speciale A, Cutrona
G, Fais F, Taiana E, Neri A, Bomben R, Gentile M, Gattei V, et al:
Heterogeneity of TP53 mutations and P53 protein residual function
in cancer: Does it matter? Front Oncol. 10:5933832020. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Huang MF, Wang YX, Chou YT and Lee DF:
Therapeutic strategies for RB1-deficient cancers: Intersecting gene
regulation and targeted therapy. Cancers (Basel). 16:15582024.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Koo N, Sharma AK and Narayan S:
Therapeutics targeting p53-MDM2 interaction to induce cancer cell
death. Int J Mol Sci. 23:50052022. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Synoradzki KJ, Bartnik E, Czarnecka AM,
Fiedorowicz M, Firlej W, Brodziak A, Stasinska A, Rutkowski P and
Grieb P: TP53 in biology and treatment of osteosarcoma. Cancers
(Basel). 13:42842021. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Mannheimer JD, Tawa G, Gerhold D, Braisted
J, Sayers CM, McEachron TA, Meltzer P, Mazcko C, Beck JA and
LeBlanc AK: Transcriptional profiling of canine osteosarcoma
identifies prognostic gene expression signatures with translational
value for humans. Commun Biol. 6:8562023. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Nie JJ, Zhang B, Luo P, Luo M, Luo Y, Cao
J, Wang H, Mao J, Xing Y, Liu W, et al: Enhanced pyroptosis
induction with pore-forming gene delivery for osteosarcoma
microenvironment reshaping. Bioact Mater. 38:455–471.
2024.PubMed/NCBI
|
|
25
|
Jin J, Cong J, Lei S, Zhang Q, Zhong X, Su
Y, Lu M, Ma Y, Li Z, Wang L, et al: Cracking the code: Deciphering
the role of the tumor microenvironment in osteosarcoma metastasis.
Int Immunopharmacol. 121:1104222023. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Luo ZW, Liu PP, Wang ZX, Chen CY and Xie
H: Macrophages in osteosarcoma immune microenvironment:
Implications for immunotherapy. Front Oncol. 10:5865802020.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ichikawa J, Schoenecker JG, Tatsuno R,
Kawasaki T, Suzuki-Inoue K and Haro H: Advancing tissue
factor-targeted therapy for osteosarcoma via understanding its role
in the tumor microenvironment. Curr Pharm Des. 29:1009–1012. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Tatsuno R, Ichikawa J, Komohara Y, Pan C,
Kawasaki T, Enomoto A, Aoki K, Hayakawa K, Iwata S, Jubashi T and
Haro H: Pivotal role of IL-8 derived from the interaction between
osteosarcoma and tumor-associated macrophages in osteosarcoma
growth and metastasis via the FAK pathway. Cell Death Dis.
15:1082024. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Cui J, Dean D, Hornicek FJ, Chen Z and
Duan Z: The role of extracelluar matrix in osteosarcoma progression
and metastasis. J Exp Clin Cancer Res. 39:1782020. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhou Y, Yang Q, Dong Y, Ji T, Zhang B,
Yang C, Zheng S, Tang L, Zhou C, Qian G, et al:
First-in-maintenance therapy for localized high-grade osteosarcoma:
An open-label phase I/II trial of the anti-PD-L1 antibody ZKAB001.
Clin Cancer Res. 29:764–774. 2023. View Article : Google Scholar
|
|
31
|
Sun Y, Zhang C, Fang Q, Zhang W and Liu W:
Abnormal signal pathways and tumor heterogeneity in osteosarcoma. J
Transl Med. 21:992023. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Rajan S, Franz EM, McAloney CA, Vetter TA,
Cam M, Gross AC, Taslim C, Wang M, Cannon MV, Oles A and Roberts
RD: Osteosarcoma tumors maintain intra-tumoral transcriptional
heterogeneity during bone and lung colonization. BMC Biol.
21:982023. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Tian H, Cao J, Li B, Nice EC, Mao H, Zhang
Y and Huang C: Managing the immune microenvironment of
osteosarcoma: The outlook for osteosarcoma treatment. Bone Res.
11:112023. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Lilienthal I and Herold N: Targeting
molecular mechanisms underlying treatment efficacy and resistance
in osteosarcoma: A review of current and future strategies. Int J
Mol Sci. 21:68852020. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Liao Y, Yi Q, He J, Huang D, Xiong J and
Sun W and Sun W: Extracellular vesicles in tumorigenesis,
metastasis, chemotherapy resistance and intercellular communication
in osteosarcoma. Bioengineered. 14:113–128. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Jia C, Liu M, Yao L, Zhao F, Liu S, Li Z
and Han Y: Multi-omics analysis reveals cuproptosis and
mitochondria-based signature for assessing prognosis and immune
landscape in osteosarcoma. Front Immunol. 14:12809452024.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Schott C, Shah AT and Sweet-Cordero EA:
Genomic complexity of osteosarcoma and its implication for
preclinical and clinical targeted therapies. Adv Exp Med Biol.
1258:1–19. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Li L, Sun M, Wang J and Wan S: Multi-omics
based artificial intelligence for cancer research. Adv Cancer Res.
163:303–356. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Meijer DM, Ruano D, Briaire-de Bruijn IH,
Wijers-Koster PM, van de Sande MAJ, Gelderblom H, Cleton-Jansen AM,
de Miranda NFCC, Kuijjer ML and Bovée JVMG: The variable genomic
landscape during osteosarcoma progression: Insights from a
longitudinal WGS analysis. Genes Chromosomes Cancer. 63:e232532024.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Bagger FO, Borgwardt L, Jespersen AS,
Hansen AR, Bertelsen B, Kodama M and Nielsen FC: Whole genome
sequencing in clinical practice. BMC Med Genomics. 17:392024.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Li Y and Yang S, Liu Y and Yang S:
Deletion of Trp53 and Rb1 in Ctsk-expressing cells drives
osteosarcoma progression by activating glucose metabolism and YAP
signaling. MedComm (2020). 3:e1312022. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Feng W, Wang Z, Feng D, Zhu Y, Zhang K and
Huang W: The effects of common variants in MDM2 and GNRH2 genes on
the risk and survival of osteosarcoma in Han populations from
Northwest China. Sci Rep. 10:159392020. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
He X, Pang Z, Zhang X, Lan T, Chen H, Chen
M, Yang H, Huang J, Chen Y, Zhang Z, et al: Consistent
amplification of FRS2 and MDM2 in low-grade osteosarcoma: A genetic
study of 22 cases with clinicopathologic analysis. Am J Surg
Pathol. 42:1143–1155. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Limbach AL, Lingen MW, McElherne J, Mashek
H, Fitzpatrick C, Hyjek E, Mostofi R and Cipriani NA: The utility
of MDM2 and CDK4 immunohistochemistry and MDM2 FISH in craniofacial
osteosarcoma. Head Neck Pathol. 14:889–898. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Kaur H, Kala S, Sood A, Mridha AR, Kakkar
A, Yadav R, Mishra S and Mishra D: Role of MDM2, CDK4, BCL2,
parafibromin and galectin 1 in differentiating osteosarcoma from
its benign fibro-osseous lesions. Head Neck Pathol. 16:728–737.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Ren C, Pan R, Hou L, Wu H, Sun J, Zhang W,
Tian X and Chen H: Suppression of CLEC3A inhibits osteosarcoma cell
proliferation and promotes their chemosensitivity through the
AKT1/mTOR/HIF1α signaling pathway. Mol Med Rep. 21:1739–1748.
2020.PubMed/NCBI
|
|
47
|
Wang J, Ni J, Song D, Ding M, Huang J, Li
W and He G: MAT1 facilitates the lung metastasis of osteosarcoma
through upregulation of AKT1 expression. Life Sci. 234:1167712019.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Madda R, Chen CM, Chen CF, Wang JY, Wu HY,
Wu PK and Chen WM: Analyzing BMP2, FGFR, and TGF beta expressions
in high-grade osteosarcoma untreated and treated autografts using
proteomic analysis. Int J Mol Sci. 23:74092022. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Huang YC, Chen WC, Yu CL, Chang TK, I-Chin
Wei A, Chang TM, Liu JF and Wang SW: FGF2 drives osteosarcoma
metastasis through activating FGFR1-4 receptor pathway-mediated
ICAM-1 expression. Biochem Pharmacol. 218:1158532023. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Kim JA, Berlow NE, Lathara M, Bharathy N,
Martin LR, Purohit R, Cleary MM, Liu Q, Michalek JE, Srinivasa G,
et al: Sensitization of osteosarcoma to irradiation by targeting
nuclear FGFR1. Biochem Biophys Res Commun. 621:101–108. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Cui J, Wang W, Li Z, Zhang Z, Wu B and
Zeng L: Epigenetic changes in osteosarcoma. Bull Cancer.
98:E62–E68. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Makise N, Sekimizu M, Kubo T, Wakai S,
Watanabe SI, Kato T, Kinoshita T, Hiraoka N, Fukayama M, Kawai A,
et al: Extraskeletal osteosarcoma: MDM2 and H3K27me3 analysis of 19
cases suggest disease heterogeneity. Histopathology. 73:147–156.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Saba KH, Cornmark L, Rissler M, Fioretos
T, Åström K, Haglund F, Rosenberg AE, Brosjö O and Nord KH: Genetic
profiling of a chondroblastoma-like osteosarcoma/malignant
phosphaturic mesenchymal tumor of bone reveals a homozygous
deletion of CDKN2A, intragenic deletion of DMD, and a targetable
FN1-FGFR1 gene fusion. Genes Chromosomes Cancer. 58:731–736. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Wu CC and Livingston JA: Genomics and the
immune landscape of osteosarcoma. Adv Exp Med Biol. 1258:21–36.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Yang S, Tian Z, Feng Y, Zhang K, Pan Y, Li
Y, Wang Z, Wei W, Qiao X, Zhou R, et al: Transcriptomics and
metabolomics reveal changes in the regulatory mechanisms of
osteosarcoma under different culture methods in vitro. BMC Med
Genomics. 15:2652022. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Yoon H, Liyanarachchi S, Wright FA,
Davuluri R, Lockman JC, de la Chapelle A and Pellegata NS: Gene
expression profiling of isogenic cells with different TP53 gene
dosage reveals numerous genes that are affected by TP53 dosage and
identifies CSPG2 as a direct target of p53. Proc Natl Acad Sci USA.
99:15632–15637. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Das S, Idate R, Regan DP, Fowles JS, Lana
SE, Thamm DH, Gustafson DL and Duval DL: Immune pathways and TP53
missense mutations are associated with longer survival in canine
osteosarcoma. Commun Biol. 4:11782021. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Luo M, Huang M, Yang N, Zhu Y, Huang P, Xu
Z, Wang W and Cai L: Impairment of rigidity sensing caused by
mutant TP53 gain of function in osteosarcoma. Bone Res. 11:282023.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Feng W, Dean DC, Hornicek FJ, Spentzos D,
Hoffman RM, Shi H and Duan Z: Myc is a prognostic biomarker and
potential therapeutic target in osteosarcoma. Ther Adv Med Oncol.
12:17588359209220552020. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Akkawi R, Hidmi O, Haj-Yahia A, Monin J,
Diment J, Drier Y, Stein GS and Aqeilan RI: WWOX promotes
osteosarcoma development via upregulation of Myc. Cell Death Dis.
15:132024. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Ma L, Xue W and Ma X: GATA3 is
downregulated in osteosarcoma and facilitates EMT as well as
migration through regulation of slug. Onco Targets Ther.
11:7579–7589. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Wu H, Zhang J, Dai R, Xu J and Feng H:
Transferrin receptor-1 and VEGF are prognostic factors for
osteosarcoma. J Orthop Surg Res. 14:2962019. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Gu J, Ji Z, Li D and Dong Q: Proliferation
inhibition and apoptosis promotion by dual silencing of VEGF and
survivin in human osteosarcoma. Acta Biochim Biophys Sin
(Shanghai). 51:59–67. 2019. View Article : Google Scholar
|
|
64
|
Quan B, Li Z, Yang H, Li S, Yan X and Wang
Y: The splicing factor YBX1 promotes the progression of
osteosarcoma by upregulating VEGF165 and downregulating
VEGF165b. Heliyon. 9:e187062023. View Article : Google Scholar
|
|
65
|
Luo P, Zhang YD, He F, Tong CJ, Liu K, Liu
H, Zhu SZ, Luo JZ and Yuan B: HIF-1α-mediated augmentation of
miRNA-18b-5p facilitates proliferation and metastasis in
osteosarcoma through attenuation PHF2. Sci Rep. 12:103982022.
View Article : Google Scholar
|
|
66
|
Zeng X, Liu S, Yang H, Jia M, Liu W and
Zhu W: Synergistic anti-tumour activity of ginsenoside Rg3 and
doxorubicin on proliferation, metastasis and angiogenesis in
osteosarcoma by modulating mTOR/HIF-1α/VEGF and EMT signalling
pathways. J Pharm Pharmacol. 75:1405–1417. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Liao YX, Zhou CH, Zeng H, Zuo DQ, Wang ZY,
Yin F, Hua YQ and Cai ZD: The role of the CXCL12-CXCR4/CXCR7 axis
in the progression and metastasis of bone sarcomas (Review). Int J
Mol Med. 32:1239–1246. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Zhang P, Dong L, Yan K, Long H, Yang TT,
Dong MQ, Zhou Y, Fan QY and Ma BA: CXCR4-mediated osteosarcoma
growth and pulmonary metastasis is promoted by mesenchymal stem
cells through VEGF. Oncol Rep. 30:1753–1761. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Li Z, Lu H, Zhang Y, Lv J, Zhang Y, Xu T,
Yang D, Duan Z, Guan Y, Jiang Z, et al: Blocking CXCR4-CARM1-YAP
axis overcomes osteosarcoma doxorubicin resistance by suppressing
aerobic glycolysis. Cancer Sci. 115:3305–3319. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Gong C, Sun K, Xiong HH, Sneh T, Zhang J,
Zhou X, Yan P and Wang JH: Expression of CXCR4 and MMP-2 is
associated with poor prognosis in patients with osteosarcoma.
Histol Histopathol. 35:863–870. 2020.PubMed/NCBI
|
|
71
|
Liu JF, Chen PC, Chang TM and Hou CH:
Monocyte chemoattractant protein-1 promotes cancer cell migration
via c-Raf/MAPK/AP-1 pathway and MMP-9 production in osteosarcoma. J
Exp Clin Cancer Res. 39:2542020. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Harrison BM and Loukopoulos P: Genomics
and transcriptomics in veterinary oncology. Oncol Lett. 21:3362021.
View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Feleke M, Feng W, Song D, Li H, Rothzerg
E, Wei Q, Kõks S, Wood D, Liu Y and Xu J: Single-cell RNA
sequencing reveals differential expression of EGFL7 and VEGF in
giant-cell tumor of bone and osteosarcoma. Exp Biol Med (Maywood).
247:1214–1227. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Chen T, Chen Z, Lian X, Wu W, Chu L, Zhang
S and Wang L: MUC 15 promotes osteosarcoma cell proliferation,
migration and invasion through livin, MMP-2/MMP-9 and Wnt/β-catenin
signal pathway. J Cancer. 12:467–473. 2021. View Article : Google Scholar :
|
|
75
|
Li AA, Zhang Y, Li F, Zhou Y, Liu ZL and
Long XH: The mechanism of VCP-mediated metastasis of osteosarcoma
based on cell autophagy and the EMT pathway. Clin Transl Oncol.
25:653–661. 2023. View Article : Google Scholar
|
|
76
|
Zheng X, Liu X, Zhang X, Zhao Z, Wu W and
Yu S: A single-cell and spatially resolved atlas of human
osteosarcomas. J Hematol Oncol. 17:712024. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Sirikaew N, Pruksakorn D, Chaiyawat P and
Chutipongtanate S: Mass spectrometric-based proteomics for
biomarker discovery in osteosarcoma: Current status and future
direction. Int J Mol Sci. 23:97412022. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Wu C, Gong S, Duan Y, Deng C, Kallendrusch
S, Berninghausen L, Osterhoff G and Schopow N: A tumor
microenvironment-based prognostic index for osteosarcoma. J Biomed
Sci. 30:232023. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Chang TY, Lan KC, Wu CH, Sheu ML, Yang RS
and Liu SH: Nε-(1-carboxymethyl)-L-lysine/RAGE signaling drives
metastasis and cancer stemness through ERK/NFκB axis in
osteosarcoma. Int J Biol Sci. 20:880–896. 2024. View Article : Google Scholar :
|
|
80
|
Jiang K, Li J, Zhang J, Wang L, Zhang Q,
Ge J, Guo Y, Wang B, Huang Y, Yang T, et al: SDF-1/CXCR4 axis
facilitates myeloid-derived suppressor cells accumulation in
osteosarcoma microenvironment and blunts the response to anti-PD-1
therapy. Int Immunopharmacol. 75:1058182019. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Liao YX, Lv JY, Zhou ZF, Xu TY, Yang D,
Gao QM, Fan L, Li GD, Yu HY and Liu KY: CXCR4 blockade sensitizes
osteosarcoma to doxorubicin by inducing autophagic cell death via
PI3K-Akt-mTOR pathway inhibition. Int J Oncol. 59:492021.
View Article : Google Scholar :
|
|
82
|
Zhang L, Pan Y, Pan F, Huang S, Wang F,
Zeng Z, Chen H and Tian X: MATN4 as a target gene of HIF-1α
promotes the proliferation and metastasis of osteosarcoma. Aging
(Albany NY). 16:10462–10476. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Shi X and Wang X, Yao W, Shi D, Shao X, Lu
Z, Chai Y, Song J, Tang W and Wang X: Mechanism insights and
therapeutic intervention of tumor metastasis: latest developments
and perspectives. Signal Transduct Target Ther. 9:1922024.
View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Gao Y, Qu Y, Zhou Q and Ma Y: SIRT6
inhibits proliferation and invasion in osteosarcoma cells by
targeting N-cadherin. Oncol Lett. 17:1237–1244. 2019.PubMed/NCBI
|
|
85
|
Masuelli L, Benvenuto M, Izzi V, Zago E,
Mattera R, Cerbelli B, Potenza V, Fazi S, Ciuffa S, Tresoldi I, et
al: In vivo and in vitro inhibition of osteosarcoma growth by the
pan Bcl-2 inhibitor AT-101. Invest New Drugs. 38:675–689. 2020.
View Article : Google Scholar
|
|
86
|
Esperança-Martins M, Fernandes I, Soares
do Brito J, Macedo D, Vasques H, Serafim T, Costa L and Dias S:
Sarcoma metabolomics: Current horizons and future perspectives.
Cells. 10:14322021. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Jiang Y, Li F, Gao B, Ma M, Chen M, Wu Y,
Zhang W, Sun Y, Liu S and Shen H: KDM6B-mediated histone
demethylation of LDHA promotes lung metastasis of osteosarcoma.
Theranostics. 11:3868–3881. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Mei Z, Shen Z, Pu J, Liu Q, Liu G, He X,
Wang Y, Yue J, Ge S, Li T, et al: NAT10 mediated ac4C acetylation
driven m6A modification via involvement of
YTHDC1-LDHA/PFKM regulates glycolysis and promotes osteosarcoma.
Cell Commun Signal. 22:512024. View Article : Google Scholar
|
|
89
|
Xia K, Zheng D, Wei Z, Liu W and Guo W:
TRIM26 inhibited osteosarcoma progression through destabilizing
RACK1 and thus inactivation of MEK/ERK signaling. Cell Death Dis.
14:5292023. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Liu W, Zhao Y, Wang G, Feng S, Ge X, Ye W,
Wang Z, Zhu Y, Cai W, Bai J and Zhou X: TRIM22 inhibits
osteosarcoma progression through destabilizing NRF2 and thus
activation of ROS/AMPK/mTOR/autophagy signaling. Redox Biol.
53:1023442022. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Wei Z, Xia K, Zhou B, Zheng D and Guo W:
Zyxin inhibits the proliferation, migration, and invasion of
osteosarcoma via Rap1-mediated inhibition of the MEK/ERK signaling
pathway. Biomedicines. 11:23142023. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Li G, Stampas A, Komatsu Y, Gao X, Huard J
and Pan S: Proteomics in orthopedic research: Recent studies and
their translational implications. J Orthop Res. 42:1631–1640. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Dean DC, Shen S, Hornicek FJ and Duan Z:
From genomics to metabolomics: Emerging metastatic biomarkers in
osteosarcoma. Cancer Metastasis Rev. 37:719–731. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Yin Z, Shen G, Fan M and Zheng P: Lipid
metabolic reprogramming and associated ferroptosis in osteosarcoma:
From molecular mechanisms to potential targets. J Bone Oncol.
51:1006602025. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Hu X, Zhou X, Zhang J and Li L:
Sphingolipid metabolism is associated with osteosarcoma metastasis
and prognosis: Evidence from interaction analysis. Front Endocrinol
(Lausanne). 13:9836062022. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Cai F, Liu L, Bo Y, Yan W, Tao X, Peng Y,
Zhang Z, Liao Q and Yi Y: LncRNA RPARP-AS1 promotes the progression
of osteosarcoma cells through regulating lipid metabolism. BMC
Cancer. 24:1662024. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Bispo DSC, Correia M, Carneiro TJ, Martins
AS, Reis AAN, Carvalho ALMB, Marques MPM and Gil AM: Impact of
conventional and potential new metal-based drugs on lipid
metabolism in osteosarcoma MG-63 cells. Int J Mol Sci.
24:175562023. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Fritsche-Guenther R, Gloaguen Y, Kirchner
M, Mertins P, Tunn PU and Kirwan JA: Progression-dependent altered
metabolism in osteosarcoma resulting in different nutrient source
dependencies. Cancers (Basel). 12:13712020. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Shen S, Xu Y, Gong Z, Yao T, Qiao D, Huang
Y, Zhang Z, Gao J, Ni H, Jin Z, et al: Positive feedback regulation
of circular RNA Hsa_circ_0000566 and HIF-1α promotes osteosarcoma
progression and glycolysis metabolism. Aging Dis. 14:529–547.
2023.PubMed/NCBI
|
|
100
|
Li G, Li Y and Wang DY: Overexpression of
miR-329-3p sensitizes osteosarcoma cells to cisplatin through
suppression of glucose metabolism by targeting LDHA. Cell Biol Int.
45:766–774. 2021. View Article : Google Scholar
|
|
101
|
Wang B, Zhou Y, Zhang P, Li J and Lu X:
Solasonine inhibits cancer stemness and metastasis by modulating
glucose metabolism via Wnt/β-catenin/snail pathway in osteosarcoma.
Am J Chin Med. 51:1293–1308. 2023. View Article : Google Scholar
|
|
102
|
Ren J, Zhao C, Sun R, Sun J, Lu L, Wu J,
Li S and Cui L: Augmented drug resistance of osteosarcoma cells
within decalcified bone matrix scaffold: The role of glutamine
metabolism. Int J Cancer. 154:1626–1638. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Wang H, Tao Y, Han J, Shen J, Mu H, Wang
Z, Wang J, Jin X, Zhang Q, Yang Y, et al: Disrupting YAP1-mediated
glutamine metabolism induces synthetic lethality alongside ODC1
inhibition in osteosarcoma. Cell Oncol (Dordr). 47:1845–1861. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Lin S, Miao Y, Zheng X, Dong Y, Yang Q,
Yang Q, Du S, Xu J, Zhou S and Yuan T: ANGPTL4 negatively regulates
the progression of osteosarcoma by remodeling branched-chain amino
acid metabolism. Cell Death Discov. 8:2252022. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Qian H, Lei T, Hu Y and Lei P: Expression
of lipid-metabolism genes is correlated with immune
microenvironment and predicts prognosis in osteosarcoma. Front Cell
Dev Biol. 9:6738272021. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Wu C, Tan J, Shen H, Deng C, Kleber C,
Osterhoff G and Schopow N: Exploring the relationship between
metabolism and immune microenvironment in osteosarcoma based on
metabolic pathways. J Biomed Sci. 31:42024. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Li Q, Fang J, Liu K, Luo P and Wang X:
Multi-omic validation of the cuproptosis-sphingolipid metabolism
network: Modulating the immune landscape in osteosarcoma. Front
Immunol. 15:14248062024. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Lin Z, He Y, Wu Z, Yuan Y, Li X and Luo W:
Comprehensive analysis of copper-metabolism-related genes about
prognosis and immune microenvironment in osteosarcoma. Sci Rep.
13:150592023. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Chen C, Wang J, Pan D, Wang X, Xu Y, Yan
J, Wang L, Yang X, Yang M and Liu GP: Applications of multi-omics
analysis in human diseases. MedComm (2020). 4:e3152023. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Lin Y, Yang Y, Yuan K, Yang S, Zhang S, Li
H and Tang T: Multi-omics analysis based on 3D-bioprinted models
innovates therapeutic target discovery of osteosarcoma. Bioact
Mater. 18:459–470. 2022.PubMed/NCBI
|
|
111
|
Zhou Y, Yang D, Yang Q, Lv X, Huang W,
Zhou Z, Wang Y, Zhang Z, Yuan T, Ding X, et al: Single-cell RNA
landscape of intratumoral heterogeneity and immunosuppressive
microenvironment in advanced osteosarcoma. Nat Commun. 11:63222020.
View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Jia C, Yao X, Dong Z, Wang L, Zhao F, Gao
J and Cai T: Molecular landscape and prognostic value in the
post-translational ubiquitination, SUMOylation and neddylation in
osteosarcoma: A transcriptome study. J Inflamm Res. 17:4315–4330.
2024. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Truong DD, Weistuch C, Murgas KA, Admane
P, King BL, Chauviere Lee J, Lamhamedi-Cherradi SE, Swaminathan J,
Daw NC, Gordon N, et al: Mapping the single-cell differentiation
landscape of osteosarcoma. Clin Cancer Res. 30:3259–3272. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Heo YJ, Hwa C, Lee GH, Park JM and An JY:
Integrative multi-omics approaches in cancer research: From
biological networks to clinical subtypes. Mol Cells. 44:433–443.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Wei Z, Xia K, Zheng D, Gong C and Guo W:
RILP inhibits tumor progression in osteosarcoma via Grb10-mediated
inhibition of the PI3K/AKT/mTOR pathway. Mol Med. 29:1332023.
View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Zhang X, Wen Z, Wang Q, Ren L and Zhao S:
A novel stratification framework based on anoikis-related genes for
predicting the prognosis in patients with osteosarcoma. Front
Immunol. 14:11998692023. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Fan J, Jahed V and Klavins K: Metabolomics
in bone research. Metabolites. 11:4342021. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Jiang Z, Han K, Min D, Kong W, Wang S and
Gao M: Identification of the methotrexate resistance-related
diagnostic markers in osteosarcoma via adaptive total variation
netNMF and multi-omics datasets. Front Genet. 14:12880732023.
View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Audinot B, Drubay D, Gaspar N, Mohr A,
Cordero C, Marec-Bérard P, Lervat C, Piperno-Neumann S, Jimenez M,
Mansuy L, et al: ctDNA quantification improves estimation of
outcomes in patients with high-grade osteosarcoma: A translational
study from the OS2006 trial. Ann Oncol. 35:559–568. 2024.
View Article : Google Scholar
|
|
120
|
Schaafsma E, Takacs EM, Kaur S, Cheng C
and Kurokawa M: Predicting clinical outcomes of cancer patients
with a p53 deficiency gene signature. Sci Rep. 12:13172022.
View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Chen Z, Guo J, Zhang K and Guo Y: TP53
mutations and survival in osteosarcoma patients: A meta-analysis of
published data. Dis Markers. 2016:46395752016. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Ru JY, Cong Y, Kang WB, Yu L, Guo T and
Zhao JN: Polymorphisms in TP53 are associated with risk and
survival of osteosarcoma in a Chinese population. Int J Clin Exp
Pathol. 8:3198–3203. 2015.PubMed/NCBI
|
|
123
|
Jeon DG, Koh JS, Cho WH, Song WS, Kong CB,
Cho SH and Lee SY and Lee SY: Clinical outcome of low-grade central
osteosarcoma and role of CDK4 and MDM2 immunohistochemistry as a
diagnostic adjunct. J Orthop Sci. 20:529–537. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Iwata S, Tatsumi Y, Yonemoto T, Araki A,
Itami M, Kamoda H, Tsukanishi T, Hagiwara Y, Kinoshita H, Ishii T,
et al: CDK4 overexpression is a predictive biomarker for resistance
to conventional chemotherapy in patients with osteosarcoma. Oncol
Rep. 46:1352021. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Schubert NA, Chen CY, Rodríguez A, Koster
J, Dowless M, Pfister SM, Shields DJ, Stancato LF, Vassal G, Caron
HN, et al: Target actionability review to evaluate CDK4/6 as a
therapeutic target in paediatric solid and brain tumours. Eur J
Cancer. 170:196–208. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Sun R, Shen J, Gao Y, Zhou Y, Yu Z,
Hornicek F, Kan Q and Duan Z: Overexpression of EZH2 is associated
with the poor prognosis in osteosarcoma and function analysis
indicates a therapeutic potential. Oncotarget. 7:38333–38346. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Yang X, Xu L and Yang L: Recent advances
in EZH2-based dual inhibitors in the treatment of cancers. Eur J
Med Chem. 256:1154612023. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Nazarizadeh A, Alizadeh-Fanalou S,
Hosseini A, Mirzaei A, Salimi V, Keshipour H, Safizadeh B, Jamshidi
K, Bahrabadi M and Tavakoli-Yaraki M: Evaluation of local and
circulating osteopontin in malignant and benign primary bone
tumors. J Bone Oncol. 29:1003772021. View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Barris DM, Weiner SB, Dubin RA, Fremed M,
Zhang X, Piperdi S, Zhang W, Maqbool S, Gill J, Roth M, et al:
Detection of circulating tumor DNA in patients with osteosarcoma.
Oncotarget. 9:12695–12704. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Kathiresan N, Selvaraj C, Pandian S,
Subbaraj GK, Alothaim AS, Safi SZ and Kulathaivel L: Proteomics and
genomics insights on malignant osteosarcoma. Adv Protein Chem
Struct Biol. 138:275–300. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Zhao D, Jia P, Wang W and Zhang G:
VEGF-mediated suppression of cell proliferation and invasion by
miR-410 in osteosarcoma. Mol Cell Biochem. 400:87–95. 2015.
View Article : Google Scholar
|
|
132
|
Tawbi HA, Burgess M, Bolejack V, Van Tine
BA, Schuetze SM, Hu J, D'Angelo S, Attia S, Riedel RF, Priebat DA,
et al: Pembrolizumab in advanced soft-tissue sarcoma and bone
sarcoma (SARC028): A multicentre, two-cohort, single-arm,
open-label, phase 2 trial. Lancet Oncol. 18:1493–1501. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Assi T, Watson S, Samra B, Rassy E, Le
Cesne A, Italiano A and Mir O: Targeting the VEGF pathway in
osteosarcoma. Cells. 10:12402021. View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Mickymaray S, Alfaiz FA, Paramasivam A,
Veeraraghavan VP, Periadurai ND, Surapaneni KM and Niu G:
Rhaponticin suppresses osteosarcoma through the inhibition of
PI3K-Akt-mTOR pathway. Saudi J Biol Sci. 28:3641–3649. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Lu DG, Tang QL, Wei JH, He FY, Lu L and
Tang YJ: Targeting EZH2 by microRNA-449a inhibits osteosarcoma cell
proliferation, invasion and migration via regulation of PI3K/AKT
signaling pathway and epithelial-mesenchymal transition. Eur Rev
Med Pharmacol Sci. 24:1656–1665. 2020.PubMed/NCBI
|
|
136
|
Chen ZY, Huang HH, Li QC, Zhan FB, Wang
LB, He T, Yang CH, Wang Y, Zhang Y and Quan ZX: Capsaicin reduces
cancer stemness and inhibits metastasis by downregulating SOX2 and
EZH2 in osteosarcoma. Am J Chin Med. 51:1041–1066. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Liu X, He S, Wu H, Xie H, Zhang T and Deng
Z: Blocking the PD-1/PD-L1 axis enhanced cisplatin chemotherapy in
osteosarcoma in vitro and in vivo. Environ Health Prev Med.
24:792019. View Article : Google Scholar : PubMed/NCBI
|
|
138
|
Yoshida K, Okamoto M, Sasaki J, Kuroda C,
Ishida H, Ueda K, Okano S, Ideta H, Kamanaka T, Sobajima A, et al:
Clinical outcome of osteosarcoma and its correlation with
programmed death-ligand 1 and T cell activation markers. Onco
Targets Ther. 12:2513–2518. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
139
|
Davis KL, Fox E, Merchant MS, Reid JM,
Kudgus RA, Liu X, Minard CG, Voss S, Berg SL, Weigel BJ and Mackall
CL: Nivolumab in children and young adults with relapsed or
refractory solid tumours or lymphoma (ADVL1412): A multicentre,
openlabel, single-arm, phase 1-2 trial. Lancet Oncol. 21:541–550.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Zhou Y, Shen JK, Yu Z, Hornicek FJ, Kan Q
and Duan Z: Expression and therapeutic implications of
cyclin-dependent kinase 4 (CDK4) in osteosarcoma. Biochim Biophys
Acta Mol Basis Dis. 1864:1573–1582. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Oshiro H, Tome Y, Miyake K, Higuchi T,
Sugisawa N, Kanaya F, Nishida K and Hoffman RM: Combination of
CDK4/6 and mTOR inhibitors suppressed doxorubicin-resistant
osteosarcoma in a patient-derived orthotopic xenograft mouse model:
A translatable strategy for recalcitrant disease. Anticancer Res.
41:3287–3292. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
142
|
Athieniti E and Spyrou GM: A guide to
multi-omics data collection and integration for translational
medicine. Comput Struct Biotechnol J. 21:134–149. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
143
|
Downing JR, Wilson RK, Zhang J, Mardis ER,
Pui CH, Ding L, Ley TJ and Evans WE: The pediatric cancer genome
project. Nat Genet. 44:619–622. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
144
|
Freeberg MA, Fromont LA, D'Altri T, Romero
AF, Ciges JI, Jene A, Kerry G, Moldes M, Ariosa R, Bahena S, et al:
The European genome-phenome archive in 2021. Nucleic Acids Res.
50(D1): D980–D987. 2022. View Article : Google Scholar :
|
|
145
|
Jovic D, Liang X, Zeng H, Lin L, Xu F and
Luo Y: Single-cell RNA sequencing technologies and applications: A
brief overview. Clin Transl Med. 12:e6942022. View Article : Google Scholar : PubMed/NCBI
|
|
146
|
Liu W, Hu H, Shao Z, Lv X, Zhang Z, Deng
X, Song Q, Han Y, Guo T, Xiong L, et al: Characterizing the tumor
microenvironment at the single-cell level reveals a novel immune
evasion mechanism in osteosarcoma. Bone Res. 11:42023. View Article : Google Scholar : PubMed/NCBI
|
|
147
|
Huang X, Wang L, Guo H, Zhang W and Shao
Z: Single-cell transcriptomics reveals the regulative roles of
cancer associated fibroblasts in tumor immune microenvironment of
recurrent osteosarcoma. Theranostics. 12:5877–5887. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
148
|
Liu Y, Feng W, Dai Y, Bao M, Yuan Z, He M,
Qin Z, Liao S, He J, Huang Q, et al: Single-cell transcriptomics
reveals the complexity of the tumor microenvironment of
treatment-naive osteosarcoma. Front Oncol. 11:7092102021.
View Article : Google Scholar : PubMed/NCBI
|
|
149
|
Fu Y, Xu Y, Liu W, Zhang J, Wang F, Jian
Q, Huang G, Zou C, Xie X, Kim AH, et al: Tumor-informed deep
sequencing of ctDNA detects minimal residual disease and predicts
relapse in osteosarcoma. EClinicalMedicine. 73:1026972024.
View Article : Google Scholar : PubMed/NCBI
|
|
150
|
Landuzzi L, Manara MC, Lollini PL and
Scotlandi K: Patient derived xenografts for genome-driven therapy
of osteosarcoma. Cells. 10:4162021. View Article : Google Scholar : PubMed/NCBI
|
|
151
|
He A, Huang Y, Cheng W, Zhang D, He W, Bai
Y, Gu C, Ma Z, He Z, Si G, et al: Organoid culture system for
patient-derived lung metastatic osteosarcoma. Med Oncol.
37:1052020. View Article : Google Scholar : PubMed/NCBI
|