Introduction
Matrine is one of the main components extracted from
Sophora flavescens (1), and
matrine has been widely applied in the treatment of a variety of
tumors including hepatocellular carcinoma (HCC) (2). It has been indicated that matrine can
inhibit proliferation and then induce apoptosis and autophagy of
certain types of tumor cells (3,4),
e.g., C6 glioma cells and hepatoma G2 cells (5,6).
Autophagy is a catabolic process involved in degradation of the
cellular components such as long-lived proteins and organelles,
through the lysosomal machinery (7). Although initially identified as a
process induced by cellular starvation, autophagy is currently
recognized as the cellular adaptation to a variety of stimulations
(8). However, the role of
autophagy in cancer is not clear. In some settings, autophagy
delays apoptosis in response to chemotherapy (9). Inhibition of autophagy enhances
drug-induced apoptosis in human cancer lines (10–14).
In other settings, antitumor agents augment autophagic cell
death.
The tumor suppressor p53 can maintain the integrity
of genome DNA, regulate cell cycle progression and cell death, and
thus, play a central role in response to stress (15). Autophagy, like apoptosis and cell
cycle progression, is associated with p53 (15). The tumor suppressor p53 activates
autophagy through its downstream targets. It has been shown that
p53 interacts with mammalian target of rapamycin (mTOR) through
AMP-activated protein kinase (AMPK) (16,17).
However, the role of p53 signaling in regulation of autophagy
remains to be studied.
The molecular mechanism of autophagy induction by
matrine is not well studied so far. In this study, we found
matrine-induced autophagy was mediated by p53/AMPK signaling
pathway. Furthermore, for the first time, we demonstrated that p53
isoforms and the interferon (IFN)-inducible genes, i.e., IFI27 and
IFITM1, might be regulated by the p53-mediated autophagy induction
by matrine. The present study shed new insight into the mechanism
of autophagy induction by matrine.
Materials and methods
Reagents
3-Methyladenine (3MA), rapamycin, dimethylsulfoxide
(DMSO), compound c and antibody against microtubule-associated
protein 1 light chain 3 (LC3) were purchased from Sigma Chemical
Co. (St. Louis, MO, USA). Antibodies against p70S6 (Thr389), AMPK
(Thr172), p-ACC (Ser79), p-Akt (Thr473) and p-Erk1/2
(Thy202/Tyr204) were provided commercially by Cell Signaling
Technology, Inc. (Beverly, MA, USA), and p62, β-actin and p53 by
Santa Cruz Biotechnology (Santa Cruz, CA, USA). Z-VAD-fmk was
purchased from Promega (Madison, WI, USA). Constitutively active
p53 was kindly donated by Dr Zhixin Zheng (Peking University School
of Medicine, Peking, China). GFP-LC3 plasmid was kindly donated by
Dr Mengqiang Li (Peking University School of Medicine, Peking,
China). Matrine was purchased from the National Institute for the
Control of Pharmaceutical and Biological Products (Peking,
China).
Cell line and cell culture
HepG2 and SMMC-7721 cells were purchased from ATCC,
and cultured in a 25-cm3 flask with Dulbecco’s modified
Eagle’s medium (Invitrogen, Carlsbad, CA, USA) supplemented with
10% (v/v) fetal bovine serum (FBS) (Invitrogen), 100 U/l penicillin
(Invitrogen), and 100 mg/l streptomycin (Invitrogen). The cells
were treated with matrine for 48 h, and collected for further study
thereafter.
Confocal microscopy
Cells were incubated with monodansylcadaverine (MDC,
0.05 mM), which was purchased from Sigma Chemical Co. After 1-h
incubation at 37°C, the cells were fixed in 4% paraformaldehyde for
15 min and immediately analyzed using a scanning confocal
microscope (Olympus, Japan).
Cells at ~60% confluence were transfected with
GFP-LC3 using Vigofect (Vigorous, Peking, China), 24 h later, and
cells were subjected to various treatments and then detected by a
scanning confocal microscope (Olympus, Japan). Percentages of
punctuate distribution of GFP-LC3 dots were calculated in five
non-overlapping fields.
Electron microscopy
The trypsinized cells were fixed with ice-cold
glutaraldehyde (3% in 0.1 M cacodylate buffer, pH 7.4) for 30 min.
After fixation in OsO4, the cells were embedded in Epon,
and stained with uranyl acetate/lead citrate (Fluka, Buchs,
Switzerland). Observation was performed on a JEM1230 electron
microscope (JEOL, Japan).
Flow cytometry analysis
Cells were treated with the indicated concentrations
of chemotherapeutic agents for 48 h, followed by staining with
Annexin V-fluorescein isothiocyanate (FITC) (BD Pharmingen, CA,
USA) and propidium iodide (PI) (BD Pharmingen), and subsequently,
analyzed by flow cytometry (FACSAria, Becton-Dickinson, USA).
Isolation of RNA and cDNA microarray
assay
Total RNA was extracted from matrine-treated HepG2
cells using the TRIzol reagents (Invitrogen). The cDNA microarray
analysis was performed by CapitalBio Co. (Peking, China) according
to the standardized protocol. A Lux-Scan 10KA dual pathways laser
scanner (CapitalBio) and a GenePix Pro 4.0 image analysis software
(Axon Instruments, Inc., Union City, CA, USA) was applied to scan
the chips and analyze the images, respectively. The raw image files
of all the data were normalized and analyzed. Annotation was in
correspondence with Unigene database http://www.ncbi.nlm.nih.gov/unigene, including gene
number and gene symbol. Ratio values >2-fold upregulation or
downregulation (p<0.05) was regarded as significantly expressed
genes.
Reverse transcription-polymerase chain
reaction (RT-PCR) and real-time PCR
One microgram of total RNA was reverse-transcribed
into cDNA using cDNA reverse transcription kits (Invitrogen).
Hot-start PCR was then performed (Eppendorf, Germany). The PCR
results were verified by varying the number of PCR cycles for each
cDNA and set of primers. The target gene primer pairs were as
follows: IFI27 upper primer: GGCCAGGATTGCTACAGTTGTGATT; lower
primer: GCGGACATCATCTTGGCTGCTAT; IFITM1 upper primer:
TAGCATTCGCCTACTCCGTGAA, lower primer: AGCCGA ATACCAGTAACAGGATGAA;
CASP1 upper primer: TGAAGGACAAACCGAAGGTGA, lower primer: TGTGGA
AGAGCAGAAAGCGATA; p53 upper primer: CTCCAG CCACCTGAAGTC, lower
primer: GTCAGTGGGG AACAAGAAG; p53β upper primer: ATGGAGGAGCC
GCAGTCAGAT, lower primer: TTGAAAGCTGGTCTGG TCCTGA; p53γ upper
primer: ATGGAGGAGCCGCA GTCAGAT, lower primer: TCGTAAGTCAAGTAGCATCT
GAAGG; Δ133p53 upper primer: TGGGTTGCAGGAGG TGCTTA, lower primer:
CTCACGCCCACGGATCTGA; Δ133p53β upper primer: TGGGTTGCAGGAGGTGCTTA,
lower primer: TGGGTTGCAGGAGGTGCTTA; Δ133p53γ upper primer:
TGGGTTGCAGGAGGTGCTTA, lower primer: TCGTAAGTCAAGTAGCATCTGAAGG
(20); GAPDH upper primer:
ACGGATTTGGTCGTATTGGG, lower primer: TGATTTTGGA GGGATCTCGC. The
amplified products were separated on 2 and 1.5% agarose gels and
visualized under ultraviolet transillumination.
Real-time PCR was performed by pre-denaturation at
95°C for 10 min, followed by 40 cycles of 95°C for 15 sec and 60°C
for 1 min. The data were analyzed with ΔCt method according to the
manufacturer’s instructions (Applied Biosystems, CA, USA). The data
were normalized to the housekeeping gene GAPDH. Changes in gene
expression were illustrated as a fold increase/decrease. The
experiments were repeated three times.
RNA interference
Cells were seeded at 30–50% confluence per well in
6-well plates overnight and transfected with target gene specific
siRNA or control siRNA duplex (Santa Cruz Biotechnology, Inc.), and
then cultured with various treatments. Blockage of target gene was
successfully examined by immunoblotting analysis.
Western blot analysis
Proteins obtained from cell lysates (40 μg
protein/lane) were separated on SDS-PAGE by electrophoresis, and
transferred to nitrocellulose membranes. The membranes were
incubated with the primary antibodies, and then probed with
secondary antibodies. The specific bands were visualized using an
enhanced chemiluminescence system (Pierce Biotechnology, Inc.,
Rockford, IL, USA).
Statistical analysis
Data are expressed as mean ± SEM. The number of
individual experiments is described in the figure legends.
Statistical analysis was performed using SPSS 17.0 statistics
software (SPSS, Inc., Chicago, IL, USA). P-values <0.05 were
considered as statistically significant.
Results
Matrine inhibits proliferation and
induces apoptosis of human hepatoma cells
Matrine has been reported to promote apoptosis in
previous in vitro studies (21,22).
To study the effect of matrine on proliferation and apoptosis of
human hepatoma cells, we treated HepG2 cells and SMMC-7721 cells
with different doses of matrine for 48 h, and detected apoptosis of
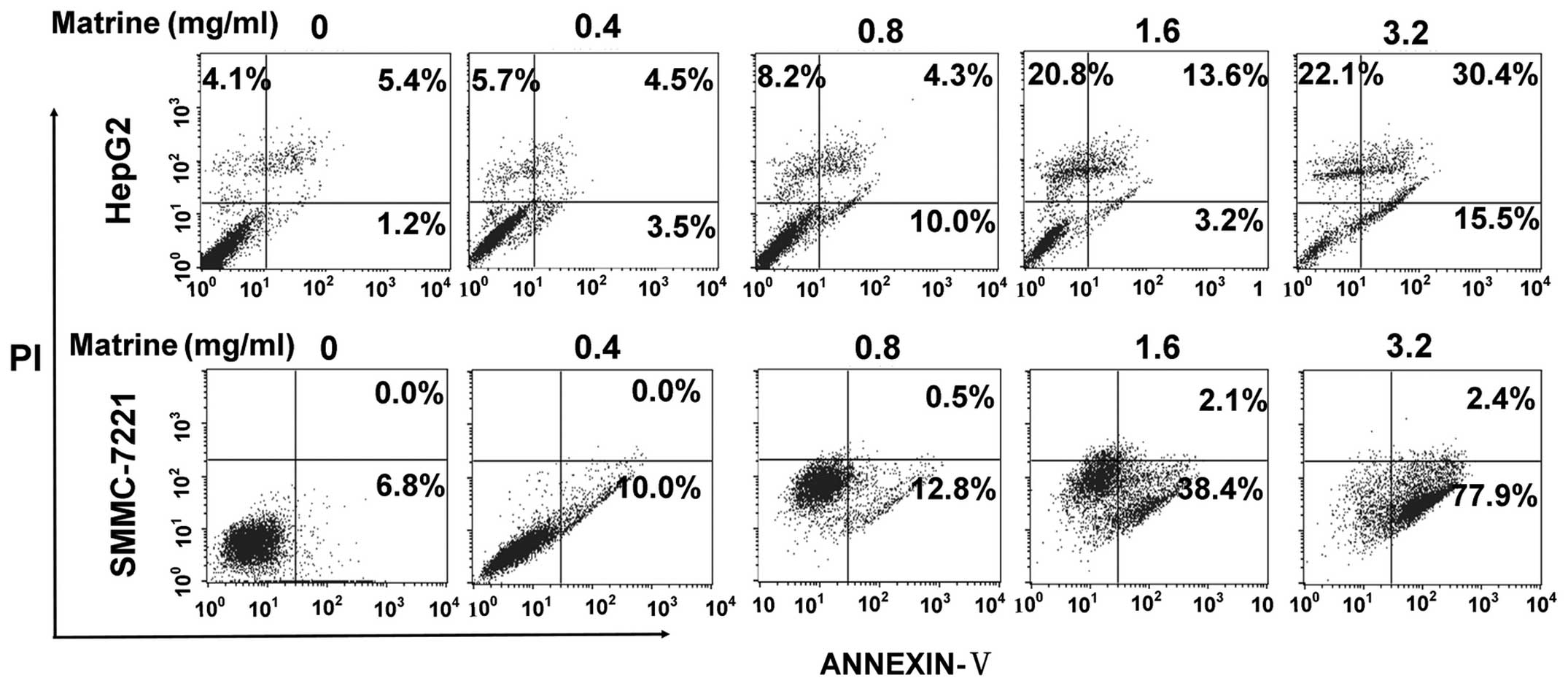
the cells, respectively. As shown in Fig. 1, at low dose of 0.8 mg/ml, matrine
induced a slight increase of apoptosis of HepG2 cells and SMMC-7721
cells as compared with the untreated cells, while matrine induced a
significant elevation in apoptosis level at median dose of 1.6
mg/ml and at large dose of 3.2 mg/ml.
Matrine stimulates autophagy in human
hepatoma cells
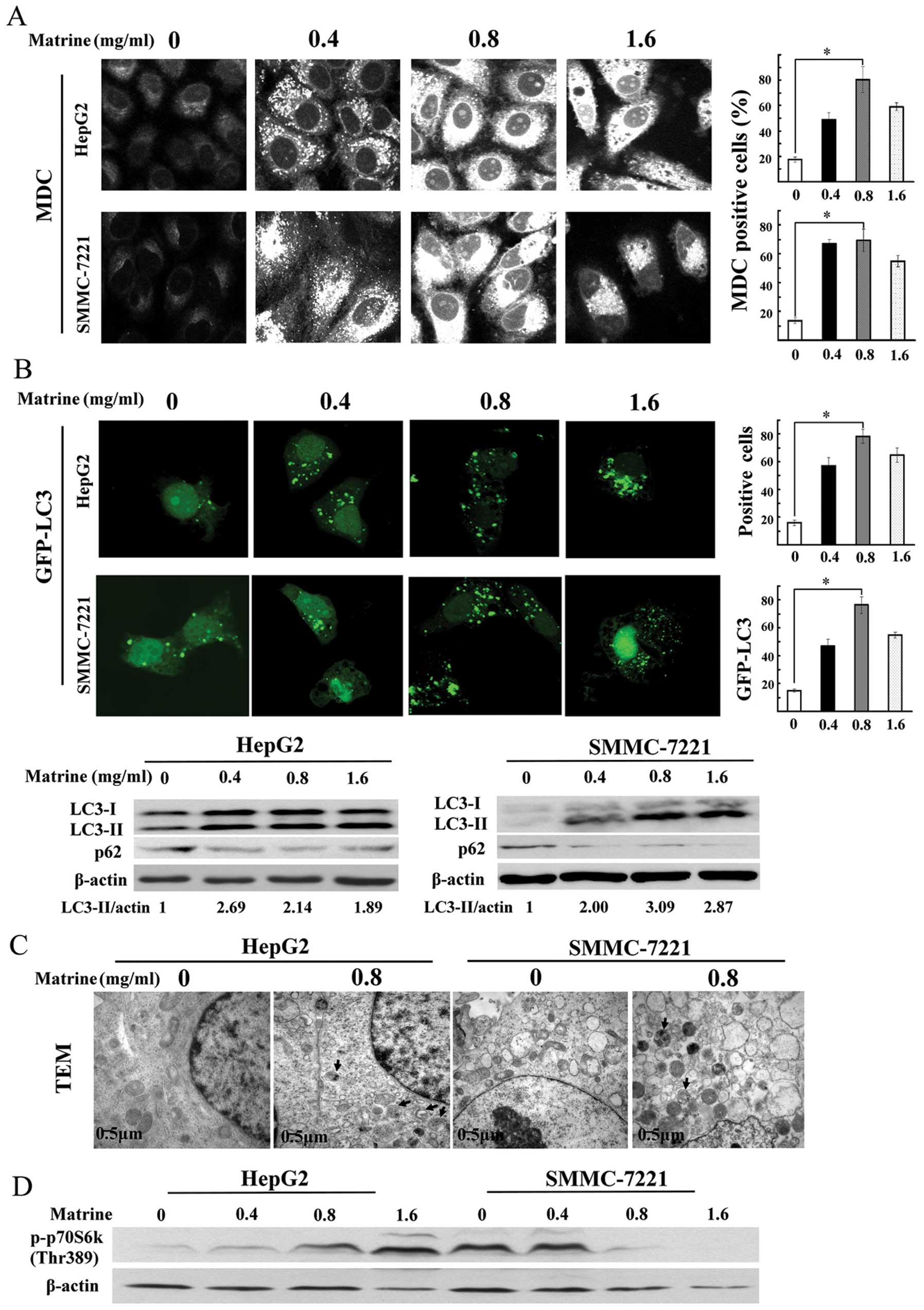
The number of MDC-stained punctuate structure, an
indicator of autophagosomes, was significantly increased in human
hepatoma cell lines after exposure to matrine at low dose of 0.4
mg/ml, and reached a peak at 0.8 mg/ml, then a slight decrease at
1.6 mg/ml (Fig. 2A). Based on
these results, treatment with 0.8 mg/ml of matrine for 48 h was
used for further studies on autophagy in hepatoma cells.
LC3, a mammalian homologue of Atg8, is widely used
as a specific marker for detecting autophagy (23). As shown in Fig. 2B, confocal microscopy assay showed
the significant increase of the punctuate GFP-LC3 in
matrine-treated cells, which was in line with MDC staining
analysis. Similar results were observed through immunoblotting
assay. P62/SQSTM1, as a substrate of autophagy, was selectively
degraded by autolysosomes. Autophagy inhibition promoted the
accumulation of p62/SQSTM1. The degradation of p62/SQSTM1 was
detected in matrine-treated cells through western blot analysis
(Fig. 2B). Transmission electron
microscopy (TEM), one of the most reliable methods for detecting
autophagy (24), demonstrated that
autophagic vacuoles were easily observed in both hepatoma cell
types exposed to matrine at 0.8 mg/ml (Fig. 2C). These results indicated that
autophagy was activated by matrine in human hepatoma cells.
Furthermore, we analyzed whether matrine-induced autophagy is
regulated by mTOR kinase, a central regulator for autophagy.
Phosphorylation of p70S6K at Thr389, a downstream effector of mTOR
and widely considered to reflect mTOR activity, was downregulated
in matrine-treated SMMC-7721 cells, but upregulated prominently in
matrine-treated HepG2 cells, compared with untreated cells,
suggesting that induction of autophagy by matrine was through an
mTOR-dependent manner in SMMC-7221 cells, but an mTOR-independent
manner in HepG2 cells (Fig.
2D).
Matrine-induced autophagy involves p53
inactivation via AMPK signaling pathway
We then investigated the molecular mechanism of
matrine-induced autophagy in HepG2 cells. We first examined
PI3K-Akt and MAPK/ERK signaling pathways, which are associated with
autophagy. A decrease of Akt and ERK phosphorylation was detected
in matrine-treated cells as compared with control (Fig. 3A). Then, we analyzed the AMPK
activity, which plays an important role in regulating autophagic
activities. A slight elevation on AMPK phosphorylation at Thr172
was observed after matrine treatment at the dose of 0.8 and 1.6
mg/ml. Consistently, similar results were obtained from analysis of
phosphorylation of acetyl-CoA carboxylase (ACC), a substrate of
AMPK. This suggests that AMPK signaling pathway participates in
matrine-induced autophagy (Fig.
3B). Accumulating evidence has shown that the p53 tumor
suppressor gene is implicated in autophagic activity via the AMPK
signaling pathway (17).
Furthermore, we investigated whether p53 regulates AMPK and thus
activates matrine-induced autophagy. Immunoblotting analysis showed
that p53 suppression was detected in HepG2 cells under matrine
treatment (Fig. 3C). Subsequently,
after transfecting matrine-treated cells with p53 overexpression
plasmid, a remarkable increase of p53 was observed. The ratio of
LC3-II/β-actin and AMPK phosphorylation at Thr172 was reversely
alleviated as compared with cells transfected with the pcDNA 3.1
empty vector (Fig. 3D). These
results suggest that matrine-induced autophagy is negatively
regulated by p53 through AMPK signaling transduction.
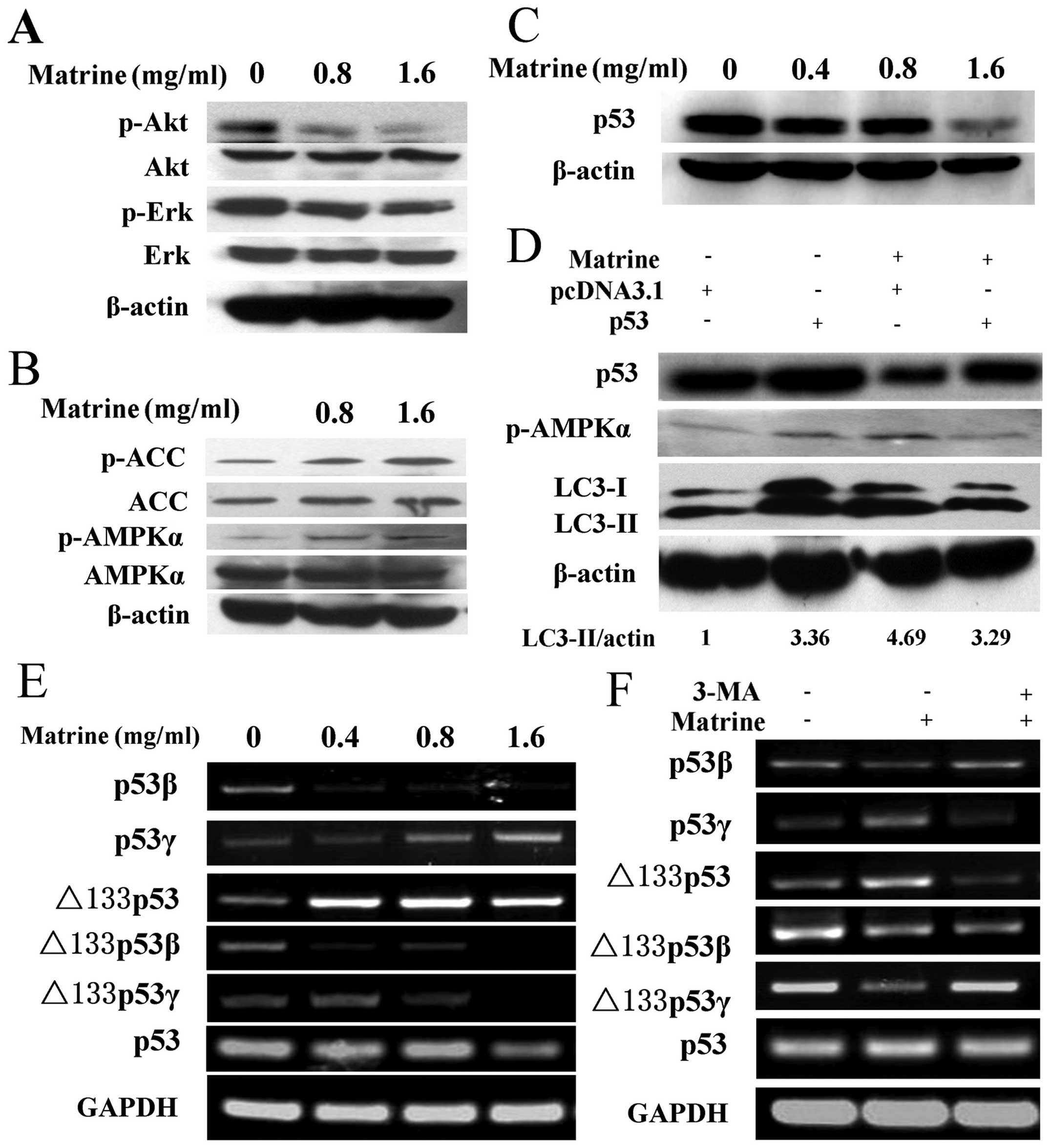 | Figure 3Matrine-induced autophagy is via
p53/AMPK pathway, and p53 variants were converted by autophagy
inhibition. (A) Immunoblotting analysis of the expression of p-Akt
(Thr473) and p-ERK (Thy202/Tyr204) in HepG2 cells after exposure to
matrine with 0.8 and 1.6 mg/ml, as compared with control. (B)
Western blot analysis of the expression of p-AMPKα (Thr172) and
p-ACC (Ser79) in treated cells, as compared with control. (C) After
exposure to matrine with 0.4, 0.8 and 1.6 mg/ml, western blot
analysis of p53 levels in matrine-treated cells in comparison with
control. (D) The cells initially were transiently transfected with
p53 expression vector for 24 h followed by exposure to matrine at
the dose of 0.8 mg/ml for 48 h. Western blot analysis of p53, LC3
and p-AMPKα in matrine-treated cells containing p53 overexpression
vector in comparison with control vector. (E) RT-PCR analysis of
p53 and its isoforms-p53β, p53γ, Δ133p53, Δ133p53β and Δ133p53γ in
the cells under matrine treatment at different dose, in contrast to
control. (F) Matrine at the dose of 0.8 mg/ml in combination with 5
mM of 3-MA is presented. RT-PCR analysis of p53 and its splice
variants between matrine alone and matrine cotreated with 3-MA. |
The p53β and Δ133p53γ isoforms are
upregulated by 3-MA, whereas the p53γ and Δ133p53 variants are
downregulated by 3-MA
Moreover, to determine whether the human p53 mRNA
splice variants are linked to the matrine-induced autophagy, we
amplified splice variants of p53 mRNA, including p53β, p53γ,
Δ133p53, Δ133p53β and Δ133p53γ, by RT-PCR. As shown in Fig. 3E, the expression of p53β and
Δ133p53γ isoform was downregulated in matrine-treated cells
compared with control. However, the expression of p53β and Δ133p53γ
isoform was upregulated in cells treated with matrine and
3-methyladenine (3-MA), a commonly used autophagy inhibitor.
Conversely, the p53γ and Δ133p53 mRNA variant was upregulated in
matrine-treated cells and downregulated in cells treated with
matrine and 3-MA. The expression of Δ133p53β mRNA variant was
attenuated in the cells exposed to matrine. However, no significant
difference was observed in Δ133p53β mRNA variant expression between
cells treated with matrine and 3-MA and cells treated with matrine
alone (Fig. 3F). These results
imply that the autophagic process induced by matrine might
contribute to the conversion of p53 splice variants, i.e., p53β,
p53γ, Δ133p53 and Δ133p53γ expression.
AMPK inhibition switches matrine-induced
autophagy to apoptosis
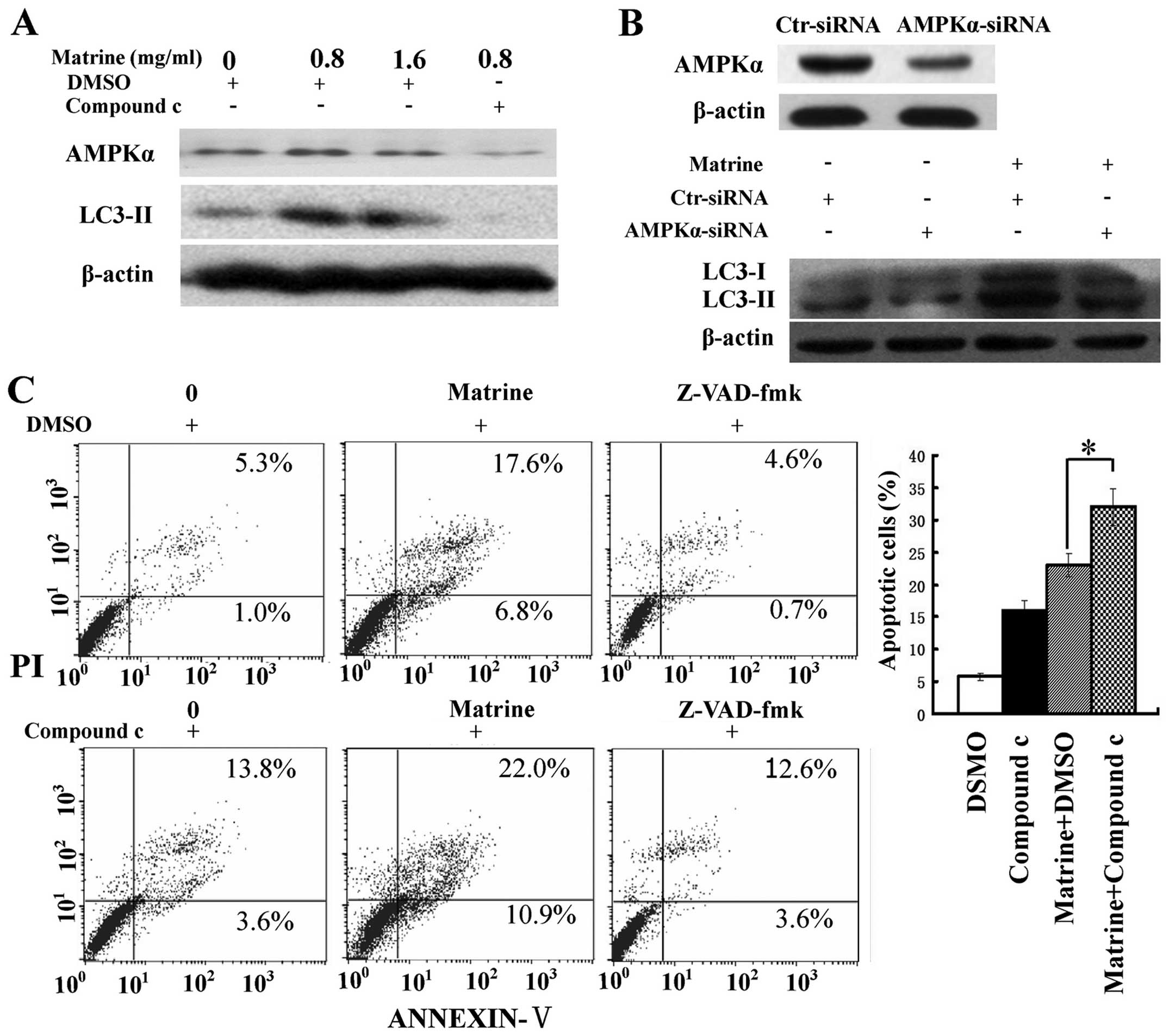
We assessed the role of AMPK in matrine-induced
autophagy and apoptosis. Inactivation of AMPK by compound c, a
specific inhibitor of AMPK, and RNA interference (RNAi) was
conducted. The increase of LC3-II/β-actin ratio after matrine
treatment was attenuated in cells treated with matrine and compound
c, compared with cells treated with matrine alone. Similar results
were obtained when blocking AMPK expression with AMPK-α siRNA
(Fig. 4A and B), indicating that
AMPK signaling was implicated in matrine-promoted autophagy in
HepG2 cells. Moreover, there was an increase of apoptotic cells in
matrine-treated cells in combination with compound c than matrine
alone. In addition, this elevation in apoptosis level was
downregulated by Z-VAD-fmk, a pan-caspase inhibitor (25) (Fig.
4C). Altogether, these results indicate AMPK inactivation
triggers apoptosis through autophagy inhibition in HepG2 cells.
The IFN-signaling transduction is
alleviated by autophagy inhibition and p53 activation,
respectively
We further investigated the molecular mechanism of
matrine-induced autophagy through cDNA array analysis. The
expression of 280 genes were upregulated ≥2-fold, and the
expression of 58 genes were downregulated ≥2-fold in
matrine-treated cells at the dose of 0.8 mg/ml. The 3-fold
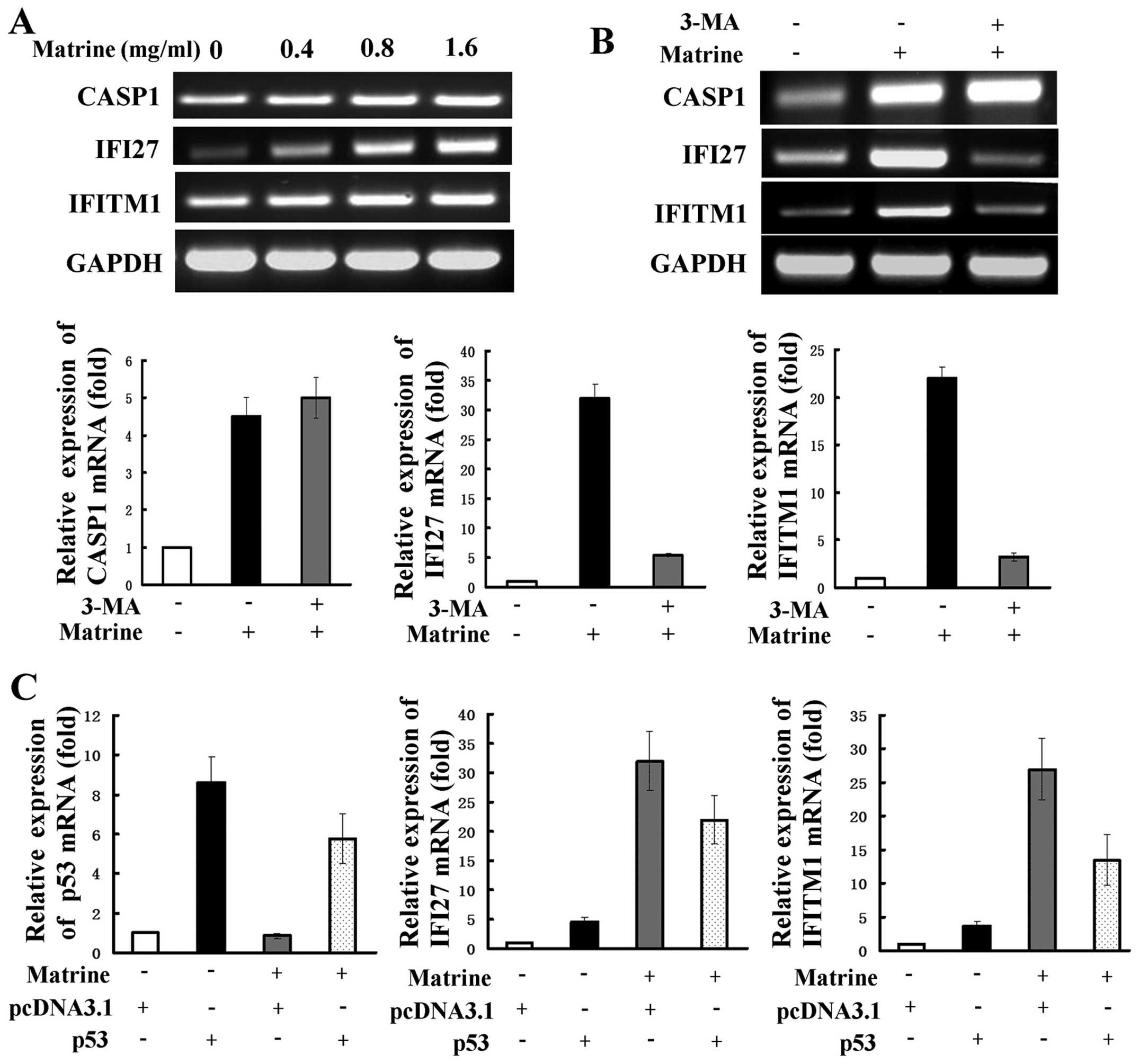
downregulated or 4-fold upregulated genes are shown in Table I. RT-PCR was used to examine
expression of several representative genes, including interferon
α-inducible protein 27 (IFI27), interferon induced transmembrane
protein 1 (IFITM1), and caspase 1 (CASP1). IFI27 and IFITM1 are two
IFN-inducible genes and are reported to be related to autophagy
(26). CASP1 is one of the
apoptosis-related genes. The expression levels of IFI27, IFITM1 and
CASP1 as revealed by RT-PCR analysis were consistent with those by
cDNA array analysis (Fig. 5A).
Next, 3-MA was used to detect whether the selected genes are
related to matrine-induced autophagy. The increased expression of
IFI27 and IFITM1 were significantly attenuated, but CASP1
expression were slightly strengthened after co-treatment with 3-MA,
as shown by RT-PCR and real-time PCR (Fig. 5B). This suggests that the two
IFN-inducible genes of IFI27 and IFITM1, but not CASP1, may be
positively regulated by matrine-induced autophagy. We evaluated
whether the role of IFN-mediated signaling transduction in hepatoma
cells is modulated by p53. Real-time PCR showed that cells
transfected with p53 overexpression plasmid had increased levels of
IFI27 and IFITM1 expression in comparison with cells transfected
with the control plasmid. After subjecting the cells transfected
with the control plasmid to matrine, a more remarked increase of
the two genes was observed and this increase was reversely
inhibited by transfection with the p53 overexpressing plasmid
(Fig. 5C).
 | Table IChanges of gene expression induced by
matrine at 0.8 mg/ml in HepG2 cells. |
Table I
Changes of gene expression induced by
matrine at 0.8 mg/ml in HepG2 cells.
| Gene symbol | Fold change |
|---|
| IFI27 | 24.23 |
| HRASLS2 | 10.72 |
| CFHR1 | 8.60 |
| TNFSF10 | 8.00 |
| ISG20 | 7.87 |
| TNFSF10 | 7.79 |
| IFITM1 | 7.38 |
| LAMP3 | 7.34 |
| ISG15 | 7.25 |
| C4A | 6.92 |
| CFH | 6.21 |
| CCL5 | 6.14 |
| SECTM1 | 5.97 |
| ATF3 | 5.96 |
| OAS3 | 5.90 |
| OAS1 | 5.81 |
| SP110 | 5.54 |
| RARRES3 | 5.51 |
| CTSD | 5.39 |
| DDX60L | 5.24 |
| SOD2 | 5.02 |
| STAT2 | 4.96 |
| CFB | 4.83 |
| BDKRB2 | 4.79 |
| HLA-E | 4.78 |
| C4BPA | 4.52 |
| ANG | 4.43 |
| PTGS1 | 4.34 |
| ARNT2 | 4.23 |
| CARD16 | 4.19 |
| CASP1 | 4.06 |
| RBM3 | −3.11 |
| HEATR1 | −3.42 |
| GKN2 | −3.73 |
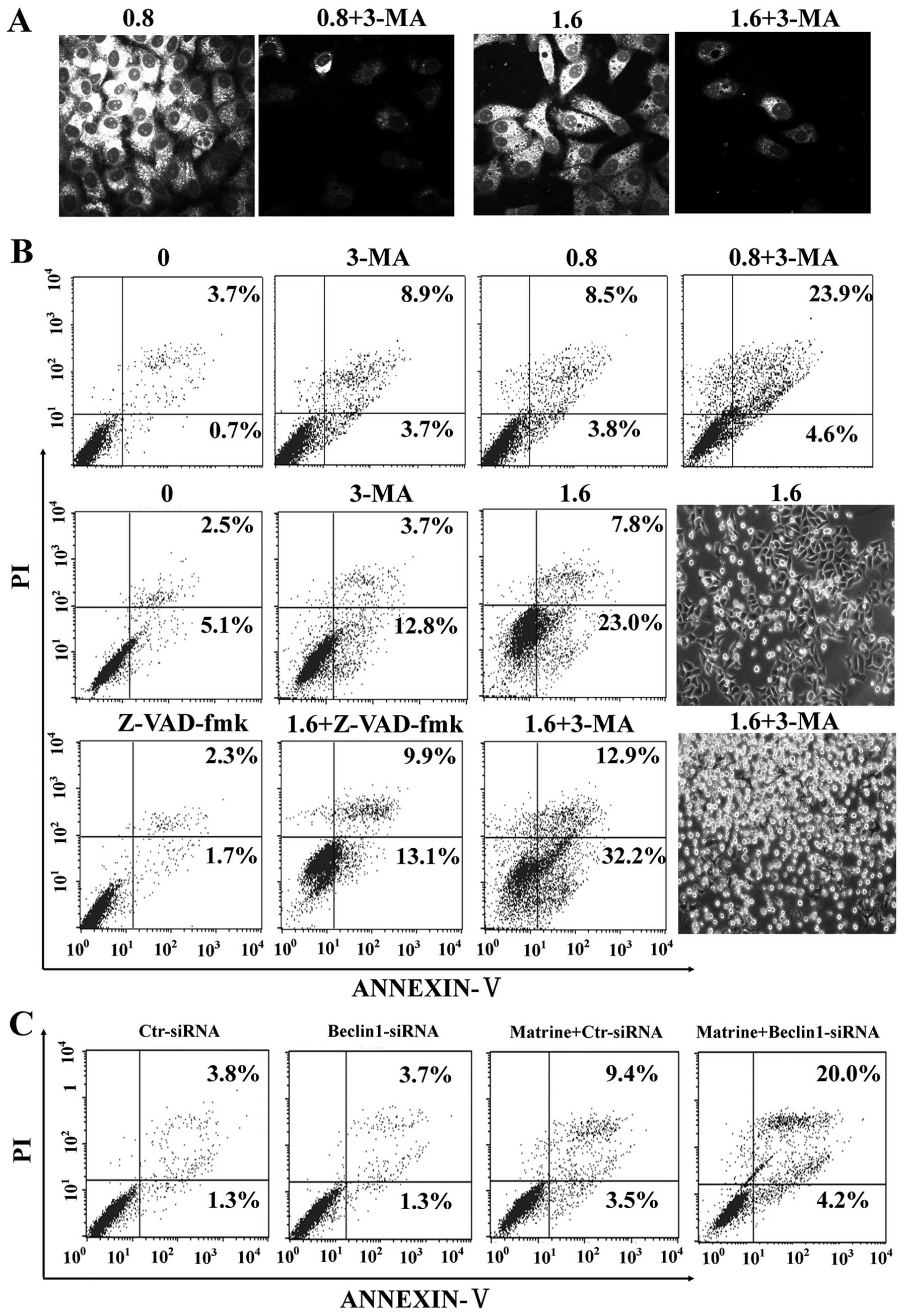
Matrine-induced autophagy protects
against death of HepG2 cells
It is unclear whether autophagy induction by matrine
is protective or toxic. Initially, autophagy inhibition by 3-MA, a
commonly used autophagy inhibitor, was confirmed by MDC staining in
HepG2 cells (Fig. 6A).
Subsequently, our study showed that 3-MA significantly enhanced
matrine-induced apoptosis as showed by flow cytometry, which was
alleviated by Z-VAD-fmk (Fig. 6B).
Additionally, we explored whether blockage of autophagy by RNA
interference (RNAi) enhanced matrine-induced cell death or not.
After matrine treatment, beclin-1-siRNA transfectants had 24.2%
increase of apoptotic cell death in comparison with control-siRNA
transfectants (12.9%) (Fig. 6C).
These results indicated that matrine-induced autophagy has
protective effects on hepatoma cells.
Discussion
Matrine has been reported to induce apoptosis and
inhibit proliferation of cancer cells. However, the exact mechanism
is still unknown. The present study demonstrates that matrine was
able to affect both autophagy and apoptosis in human hepatoma
cells. Our results were consistent with previous studies in C6
glioma cells and hepatoma cells (5,6).
However, our study showed that there was inverse correlation
between apoptosis and autophagy in matrine-treated cells. Another
finding of this study was that autophagy, but not apoptosis,
predominated at low dose of matrine treatment. LC3 expression
further confirmed this finding, suggesting that the initiation of
apoptosis may be blocked by the autophagic system. Autophagy is
usually activated by antineoplastic agents, yet it is not clear
whether this is a futile effort for cellular preservation or a
death mechanism. It has been reported that in matrine-treated C6
glioma cells, besides apoptosis, autophagy is another process of
cell death whereas other studies demonstrate that autophagy
inhibits apoptosis (11–14). What is the relationship between
autophagy and apoptosis in matrine-treated hepatoma cells? To
clarify this question, 3-MA and beclin-1 siRNA were administered to
inhibit autophagy respectively, resulting in increment of apoptosis
of HepG2 cells. We therefore conclude that matrine-induced
autophagy may protect the cells against apoptosis.
The tumor suppressor p53 plays a dual role in
autophagy, i.e., turning autophagy either on or off (15). It has been demonstrated that p53
inactivation by deletion, depletion or inhibition exerts its
ability to stimulate autophagy (8). Consistent with these observations,
our studies demonstrated that p53 inactivation was observed in
HepG2 cells under matrine treatment, whereas p53 activation
reversely suppressed the increased autophagic process induced by
matrine. It is conceivable that p53 may negatively regulate
matrine-induced autophagy in HCC cells. Inactivation of p53 has
been recently shown to negatively regulate autophagy via AMPK
signaling pathway, which is the same to autophagy induction by p53
activation (27). Similarly, we
found that p53 overexpression lead to AMPK dephosphorylation and
LC3-II conversion, suggesting that p53 negatively regulating
autophagy may be via AMPK signaling pathway. Accumulating data have
provided evidence that AMPK plays a key role in autophagy. Our
earlier report showed that AMPK activation contributes to DNA
damaged-induced autophagy in HepG2 cells (28). Consistently, in this study, we
showed that AMPK was activated in matrine-treated HepG2 cells.
Moreover, inactivation of AMPK by compoud c caused the conversion
of LC3-II, implying that AMPK signaling may be involved in
autophagy induction by matrine. These results suggest that AMPK
activation is implicated in matrine-induced autophagy through p53
suppression. Then inhibition of AMPK by compound c clearly enhanced
matrine-induced apoptosis in HepG2 cells. The disruption of AMPK
may contribute to the switch from autophagy to apoptosis.
Presumably, AMPK signaling transduction is a control switch from
autophagy to apoptosis. Conceivably, increase of ATP levels in
matrine-treated cells triggers AMPK loss and thereby switch
autophagy which functions as maintaining cell survival to apoptotic
cell death. AMPK initiates a series of downstream phosphorylation
events and one major downstream event is to induce autophagy
through inhibition of mTOR, a pivotal regulator for autophagy
(29). Interestingly, we found
that matrine induced autophagy may be independent of mTOR
inhibition in HepG2 cells, but dependent of mTOR inhibition in
SMMC-7721 cells. A similar result was observed where
fangchinoline-induced autophagy is mediated by p53/AMPK signaling
pathway in an mTOR-independent manner in both HepG2 and PLC/PRF/5
cells (30). Some small molecules,
such as intracellular inositol or inositol 1,4,5-trisphosphate
(IP3), have also been shown to activate mTOR-independent autophagy
in vitro (31,32). These results indicate that
autophagy induction by matrine in HepG2 cells may not act through
the mTOR signaling pathway, but rather through stimulating an
unknown signaling pathway.
The p53 isoforms, including p53β, p53γ, Δ133p53,
Δ133p53β and Δ133p53γ, are generated by alternative splicing of
intron 9 and have been reported to be encoded in human p53 gene
(20), and implicated in apoptosis
(20). The Δ133p53 and p53β splice
variants have also been demonstrated to function as endogenous
modulators of cellular senescence (33). However, it is unclear whether the
p53 splice variants function in the increased autophagy in
context-specific ways. In the present study, matrine treatment
caused a decline of p53β and Δ133p53γ, but an elevation of p53γ and
Δ133p53. However, chemical inhibition of autophagy by 3-MA reversed
the effects of these isoforms induced by matrine. This study, for
the first time, suggests that autophagy induction by matrine may
influence the splice variants. The p53β and Δ133p53γ variants may
be negatively controlled by autophagy, while the p53γ and Δ133p53
isoforms may be positively controlled by autophagy. Nonetheless,
chemical inhibition of autophagy had no effect on Δ133p53β levels,
implying that Δ133p53β may not be regulated by matrine-induced
autophagy. In this regard, p53 variants perhaps was modulated by
autophagy within a broader network. The interplay between autophagy
and their role in p53 isoforms is still under investigation because
our preliminary results are merely able to offer a clue at mRNA
levels.
Autophagy has been implicated in the immune system.
The induction of autophagy in immune cells serves as an innate
immune response to defend against intruding pathogens (34,35).
In addition to regulation of the immune system, interferon
(IFN)-inducible genes also play important roles in proliferation
and tumorigenesis of tumor cells. IFITM1, a member of the
interferon-induced transmembrane protein family, has been observed
to induce tumor cell proliferation, and be upregulated in many
kinds of tumors (36). IFI27,
among the interferon α-inducible proteins, is also a mitochondrial
protein resistant to apoptosis. IFI27 may influence the innate
immune responses of IFNs, and thus may be a novel marker of
proliferation and cancer (37).
Given that autophagy functions to promote cell survival and to
resist apoptosis, what is the relationship between the
IFN-inducible proteins and autophagy? Our results revealed a
positive role for autophagy induction by matrine on the
IFN-mediated signaling transduction. Conversely, a recent study
indicates that HCV-induced autophagy inhibits the expression of
IFN-stimulated genes, including IFI27, and that suppression of
autophagy induces the IFN signaling pathway and apoptosis in
HCV-infected hepatocytes (26).
This study has demonstrated that autophagy can negate the IFN
signaling pathway and serve as a defensive mechanism. This
discrepancy may be partly explained by the distinct effect of
IFN-stimulated genes on cell types. We further demonstrate that the
IFN signaling transduction might be positively modulated by p53.
Matrine treatment caused a significant elevation of the
IFN-inducible genes, which were reversely downregulated by p53
activation. In addition to the central p53/AMPK pathway of
autophagy induced by matrine, p53 mediated-autophagy is at least in
part through the IFN-inducible genes in human hepatoma cells under
matrine treatment.
In conclusion, matrine induces autophagy through
p53/AMPK signaling. In addition, matrine-induced autophagy might
contribute to the regulation of the p53 splice variants and the
IFN-inducible genes in human hepatoma cells. Pharmacological
regulation of autophagy may improve the therapeutic effect of
matrine against HCC and thereby improve survival of patients with
malignancy.
Acknowledgements
This study was supported by National Natural Science
Foundation of China (grant nos. 307728598 and 81273975). The
authors thank Professor Hongchun Liu and Aiping Bai for
professional support and help.
References
|
1
|
Luo C, Zhu Y, Jiang T, Lu X, Zhang W, Jing
Q, Li J, Pang L, Chen K, Qiu F, et al: Matrine induced gastric
cancer MKN45 cells apoptosis via increasing pro-apoptotic molecules
of Bcl-2 family. Toxicology. 229:245–252. 2007. View Article : Google Scholar
|
|
2
|
Wan XY, Luo M, Li XD and He P:
Hepatoprotective and anti-hepatocarcinogenic effects of
glycyrrhizin and matrine. Chem Biol Interact. 181:15–19. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Guo D, Chen NN, Zhou P, Pan B and Hou LB:
Suppressive effect of matrine on cell growth and decreases
beta-catenin-dependent transcriptional activity in hepatoma cell
line Hep3B. Zhong Yao Cai. 33:778–781. 2010.(In Chinese).
PubMed/NCBI
|
|
4
|
Zhang Y, Zhang H, Yu P, Liu Q, Liu K, Duan
H, Luan G, Yagasaki K and Zhang G: Effects of matrine against the
growth of human lung cancer and hepatoma cells as well as lung
cancer cell migration. Cytotechnology. 59:191–200. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhang S, Qi J, Sun L, Cheng B, Pan S, Zhou
M and Sun X: Matrine induces programmed cell death and regulates
expression of relevant genes based on PCR array analysis in C6
glioma cells. Mol Biol Rep. 36:791–799. 2009. View Article : Google Scholar
|
|
6
|
Zhang JQ, Li YM, Liu T, He WT, Chen YT,
Chen XH, Li X, Zhou WC, Yi JF and Ren ZJ: Antitumor effect of
matrine in human hepatoma G2 cells by inducing apoptosis and
autophagy. World J Gastroenterol. 16:4281–4290. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Mizushima N: Autophagy: process and
function. Genes Dev. 21:2861–2873. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tasdemir E, Chiara Maiuri M, Morselli E,
Criollo A, D’Amelio M, Djavaheri-Mergny M, Cecconi F, Tavernarakis
N and Kroemer G: A dual role of p53 in the control of autophagy.
Autophagy. 4:810–814. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kaushal GP, Kaushal V, Herzog C and Yang
C: Autophagy delays apoptosis in renal tubular epithelial cells in
cisplatin cytotoxicity. Autophagy. 4:710–712. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Huang S and Sinicrope FA:
Celecoxib-induced apoptosis is enhanced by ABT-737 and by
inhibition of autophagy in human colorectal cancer cells.
Autophagy. 6:256–269. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lee SB, Tong SY, Kim JJ, Um SJ and Park
JS: Caspase-independent autophagic cytotoxicity in
etoposide-treated CaSki cervical carcinoma cells. DNA Cell Biol.
26:713–720. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Harhaji-Trajkovic L, Vilimanovich U,
Kravic-Stevovic T, Bumbasirevic V and Trajkovic V: AMPK-mediated
autophagy inhibits apoptosis in cisplatin-treated tumor cells. J
Cell Mol Med. 13:3644–3654. 2009. View Article : Google Scholar
|
|
13
|
Nishikawa T, Tsuno NH, Okaji Y, Shuno Y,
Sasaki K, Hongo K, Sunami E, Kitayama J, Takahashi K and Nagawa H:
Inhibition of autophagy potentiates sulforaphane-induced apoptosis
in human colon cancer cells. Ann Surg Oncol. 17:592–602. 2010.
View Article : Google Scholar
|
|
14
|
Abedin MJ, Wang D, McDonnell MA, Lehmann U
and Kelekar A: Autophagy delays apoptotic death in breast cancer
cells following DNA damage. Cell Death Differ. 14:500–510. 2007.
View Article : Google Scholar
|
|
15
|
Levine B and Abrams J: p53: the Janus of
autophagy? Nat Cell Biol. 10:637–639. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Meijer AJ and Codogno P: Signalling and
autophagy regulation in health, aging and disease. Mol Aspects Med.
27:411–425. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Feng Z, Zhang H, Levine AJ and Jin S: The
coordinate regulation of the p53 and mTOR pathways in cells. Proc
Natl Acad Sci USA. 102:8204–8209. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang Q, Zhao XH and Wang ZJ: Cytotoxicity
of flavones and flavonols to a human esophageal squamous cell
carcinoma cell line (KYSE-510) by induction of G2/M arrest and
apoptosis. Toxicol In Vitro. 23:797–807. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yang YH, Dudoit S, Luu P, Lin DM, Peng V,
Ngai J and Speed TP: Normalization for cDNA microarray data: a
robust composite method addressing single and multiple slide
systematic variation. Nucleic Acids Res. 30:e152002. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Bourdon JC, Fernandes K, Murray-Zmijewski
F, Liu G, Diot A, Xirodimas DP, Saville MK and Lane DP: p53
isoforms can regulate p53 transcriptional activity. Genes Dev.
19:2122–2137. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Liu T, Song Y, Chen H, Pan S and Sun X:
Matrine inhibits proliferation and induces apoptosis of pancreatic
cancer cells in vitro and in vivo. Biol Pharm Bull. 33:1740–1745.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Han Y, Zhang S, Wu J, Yu K, Zhang Y, Yin L
and Bi L: Matrine induces apoptosis of human multiple myeloma cells
via activation of the mitochondrial pathway. Leuk Lymphoma.
51:1337–1346. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kabeya Y, Mizushima N, Ueno T, Yamamoto A,
Kirisako T, Noda T, Kominami E, Ohsumi Y and Yoshimori T: LC3, a
mammalian homologue of yeast Apg8p, is localized in autophagosome
membranes after processing. EMBO J. 19:5720–5728. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Mizushima N: Methods for monitoring
autophagy. Int J Biochem Cell Biol. 36:2491–2502. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Slee EA, Zhu H, Chow SC, MacFarlane M,
Nicholson DW and Cohen GM: Benzyloxycarbonyl-Val-Ala-Asp (OMe)
fluoromethylketone (Z-VAD.FMK) inhibits apoptosis by blocking the
processing of CPP32. Biochem J. 315:21–24. 1996.PubMed/NCBI
|
|
26
|
Shrivastava S, Raychoudhuri A, Steele R,
Ray R and Ray RB: Knockdown of autophagy enhances the innate immune
response in hepatitis C virus-infected hepatocytes. Hepatology.
53:406–414. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Tasdemir E, Maiuri MC, Galluzzi L, Vitale
I, Djavaheri-Mergny M, D’Amelio M, Criollo A, Morselli E, Zhu C,
Harper F, et al: Regulation of autophagy by cytoplasmic p53. Nat
Cell Biol. 10:676–687. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
28
|
Xie BS, Zhao HC, Yao SK, Zhuo DX, Jin B,
Lv DC, Wu CL, Ma DL, Gao C, Shu XM, et al: Autophagy inhibition
enhances etoposide-induced cell death in human hepatoma G2 cells.
Int J Mol Med. 27:599–606. 2011.PubMed/NCBI
|
|
29
|
Meley D, Bauvy C, Houben-Weerts JH,
Dubbelhuis PF, Helmond MT, Codogno P and Meijer AJ: AMP-activated
protein kinase and the regulation of autophagic proteolysis. J Biol
Chem. 281:34870–34879. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang N, Pan W, Zhu M, Zhang M, Hao X,
Liang G and Feng Y: Fangchinoline induces autophagic cell death via
p53/sestrin2/AMPK signaling in human hepatocellular carcinoma
cells. Br J Pharmacol. 164:731–742. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Rubinsztein DC, Gestwicki JE, Murphy LO
and Klionsky DJ: Potential therapeutic applications of autophagy.
Nat Rev Drug Discov. 6:304–312. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
32
|
Botti J, Djavaheri-Mergny M, Pilatte Y and
Codogno P: Autophagy signaling and the cogwheels of cancer.
Autophagy. 2:67–73. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Fujita K, Mondal AM, Horikawa I, Nguyen
GH, Kumamoto K, Sohn JJ, Bowman ED, Mathe EA, Schetter AJ, Pine SR,
et al: p53 isoforms Delta133p53 and p53beta are endogenous
regulators of replicative cellular senescence. Nat Cell Biol.
11:1135–1142. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
34
|
Weidberg H and Elazar Z: TBK1 mediates
crosstalk between the innate immune response and autophagy. Sci
Signal. 4:pe392011.PubMed/NCBI
|
|
35
|
Deretic V: Autophagy in innate and
adaptive immunity. Trends Immunol. 26:523–528. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yu F, Ng SS, Chow BK, Sze J, Lu G, Poon
WS, Kung HF and Lin MC: Knockdown of interferon-induced
transmembrane protein 1 (IFITM1) inhibits proliferation, migration,
and invasion of glioma cells. J Neurooncol. 103:187–195. 2011.
View Article : Google Scholar :
|
|
37
|
Suomela S, Cao L, Bowcock A and
Saarialho-Kere U: Interferon alpha-inducible protein 27 (IFI27) is
upregulated in psoriatic skin and certain epithelial cancers. J
Invest Dermatol. 122:717–721. 2004. View Article : Google Scholar : PubMed/NCBI
|