Introduction
FOXM1 is a member of the Forkhead family of
transcription factors, previously known as Trident (in mouse),
HFH-11 (in human), WIN or INS-1 (in rat), MPP-2 (partial human
cDNA) and FKHL-16 (1). FOXM1 is a
potent oncogene whose expression is frequently upregulated during
cancer initiation. FOXM1 expression increases at the entry of the
S-phase of the cell cycle, remains stable during G2/M phase, before
being degraded at the mitotic exit (2,3).
FOXM1 also controls mitosis through the transcriptional regulation
of mitotic regulatory genes, including PLK, Cyclin B1, Aurora A and
B kinases (4), and in addition, it
plays a major role in maintaining chromosome stability (2). FOXM1 has an important role in cell
cycle progression and cell proliferation. As such, its expression
levels correlate with the proliferative state of a cell. Not
surprisingly, FOXM1 is highly expressed in all embryonic tissues,
particularly in proliferating cells of epithelial and mesenchymal
origin (5). Its overexpression has
also been detected in numerous human cancer cell lines and has been
associated with the development and progression of many
malignancies (6,7), with high cell proliferation rates,
drug resistance (8–10) and poor prognosis in many cancer
types (11–15).
B-lymphoblastic leukemia (B-ALL) is a malignant
disorder which derives from clonal proliferation of lymphoid
precursors with arrested maturation. In recent years the role and
involvement of FOXM1 in B-ALL and other hematological malignancies
has become increasingly important. A previous study by Nakamura
et al has examined the role of FOXM1 in cell proliferation
in myeloid leukemia, showing its capability to promote cell cycle
progression (16). Other studies
have also demonstrated that FOXM1 downregulation causes the
inhibition of cell proliferation in B-lymphoma (17). A different report by Uddin et
al has instead described the involvement of FOXM1 in B-cell
lymphoma migration and invasion (18), and recently it has been pointed out
that FOXM1 pathway could be a potential therapeutic target in B
cell malignancy (19,20). The role of FOXM1 as an oncogene and
its upregulation in relapsed B-ALL patients (21), prompted us to investigate whether
FOXM1 has a potential role in B-ALL cell proliferation, with
particular focus on whether it can become a target that would
increase the efficiency of chemotherapeutic treatment, and allow us
to overcome drug resistance in this hematological malignancy.
Materials and methods
Primary leukemia cell cultures
The mRNA and protein samples of PBMC from healthy
donors were obtained from cells separated by Ficoll-Paque
centrifugation, while healthy B-cells (mainly CD19+)
were obtained by cell sorting of bone marrows from healthy
volunteers. Diagnostic RNA samples of bone marrow (BM) aspirates of
B-leukaemic patients with a blast count of 80–95% were kindly
allowed from the Cell Bank of the Dipartimento di Salute della
Donna e del Bambino, University of Padova, Italy. B-ALL patient
samples were obtained after informed consent following the tenets
of the Declaration of Helsinki. The study was approved by the
Italian Association of Pediatric Onco-Hematology (AIEOP). Written
consent was obtained from participants. All analyzed B-ALL samples
were obtained at the time of diagnosis before treatment, after
Ficoll-Hypaque (Pharmacia, Uppsala, Sweden) separation of
mononuclear cells as described previously (22). The percentage of CD19+
cells ranged from 80 to 95%. Human B-leukemia cell lines, REH, SEM,
MHH-CALL2, RS4;11 and NALM-6, were grown in RPMI-1640 medium
(Gibco, Milan, Italy) all supplemented with 115 U/ml penicillin G
(Gibco), 115 μg/ml streptomycin (Invitrogen), 10% fetal bovine
serum (Invitrogen), and maintained at 37°C in a humidified
atmosphere with 5% CO2.
Quantitative real-time PCR
Total RNA was isolated from frozen cell pellets
using the RNeasy Mini kit (Qiagen, UK) according to the
manufacturer's instructions and RNA purity and concentration were
determined by measuring the spectrophotometric absorption at 260 nm
and 280 nm on NanoDrop ND-1000. Total RNA (1 μg) was reverse
transcribed into first strand cDNA using Superscript III first
stand cDNA synthesis (Life Technologies, UK) Briefly, 1 μl of 50 μM
oligo(dT)20 and 1 μl of 10 mM dNTPs mix were added to the RNA
before the volume was adjusted to 11 μl using RNase-free water.
Samples were denaturated at 65°C for 5 min and then quickly chilled
on ice for 1 min. Subsequently, 1 μl of the reverse transcriptase
Superscript III (200 U/μl) was added, along with 1 μl 0.1 M DTT, 1
μl RNaseOUT Recombinase Inhibitor and 1X first stand buffer. The
solution was incubated at 25°C for 5 min then heated at 50°C for 50
min. The reaction was inactivated by heating at 70°C for 15 min.
For real-time quantitative PCR, 1 μl of cDNA was used as template
in a 24-μl reaction carried out with Power SYBR Green kit (Applied
Biosystems, UK) with ABI 7800 system (Applied Biosystems). The mRNA
levels of target genes were calculated relative to the expression
of L19 mRNA levels using the ΔCt method. Primers used:
FOXM1-fwd, 5′-TGCAGCTAGGGATGTGAATCTTC-3′; FOXM1-rv,
5′-G GAG CCCAGTCCATCAGA ACT-3′; CCNB1-fwd,
5′-CAGTTATGCAGCACCTGGCTAAG-3′; CCNB1-rv,
5′-TGTGGTAGAGTGCTGATCTTAGCAT-3′; AURKB-fwd,
5′-AGTGGGACACCCGACATC-3′; AURKB-rv, 5′-G CCCA ATCTCA A AGTCATCA AT
T-3′; L19-fwd, 5′-GCGGAAGGGTACAGCCAAT-3′; L19-rv,
5′-GCAGCCGGCGCAAA-3′.
Western blot analysis
REH, SEM, MHH-CALL2, RS4;11 and NALM-6, after
experimental conditions, were collected, centrifuged, and washed
two times with ice cold phosphate-buffered saline (PBS). For
western blot analysis cells were lysed as described previously
(23). Proteins were resolved by
sodium dodecyl sulphate-polyacrylamide gel electrophoresis
(SDS–PAGE) (using 7 or 10% acrylamide gels), transferred to PVDF
Hybond-P membrane (GE Healthcare) and immunoblotted with primary
antibodies against, FOXM1 (Santa Cruz, C-20), β-tubulin (Santa
Cruz), Aurora B (Cell Signaling), Cyclin B1 (Santa Cruz). The
membranes were washed four times with Tris-buffered saline and
Tween-20 (TBS-T) for 15 min prior to incubation with the respective
peroxidise (HRP)-conjugated secondary antibody (Dako, Ely, UK), at
1:2,000 dilution for 30 min at room temperature and again washed
four times with TBST-T for 20 min. Protein were visualised using
enhanced chemiluminescence (ECL) detection system (Perkin-Elmer,
Seer Green, UK) with Amsterdam Hyperfilm ECL (GE Helthcare, Little
Chalfont, UK) and signal was detected using the SRX-101A X-ray
developer (Konica Minolta, Tokyo, Japan).
RNA interference with small interfering
RNAs (siRNAs)
For FOXM1 silencing, REH or NALM-6 cells were
transiently transfected with siRNA SMARTpool reagents purchased
from Thermo Scientific Dharmacon (Lafayette, CO, USA) using the
transfection reagent Oligofectamine (Life Technologies, UK)
according to the manufacturer's instructions. SMARTpool siRNAs used
were: siRNA FOXM1 (l-009762-00) and the non-specific (NS) control
siRNA, ON-TARGETplus Non-Targeting pool (D-001810-10) confirmed to
have minimal targeting of known genes. All siRNA pools were
resuspended to 20 μM in 1X siRNA buffer.
Annexin V assay
Surface exposure of phosphatidylserine on apoptotic
cells was measured by flow cytometry with a Coulter Cytomics FC500
(Beckman Coulter) by adding Annexin V conjugated to fluorescein
isothiocyanate (FITC) to cells according to the manufacturer's
instructions (Annexin V Fluos, Roche Diagnostic). Simultaneously,
the cells were stained with PI. Excitation was set at 488 nm, and
the emission filters were at 525 and 585 nm, respectively, for FITC
and PI.
Flow cytometric analysis of cell cycle
distribution
For flow cytometric analysis of DNA content,
5×105 of REH, and NALM-6 cells were either treated with
0.5 and 1 μM of thiostrepton or knocked-down for FOXM1 and after
24, 48 and 72 h of treatment or knockdown, cell cycle analysis was
performed as previously described (24).
Drug combination studies
Cell proliferation was assessed by MTT
(3-4,5-dimethylthiazol-2-yl-2,5-diphenyl tetrazolium bromide) assay
after treatment. Equal concentrations of cells were plated in
triplicate in a 96-well plate and incubated with 10 μl of MTT
(Sigma-Aldrich, St. Louis, MO, USA) for 4 h. Absorbance was
measured at 562 nm using Victor3™ 1420 Multilabel Counter
(Perkin-Elmer, Waltham, MA, USA). Cells were treated for 48 h using
scalar dilutions of thiostrepton (Sigma-Aldrich), combined with
cytarabine (Aractyn, Pfizer), daunorubicin (Pfizer), vincristine,
dexamethasone (Sigma-Aldrich). Thiostrepton was also added to drug
solutions at fixed combination ratios. The effectiveness of various
drug combinations was analyzed by Calcusyn Version 2.1 software
(Biosoft). The combination index (CI) was calculated according to
the Chou-Talalay method (25). A
combination index of 1 indicates an additive effect of the 2 drugs.
Combination index values <1 indicate synergy, and combination
index values >1 indicate antagonism.
Statistical analysis
Results are presented as the mean ± SEM. The
differences between different conditions were analyzed using the
two-sided Student's t-test. P-values <0.05 were considered
statistically significant.
Results
FOXM1 is overexpressed in B lymphoblastic
leukaemic patients
To investigate whether FOXM1 regulates B
lymphoblastic leukemia (B-ALL) proliferation, we analyzed FOXM1
mRNA levels in ten B-ALL pediatric patients comparing them to
peripheral blood mononuclear cells (PBMC) from healthy donors. For
these experiments, the mRNA was extracted from cell pellets of
patients recruited at the time of diagnosis. The first two selected
patients were characterized by chromosomal translocations at
chromosome 12 and 21 [t(12;21)]; patients 3, 4 and 5 carried the
translocation between the chromosome 9 and 22 [t(9;22)], whereas
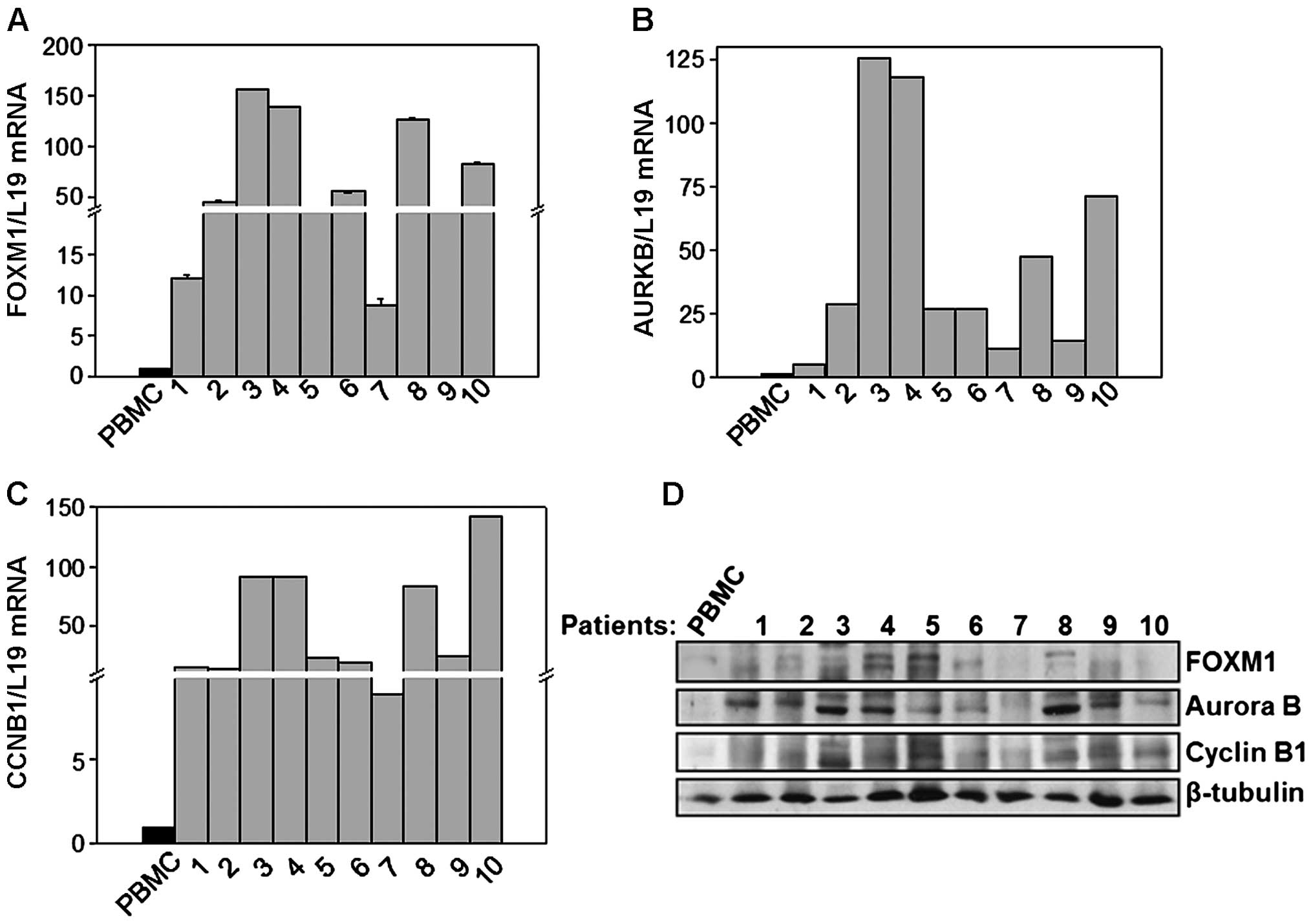
the other five patients were without translocations (Table I). As shown in Fig. 1A, FOXM1 mRNA levels were
significantly higher in patients when compared to the samples of
healthy donors. Interestingly, a significant increment on Cyclin B1
and Aurora B mRNA levels, two genes whose expression is directly
regulated by FOXM1 was also observed (Fig. 1B and C). In addition to mRNA
levels, protein expression was also examined by western blot
analysis. As shown in Fig. 1D, the
expression of FOXM1 and of its targets Aurora B and Cyclin B1 was
generally higher in patients compared to healthy cells, confirming
the upregulated FOXM1 activity. Similar results were obtained when
the mRNA or protein levels of B-ALL patients were compared to those
of CD19-positive cells isolated from bone marrows of healthy donors
(data not shown). Taken together, these data showed that the
expression levels and activity of FOXM1 are increased in B-ALL
patient samples when compared to healthy lymphocytes and normal
B-cells, suggesting that FOXM1 could plays a key function in
supporting the cell proliferation of B-ALL.
 | Table ICharacteristics of patients
samples. |
Table I
Characteristics of patients
samples.
| B-ALL sample | Gender | Age at
diagnosis | % of blast in
BM | Cytogenetics |
|---|
| #1 | F | 6 | 82 | 9q22 |
| #2 | M | 16 | 91 | 9q22 |
| #3 | F | 5 | 77 | 12q21 |
| #4 | M | 3 | 90 | 12q21 |
| #5 | F | 3 | 63 | 12q21 |
| #6 | F | 8 | 97 | Normal |
| #7 | M | 1 | 89 | Normal |
| #8 | F | 8 | 90 | Normal |
| #9 | M | 4 | 90 | Normal |
| #10 | M | 2 | 89 | Normal |
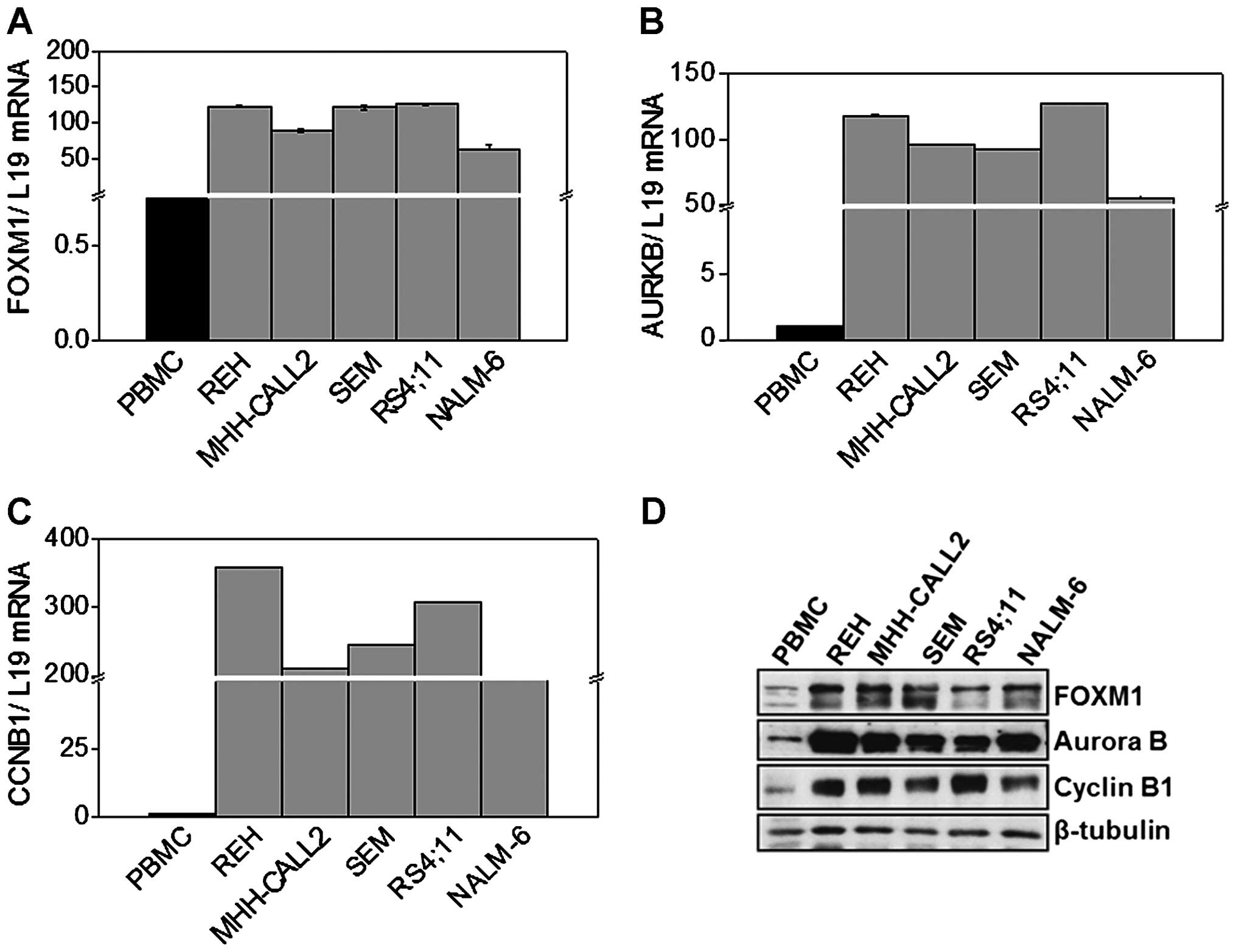
FOXM1 is overexpressed in B-ALL cell
lines
Next, we analyzed FOXM1 expression levels in five
different B-ALL cell lines. More specifically, we quantified FOXM1
mRNA levels, at basal conditions, comparing five different B-ALL
cell lines (REH, MHH-CALL2, SEM, NALM6, RS4;11) with mRNA samples
of PBMC obtained from healthy donors. RT-qPCR analysis showed in
Fig. 2A reveals that FOXM1 mRNA
levels were significantly higher in B-ALL cell lines when compared
to normal lymphocytes. This pattern was again accompanied by
increased transcriptional levels of Cyclin B1 and Aurora B
(Fig. 2B and C). In agreement with
the mRNA expression patterns, western blot analysis revealed that
FOXM1 expression was generally higher in leukaemic cells compared
to normal lymphocytes. This was again associated with Cyclin B1 and
Aurora B upregulation in leukaemic cells (Fig. 2D). Similar results were obtained
comparing the FOXM1 mRNA and protein expression of B-ALL cell lines
with healthy CD19+ cells (data not shown).
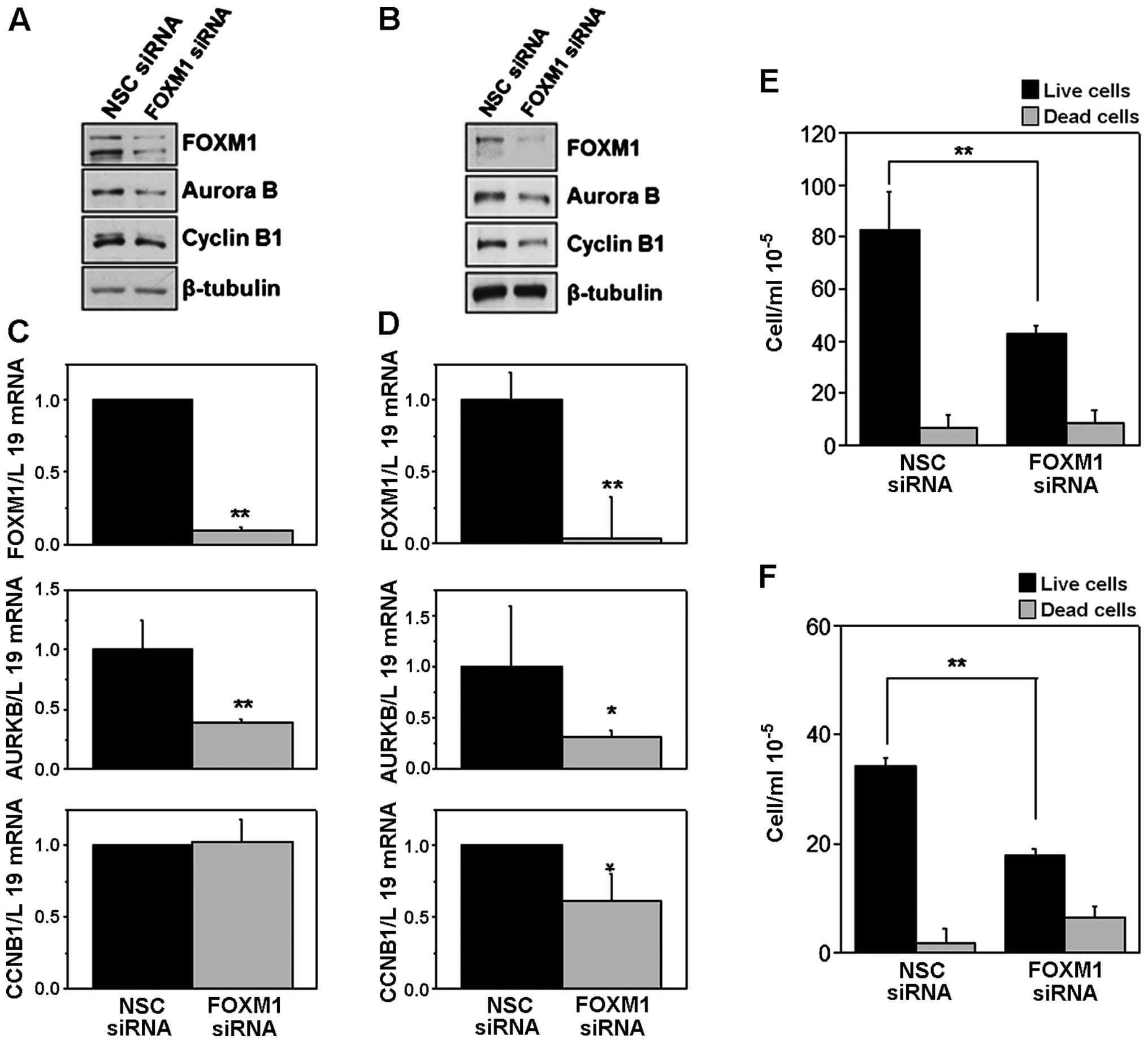
FOXM1 silencing decreases cell
proliferation in B-ALL cell lines and induces a G2/M cell cycle
arrest
To test if FOXM1 has a role in promoting B-ALL
proliferation, we knocked down its expression in two B-ALL cell
lines REH and NALM-6 using a specific FOXM1 siRNA pool and assessed
the cell viability at 24, 48, and 72 h post-transfection. FOXM1
silencing was confirmed by western blot and RT-qPCR analysis
(Fig. 3A–D). Aurora B and Cyclin
B1 protein levels were clearly downregulated in FOXM1-depleted
NALM-6 and REH cells compared to cells transfected with
non-specific (NSC) control siRNA pool (Fig. 3A and B). Furthermore, in both REH
and NALM-6 cell lines, RT-qPCR analysis revealed a significant
decrease in Aurora B mRNA levels, although Cyclin B1 mRNA levels
appeared significantly decreased only in REH and not in NALM-6
cells (Fig. 3C and D).
Cell proliferation analysis assessed by trypan blue
exclusion assay, revealed that upon FOXM1 depletion, there is a
significant decrease in cell proliferation after 72 h of
transfection in NALM-6 (Fig. 3E)
and after 48 h in REH cells (Fig.
3F), suggesting that FOXM1 plays an important role in B-ALL
cell proliferation. Trypan blue-negative population (live cells)
was strongly decreased in cells that were silenced for FOXM1, with
no significant changes in the levels of dead cells (trypan
blue-positive) in NALM-6, and only with a slight increase in the
percentage of dead cells in REH. These results pointed out that
FOXM1 plays an important role in B-ALL cell proliferation.
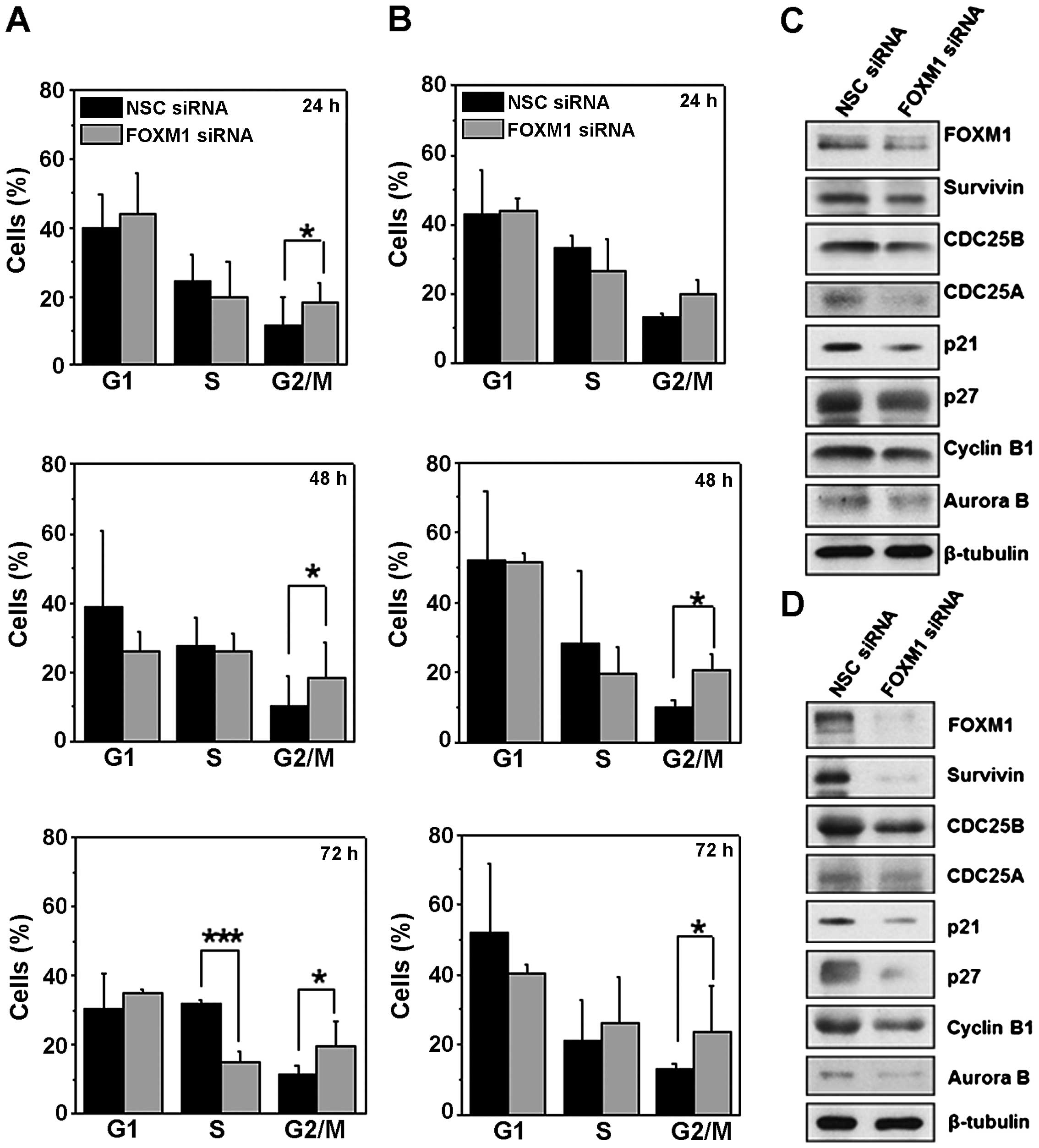
To further analyze the role of FOXM1 on cell cycle
progression in B-ALL, we analyzed the cell cycle phase distribution
in NALM-6 and REH cells following FOXM1 knockdown. As shown in
Fig. 4A and B, there was a
significant increase on the percentage of cells in G2/M in both
cell lines, and a corresponding decrease of cells in the S phase
compared with non-specific (NSC) control cells, only in NALM-6
cells at 72 h. The G2/M cell cycle arrest following FOXM1 silencing
by siRNA is confirmed by the consistent downregulation of the
expression of the G2/M regulators, Cyclin B1 and Aurora B (Fig. 4C and D). This further supports the
role of FOXM1 in cell cycle progression in B-ALL, particularly at
the G2/M transition.
FOXM1 silencing reduces the expression of
cell cycle regulators
We next evaluated if FOXM1 knockdown could modulate
the expression of proteins involved in late phase cell cycle
regulation. As depicted in Fig. 4,
the FOXM1 silencing in both NALM-6 (Fig. 4C) and in REH cells (Fig. 4D), caused downregulation of cell
cycle regulatory proteins such as Cyclin B1, Aurora B, Survivin,
and Cdc25b, involved in mitotic progression. The expression of the
S-phase promoting Cdc25a phosphatase, which plays a major role in
G1/S progression, dephosphorylating Cdk2 and activating CDK2-cyclin
E activity (26,27), was strongly reduced. Interestingly,
both p27Kip1 and p21Cip/Waf1, were also
downregulated. These two cyclin-dependent kinase inhibitor (CKI)
proteins also play a role in the assembly of cyclin-CDK complexes.
Altogether, these results indicate that FOXM1 is strongly involved
in modulating the expression of cell cycle regulatory proteins at
both the G1/S and at the G2/M cell cycle transitions.
Thiostrepton treatment induces G2/M
arrest and decreased cell viability
To further confirm the role of FOXM1 on B-ALL cell
proliferation, we treated cells with the thiazole ring containing
antibiotic, thiostrepton, a well-established specific FOXM1
inhibitor (28,29). This drug causes growth inhibition
in a small panel of B-ALL cell lines with GI50 in the
micromolar and sub-micromolar range (GI50= 0.4–1.4 μM).
In this context, we treated both NALM-6 and REH cell lines with two
different concentrations of thiostrepton, 0.5 and 1 μM,
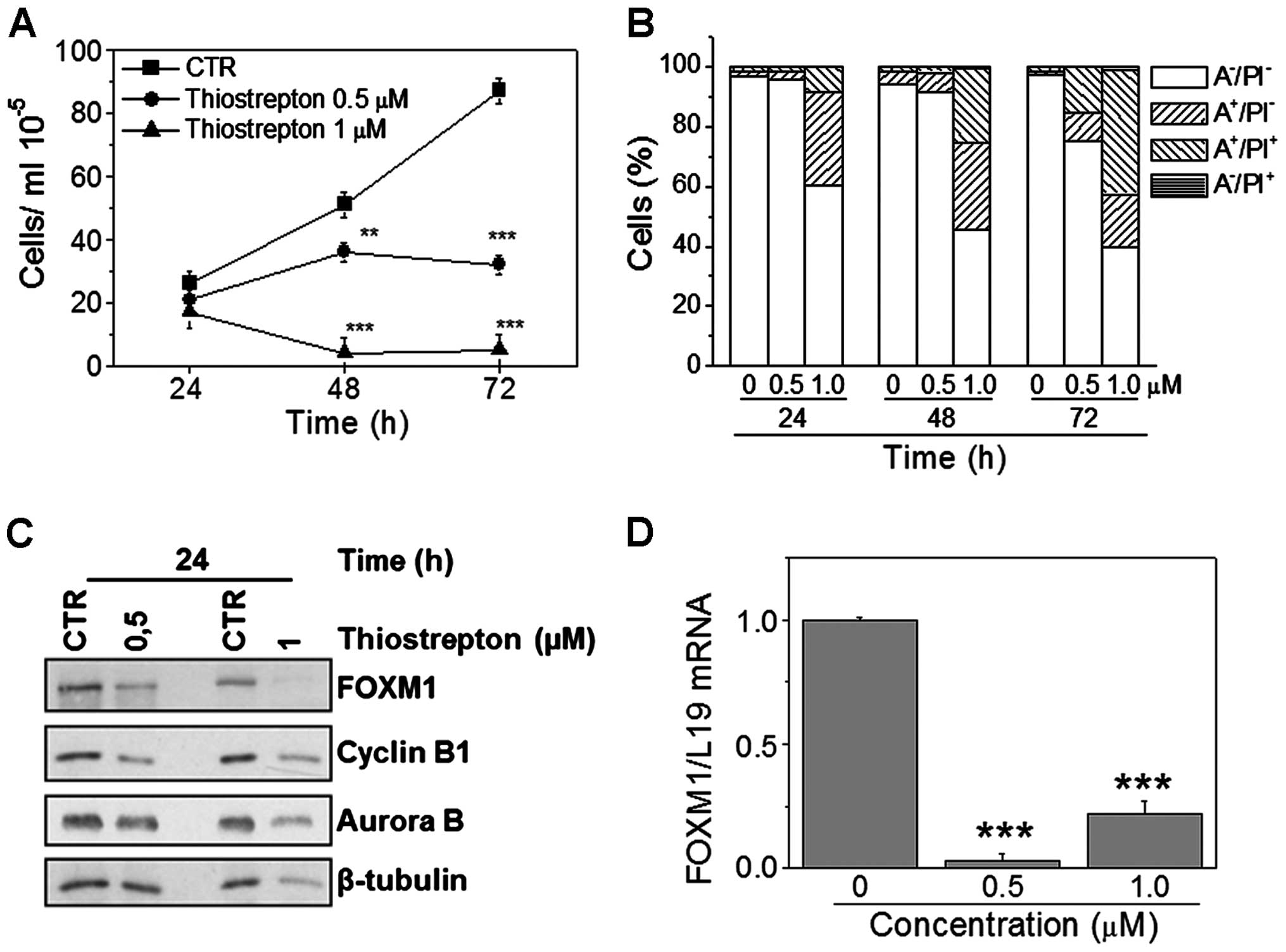
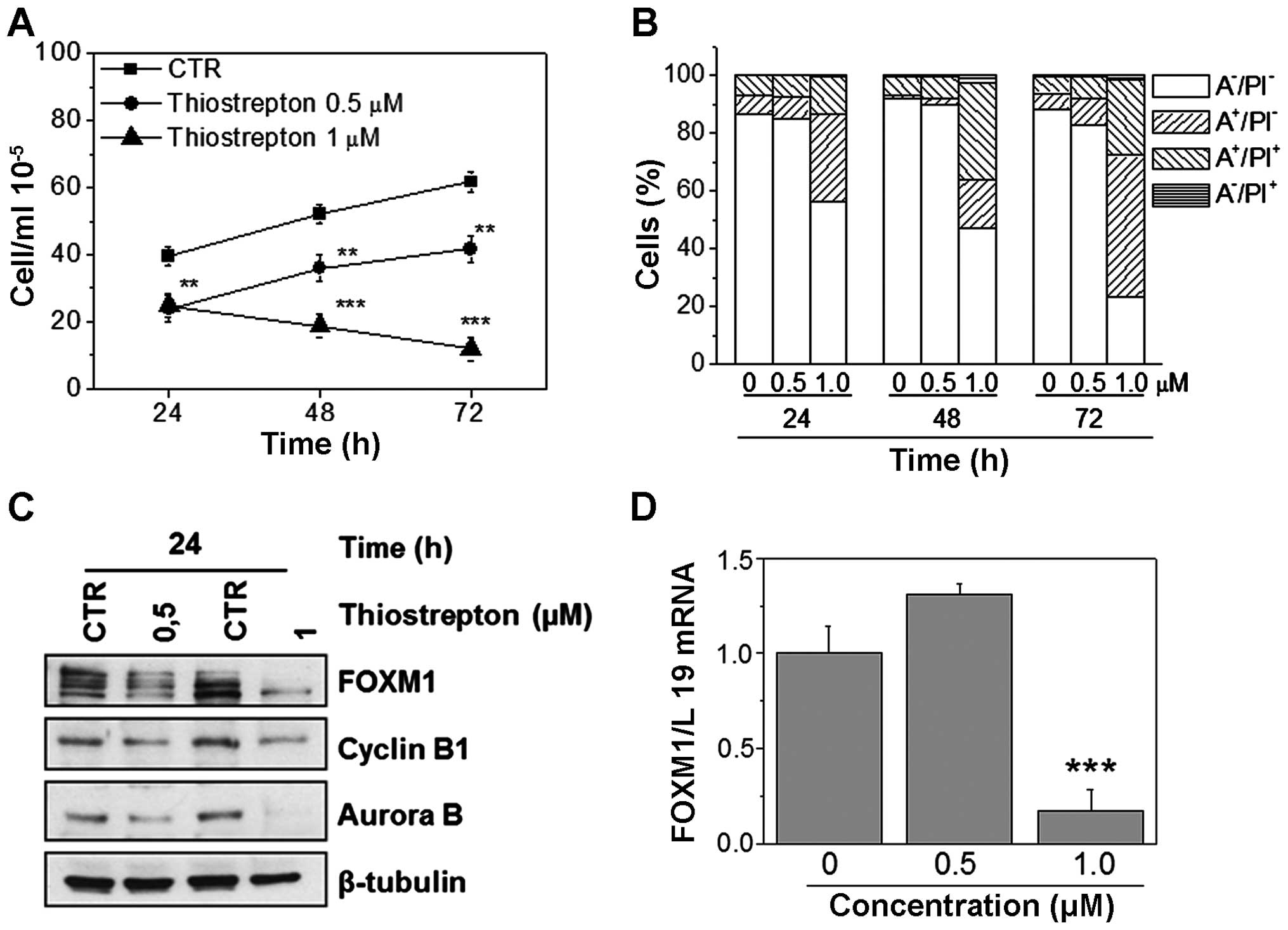
respectively for 24, 48, and 72 h. Results show a significant
reduction in cell viability, revealed by the considerable low
numbers of trypan blue-negative cells in both NALM-6 and REH cell
lines (Figs. 5A and 6A). Moreover, flow cytometric analysis
carried out following different treatment times indicates that
thiostrepton induces apoptosis, demonstrated by the appearance of a
large percentage of Annexin V-positive cells in both cell lines
(Figs. 5B and 6B). Importantly, apoptosis occurs in a
concentration- and time-dependent manner. In each assay, FOXM1
downregulation was confirmed by both western blot analysis
(Figs. 5C and 6C) and by RT-qPCR (Figs. 5D and 6D). Note that in both cell lines, FOXM1
and its downstream target protein levels were decreased following
thiostrepton treatment, in a manner similar to that observed for
FOXM1 silencing.
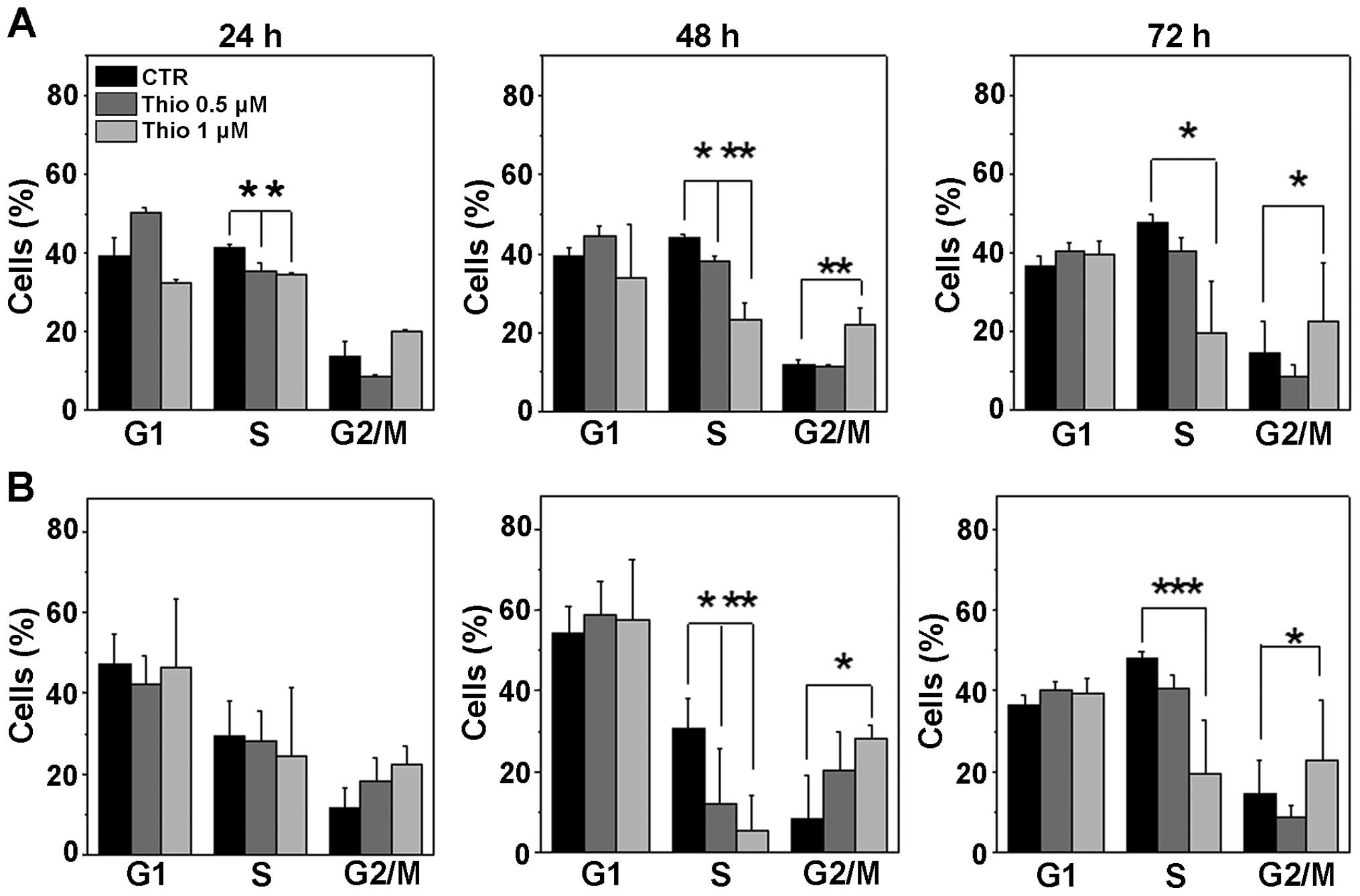
We then analyzed the cell cycle phase distribution
in REH and NALM-6 cells following treatment with both 0.5 and 1 μM
concentrations of thiostrepton at 24, 48, and 72 h. Results
(Fig. 7A and B) show that
similarly to FOXM1 knockdown experiments, thiostrepton treatment
induces a cellular arrest at the G2/M cell cycle phase as well as a
decrease of cells in the S phase.
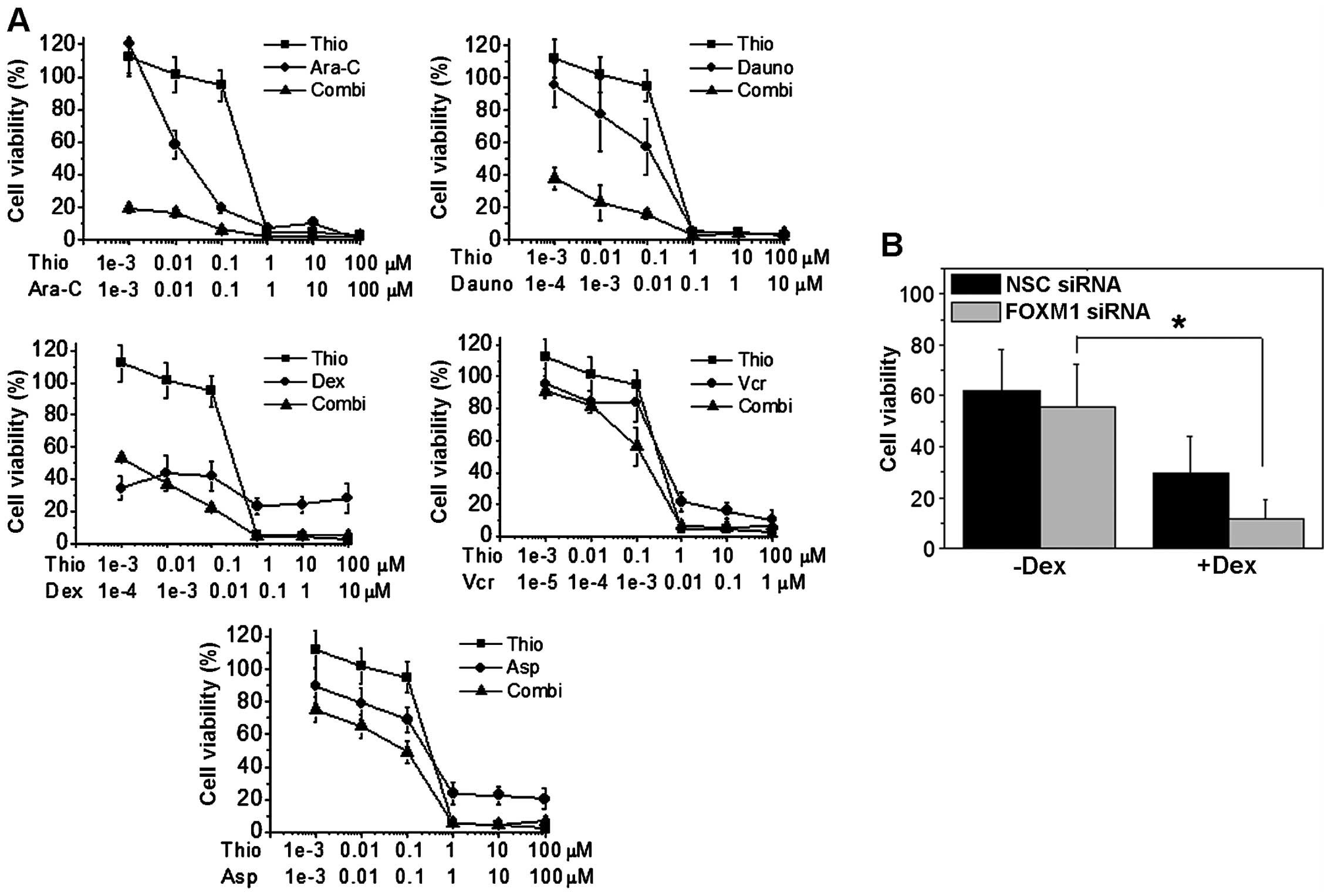
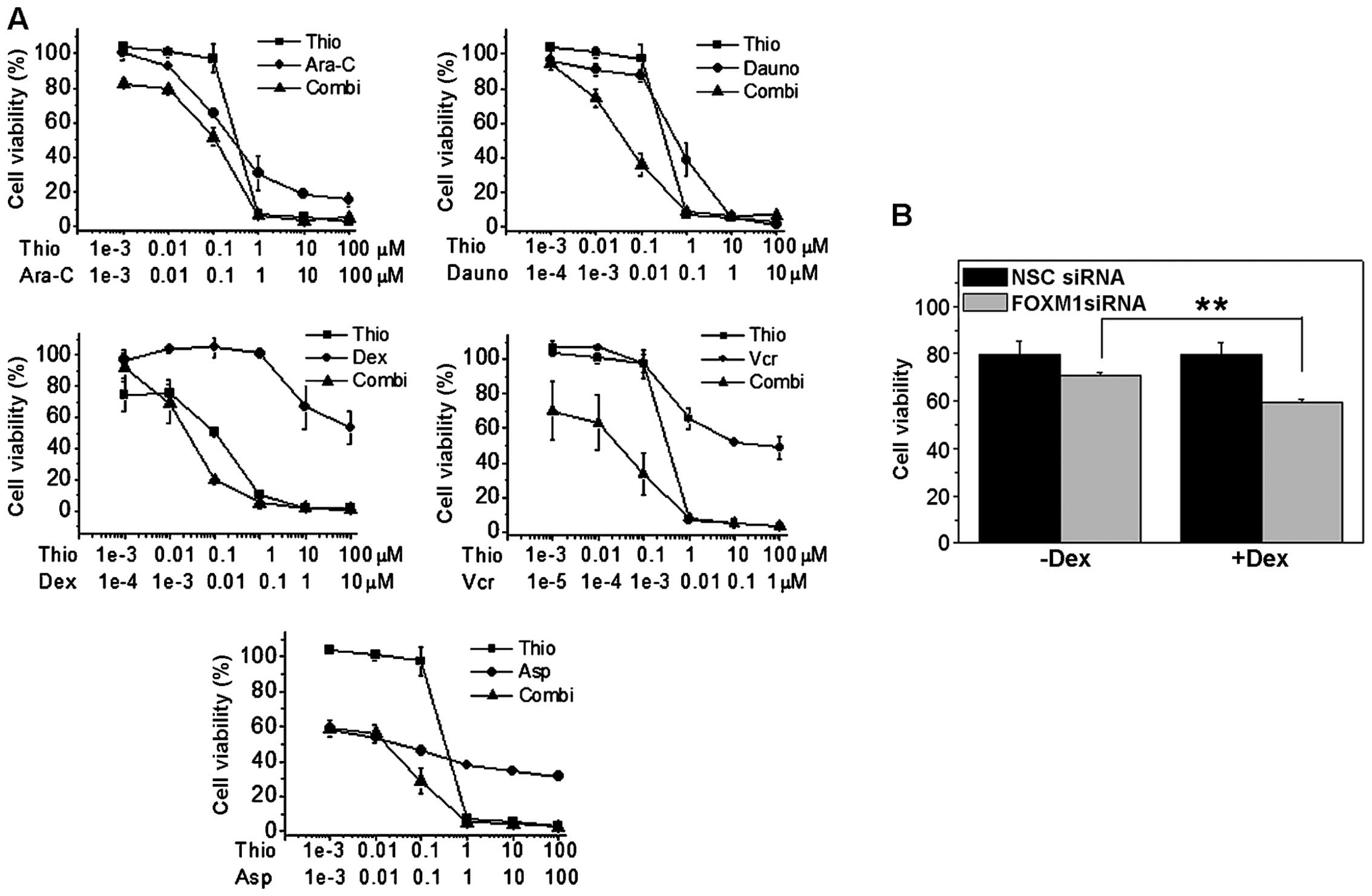
Thiostrepton synergises with conventional
chemotherapeutic agents to inhibit B-ALL proliferation
As FOXM1 down-regulation led to a decrease on B-ALL
cell proliferation, we next-tested if thiostrepton could be used in
combination with the most commonly used chemotherapeutics in B-ALL
treatment. To this end, four different B-ALL cell lines, two
glucocorticoid-resistant (REH, SEM) and two
glucocorticoid-sensitive (NALM-6, RS4;11), were treated for 48 h
with thiostrepton in combination with chemotherapeutic agents
(i.e., dexamethasone, asparaginase, daunorubicin, vincristine and
Ara-C) normally used to treat pediatric B-ALL patients. More
specifically, thiostrepton was combined with different drugs at
fixed molar combination ratios, and cell viability analyzed by MTT
assay (Figs. 8A and 9A). As described above, thiostrepton has
a significant cytotoxicity when used as single agent. Notably, when
thiostrepton was used in combination with chemotherapic drugs, we
observed a synergistic increase in the cytotoxicity as demonstrated
by the values of combination index (CI) according to Chou and
Talalay (25,30). As depicted in Table II, reporting CI values calculated
at GI50, GI75 and GI90, in almost
all cell lines tested, thiostrepton and chemotherapeutic drugs act
in a synergistic fashion (CI<1). Our results therefore show
that, in general, the pharmacological downregulation of FOXM1
caused by thiostrepton, can significantly increase the cell death
induced by the treatment with conventional chemotherapeutic
agents.
 | Table IICombination index values (CI) in
B-ALL cell lines treated with thiostrepton in combination with
chemotherapic drugs. |
Table II
Combination index values (CI) in
B-ALL cell lines treated with thiostrepton in combination with
chemotherapic drugs.
|
GI50b |
GI75b |
GI90b |
|---|
| Dauno
(1:10)a |
| RS4;11 | 2.4 | 0.17 | 0.1 |
| SEM | 0.27 | 0.28 | 0.35 |
| NALM-6 | 0.06 | 0.12 | 0.25 |
| REH | 0.84 | 1.1 | 1.1 |
| Ara-C (1:1)a |
| RS4;11 | 0.07 | 0.2 | 0.006 |
| SEM | 0.33 | 0.35 | 0.42 |
| NALM-6 | 0.0017 | 0.001 | 0.09 |
| REH | 0.16 | 0.29 | 0.54 |
| Dex (1:10)a |
| RS4;11 | 0.10 | 0.9 | 8.0 |
| SEM | 0.18 | 0.15 | 0.13 |
| NALM-6 | 0.8 | 0.044 | 0.24 |
| REH | 1.1 | 0.97 | 0.85 |
| Vcr
(1:1,000a |
| RS4;11 | 0.04 | 0.02 | 9.5 |
| SEM | 1.1 | 0.75 | 0.6 |
| NALM-6 | 0.64 | 0.44 | 0.74 |
| REH | 0.09 | 0.02 | 0.01 |
| Asp (1:1)a |
| RS4;11 | 0.5 | 0.2 | 0.1 |
| SEM | 1.6 | 0.8 | 0.4 |
| NALM-6 | 0.10 | 0.13 | 0.5 |
| REH | 1.3 | 0.05 | 0.15 |
To prove the above further, we knocked down FOXM1 in
REH and NALM-6 cells using siRNA, and then treated them with
dexamethasone. After 48 h of treatment we analyzed cell viability
by flow cytometry staining with Annexin V and propidium iodide
(PI). Results showed a significant decrease in cell viability after
48 h of treatment in FOXM1 silenced cells when compared to its
controls, this was particularly noted in glucocorticoid resistant
cells, indicating that FOXM1 can be an important therapeutic target
for overcoming glucocorticoid resistance in B-ALL (Figs. 8B and 9B).
Discussion
FOXM1 is an important cell cycle regulator and plays
a crucial role in tumorigenesis and its overexpression has been
found in many different human cancers. However, very little is
known about its function in hematological malignancies. Our aim was
to investigate the role of FOXM1 in B-ALL, the most common
pediatric leukemia. RT-PCR analysis performed on ten B-ALL patient
samples showed that FOXM1 mRNA is highly overexpressed in
comparison to FOXM1 mRNA from lymphocytes of healthy donors and
more importantly also in comparison with normal B-cells
(CD19+). These results are in excellent agreement with
Buchner et al (20) who
recently showed that FOXM1 is highly overexpressed in B-ALL
irrespectively of different B-ALL subsets. It is important to note
that our primers, irrespectively detect both B and C isoforms of
FOXM1 which represent the active forms since the isoform FOXM1A, is
not translated.
The pattern of FOXM1 is mirrored by Cyclin B1 and
Aurora B, two G2/M phase regulators directly regulated by FOXM1.
Immunoblot analysis also demonstrated similar over-expression of
FOXM1 and its downstream targets in samples from blast patients,
suggesting that FOXM1 is also aberrantly overexpressed in B-ALL as
in many other cancer types. In agreement, the five B-ALL cell lines
analyzed also overexpressed FOXM1 and its downstream targets,
confirming the results obtained in patients. Importantly, our study
revealed that the FOXM1 depletion by siRNA causes a significant
reduction in the proliferation rate of B-ALL cell lines, suggesting
a role for FOXM1 in the oncogenesis of B-ALL. Moreover, FOXM1
silencing led to cell cycle arrest in G2/M, suggesting that FOXM1
promotes cell proliferation through cell-cycle modulation,
previously reported by Nakamura et al in acute myeloid
leukemia cells (16). Consistent
with previous data (16), FOXM1
silencing in B-ALL cell lines led to a down-regulation of proteins
involved in the regulation of mitotic progression. We also showed
that FOXM1 silencing is accompanied by a decrease on expression
levels of p27Kip1 and p21Cip1/Waf1 proteins,
which are known to assemble different Cyclin/Cdk complexes.
p27Kip1 and p21Cip1/Waf1 proteins are
phosphorylated by the Cdk2-cyclin E complex, to be recognized by
the specificity subunits Skp2 and Cks1 of the SCF ubiquitin ligase
complex, which targets them for ubiquitin-mediated proteasome
degradation (31–33). It could therefore be speculated
that the noted decrease in their expression could reflect an
accelerated ubiquitinylation and subsequent degradation.
Thiostrepton is a thiazole antibiotic that inhibits
the transcriptional activity of FOXM1. It also downregulates FOXM1
mRNA expression, since FOXM1 can positively auto-regulate its own
transcription (28,34). Indeed, we found that both the mRNA
and protein expression of FOXM1 were downregulated by thiostrepton
in B-ALLs. In our study, we also found that thiostrepton remarkably
reduces the cell viability of different B-ALL cell lines causing
apoptosis in a concentration- and time-dependent manner, as well as
inducing a G2/M arrest of the cell cycle, consistent with the
result obtained with the siRNA-mediated knockdown of FOXM1.
The standard treatment option for newly diagnosed
childhood B-ALL is predominantly chemotherapy. Patients that
respond poorly to chemotherapy are predicted to undergo a future
relapse. Understanding the biological mechanisms which underlie
poor responsiveness is therefore crucial for the development of
more effective therapies. Consistent with this, Bhatla et al
have identified in a cohort of relapsed B-ALL pediatric patients a
series of upregulated genes which also includes FOXM1 (21).
In light of these results we examined if combining
thiostrepton with the chemotherapeutics used in B-ALL therapy could
increase their efficacy. Our results clearly indicate a strong
synergistic effect (CI<1) between thiostrepton and drugs with
different mechanisms of action in the four B-ALL cell lines tested.
Our results are in good agreement with that reported by Uddin et
al who reported a synergistic interaction of thiostrepton with
bortezomib in diffuse large B-cell lymphoma (18). It is also important to note that
thiostrepton is able to partially reverse the glucocorticoid
resistance in REH cells. These results were further confirmed, in a
more specific way, in the same cell lines with silenced FOXM1
(Figs. 8B and 9B). Given that patients that respond
poorly to glucocorticoid therapy at diagnosis are usually predicted
to undergo relapse in the future, our findings suggest that FOXM1
inhibition could be a potential useful strategy in clinical therapy
to optimize the efficacy of existing therapeutics for B-ALL,
although further studies are needed to better understand the
molecular mechanism(s) involved in these synergistic effects.
Indeed, very recently Buchner et al (20) reported the efficacy of thiostrepton
also in vivo in a mouse xenograft model of B-ALL. In
conclusion, we show that FOXM1 has a role in both oncogenesis and
the development of drug resistance in B-ALL, and the targeting of
FOXM1 could be a useful means for treating B-ALL and for overcoming
drug resistance.
References
|
1
|
Lam EW-F, Brosens JJ, Gomes AR and Koo
C-Y: Forkhead box proteins: Tuning forks for transcriptional
harmony. Nat Rev Cancer. 13:482–495. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Laoukili J, Kooistra MRH, Brás A, Kauw J,
Kerkhoven RM, Morrison A, Clevers H and Medema RH: FoxM1 is
required for execution of the mitotic programme and chromosome
stability. Nat Cell Biol. 7:126–136. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Laoukili J, Alvarez M, Meijer LA, Stahl M,
Mohammed S, Kleij L, Heck AJ and Medema RH: Activation of FoxM1
during G2 requires cyclin A/Cdk-dependent relief of autorepression
by the FoxM1 N-terminal domain. Mol Cell Biol. 28:3076–3087. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wang I-C, Chen Y-J, Hughes D, Petrovic V,
Major ML, Park HJ, Tan Y, Ackerson T and Costa RH: Forkhead box M1
regulates the transcriptional network of genes essential for
mitotic progression and genes encoding the SCF (Skp2-Cks1)
ubiquitin ligase. Mol Cell Biol. 25:10875–10894. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gemenetzidis E, Elena-Costea D, Parkinson
EK, Waseem A, Wan H and Teh M-T: Induction of human epithelial
stem/progenitor expansion by FOXM1. Cancer Res. 70:9515–9526. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Huynh KM, Soh J-W, Dash R, Sarkar D,
Fisher PB and Kang D: FOXM1 expression mediates growth suppression
during terminal differentiation of HO-1 human metastatic melanoma
cells. J Cell Physiol. 226:194–204. 2011. View Article : Google Scholar
|
|
7
|
Wang Z, Park HJ, Carr JR, Chen YJ, Zheng
Y, Li J, Tyner AL, Costa RH, Bagchi S and Raychaudhuri P: FoxM1 in
tumorigenicity of the neuroblastoma cells and renewal of the neural
progenitors. Cancer Res. 71:4292–4302. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Monteiro LJ, Khongkow P, Kongsema M,
Morris JR, Man C, Weekes D, Koo CY, Gomes AR, Pinto PH, Varghese V,
et al: The Forkhead Box M1 protein regulates BRIP1 expression and
DNA damage repair in epirubicin treatment. Oncogene. 32:4634–4645.
2013. View Article : Google Scholar
|
|
9
|
Khongkow P, Karunarathna U, Khongkow M,
Gong C, Gomes AR, Yagüe E, Monteiro LJ, Kongsema M, Zona S, Man EP,
et al: FOXM1 targets NBS1 to regulate DNA damage-induced senescence
and epirubicin resistance. Oncogene. 33:4144–4155. 2014. View Article : Google Scholar :
|
|
10
|
Li X, Qiu W, Liu B, Yao R, Liu S, Yao Y
and Liang J: Forkhead box transcription factor 1 expression in
gastric cancer: FOXM1 is a poor prognostic factor and mediates
resistance to docetaxel. J Transl Med. 11:2042013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chu X-Y, Zhu Z-M, Chen L-B, Wang JH, Su
QS, Yang JR, Lin Y, Xue LJ, Liu XB and Mo XB: FOXM1 expression
correlates with tumor invasion and a poor prognosis of colorectal
cancer. Acta Histochem. 114:755–762. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Martin KJ, Patrick DR, Bissell MJ and
Fournier MV: Prognostic breast cancer signature identified from 3D
culture model accurately predicts clinical outcome across
independent datasets. PLoS One. 3:e29942008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bektas N, Haaf A, Veeck J, Wild PJ,
Lüscher-Firzlaff J, Hartmann A, Knüchel R and Dahl E: Tight
correlation between expression of the Forkhead transcription factor
FOXM1 and HER2 in human breast cancer. BMC Cancer. 8:422008.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Xia L, Huang W, Tian D, Zhu H, Zhang Y, Hu
H, Fan D, Nie Y and Wu K: Upregulated FoxM1 expression induced by
hepatitis B virus X protein promotes tumor metastasis and indicates
poor prognosis in hepatitis B virus-related hepatocellular
carcinoma. J Hepatol. 57:600–612. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liu M, Dai B, Kang S-H, Ban K, Huang FJ,
Lang FF, Aldape KD, Xie TX, Pelloski CE, Xie K, et al: FoxM1B is
overexpressed in human glioblastomas and critically regulates the
tumorigenicity of glioma cells. Cancer Res. 66:3593–3602. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Nakamura S, Hirano I, Okinaka K, Takemura
T, Yokota D, Ono T, Shigeno K, Shibata K, Fujisawa S and Ohnishi K:
The FOXM1 transcriptional factor promotes the proliferation of
leukemia cells through modulation of cell cycle progression in
acute myeloid leukemia. Carcinogenesis. 31:2012–2021. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang Z, Zheng Y, Park HJ, Li J, Carr JR,
Chen YJ, Kiefer MM, Kopanja D, Bagchi S, Tyner AL, et al: Targeting
FoxM1 effectively retards p53-null lymphoma and sarcoma. Mol Cancer
Ther. 12:759–767. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Uddin S, Hussain AR, Ahmed M, Siddiqui K,
Al-Dayel F, Bavi P and Al-Kuraya KS: Overexpression of FoxM1 offers
a promising therapeutic target in diffuse large B-cell lymphoma.
Haematologica. 97:1092–1100. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhao B, Barrera LA, Ersing I, Willox B,
Schmidt SC, Greenfeld H, Zhou H, Mollo SB, Shi TT, Takasaki K, et
al: The NF-κB genomic landscape in lymphoblastoid B cells. Cell
Rep. 8:1595–1606. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Buchner M, Park E, Geng H, Klemm L, Flach
J, Passegué E, Schjerven H, Melnick A, Paietta E, Kopanja D, et al:
Identification of FOXM1 as a therapeutic target in B-cell lineage
acute lymphoblastic leukaemia. Nat Commun. 6:64712015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Bhatla T, Wang J, Morrison DJ, Raetz EA,
Burke MJ, Brown P and Carroll WL: Epigenetic reprogramming reverses
the relapse-specific gene expression signature and restores
chemosensitivity in childhood B-lymphoblastic leukemia. Blood.
119:5201–5210. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Accordi B, Galla L, Milani G, Curtarello
M, Serafin V, Lissandron V, Viola G, te Kronnie G, De Maria R,
Petricoin EF III, et al: AMPK inhibition enhances apoptosis in
MLL-rearranged pediatric B-acute lymphoblastic leukemia cells.
Leukemia. 27:1019–1027. 2013. View Article : Google Scholar
|
|
23
|
Hui RC-Y, Gomes AR, Constantinidou D,
Costa JR, Karadedou CT, Fernandez de Mattos S, Wymann MP, Brosens
JJ, Schulze A and Lam EW: The forkhead transcription factor FOXO3a
increases phosphoinositide-3 kinase/Akt activity in drug-resistant
leukemic cells through induction of PIK3CA expression. Mol Cell
Biol. 28:5886–5898. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Bortolozzi R, Viola G, Porcù E, et al: A
novel copper (I) complex induces ER-stress-mediated apoptosis and
sensitizes B-acute lymphoblastic leukemia cells to chemotherapeutic
agents. Oncotarget. 5:5978–5991. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chou T-C: Drug combination studies and
their synergy quantification using the Chou-Talalay method. Cancer
Res. 70:440–446. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Nilsson I and Hoffmann I: Cell cycle
regulation by the Cdc25 phosphatase family. Prog Cell Cycle Res.
4:107–114. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Bertoli C, Skotheim JM and de Bruin RAM:
Control of cell cycle transcription during G1 and S phases. Nat Rev
Mol Cell Biol. 14:518–528. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kwok JM-M, Myatt SS, Marson CM, Coombes
RC, Constantinidou D and Lam EW-F: Thiostrepton selectively targets
breast cancer cells through inhibition of forkhead box M1
expression. Mol Cancer Ther. 7:2022–2032. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hegde NS, Sanders DA, Rodriguez R and
Balasubramanian S: The transcription factor FOXM1 is a cellular
target of the natural product thiostrepton. Nat Chem. 3:725–731.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chou TC: Theoretical basis, experimental
design, and computerized simulation of synergism and antagonism in
drug combination studies. Pharmacol Rev. 58:621–681. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Carrano AC, Eytan E, Hershko A and Pagano
M: SKP2 is required for ubiquitin-mediated degradation of the CDK
inhibitor p27. Nat Cell Biol. 1:193–199. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hara T, Kamura T and Nakayama K, Oshikawa
K, Hatakeyama S and Nakayama K: Degradation of p27(Kip1) at the
G(0)–G(1) transition mediated by a Skp2-independent ubiquitination
pathway. J Biol Chem. 276:48937–48943. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lu Z and Hunter T: Ubiquitylation and
proteasomal degradation of the p21(Cip1), p27(Kip1) and p57(Kip2)
CDK inhibitors. Cell Cycle. 9:2342–2352. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Koo C-Y, Muir KW and Lam EW-F: FOXM1: From
cancer initiation to progression and treatment. Biochim Biophys
Acta. 1819.28–37. 2012.
|