Introduction
Out of the various types of gynecological malignant
tumors, ovarian cancer is considered the leading cause of fatality
among women worldwide (1). This is
a result of two factors. Firstly, lack of reliable and accurate
methods for early diagnosis results in the confirmation of the
late-stage ovarian cancer in >70% of patients (2,3).
Secondly, the multidrug resistance (MDR) phenotype can develop
post-chemotherapy treatment, which typically results in a high
recurrence rate (>50%) and poor prognosis for these patients
(4).
A great effort has been made to understand the
molecular mechanisms of the MDR phenotype, and several models have
been proposed, including the upregulated expression of MDR proteins
(which efflux the anti-cancer agents out of cancer cells), the
evasion of cell apoptosis through the overexpression of
anti-apoptotic proteins, the increased recovery capability of the
DNA-damage repairing system and the accelerated metabolism of
anti-cancer agents (5–7). Various proteins affect cell growth
and cytokine signaling pathways, and cell cycle behavior also
serves important roles in the development of the MDR phenotype
(8–10). Since MDR has become a more serious
concern in the clinic, the development of novel strategies to
overcome the MDR phenotype is of importance.
ERp57 was first reported to be associated with the
chemotherapy resistance of ovarian cancer by Bernardini et
al (11) in 2005. Their array
comparative genomic hybridization and microarray expression
profiling result identified a novel class of genes associated with
in vivo drug response in patients with. ovarian cancer.
ERp57 has been indicated to form a nuclear complex that is
associated with resistance to DNA conformation-altering
chemotherapeutic agents in in vitro systems. Cicchillitti
et al (12) used a
comparative proteomic approach to analyze the paclitaxel
sensitivity of A2780 epithelial ovarian cancer cells and identified
that ERp57 is a protein that is altered in paclitaxel-resistant
cell lines when compared with paclitaxel-sensitive cell lines
(12). ERp57 interacts with class
III β-tubulin (TUBB3) in paclitaxel-resistance ovarian cells, and
this ERp57/TUBB3 interaction occurs in a novel location of the
cytoskeleton rather than the nuclear compartment (12). These results indicate that ERp57
may serve an important role in the chemoresistance of ovarian
cancer by modulating the attachment of microtubules to chromosomes
following paclitaxel treatment through its interaction with TUBB3.
However, the biological roles of ERp57 in the chemoresistance of
ovarian cancer remain unknown, and no studies have explored the
effects of ERp57 downregulation on the improvement of the
paclitaxel sensitivity of chemoresistant ovarian cancer.
In the present study, the expression levels of ERp57
were compared in SKOV3 ovarian cancer cells and
paclitaxel-resistant SKOV3/tax cells. The small interfering (si)RNA
approach was used to inhibit the expression of ERp57. Furthermore,
the biological effects of ERp57-siRNA silencing on the possible MDR
reversal of SKOV3/tax cells were examined. Bioinformatics analysis
was also performed to identify the biological processes and
pathways associated with ERp57 and chemoresistant ovarian
cancer.
Materials and methods
Chemicals and reagents
All chemicals were obtained from Shenyang Sinopharm
Group (Shenyang, China) unless otherwise stated. ERp57 inhibitor
DNTB was obtained from Sigma-Aldrich; Merck KGaA (Darmstadt,
Germany).
Cell lines and cell cultures
Human epithelial ovarian cancer cells SKOV3 were
purchased from Beijing Shijitan Hospital (Beijing, China). Cells
were cultured in RPMI-1640 medium (Hyclone; GE Heathcare Life
Sciences, Logan, UT, USA) supplemented with L-glutamine and 10%
fetal bovine serum (TBD, Tianjin, China) in a humidified incubator
(5% CO2 at 37°C). Cell lines grew in a monolayer and
were passaged when cultures were 70–80% confluent.
Establishment of SKOV3/tax cells
The paclitaxel-resistant SKOV3 (SKOV3/tax) were
prepared following a standard stepwise selection procedure. SKOV3
cells were cultured in RPMI-1640 medium containing paclitaxel at a
concentration of 0.1 nM for 24 h. Cells that survived were selected
for the next survival selection step using a higher paclitaxel
concentration. This cell culture process was repeated for several
steps with an increment of 0.5 nM at each step until all cells
could survive at the paclitaxel concentration of 10 nM. The
survived cells were able to maintain the paclitaxel-resistance
phenotype in the absence of the selection pressure and were named
SKOV3/tax (13).
Giemsa staining
SKOV3 and SKOV3/tax cells were seeded on 6-well
plates at the cell density of 2×106 cells/ml. Following
24 h of incubation, cells were washed with PBS 3 times and fixed
with methanol for 30 min at room temperature. Subsequently, cells
were stained with Giemsa dye (Beyotime Institute of Biotechnology,
Shanghai, China) for 15 min at room temperature. Cells were washed
with PBS once and with deionized water three times. Once they were
dried, SKOV3 and SKOV3/tax cells were examined under an optic
microscope (at magnifications, ×100 and ×200, respectively).
Cell viability analysis
SKOV3 and SKOV3/tax cells were seeded on 96-well
plates at a cell density of 5×103 cells/well or on
6-well plates at the cell density of 1×106 cells/well,
respectively. Following 24 h of incubation, cell culture medium
(RPMI-1640) was aspirated and replaced with fresh culture medium
containing different concentrations of paclitaxel (0.01, 0.1, 1.0,
10, 100 and 1,000 nM). Following 48 or 72 h incubation, cell
viability was assessed using MTT assay. Each well was aspirated and
incubated with 5 mg/ml MTT reagent (in 0.01 M PBS, pH 7.4). 4 h
later, extraction buffer was added to each well to resolve the MTT
crystals and the optic absorbance at 570 nm was measured using an
Infinite M200 PRO multiplate reader (Tecan Group Ltd., Männedorf,
Switzerland). Cell viability was calculated based on the optic
absorbance. Untreated cells were used as a blank control
(considered as 100% viable). IC50 values were estimated
from concentration-viability curves.
ERp57 overexpression induced by
lentiviral particle infection
Lentiviruses carrying ERp57 expression vectors were
obtained from GeneChem (Shanghai, China). ERp57 overexpression was
conducted according to the company’s instructions. Briefly, cells
(0.5×105 cells/well) were seeded in a 12-well plate and
treated with lentiviral particles to establish ERp57 overexpression
[40 µl polybrene and 2.5 µl/well containing
1×108 infectious units (IFU) of ERp57 overexpression
virus] and scramble control (40 µl polybrene and 2.5
µl/well containing 1×108 IFU scramble virus)
groups. The blank group consisted of SKOV3 cells without
lentiviruses transfection. Fresh medium (RPMI-1640) without
polybrene was placed on each infected well following 24 h of
incubation.
Silencing ERp57 with siRNA
ERp57-siRNA was used to target the ERp57 gene
(nucleotide sequence, 5′-GGGCAAGGACUUACUUAUU-3′). For comparison, a
random nucleotide sequence of 5′-UUCUCCGAACGUGUCACGU-3′ was used as
a negative control (NC-siRNA). Transfection of NC-siRNA and
ERp57-siRNA was carried out, respectively, in a final concentration
of 50 nM using Lipofectamine 2000 (Invitrogen; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) according to the manufacturer’s
protocol. The blank group was SKOV3/tax cells without siRNA
transfection. After 48 h of incubation, the transfected cells were
collected for cell apoptosis analysis. Total mRNAs and proteins of
these cells were isolated for reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) and western blot analysis,
respectively.
Western blot analysis
SKOV3/tax cells were treated with ERp57-siRNA for 48
h, followed by 10 nM paclitaxel (the dosage commonly used for
treatment of SKOV3 cells) for 48 h and 100 nM paclitaxel (the
concentration near the IC50 value of SKOV3/tax cells)
for 48 h, respectively. Subsequently, cells were pelleted by
centrifugation (500 x g for 5 min at 4°C) and washed with cold PBS.
Cell pellets were resuspended in the radioimmunoprecipitation assay
buffer (Beyotime Institute of Biotechnology) containing protease
inhibitor (protease inhibitor cocktail; Roche Diagnostics, Basel,
Switzerland), and lysed by bath sonication (4 times for 30 sec
on/off). Lysates were centrifuged (15,000 x g for 30 min at 4°C)
and the concentration of proteins were diluted to 3
µg/µl with 5X sample loading buffer (as determined by
BCA assay). Samples were boiled at 100°C for 5 min. Following this,
the extracted proteins (30 µg per lane) were separated by
SDS-PAGE on Bis-Tris 4-12% gradient gels, transferred to
polyvinylidene difluoride membranes and detected using specific
antibodies. The membranes were blocked using 5% skimmed milk at
room temperature for 2 h and incubated with appropriate primary
antibodies at 4°C overnight. The following antibodies were used:
Monoclonal anti-ERp57 (cat. no. sc80648), anti-TUBB3 (cat. no.
sc-69966), anti-STAT3 (cat. no. sc8019) and anti-phospho(p)-STAT3
(Tyr705) antibodies (cat. no. sc81523) from Santa Cruz
Biotechnology Inc. (Santa Cruz, CA, USA); monoclonal
anti-phospho-glycoprotein (P-gp) antibodies (cat. no. ab170904)
from Abcam (Cambridge, UK); polyclonal anti-p53 (cat. no.
10442-1-AP), anti-nucleolin (cat. no. 10556-1-AP) and monoclonal
anti-GAPDH antibodies (cat. no.60004-1-lg) from ProteinTech Group
Inc. (Wuhan, China); polyclonal anti-proliferating cell nuclear
antigen (PCNA; cat. no. WL01804), anti-B-cell lymphoma B-cell
lymphoma-extra large (Bcl-xl; cat. no. WL01558), anti-matrix
metalloproteinase (MMP)1 (cat. no. WL01201), anti-MMP2 (cat. no.
WL01579a), anti-MMP9 (cat. no. WL01580) and anti-vimentin
antibodies (cat. no. WL00742) from Wanlei Biotechnology Inc.
(Shenyang, China); and anti-B-cell lymphoma-2 (Bcl-2; cat. no.
D260117) and anti-Bcl-2-associated X protein (Bax; cat. no.
D120073) from Sangon Biotechnology Inc. (Shanghai, China). The
primary antibodies were diluted to 1:800. Thereafter, the membranes
were incubated with the secondary antibodies (anti-rabbit cat. no.
ZB-2301 or anti-mouse cat. no. ZB-2305; ZSGB-Bio, Beijing, China)
for 1 h at room temperature. The secondary antibodies were diluted
to 1:10,000. Finally, the immune reactive proteins were detected
using an enhanced chemiluminescence kit (cat. no. WLA003a; Wanlei
Life Science, Shenyang, China) and the enhanced chemiluminescence
detection system (Tanon-5200; Tanon Science and Technology Co.,
Ltd., Shanghai, China).
RT-qPCR
SKOV3 and SKOV3/tax cells were incubated at the
density of 2×106 cells/well in 4 ml of RPMI-1640
supplemented with 10% fetal bovine serum for 48 h. Once cells were
collected and washed with PBS, total RNA was extracted using TRIzol
reagent (Invitrogen; Thermo Fisher Scientific, Inc.) and cDNA was
obtained with RT using a PrimeScript RT Reagent kit (Takara Bio,
Inc., Otsu, Japan). qPCR was performed using SYBR Premix Ex Taq II
(Takara Bio, Inc.) according to the manufacturer’s protocols. The
following primers were used in the present study: ERp57, forward
5′-GAGCAATGATGGGCC TGTGA-3′ and reverse 5′-TGACGATATTTGGGTCTTTGC
TGA-3′; and GAPDH, forward 5′-TGCACCACCAACTGCTT AGC-3′ and reverse
5′-GGCATGGACTGTGGTCATGAG-3′. The PCR process was performed using an
ABI PRISM 7500 system (Applied Biosystems; Thermo Fisher
Scientific, Inc.). PCR was performed as follows: 95°C for 30 sec;
followed by 40 cycles of 95°C for 5 sec and 60°C for 34 sec; and 1
cycle of 95°C for 15 sec, 60°C for 1 h and 95°C for 15 sec. RT-qPCR
data were normalized using GADPH as an internal standard and
analyzed using the 2−ΔΔCq method (14).
Clonogenic assay
SKOV3 and SKOV3/tax cells were transfected with
ERp57-siRNA, respectively, for 48 h, then harvested and washed with
PBS. Similarly, SKOV3 and SKOV3/tax cells were also transfected
with NC-siRNA for comparison. A total of 500 cells/well were plated
for 1-2 weeks at 37°C. Cells were fixed with 10% paraformaldehyde
at room temperature for 30 min. Colonies were stained with 0.25% of
crystal violet at room temperature for 30 min and counted using
ImageJ (version 1.46r) software (National Institutes of Health,
Bethesda, MD, USA).
Cell migration analysis
The cell migration capability was examined using the
Transwell assay (24-well insert; pore size, 8 µm; Corning
Inc., NY, USA). Cells transfected with ERp57-siRNA or NC-siRNA for
48 h were harvested, suspended (5×104 cells/well) in 100
µl serum-free RPMI-1640 medium, and loaded on the upper
chamber. A total of 500 µl complete RPMI-1640 medium
(containing 10% fetal serum albumin) was added in the lower
chamber. Following 24 h of incubation, cells were fixed with 10%
paraformaldehyde at room temperature for 30 min and free cells were
removed carefully from the upper surface of the filter with a
cotton swab. Migrated cells on the lower side of the filter were
stained with 0.5% crystal violet for 1 h at room temperature and
counted from five random fields under a optic microscope
(magnification, ×200) using ImageJ (version 1.46r) software.
Apoptosis assay
Cells were subjected to paclitaxel treatment and
compared with those without this treatment. In the non-treatment
group, SKOV3/tax cells were transfected with NC-siRNA, ERp57-siRNA
and blank for 48 h, respectively, and then harvested directly
without paclitaxel treatment. In the paclitaxel-treated group,
SKOV3/tax cells were treated with NC-siRNA, ERp57-siRNA and blank
for 48 h, respectively, followed by treatment with 10 nM paclitaxel
for 24 h, and then harvested. For comparison, one extra sample was
prepared: SKOV3/tax cells were treated with 1 mM DNTB for 48 h,
followed by 10 nM paclitaxel treatment for 24 h, and then
harvested. After the cells were harvested, all the samples were
resuspended with 100 µl binding buffer (140 mmol/l NaCl, 5
mmol/l CaCl2 and 10 mmol/l HEPES buffer) and washed
three times with PBS (pH 7.4). A total of 5 µl Annexin
V-fluorescein isothiocyanate and 10 µl propidium iodide
(Beijing Biosea Biotechnology, Co., Ltd., Beijing, China) were
added and the cell suspension was incubated at room temperature in
dark for 10 min. Following centrifugation, the cell pellet was
resus-pended in 200 µl binding buffer and analyzed using a
FACSort flow cytometer (BD Biosciences, San Jose, CA, USA). The
percentage of apoptotic and necrotic cells was determined using FCS
express software (version 3.0; DeNovo Software, Los Angeles, CA,
USA).
Bioinformatics analysis
The protein-protein interaction (PPI) network was
established using the online tool STRING (string-db.org) (15).
The gene/protein-gene/protein interaction network was generated
with GeneMANIA (genemania.org) (16). Biological process and gene
co-occurrence analysis was performed using COREMINE
(coremine/medical) (17). Pathway
enrichment analysis was performed using DAVID (david.abcc.ncifcrf.gov) (18).
Statistical methods
Data analysis was performed using SPSS software 17.0
(SPSS, Inc., Chicago, IL, USA). All experimental data were
expressed as the mean ± standard deviation and statistically
analyzed. The statistical significance of the results was assessed
using one-way analysis of variance followed by Tukey’s post hoc
multiple comparison tests. P<0.05 was considered to indicate a
statistically significant difference. All the measurements were
repeated at least three times.
Results
Characterization of paclitaxel-resistant
ovarian cancer cell lines
SKOV3/tax cells were characterized. The difference
in cell morphology between SKOV3 and SKOV3/tax cells was clarified
under an optic microscope and with Giemsa staining (Fig. 1A and B). Notably, more vesicles and
vacuoles were observed in SKOV3/tax cells compared with SKOV3
cells.
Cell proliferation was examined using the clonogenic
assay. Equal numbers of SKOV3 and SKOV3/tax cells were cultured for
2 weeks and colony numbers were subsequently counted. SKOV3/tax
cells grew significantly slower compared with SKOV3 cells
(P<0.05; Fig. 1C). Cell
population doubling times were estimated to be ~22 h for SKOV3
cells and 36 h for SKOV3/tax cells, respectively (by a factor of
1.6; Fig. 1D). The IC50
values of SKOV3 and SKOV3/tax cells to paclitaxel were determined
based on the growth curves in Fig.
1E to be 3.24±0.03 and 101.06±0.99 nM, respectively. The
drug-resistance index of SKOV3/tax to SKOV3 was calculated to be
>30-fold. These data confirmed the paclitaxel-resistant
characteristics of SKOV3/tax cells.
Furthermore, several proteins associated with
apoptosis (Bcl-2, Bax, Bcl-xl and p53), migration (MMP1, MMP2, MMP9
and vimentin), cell proliferation (PCNA and nucleolin) and
drug-resistance (P-gp and TUBB3) were compared in SKOV3 and
SKOV3/tax cells using western blot analysis to assess
paclitaxel-resistant behavior (Fig.
1F). The expression levels of MDR phenotype biomarkers P-gp and
TUBB3 were increased in SKOV3/tax cells compared with SKOV3 cells.
Protein PCNA and nucleolin expression levels were considered
biomarkers for cell proliferation. Notably, PCNA expression levels
were reduced in SKOV3/tax cells compared with SKOV3 cells. By
contrast, nucleolin expression levels were similar in SKOV3/tax and
SKOV3 cells. Weak p-STAT3 expression was indicated in SKOV3 cells;
however, p-STAT3 protein expression levels were increased in
SKOV3/tax cells. In SKOV3/tax cells, apoptosis-inhibiting proteins
Bcl-2 and Bcl-xl were highly expressed, whereas the apoptosis
promoting protein Bax was expressed in lower levels. The SKOV3 cell
is a p53-mutant cell line that does not express p53 (19); however, p53 was highly expressed in
SKOV3/tax cells. MMP1, MMP2 and MMP9 proteins, which are associated
with metastasis (20), were
expressed in lower levels in SKOV3/tax cells compared with SKOV3
cells, suggesting a lower invading ability of SKOV3/tax cells.
Furthermore, an epithelial-mesenchymal transition (EMT) protein
marker, vimentin, was expressed to almost at the same level in
SKOV3 and SKOV3/tax cells.
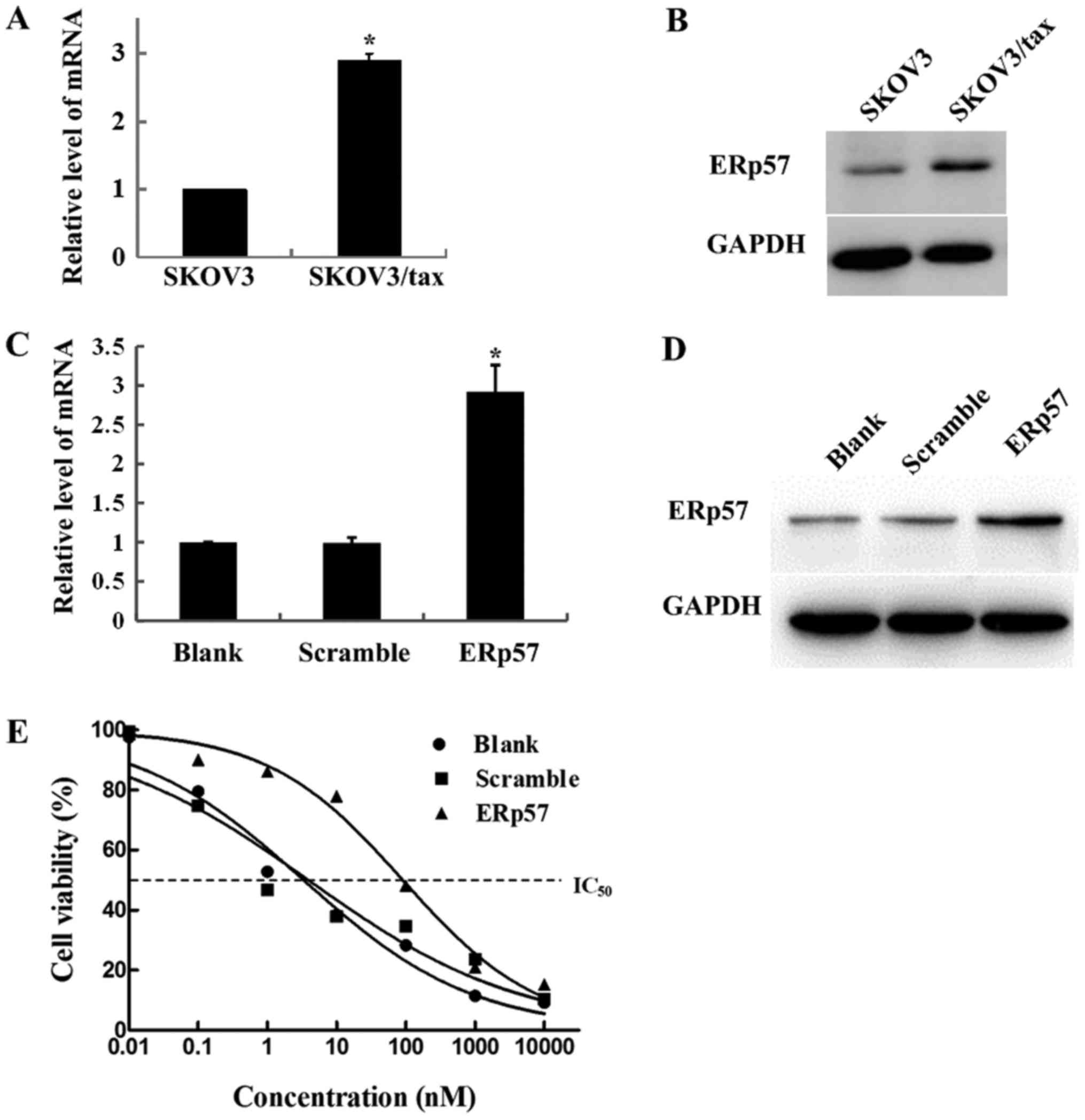
ERp57 overexpression of
paclitaxel-resistant SKOV3/tax cells
ERp57 mRNA and protein expression levels were
compared in SKOV3 and SKOV3/tax cells using RT-qPCR and western
blot analysis. As indicated in Fig.
2A, mRNA expression levels of ERp57 in SKOV3/tax cells were
>2-fold higher than that of SKOV3 cells (P<0.05). In
addition, western blot analysis results also revealed that the
expression level of ERp57 protein in SKOV3/tax cells was
upregulated (Fig. 2B). These data
were consistent with a previous report (21), which indicated that ERp57 is
strongly associated with the paclitaxel-resistant ovarian cancer
cells SKOV3/tax.
To examine the effect of ERp57 overexpression on the
paclitaxel sensitivity of SKOV3 cells, ERp57 was overexpressed in
SKOV3 cells (Fig. 2C and D). As
indicated in Fig. 2E, the
IC50 values of untransfected SKOV3 cells and scramble
control were 3.33±1.18 and 3.897±1.39 nM. However, the
IC50 value of ERp57-overexpressed SKOV3 cells was
increased to 90.59±1.13 nM. These data indicated that ERp57
overexpression could increase drug resistance of SKOV3 cells.
Paclitaxel sensitivity of SKOV3/tax
affected by ERp57-siRNA silencing
The effect of ERp57-siRNA silencing on the
sensitivity of SKOV3/tax cells to paclitaxel was examined. As
indicated in Fig. 3A, ERp57-siRNA
silencing significantly inhibited the expression levels of ERp57
mRNA (P<0.05). Similarly, western blot analysis results
demonstrated that ERp57 protein expression levels were also
inhibited (Fig. 3B). As indicated
in Fig. 3C, ERp57-siRNA silencing
significantly downregulated the viability of SKOV3/tax cells at 24,
48 and 72 h (P<0.05), and the cell viability percentages were
determined to be 87, 76 and 71% at the respective time-points. By
contrast, NC-siRNA did not significantly alter the cell viability
of SKOV3/tax cells.
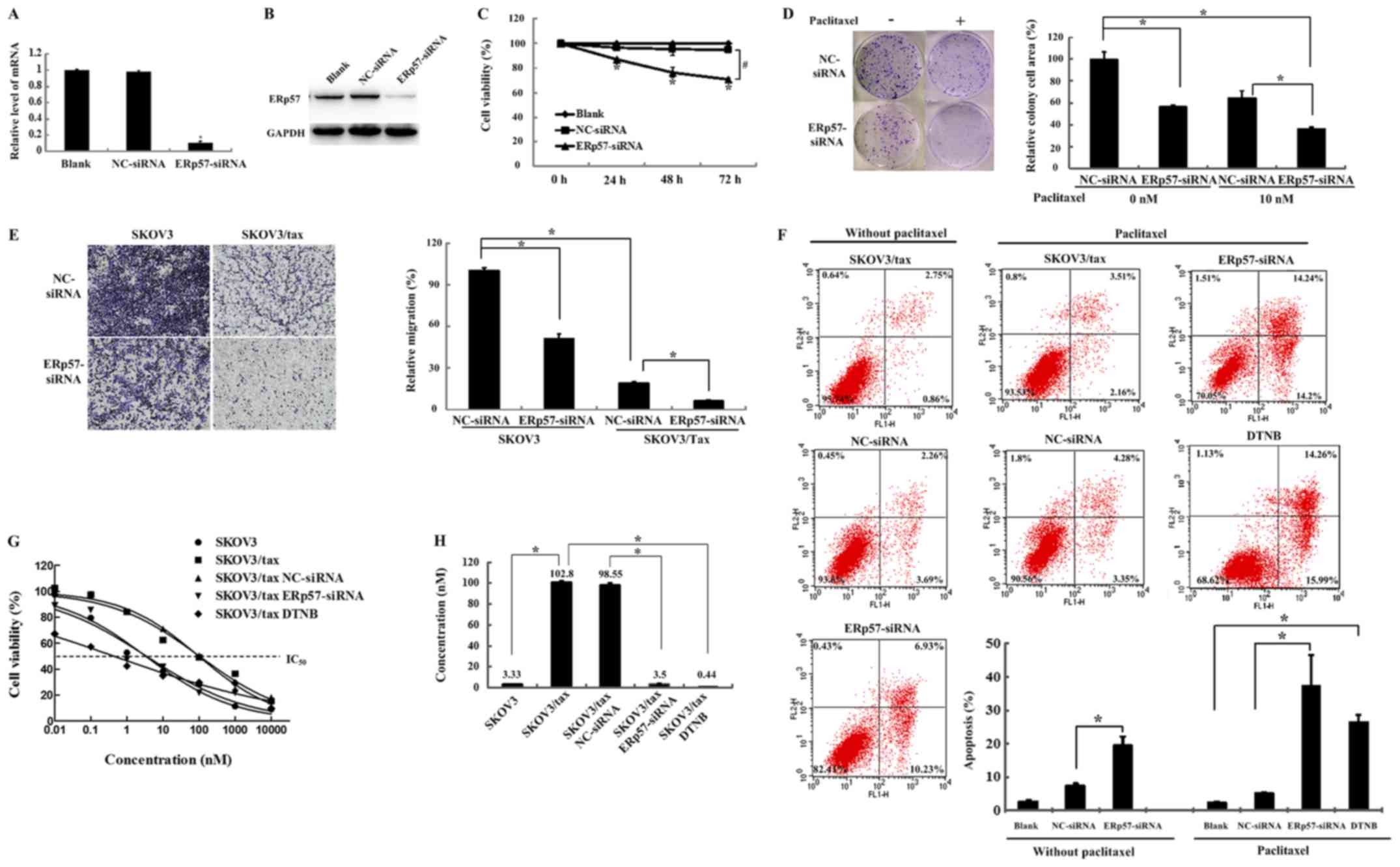 | Figure 3Cellular and molecular responses of
SKOV3/tax cells following different treatments. (A) The ERp57 mRNA
expression levels of SKOV3/tax cells treated with blank, NC-siRNA
and ERp57-siRNA were detected by reverse transcription-quantitative
polymerase chain reaction. *P<0.05 vs. NC-siRNA. (B)
ERp57 protein expression levels of SKOV3/tax cells treated with
blank, NC-siRNA and ERp57-siRNA, respectively. GAPDH was used as
the internal standard. (C) Cell viability analysis of SKOV3/tax
(treated with blank, NC-siRNA and ERp57-siRNA, respectively) for
different time periods (0, 24, 48 and 72 h) using the MTT assay.
*P<0.05 vs. NC-siRNA at 0 h; #P<0.05 as
indicated. (D) Cellular proliferation was examined using colony
formation assays. *P<0.05 as indicated. (E) Cell
migration was assessed using Transwell assays (original
magnifications, ×40). *P<0.05 as indicated. (F) Cell
apoptosis was detected by flow cytometric analyses.
*P<0.05 as indicated. (G) Cell viability analysis was
assessed by the MTT assay. (H) IC50 of SKOV3, SKOV3/tax,
SKOV3/tax treated with NC-siRNA, SKOV3/tax treated with ERp57 and
SKOV3/tax treated with ERp57 inhibitor DTNB, respectively.
*P<0.05 as indicated. |
Following ERp57-siRNA silencing, the numbers and the
size of the colonies of SKOV3/tax cells were significantly reduced
compared with the control (P<0.05; Fig. 3D, top left vs. lower left).
Furthermore, after ERp57-siRNA silencing, a total of 10 nM
paclitaxel decreased the colony formation and number of SKOV3/tax
colonies by ~60% when compared with the control with no paclitaxel
treatment (P<0.05; Fig. 3D, top
left vs. lower right). The Transwell assay was used to examine the
migration ability of SKOV3 and SKOV3/tax cells. As indicated in
Fig. 3E, the migration ability of
SKOV3 cells was significantly increased compared with SKOV3/tax
cells (P<0.05). ERp57-siRNA silencing could significantly reduce
the cell migration of SKOV3 and SKOV3/tax cells (P<0.05).
The effects of ERp57-siRNA silencing on SKOV3/tax
cell apoptosis were examined using Annexin V and PI double
staining. The apoptosis rate of the ERp57-silenced cells was
17.16%, which was significantly higher than the apoptosis rate in
the control (3.61%) and NC-siRNA cells (5.95%; P<0.05; Fig. 3F). Furthermore, in SKOV3/tax cells
treated with paclitaxel, the apoptosis rates of the control and
NC-siRNA cells were 5.67 and 7.63%, whereas the apoptosis rate of
ERp57-siRNA cells was significantly increased to ~38% (mean of the
three measurements; P<0.05). For comparison, the effect of the
ERp57 inhibitor DNTB was examined, and the results indicated that
the apoptosis rate of SKOV3/tax cells was 29.7%.
Cell viability of SKOV3/tax cells under different
conditions was assessed using the MTT assay. Fig. 3G and H revealed the growth curves
and the calculated IC50 values of the different samples.
The IC50 values of SKOV3 and SKOV3/tax cells were
~3.33±1.18 and 102.8±1.17 nM, respectively. The IC50
values of SKOV3/tax cells were reduced to 3.5±1.15 and 0.44±1.3 nM
following treatment with ERp57-siRNA or DTNB, respectively
(Fig. 3H). These data indicated
that ERp57-siRNA silencing could restore the sensitivity of
SKOV3/tax cells to paclitaxel.
The expression levels of the selected protein
biomarkers were assessed in cells treated with ERp57-siRNA and
paclitaxel. As indicated in Fig.
4, when SKOV3/tax cells were treated with ERp57-siRNA alone,
protein P-gp expression was not significantly impacted (columns 1
and 2). However, once paclitaxel was applied, P-gp expression
levels were decreased in a concentration-dependent manner (columns
4 and 6), which suggested that pre-treatment of ERp57-siRNA
effectively inhibited P-gp protein expression and increased
paclitaxel efficacy. The MDR biomarker TUBB3 also exhibited a
similar trend. The expression level of PCNA was reduced and that of
nucleolin was marginally altered in response to paclitaxel (columns
1, 3 and 5). When ERp57-siRNA was applied, PCNA and nucleolin
expression levels were further reduced in the presence of
paclitaxel in a concentration dependent-manner (columns 2, 4 and
6). Furthermore, western blot analysis revealed that co-treatment
of ERp57-siRNA and paclitaxel reduced the p-STAT3 expression levels
in a dose-dependent manner (columns 2, 4 and 6). Additionally,
Bcl-2 and Bcl-xl expression levels were downregulated when
ERp57-siRNA and 100 nM paclitaxel were used (columns 4 and 6),
whereas the apoptosis promoting protein Bax was highly
expressed.
Paclitaxel treatment alone did not impact p53
expression, even at 100 nM, due to the drug-resistant phenotype of
SKOV3/tax cells (columns 1, 3 and 5). In addition, p53 expression
was not affected by ERp57-siRNA alone (column 2). However, when
ERp57-siRNA was combined with 10 nM paclitaxel, the p53 band was
markedly reduced, and its isoform p53/p47 became merged (column 4).
When ERp57-siRNA was combined with 100 nM paclitaxel, the p53 band
was further reduced and the p53/p47 band was highly expressed
(column 6). Notably, MMP1 and MMP2 protein expression levels were
reduced when paclitaxel and/or ERp57-siRNA were applied. However,
MMP9 protein expression was unchanged with ERp57-siRNA treatment
alone (columns 1 and 2) and a minor reduction in expression was
observed with treatment of paclitaxel alone (10 or 100 nM) (columns
1, 3 and 5). When ERp57-siRNA was combined with paclitaxel, MMP9
expression was markedly reduced with 10 nM paclitaxel and
completely eliminated with 100 nM paclitaxel (columns 4 and 6). No
notable changes were indicated with regards to the expression of
the EMT marker vimentin with paclitaxel and/or ERp57-siRNA
treatment; however, a reduction was observed with 100 nM paclitaxel
and ERp57-siRNA treatment (column 6).
Bioinformatics analyses of ERp57 and drug
resistance in ovarian cancer
In order to understand the underlying mechanisms of
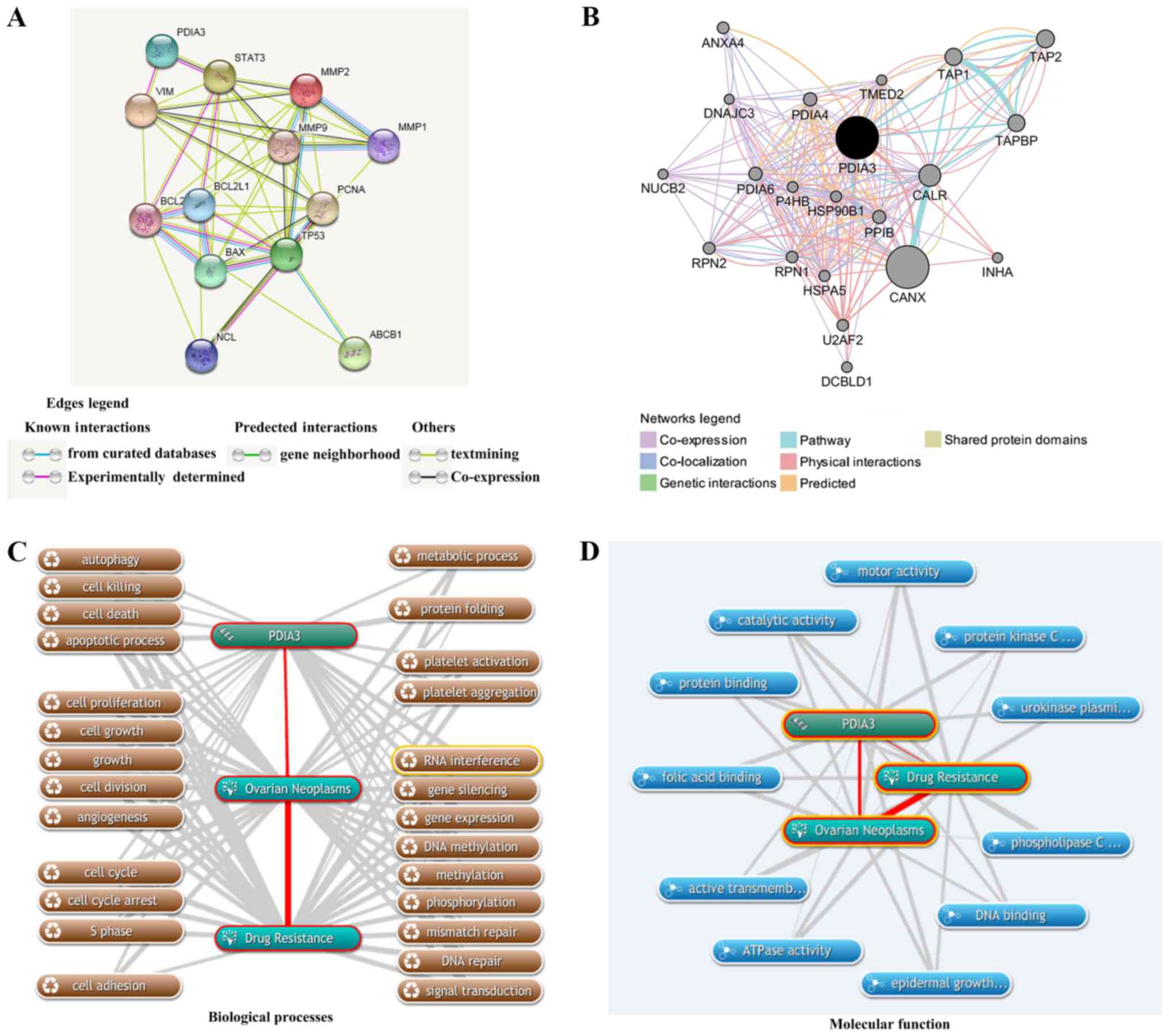
ERp57 in drug-resistant ovarian cancer, STRING was used to
construct a PPI network between ERp57 (PDIA3) and STAT3, P-gp
(ABCB1), TUBB3, p53 (TP53), Bcl-2, Bax, Bcl-xl (BCL2L1), vimentin,
PCNA, nucleolin, MMP1, MMP2 and MMP9. As indicated in Fig. 5A, ERp57 (referred as PDIA3 in
STRING) has direct interactions with STAT3 and vimentin. ERp57 can
have the indirect interactions with p53, Bcl-2, Bax, Bcl-xl, PCNA,
MMP1, MMP2 and MMP9 proteins via STAT3, and indirect interactions
with p53, Bcl-2, PCNA, MMP1, MMP2 and MMP9 via vimentin.
Furthermore, ABCB1 can be regulated indirectly by ERp57 via p53 and
Bax. GeneMANIA was used to construct a gene/protein-gene/protein
interaction network between ERp57 (PDIA3) and various other
components, including calnexin (CANX), calreticulin (CALR),
transporter 1 (TAP1), transporter 2 (TAP2), TAP binding protein
(TAPBP), protein disulfide isomerase family A member 4 (PDIA4),
protein disulfide isomerase family A member 6 (PDIA6) and heat
shock protein 90 kDa beta family member 1 (HSP90B1). As indicated
in Fig. 5B, ERp57 had strong
interactions with 20 proteins/genes. Co-expressed ERp57 had strong
physical interactions and shared a pathway with CANX and CALR.
Furthermore, CANX was associated with ATP transporter-associated
proteins/genes (ABCB1, ABCC1 and ABCC3) and apoptosis-associated
proteins/genes [Bcl-2, poly (ADP-ribose) polymerase 1 (PARP1),
caspase-3 (CASP3), p53]. ABCB1, ABCC1, ABCC3, Bcl-2, PARP1 and
CASP3 expression changes have been associated with drug resistance
(22-26). The findings suggested that
inference of ERp57 must be associated with drug resistance in
ovarian cancer.
The associated biological processes of ERp57 with
ovarian cancer and drug resistance were assessed using COREMINE.
The results indicated that a total of 25 biological processes were
associated with ERp57, ovarian cancer and drug resistance
(P<0.01), including cell death (autophagy, cell killing, cell
death and the apoptotic process), cell growth (cell proliferation,
cell growth, growth, cell division and angiogenesis), the cell
cycle (cell cycle, cell cycle arrest and S phase), gene expression
regulation (RNA interference, gene silencing, gene expression, DNA
methylation, methylation, phosphorylation, mismatch repair, DNA
repair and signal transduction), platelets (platelet activation and
platelet aggregation), cell migration, metabolism and protein
folding-associated processes (Fig.
5C). These results suggested that either ERp57 may be a
regulator of these processes or these processes contribute the
development of drug resistance phenotype of ovarian cancer.
Fig. 5D indicated
that a total of 11 molecular functions, including folic acid
binding, protein binding, catalytic activity, motor activity,
protein kinase C activity, urokinase plasminogen activator receptor
activity, phospholipase C activity, DNA binding, epidermal growth
factor binding, ATPase activity and active transmembrane
transporter activity were predicted to be associated with ERp57,
ovarian cancer and drug resistance (P<0.05). Furthermore,
pathway enrichment analysis was performed using DAVID, which
revealed a total of 31 genes (finding by Coremine Medical)
co-occurred with ERp57 and drug resistance in ovarian cancer
(Table I). Many familiar genes
were indicated, including BCL2, PARP1, STAT3, CASP3, CASP9,
vimentin, phosphatase and tensin homolog, Erb-B2 receptor tyrosine
kinase 2, heat shock protein 90 β1 and epidermal growth factor
receptor. As indicated in Table I,
ERp57 may be associated with drug resistance in ovarian cancer
through its regulation on the four pathways: The cell
death-associated pathways (apoptosis and p53 signaling pathway),
the focal adhesion signaling pathway and the cancer related
pathway.
 | Table IPathway enrichment analysis of the 31
genes which co-occurred with ERp57, drug resistance and ovarian
cancer, in accordance with DAVID. |
Table I
Pathway enrichment analysis of the 31
genes which co-occurred with ERp57, drug resistance and ovarian
cancer, in accordance with DAVID.
| Kyoto Encyclopedia
of Genes and Genomes pathway | P-value
(<0.01) | Benjamini
(<0.05) | Genes co-occurring
with ERp57, drug resistance and ovarian cancer |
|---|
| Pathway in
cancer |
6.80×10−9 |
2.00×10−7 | BCL2, BCL2L1,
CASP3, CASP9, EGFR, GSTP1, HSP90AA1, PTEN, STAT3, P53, ERBB2 |
| Apoptosis |
3.10×10−4 |
2.60×10−3 | BCL2, BCL2L1,
CASP3, CASP9, TP53 |
| p53 signaling
pathway |
2.20×10−3 |
1.40×10−2 | CASP3, CASP9, PTEN,
TP53 |
| Focal adhesion |
7.00×10−3 |
3.70×10−2 | BCL2, COL11A2,
EGFR, PTEN, ERBB2 |
Discussion
ERp57, also referred to as PDIA3, ER60, ERp60,
GRp58, Q2 and 1,25D3-MARRS receptor, is a widely expressed protein
with multiple biological functions (27). Being a member of the disulfide
isomerase family, ERp57 has been studied extensively as an
endoplasmic reticulum (ER) chaperone protein participating in the
proper folding and reshuffling of disulfide bridges of newly
synthesized proteins in ER, as well as in the assembly of major
histocompatibility complex class-I molecules (28-31).
ERp57 is considered a stress-response protein, and
its overexpression has been confirmed in various types of cancer,
including breast, uterus, lung, stomach, cervical, head and neck
cancer and laryngeal cancer (32,33).
Anti-cancer agents, particularly stress-inducing agents, can induce
ERp57 upregulation, therefore providing a protective role against
apoptosis in cancer cells under increased ER stress (34,35).
Thus, the ERp57 overexpression in chemoresistant cancer cells at
mRNA and protein levels was not unanticipated (11,12).
Choe et al (36)
demonstrated that ERp57 was upregulated in radioresistant laryngeal
cancer cells. Unfortunately, little information was available
regarding the biological effects of ERp57 on the chemoresistance
phenotype. Therefore, the present study was designed to investigate
the ERp57 roles in paclitaxel-resistant SKOV3/tax cells and the
possibility of resensitizing SKOV3/tax cells to paclitaxel by ERp57
downregulation.
The present results indicated that the expression
Bcl-2 and Bcl-xl were overexpressed and Bax expression was
significantly reduced in SKOV3/tax cells, suggesting the expression
of apoptosis-associated proteins supported the drug resistance
cellular phenotype. The apoptosis indicator, tumor repressor
protein p53, was also increased in SKOV3/tax cells. Furthermore,
the active form of STAT3, p-STAT3, was highly expressed in
SKOV3/tax cells. These results implied that the
paclitaxel-resistance of SKOV3/tax may be due to an
apoptosis-associated mechanism and the activation of STAT3.
Furthermore, the results suggested that paclitaxel-resistance was
partly associated with an effluxing mechanism involving P-gp and
the de-polymerization of TUBB3.
The present results suggested that ERp57-siRNA
silencing improved the sensitivity of SKOV3/tax cells to
paclitaxel. As confirmation, the expression levels of selected
protein markers associated with the cellular behavior of SKOV3/tax
cells were assessed. Following ERp57-siRNA silencing, P-gp and
TUBB3 were expression levels were reduced in the presence of 10 nM
paclitaxel and completely inhibited in the presence of 100 nM
paclitaxel, suggesting that ERp57-siRNA silencing could restore the
sensitivity of SKOV3/tax to paclitaxel.
With ERp57-siRNA silencing, 10 nM paclitaxel reduced
p53 expression by ~50% and the expression of its isoform p53/p47
was increased. At 100 nM paclitaxel, p53 expression was completely
eliminated and the isoform p53/p47 became further increased. These
results were consistent with a previous report (37). Transcriptionally active p53
tetramers bind to promoter regions and regulate gene products,
which prevents cancer development (37). ER stress promotes protein kinase
R-like ER kinase (PERK)-dependent induction of p53/p47 isoform
(37). Furthermore, P53/p47
induces G2 arrest but has no effect on G1
progression. It was reported that cells appear to favor
G2 arrest in response to ER-stress like paclitaxel
treatment (37). A previous study
indicated that the p53/p47 isoform was increased and H1299 and
MLS1765 cells were arrested in G2 phase with the
increase of thapsigargin dosage (38). Additionally, apoptosis-inhibiting
Bcl-2 and Bcl-xl proteins were markedly reduced with ERp57-siRNA
silencing in the present study. By contrast, the
apoptosis-promoting protein Bax was upregulated. These results were
consistent with the results that co-treatment of ERp57-siRNA and
paclitaxel could increase the apoptosis rate of SKOV3/tax
cells.
Tumorigenic STAT3 activation has been frequently
linked to malignant cancer behavior, including growth, migration,
invasion and metastasis (39).
Previous studies have revealed an association between ERp57 and
STAT3 (40,41). In M14 melanoma cells, chromatin
immunoprecipitation revealed that ERp57 binds to DNA in the
proximity of STAT3 in a subset of STAT3-regulated genes. Upon
depletion of ERp57, the quantity of p-STAT3 was reduced (41). It has been also reported that ERp57
and STAT3 are associated to α2-marroglobulin gene enhancer, when
stimulated by interleukin-6, these two proteins are bound to the
sis-inducible element sequence in HepG2 cells (42). Accumulated evidence has indicated
that STAT3 activation was also associated with tumor
chemoresistance. Gu et al (43) identified a correlation between
enhanced STAT3 expression and cisplatin-resistance in patients with
cancer, and blocking the Janus-kinase STAT3 signaling pathway could
restore cisplatin sensitivity (43). Notably, it has been demonstrated
that activating transcription factor 4 promotes the MDR phenotype
in esophageal squamous-cell carcinoma (ESCC) cells by binding
directly to the STAT3 promoter. However, inhibition of STAT3 could
reintroduce therapeutic sensitivity (44). Ryu et al (45) reported that treatment with CDDO-Me
significantly decreased the level of nuclear translocation and
phosphorylation of STAT3. A previous study revealed that the
inhibition of the STAT3 signaling pathway correlated with the
suppression of the anti-apoptotic genes Bcl-xl, survivin and MCL-1
(45). The correlation between the
STAT3 activation and the chemoresistance of cancer cells has been
documented previously (46-48).
In the present study, the high expression of the activated STAT3
and the chemoresistance of SKOV3/tax cells was confirmed, and the
downregulation of p-STAT3 by ERp57-siRNA silencing was associated
with the chemoresistance reversal of SKOV3/tax cells.
EMT is associated with drug resistance. In some
cases MMPs are overexpressed (high invading) in drug-resistant
cancer cells (49,50), and in other cases MMPs are
underexpressed (low invading) in drug-resistant cancer cells
(51,52). These findings suggest that the
association of MMPs and drug resistance varies among different
samples. Notably, the expression levels of EMT-associated proteins
MMP1, MMP2, MMP9 and vimentin were lower in SKOV3/tax cells than
that in SKOV3 cells, suggesting that SKOV3/tax cells exhibited less
metastasis than SKOV3 cells. In the present study, ERp57 was highly
expressed in less metastatic cells (SKOV3/tax), which was not in
agreement with Naiara’s observation that overexpression ERp57 was
related to bone metastasis in breast cancer cell (53).
Although the role of ERp57 as a cell protective
agent against apoptosis has been accepted, some controversial
evidence has also emerged. Xu et al (54) suggested that ERp57-siRNA could
significantly reduce hyperoxia- or tunicamycin-induced apoptosis in
human endothelial cells by the inhibition of caspase-3 activation
and stimulation of binding immunoglobulin protein/glucose-regulated
protein 78 induction (54). It was
also reported that ERp57 possesses Bcl-2 homologous
antagonist/killer-dependent proapoptotic function through inducing
mitochondrial outer membrane permeabilization (55). These discrepancies are likely due
to the differences in cellular context and tumor types as well as
upstream regulators, parallel transcription co-regulators and
downstream target genes of ERp57. Hence, these findings signify the
pivotal role of ERp57 in the coordination of complex regulatory
systems.
To further illustrate the potential association of
ERp57 with drug resistance in ovarian cancer, comprehensive
bioinformatics analyses were performed in the present study. A
network of ERp57 and other proteins was constructed. ERp57 was
predicted to directly regulate STAT3 and vimentin, and other
proteins [P-gp, p53, Bcl-2, Bax, Bcl-xl (BCL2L1), nucleolin, PCNA,
MMP1, MMP2 and MMP9] were linked with ERp57 indirectly. These
predictions were partially consistent with the present experimental
results that ERp57-siRNA silencing could directly decrease the
expression of p-STAT3; however, the results indicated that ERp57
siRNA silencing could not affect the expression of vimentin
directly in the present study. Protein/gene interaction analysis
revealed a total 20 proteins/genes interactions with ERp57, 7 of
which (CANX, TAP1, TAP2, PDIA4, PDIA6, HSP90B1 and ANXA4) were
associated with drug resistance. The biological process annotation
indicated that 25 biological processes, 11 molecular functions, 3
pathways and 36 genes co-occurred with ERp57, ovarian cancer and
drug resistance.
In conclusion, the present study produced a model to
interpret the biological role of ERp57 in paclitaxel-resistant
SKOV3/tax cells and the paclitaxel sensitivity reversal of
SKOV3/tax by siRNA silencing (Fig.
6). The findings suggested that long-term or high-dosage
paclitaxel treatment of SKOV3 ovarian cancer cells leads to high
ERp57 expression. As a result, the STAT3 signaling pathway was
activated, which promotes cell survival by evading the apoptosis
process. However, inhibition of ERp57 expression inhibited the
STAT3 signaling pathway, which caused the SKOV3/tax cells to regain
paclitaxel sensitivity. The findings of the present study provide a
novel potential strategy to overcome the chemoresistance challenge
in the clinical treatment of ovarian cancer in patients.
Funding
The present study was funded by the National Natural
Science Foundation of China (grant no. 31670821).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors’ contributions
YG and SL conceived and designed the experiments. SL
performed the majority of the experiments, XZ, SC, YL and MG
performed some of the experiments. SL and YG wrote the manuscript.
All authors reviewed the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Abbreviations:
|
si
|
small interfering
|
|
STAT3
|
signal transducer and activator of
transcription 3
|
|
p-
|
phospho
|
|
PCNA
|
proliferating cell nuclear antigen
|
|
P-gp
|
P-glycoprotein
|
|
TUBB3
|
class III β-tubulin
|
|
Bcl-2
|
B-cell lymphoma-2
|
|
Bax
|
Bcl-2-associated X protein
|
|
Bcl-xl
|
B-cell lymphoma-extra large
|
|
MMP
|
matrix metalloproteinase
|
Acknowledgments
Not applicable.
References
|
1
|
Gloss BS and Samimi G: Epigenetic
biomarkers in epithelial ovarian cancer. Cancer Lett. 342:257–263.
2014. View Article : Google Scholar
|
|
2
|
Sundar S, Wu J, Hillaby K, Yap J and
Lilford R: A systematic review evaluating the relationship between
progression free survival and post progression survival in advanced
ovarian cancer. Gynecol Oncol. 125:493–499. 2012. View Article : Google Scholar
|
|
3
|
Ziebarth AJ, Landen CN Jr and Alvarez RD:
Molecular/genetic therapies in ovarian cancer: Future opportunities
and challenges. Clin Obstet Gynecol. 55:156–172. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kipps E, Tan DS and Kaye SB: Meeting the
challenge of ascites in ovarian cancer: New avenues for therapy and
research. Nat Rev Cancer. 13:273–282. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ford JM, Bruggemann EP, Pastan I,
Gottesman MM and Hait WN: Cellular and biochemical characterization
of thioxanthenes for reversal of multidrug resistance in human and
murine cell lines. Cancer Res. 50:1748–1756. 1990.PubMed/NCBI
|
|
6
|
Ranganathan S, Benetatos CA, Colarusso PJ,
Dexter DW and Hudes GR: Altered beta-tubulin isotype expression in
paclitaxel-resistant human prostate carcinoma cells. Br J Cancer.
77:562–566. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hari M, Loganzo F, Annable T, Tan X, Musto
S, Morilla DB, Nettles JH, Snyder JP and Greenberger LM:
Paclitaxel-resistant cells have a mutation in the
paclitaxel-binding region of beta-tubulin (Asp26Glu) and less
stable microtubules. Mol Cancer Ther. 5:270–278. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Haass NK, Beaumont KA, Hill DS, Anfosso A,
Mrass P, Munoz MA, Kinjyo I and Weninger W: Real-time cell cycle
imaging during melanoma growth, invasion, and drug response.
Pigment Cell Melanoma Res. 27:764–776. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kapse-Mistry S, Govender T, Srivastava R
and Yergeri M: Nanodrug delivery in reversing multidrug resistance
in cancer cells. Front Pharmacol. 5:1592014.PubMed/NCBI
|
|
10
|
Bordelon JR and Grichnik JM: TGF-β may
control the switch between tumorigenic growth and ‘stem
cell/mesenchymal’ potentially drug-resistant states. Dermatol Ther
(Heidelb). 28:177–178. 2015. View Article : Google Scholar
|
|
11
|
Bernardini M, Lee CH, Beheshti B, Prasad
M, Albert M, Marrano P, Begley H, Shaw P, Covens A, Murphy J, et
al: High-resolution mapping of genomic imbalance and identification
of gene expression profiles associated with differential
chemotherapy response in serous epithelial ovarian cancer.
Neoplasia. 7:603–613. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cicchillitti L, Di Michele M, Urbani A,
Ferlini C, Donat MB, Scambia G and Rotilio D: Comparative proteomic
analysis of paclitaxel sensitive A2780 epithelial ovarian cancer
cell line and its resistant counterpart A2780TC1 by 2D-DIGE: The
role of ERp57. J Proteome Res. 8:1902–1912. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Duan Z, Feller AJ, Penson RT, Chabner BA
and Seiden MV: Discovery of differentially expressed genes
associated with paclitaxel resistance using cDNA array technology:
Analysis of interleukin (IL) 6, IL-8, and monocyte chemotactic
protein 1 in the paclitaxel-resistant phenotype. Clin Cancer Res.
5:3445–3453. 1999.PubMed/NCBI
|
|
14
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
15
|
Szklarczyk D, Morris JH, Cook H, Kuhn M,
Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, et al:
The STRING database in 2017: Quality-controlled protein-protein
association networks, made broadly accessible. Nucleic Acids Res.
45D:D362–D368. 2017. View Article : Google Scholar
|
|
16
|
Zuberi K, Franz M, Rodriguez H, Montojo J,
Lopes CT, Bader GD and Morris Q: GeneMANIA prediction server 2013
update. Nucleic Acids Res. 41W:W115–W22. 2013. View Article : Google Scholar
|
|
17
|
de Leeuw N, Dijkhuizen T, Hehir-Kwa JY,
Carter NP, Feuk L, Firth HV, Kuhn RM, Ledbetter DH, Martin CL, van
Ravenswaaij-Arts CM, et al: Diagnostic interpretation of array data
using public databases and internet sources. Hum Mutat. 33:930–940.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Huang W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar
|
|
19
|
Vikhanskaya F, Erba E, D’Incalci M and
Broggini M: Introduction of wild-type p53 in a human ovarian cancer
cell line not expressing endogenous p53. Nucleic Acids Res.
22:1012–1017. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Alaseem A, Alhazzani K, Dondapati P,
Alobid S, Bishayee A and Rathinavelu A: Matrix Metalloproteinases:
A challenging paradigm of cancer management. Semin Cancer Biol. Nov
16–2017.Epub ahead of print. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Cicchillitti L, Della Corte A, Di Michele
M, Donati MB, Rotilio D and Scambia G: Characterisation of a
multimeric protein complex associated with ERp57 within the nucleus
in paclitaxel-sensitive and -resistant epithelial ovarian cancer
cells: The involvement of specific conformational states of
beta-actin. Int J Oncol. 37:445–454. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Haufroid V: Genetic polymorphisms of
ATP-binding cassette transporters ABCB1 and ABCC2 and their impact
on drug disposition. Curr Drug Targets. 12:631–646. 2011.
View Article : Google Scholar
|
|
23
|
Kwak JO, Lee SH, Lee GS, Kim MS, Ahn YG,
Lee JH, Kim SW, Kim KH and Lee MG: Selective inhibition of MDR1
(ABCB1) by HM30181 increases oral bioavailability and therapeutic
efficacy of paclitaxel. Eur J Pharmacol. 627:92–98. 2010.
View Article : Google Scholar
|
|
24
|
Calastretti A, Gatti G, Quaresmini C and
Bevilacqua A: Down-modulation of Bcl-2 sensitizes PTEN-mutated
prostate cancer cells to starvation and taxanes. Prostate.
74:1411–1422. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Luo Y, Tong L, Meng H, Zhu W, Guo L, Wei T
and Zhang J: MiR-335 regulates the chemo-radioresistance of small
cell lung cancer cells by targeting PARP-1. Gene. 600:9–15. 2017.
View Article : Google Scholar
|
|
26
|
Friedrich K, Wieder T, Von Haefen C,
Radetzki S, Jänicke R, Schulze-Osthoff K, Dörken B and Daniel PT:
Overexpression of caspase-3 restores sensitivity for drug-induced
apoptosis in breast cancer cell lines with acquired drug
resistance. Oncogene. 20:2749–2760. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Turano C, Gaucci E, Grillo C and
Chichiarelli S: ERp57/GRP58: A protein with multiple functions.
Cell Mol Biol Lett. 16:539–563. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Khanal RC and Nemere I: The
ERp57/GRp58/1,25D3-MARRS receptor: Multiple functional roles in
diverse cell systems. Curr Med Chem. 14:1087–1093. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Vigneron N, Peaper DR, Leonhardt RM and
Cresswell P: Functional significance of tapasin membrane
association and disulfide linkage to ERp57 in MHC class I
presentation. Eur J Immunol. 39:2371–2376. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chapman DC and Williams DB: ER quality
control in the biogenesis of MHC class I molecules. Semin Cell Dev
Biol. 21:512–519. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Frenkel Z, Shenkman M, Kondratyev M and
Lederkremer GZ: Separate roles and different routing of calnexin
and ERp57 in endoplasmic reticulum quality control revealed by
interactions with asialoglycoprotein receptor chains. Mol Biol
Cell. 15:2133–2142. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Celli CM and Jaiswal AK: Role of GRP58 in
mitomycin C-induced DNA cross-linking. Cancer Res. 63:6016–6025.
2003.PubMed/NCBI
|
|
33
|
He Y, Shao F, Pi W, Shi C, Chen Y, Gong D,
Wang B, Cao Z and Tang K: Largescale transcriptomics analysis
suggests over-expression of BGH3, MMP9 and PDIA3 in oral squamous
vell carcinoma. PLoS One. 11:e01465302016. View Article : Google Scholar
|
|
34
|
Lovat PE, Corazzari M, Armstrong JL,
Martin S, Pagliarini V, Hill D, Brown AM, Piacentini M,
Birch-Machin MA and Redfern CP: Increasing melanoma cell death
using inhibitors of protein disulfide isomerases to abrogate
survival responses to endoplasmic reticulum stress. Cancer Res.
68:5363–5369. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Corazzari M, Lovat PE, Armstrong JL, Fimia
GM, Hill DS, Birch-Machin M, Redfern CP and Piacentini M: Targeting
homeostatic mechanisms of endoplasmic reticulum stress to increase
susceptibility of cancer cells to fenretinide-induced apoptosis:
The role of stress proteins ERdj5 and ERp57. Br J Cancer.
96:1062–1071. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Choe MH, Min JW, Jeon HB, Cho DH, Oh JS,
Lee HG, Hwang SG, An S, Han YH and Kim JS: ERp57 modulates STAT3
activity in radioresistant laryngeal cancer cells and serves as a
prognostic marker for laryngeal cancer. Oncotarget. 6:2654–2666.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Bourougaa K, Naski N, Boularan C,
Mlynarczyk C, Candeias MM, Marullo S and Fåhraeus R: Endoplasmic
reticulum stress induces G2 cell-cycle arrest via mRNA translation
of the p53 isoform p53/47. Mol Cell. 38:78–88. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Candeias MM, Powell DJ, Roubalova E,
Apcher S, Bourougaa K, Vojtesek B, Bruzzoni-Giovanelli H and
Fåhraeus R: Expression of p53 and p53/47 are controlled by
alternative mechanisms of messenger RNA translation initiation.
Oncogene. 25:6936–6947. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Carpenter RL and Lo HW: STAT3 target genes
relevant to human cancers. Cancers (Basel). 6:897–925. 2014.
View Article : Google Scholar
|
|
40
|
Guo GG, Patel K, Kumar V, Shah M, Fried
VA, Etlinger JD and Sehgal PB: Association of the chaperone
glucose-regulated protein 58 (GRP58/ER-60/ERp57) with Stat3 in
cytosol and plasma membrane complexes. J Interferon Cytokine Res.
22:555–563. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Chichiarelli S, Gaucci E, Ferraro A,
Grillo C, Altieri F, Cocchiola R, Arcangeli V, Turano C and Eufemi
M: Role of ERp57 in the signaling and transcriptional activity of
STAT3 in a melanoma cell line. Arch Biochem Biophys. 494:178–183.
2010. View Article : Google Scholar
|
|
42
|
Eufemi M, Coppari S, Altieri F, Grillo C,
Ferraro A and Turano C: ERp57 is present in STAT3-DNA complexes.
Biochem Biophys Res Commun. 323:1306–1312. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Gu F, Ma Y, Zhang Z, Zhao J, Kobayashi H,
Zhang L and Fu L: Expression of Stat3 and Notch1 is associated with
cisplatin resistance in head and neck squamous cell carcinoma.
Oncol Rep. 23:671–676. 2010.PubMed/NCBI
|
|
44
|
Zhu H, Chen X, Chen B, Chen B, Fan J, Song
W, Xie Z, Jiang D, Li Q, Zhou M, et al: Activating transcription
factor 4 mediates a multidrug resistance phenotype of esophageal
squamous cell carcinoma cells through transactivation of STAT3
expression. Cancer Lett. 354:142–152. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Ryu K, Susa M, Choy E, Yang C, Hornicek
FJ, Mankin HJ and Duan Z: Oleanane triterpenoid CDDO-Me induces
apoptosis in multidrug resistant osteosarcoma cells through
inhibition of Stat3 pathway. BMC Cancer. 10:1872010. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Gariboldi MB, Ravizza R, Molteni R, Osella
D, Gabano E and Monti E: Inhibition of Stat3 increases doxorubicin
sensitivity in a human metastatic breast cancer cell line. Cancer
Lett. 258:181–188. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Selvendiran K, Bratasz A, Kuppusamy ML,
Tazi MF, Rivera BK and Kuppusamy P: Hypoxia induces chemoresistance
in ovarian cancer cells by activation of signal transducer and
activator of transcription 3. Int J Cancer. 125:2198–2204. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Zhang F, Wang Z, Fan Y, Xu Q, Ji W, Tian R
and Niu R: Elevated STAT3 signaling-mediated upregulation of
MMP-2/9 confers enhanced invasion ability in multidrug-resistant
breast cancer cells. Int J Mol Sci. 16:24772–24790. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Yang JM, Xu Z, Wu H, Zhu H, Wu X and Hait
WN: Overexpression of extracellular matrix metalloproteinase
inducer in multidrug resistant cancer cells. Mol Cancer Res.
1:420–427. 2003.PubMed/NCBI
|
|
50
|
Colone M, Calcabrini A, Toccacieli L,
Bozzuto G, Stringaro A, Gentile M, Cianfriglia M, Ciervo A,
Caraglia M, Budillon A, et al: The multidrug transporter
P-glycoprotein: A mediator of melanoma invasion? J Invest Dermatol.
128:957–971. 2008. View Article : Google Scholar
|
|
51
|
Hikawa T, Mori T, Abe T and Hori S: The
ability in adhesion and invasion of drug-resistant human glioma
cells. Journal of experimental and clinical cancer research CR
(East Lansing Mich). 19:357–362. 2000.
|
|
52
|
Işeri OD, Kars MD, Arpaci F and Gündüz U:
Gene expression analysis of drug-resistant MCF-7 cells:
Implications for relation to extracellular matrix proteins. Cancer
Chemother Pharmacol. 65:447–455. 2010. View Article : Google Scholar
|
|
53
|
Santana-Codina N, Carretero R,
Sanz-Pamplona R, Cabrera T, Guney E, Oliva B, Clezardin P, Olarte
OE, Loza-Alvarez P, Méndez-Lucas A, et al: A transcriptome-proteome
integrated network identifies endoplasmic reticulum thiol
oxidoreductase (ERp57) as a hub that mediates bone metastasis. Mol
Cell Proteomics. 12:2111–2125. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Xu D, Perez RE, Rezaiekhaligh MH, Bourdi M
and Truog WE: Knockdown of ERp57 increases BiP/GRP78 induction and
protects against hyperoxia and tunicamycin-induced apoptosis. Am J
Physiol Lung Cell Mol Physiol. 297:L44–L51. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Zhao G, Lu H and Li C: Proapoptotic
activities of protein disulfide isomerase (PDI) and PDIA3 protein,
a role of the Bcl-2 protein Bak. J Biol Chem. 290:8949–8963. 2015.
View Article : Google Scholar : PubMed/NCBI
|