Introduction
According to the 2021 global cancer statistics,
colorectal cancer (CRC) is a common gastrointestinal malignancy
with the third highest incidence and the second highest mortality
(1,2). The failure of early diagnosis and the
lack of effective therapeutic targets contribute to the poor
prognosis of CRC (3,4). Therefore, finding new targets for the
diagnosis and therapy of CRC is necessary and urgent.
The long non-coding RNA (lncRNA), a group of novel
regulatory RNA, is a non-coding transcript >200 nucleotides in
length (5). It has been shown that
lncRNA is aberrantly expressed in numerous tumors and plays
critical roles in various tumor processes (6–8),
including tumor cell death (9),
metabolic remodeling (10),
migration and infiltration (11),
reflecting their important roles in carcinogenesis. For instance,
lncRNA GAS5 is downregulated in colon cancer and restrains the
proliferation of colon cancer by inhibiting the activation of the
Wnt/β-catenin signaling pathway (12); lncRNA NALT1 is upregulated and
induces invasion and metastasis of gastric cancer through the NOTCH
signaling pathway (10); lncRNA
MALAT1 regulates proliferation of SKOV3 cells and is overexpressed
in ovarian cancer tissues (9).
Accordingly, lncRNAs are considered novel diagnostic and
therapeutic targets for anticancer research.
Metabolic reprogramming, particularly aerobic
glycolysis, is one of the malignant features of tumors (13). Cancer cells prefer to produce
energy by glycolysis rather than via oxidative phosphorylation,
even in the presence of oxygen. This phenomenon is defined as the
‘Warburg effect’, also known as aerobic glycolysis (14,15).
Recently, an increasing number of studies suggested that lncRNAs
may play a role in tumorigenesis by reprogramming glucose
metabolism (16–18). Upregulation of lncRNA PVT1
expression in tumors has been reported to promote aerobic
glycolysis in gallbladder cancer cells by competitively binding to
endogenous miR-143 to regulate HK2 expression (19). LncRNA MACC1-AS1 promotes the
stabilization of MACC1 mRNA, which in turn activates the expression
of its downstream target genes Glut1, HK2 and LDHA, promotes
aerobic glycolysis in tumor cells and leads to malignant growth of
gastric cancer cells (20).
In a previous study by the authors, a novel lncRNA
495810 (named according to its ENST ID: ENST00000495810) expressed
in CRC cells was discovered (21).
However, its clinical significance and molecular mechanism in CRC
have not been reported. In the current study, data demonstrated
that lncRNA 495810 is significantly upregulated in colon cancer and
is associated with poor prognosis in patients with CRC. LncRNA
495810 functions as an oncogene and promotes aerobic glycolysis in
colon cancer. Mechanistically, lncRNA 495810 positively correlates
with pyruvate kinase isozyme M2 (PKM2) and regulates its
proteasomal degradation to promote the level of aerobic glycolysis
in CRC cells.
Materials and methods
Chemicals
DMEM/F12 (HAM) 1:1 medium and fetal bovine serum
(FBS) were from Biological Industries;
3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2-H-tetrazolium bromide
(MTT), dimethyl sulfoxide (DMSO) and cycloheximide (CHX) were
obtained from Beijing Solarbio Science & Technology Co., Ltd.
The RNAiso Plus was purchased from Takara Bio, Inc. All-in-one
First Strand cDNA Synthesis Kit II and 2X SYBR Green qPCR Master
Mix were from Sevenbio (http://www.7bio.com.cn/index.php). Turbofect was
obtained from Thermo Fisher Scientific, Inc. Annexin V-FITC
apoptosis detection kit was purchased from Nanjing KeyGen Biotech
Co., Ltd. Antibodies for PKM2 (cat. no. 15822-1-AP) and
glyceraldehyde-3-phosphate dehydrogenase (GAPDH; cat. no.
10494-1-AP) were obtained from Proteintech Group, Inc. ATP assay
kit (cat. no. S0026B) was obtained from Beyotime Institute of
Biotechnology. Glucose assay kit (cat. no. F006-1-1) and pyruvate
acid test kit (cat. no. A081-1-1) were purchased from Nanjing
Jiancheng Bioenigeering Institute. HiPerFect Transfection Reagent
were purchased from Qiagen GmbH. MG132 was obtained from
MedChemExpress.
Cell lines and cell culture
The CRC cell lines HT-29, DLD1, HCT116, SW620, SW480
and human colon epithelial cell line (FHC) were preserved in the
authors' laboratory.. The cells were cultured in DMEM/F12 medium
containing 10% FBS and placed in an incubator at 37°C with 5%
CO2.
Reverse transcription-quantitative PCR
(RT-qPCR) assay
Total RNA was extracted from cells with
TRIzol® reagent (Takara Bio, Inc.) and 500 ng of RNA was
reverse-transcribed. The reverse transcription was performed
according to the manufacturer's instructions. LncRNA and mRNA
levels were quantified using RT-qPCR. The PCR reaction in the
following conditions: one cycle at 94°C for 30 sec, followed by 40
cycles at 94°C for 5 sec, 60°C for 15 sec and 72°C for 10 sec. The
quantitative analysis was performed using the 2−ΔΔCq
method (22). GAPDH was used as a
reference gene. All primers used in the experiment are shown in
Table SI.
Bioinformatics
Kaplan-Meier survival analysis for CRC patients with
high or low gene expression level was performed through Gene
Expression Profiling Interactive Analysis 2 (GEPIA 2; http://gepia2.cancer-pku.cn/#index). The coding
potential of lncRNA was estimated by the coding-potential
assessment tool (CPAT; http://lilab.research.bcm.edu/). LncRNA PhyloCSF value
analysis was performed by the UCSC genome browser (http://genome.ucsc.edu/) with the PhyloCSF data hub.
The metascape database (www.metascape.org) was used for gene enrichment
analysis and the protein-protein interaction (PPI) network. The
expression data of PKM2 and the correlation analysis of two genes
in CRC tissues were performed through the GEPIA 2 (http://gepia2.cancer-pku.cn/#correlation). The
interaction between lncRNA 495810 and PKM2 was predicted via
RNA-Protein Interaction Prediction (RPISeq) (http://pridb.gdcb.iastate.edu/RPISeq/index.html).
A potential PKM2-binding region of lncRNA 495810 (401–452) was
predicted using catRAPI omics (http://s.tartaglialab.com/page/catrapid_omics_group).
Nuclear and cytoplasmic RNA
fractionation analysis
The collected cells were lysed with hypotonic buffer
(pH 7.4 25 mM Tris-HCl, 1 mM MgCl2, 5 mM KCl and 1%
NP-40), centrifuged (4°C, 5,000 g, 5 min), and the supernatant was
received to obtain cytosolic fractions. Then, the aforementioned
pellet was lysed with nuclear resuspension buffer (pH 7.9 20 mM
HEPES, 400 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1 mM DTT and 1 mM PMSF)
and centrifuged (4°C, 13,000 g, 10 min) to obtain nuclear
fractions. Nuclear and cytoplasmic RNA were extracted with TRIzol
reagent. Finally, the expression levels of GAPDH, U6, and lncRNA
495810 in the nuclei and cytoplasm of CRC cells were detected using
RT-qPCR assays.
siRNA and plasmid transfection
To assay siRNA transfection, the cells were
inoculated into a six-well plate, and 10 nM siRNA was transfected
with HiPerFect Transfection Reagent for 48 h according to the
manufacturer's protocol. For the overexpression assay, lncRNA
495810 overexpression plasmid and the control plasmid (pCDH; both
from Miiaoling Biotech Science) were transfected into colon cancer
cells using Turbofect reagent according to the manufacturer's
instructions. After transfection for 48 h, the cells were collected
for subsequent analysis or assay. The siRNA sequences (Shanghai
GenePharma Co., Ltd) were as follows: lncRNA 495810-homo-112
(sense, 5′-CCAUACCAUCAAUGGUCAUTT-3′ and antisense,
5′-AUGACCAUUGAUGGUAUGGTT-3′).
Cell viability assay
The HT-29, SW620 and HCT116 cells
(~5×103) were seeded into 96-well plates and were
incubated overnight to adhere to the wall. Cells were transfected
with si-495810 or 495810 for 48 h. Then 10 µl of MTT (0.5 mg/ml)
was added to each well for 4 h. After the formazan crystals formed,
they were dissolved in DMSO and the OD value at 570 nm was
evaluated. The cell survival rate was calculated with the following
formula: Cell survival rate (%)=(OD570 treated/OD570 control)
×100.
Cell apoptosis analysis
Cells were resuspended with 500 µl binding buffer at
the density of 1×10 5 cells/ml, and then 5 µl connexin
V-FITC and 5 µl propidium iodide were added. The cells were
incubated at room temperature in the dark for 15 min. After that,
the samples were detected and analyzed by flow cytometry (CytoFLEX;
Beckman Coulter, Inc.).
Transwell assays
The SW620 and HCT116 cells (~2×104) were
seeded into 24-well Transwell plates (cat. no. 3422; Corning, Inc.)
and 200 µl serum-free DMEM medium (Gibco; Thermo Fisher Scientific,
Inc.) in the upper compartment and 660 µl DMEM medium supplemented
with 15% FBS (Gibco; Thermo Fisher Scientific, Inc.) was added to
the lower compartment. The plate was incubated at 37°C for 48 h.
The migrated cells on the lower surface were detected.
Metabolic assays
Cells were seeded into the six-well plate, and
medium and cells were collected after transfection with si-495810
or 495810 for 48 h. The medium was used to measure the contents of
glucose and pyruvate acid, and the production of ATP was determined
after cell lysis. The methods of measuring glucose, ATP and
pyruvate acid were according to the manufacturer's instructions.
Briefly, the medium was added to the reaction solution in the
glucose and pyruvate acid kit, respectively, and its OD was
measured at 505 nm to calculate its content. The cell lysate was
added to the ATP assay solution, its fluorescence intensity was
measured, and its content was calculated.
In vitro transcription and RNA
pull-down
The templates for in vitro transcription were
obtained from PCR. The primers containing the T7 promoter of 495810
are listed in Table SI. In
vitro transcription was conducted using a TranscriptAid T7 High
Yield Transcription kit (Thermo Fisher Scientific, Inc.) following
the manufacturer's protocol and the target RNA was labeled with
biotin using Biotin RNA Labeling Mix 10X.
Cell lysates were prepared using IP lysis buffer.
Next, the labeled RNA was incubated with streptavidin magnetic
beads for 1 h at room temperature with agitation. Then, the
bead-RNA complex and RNA-protein binding reaction master were
incubated for 2 h at 4°C with rotation. Finally, the eluted
interacting proteins were obtained.
Mass spectrometry
Mass spectrometry was performed at Bioprofile. Mass
spectrometry experiments were performed on a Q Exactive HF-X mass
spectrometer that was coupled to Easy nLC (Thermo Fisher
Scientific, Inc.). The full MS scans were acquired at a resolution
of 60,000 at m/z 200, and 15,000 at m/z 200 for MS/MS scan. The
maximum injection time was set to for 50 ms for MS and 50 ms for
MS/MS. Normalized collision energy was 28 and the isolation window
was set to 1.6 Th. Dynamic exclusion duration was 60 sec.
Western blot assay
Western blotting was performed as previously
described (21). Total protein was
extracted by WBIP (Solarbio Science & Technology Co., Ltd.) and
its concentration was determined by BCA assay. The same amount (60
µg) of protein was separated by 10% SDS-PAGE and transferred to the
PVDF membrane. The membrane was blocked for 40 min at room
temperature with 5% skim milk and incubated with primary antibodies
against anti-PKM2 or GAPDH overnight at 4°C. The corresponding
secondary antibodies (anti-Rabbit, ZB-2306/anti-Mouse, ZB-2305)
were incubated for 2 h at room temperature the following day. The
expression changes of target proteins were observed. Relative
protein levels were analyzed using ImageJ software (National
Institutes of Health). Expression of GAPDH was used as the internal
control.
CHX chase assay
To observe the protein degradation process, HT-29,
HCT116 and SW620 cells (~1×105) were treated with CHX
(20 µg/ml) for 0, 2, 4, 6 and 8 h, respectively. Total protein was
extracted at indicated time points and analyzed by western
blotting.
Statistical analysis
The data are presented as the mean ± SEM, data error
bar indicating standard deviation. Single-variable comparisons were
performed by independent samples t-test. Continuous variables (N≥3
groups) were analyzed by Single factor analysis of variance
(ANOVA), followed by Tukey's post-hoc test. Cox proportional
hazards regression models were applied to determine the independent
factors that influence survival. Experimental data were executed
using GraphPad Prism 9 software (GraphPad Software, Inc.). The data
of TCGA was performed in the GEPIA 2. P<0.05 was considered to
indicate a statistically significant difference.
Results
LncRNA 495810 is upregulated in
CRC
To examine the clinical significance of lncRNA
495810 in colon cancer, the endogenous expression of lncRNA 495810
in normal colonic epithelial FHC and different CRC cell lines was
first examined. It was found that lncRNA 495810 was highly
expressed in most detected CRC cells compared with FHC (Fig. 1A). Then the expression of lncRNA
495810 was estimated in 68 paired samples of adjacent tissues and
CRCs. The results revealed that lncRNA 495810 expression level was
significantly increased in colon cancer tissues (Fig. 1B). Subsequently, the pathological
and clinical value of lncRNA 495810 were assessed. The results
revealed that high expression of lncRNA 495810 was associated with
poor survival by survival analysis (Fig. 1C). Through COX regression analysis,
it was identified that the expression level of lncRNA 495810 was a
prognostic factor for CRC (Tables
I and SII). The overall and
disease-free survival curve for CRC patients with high or low
lncRNA 495810 expression level further proved that high lncRNA
495810 expression levels indicated a poor prognosis in patients
(Fig. 1D and E), and it was
correlated with advanced clinical grade and stage (Fig. 1F).
 | Table I.Association of lncRNA 495810
expression with clinical characters and overall survival in
patients with colorectal cancer (n=68 pairs). |
Table I.
Association of lncRNA 495810
expression with clinical characters and overall survival in
patients with colorectal cancer (n=68 pairs).
| Factors | Subset | Hazard ratio (95%
confidence interval) | P-value |
|---|
| Sex | Male/Female | 0.83
(0.42–1.65) | 0.600 |
| Tumor size
(cm) | >5/≤5 | 0.26
(0.09–0.75) | 0.013 |
|
Differentiation | Poorly/Well,
moderately | 0.71
(0.35–1.45) | 0.350 |
| Tumor stage | (II/III/IV)/I | 1.38
(0.84–2.28) | 0.209 |
| Expression level of
lncRNA 495810 | High/Low | 2.26
(1.06–4.80) | 0.034 |
To determine whether 495810 is a lncRNA, the
characteristics of lncRNA 495810 were detected. The chromosomal
coordinate ranges of lncRNA 495810 were chr7:26195712-26201301, and
its size was 484 bp in length (Table
SIII). Then, the CPAT displayed that 495810 had no coding
ability (Fig. S1A). In addition,
the PhyloCSF value of 495810, which was calculated to verify the
conservation of the sequence, was minus (Fig. S1B). Nuclear and cytoplasmic
fractions in CRC cells were also isolated and RT-qPCR was
performed. The results demonstrated that 495810 was mainly
localized in the nucleus (Fig.
1G). Collectively, the data indicated that 495810 is a lncRNA
and exhibits oncogene potential.
High expression of lncRNA 495810
promotes CRC growth
The promotional effect of lncRNA 495810 on
colorectal carcinogenesis was then characterized by functional
assays. HCT116/SW620 cells with relatively lower or higher lncRNA
495810 expression were selected for lncRNA 495810 overexpression
and knockdown. The HT-29 cells with moderate expression were used
in both knockdown and overexpression experiments, which were used
as parallel experimental groups (Figs.
1A, S1C and D). The results
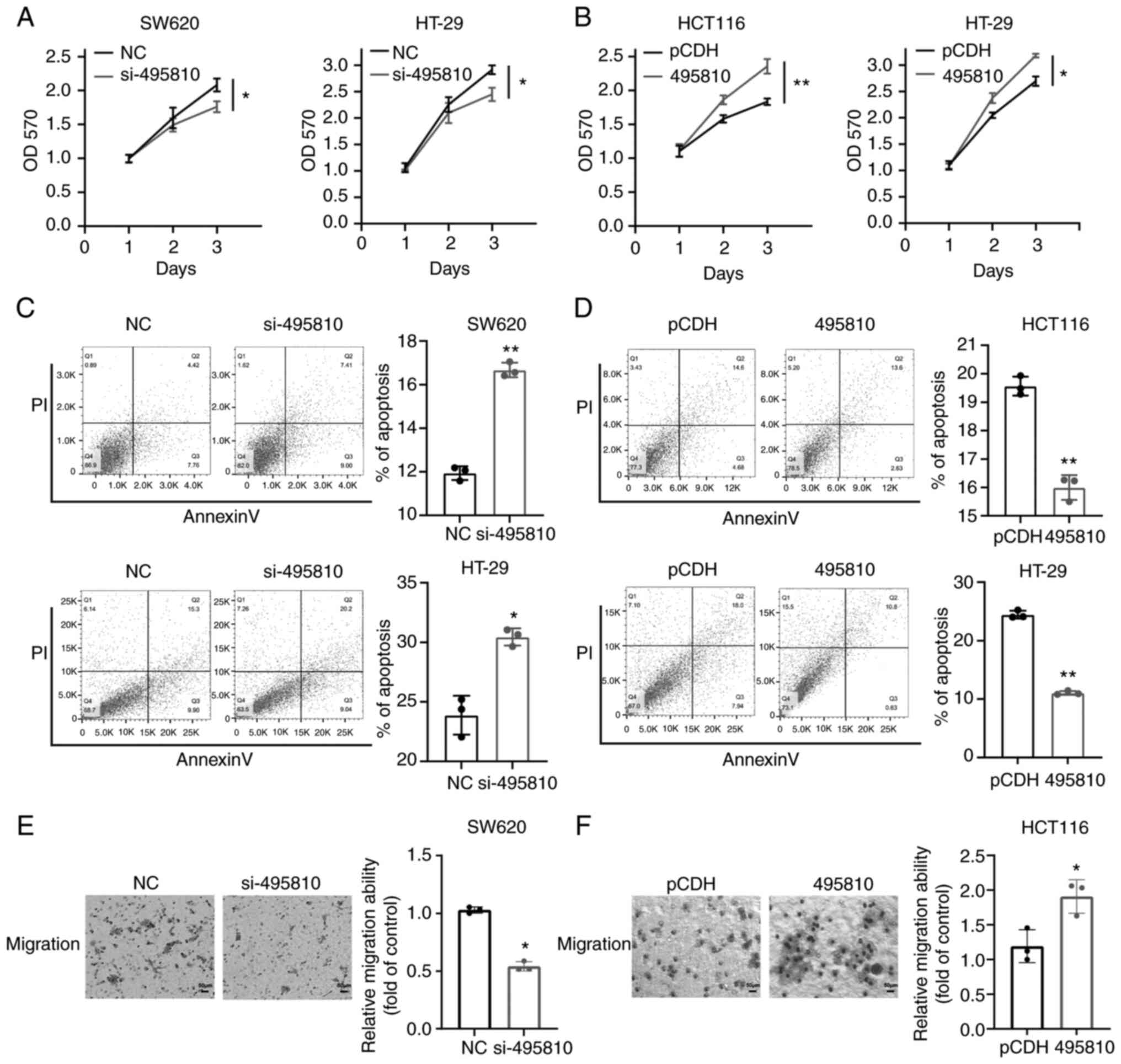
demonstrated that lncRNA 495810 knockdown significantly inhibited
cell proliferation in SW620 and HT-29 cells, whereas overexpression
of lncRNA 495810 increased CRC cell proliferation of HCT116 and
HT-29 cells (Fig. 2A and B).
Moreover, lncRNA 495810 knockdown significantly increased, whereas
overexpression of lncRNA 495810 inhibited apoptotic levels
(Fig. 2C and D).
Moreover, the migration ability of the si-495810
group was significantly reduced compared with the NC (non-specific
control) group in SW620 cells (Fig.
2E). Whereas in the gain-of-function assays, 495810
overexpression significantly increased cell migration ability of
HCT116 cells (Fig. 2F). The
aforementioned results collectively indicated that lncRNA 495810
was involved in colorectal tumorigenesis and progression as an
oncogene.
LncRNA 495810 promotes aerobic
glycolysis in CRC cells
To determine the signaling pathway affected by
lncRNA 495810 in promoting CRC, RNA pull-down combined with mass
spectrometry (Bioprofile) was used to identify targets directly
regulated by lncRNA 495810. The aforementioned target proteins were
imported into the Metascape database (www.metascape.org) for gene enrichment analysis. The
results demonstrated that the pathways closely related to lncRNA
495810 were glycolysis and gluconeogenesis (Fig. 3A-C). Furthermore, metabolic assays
revealed that silencing lncRNA 495810 expression level declined,
whereas ectopic lncRNA 495810 expression enhanced glucose
consumption, pyruvate acid production and ATP production (Fig. 3D-I). These data indicated that
495810 is a glycolysis-related lncRNA and promotes aerobic
glycolysis in CRC.
 | Figure 3.Long non-coding RNA 495810 promotes
aerobic glycolysis in colorectal cancer cells. (A-C) The 495810
target genes were imported into the Metascape database for gene
enrichment analysis; (A) Pathway and process enrichment analysis.
(B) Network of enriched terms, colored by cluster ID, where nodes
that share the same cluster ID are typically close to each other.
(C) Network of enriched terms, colored by P-value, where terms
containing more genes tend to have a more significant p-value.
(D-F) After the HCT116 cells were transfected with NC or si-495810
for 48 h, the (D) glucose consumption, (E) ATP generation (F) and
pyruvate acid production were determined. (G-I) After the HCT116
cells were transiently transfected with pCDH or 495810 for 48 h,
the (G) glucose consumption, (H) ATP generation and (I) pyruvate
acid production were determined. *P<0.05 and **P<0.01. si-,
small interfering; NC, negative control. |
PKM2 is the direct target of lncRNA
495810
To identify targets that are directly regulated by
lncRNA 495810, the proteins bound to the sense strand of lncRNA
495810 were used to construct a PPI network (Fig. 4A). At the same time, the MCODE
algorithm built into the Metascape database was used to cluster
these proteins (Table II).
Glycolysis-related enzymes, including PKM2, LDHB, ENO1, PGK1, LDHA
and TPI1, attracted the authors' attention (Fig. 4B). Among which, PKM2 was positively
correlated with lncRNA 495810 (Fig.
4C), but not other proteins (Fig.
S2A-E). Transcriptional and survival data for PKM2 were
subsequently examined in the GEPIA 2 database. Consistent with
lncRNA 495810, the expression of PKM2 was significantly upregulated
in tumor tissues (Fig. 4D). The
overall survival rates of the colon cancer patients with high
levels of PKM2 were lower than those with low expression levels of
PKM2 (Fig. 4E).
 | Table II.MCODE components identified in 495810
target genes. |
Table II.
MCODE components identified in 495810
target genes.
| MCODE | GO | Description |
Log10(P) |
|---|
| MCODE 1 | R-HSA-3371556 | Cellular response
to heat stress | −9.4 |
|
| R-HSA-5336415 | Uptake and function
of diphtheria toxin | −8.8 |
|
| GO: 0006986 | response to
unfolded protein | −8.7 |
| MCODE 2 | GO: 0006457 | protein
folding | −7.5 |
|
| CORUM: 1335 | SNW1 complex | −7.2 |
|
| R-HSA-389960 | Formation of
tubulin folding intermediates by CCT/TriC | −6.7 |
| MCODE 3 | GO:1903312 | negative regulation
of mRNA metabolic process | −6.9 |
|
| CORUM: 3055 | Nop56p-associated
pre-rRNA complex | −6.8 |
|
| WP411 | mRNA
processing | −6.5 |
| MCODE 4 | WP534 | Glycolysis and
gluconeogenesis | −8.5 |
|
| GO:0006090 | pyruvate metabolic
process | −8.1 |
|
| hsa00010 |
Glycolysis/Gluconeogenesis | −8 |
| MCODE 5 | WP4629 | Aerobic
glycolysis | −10.3 |
|
| WP1946 | Cori cycle | −9.8 |
|
| GO: 0006096 | glycolytic
process | −8.7 |
LncRNA 495810 interacts with PKM2 and
promotes its expression
Next, the interaction between lncRNA 495810 and PKM2
was predicted via RPISeq analysis. The interaction probabilities of
lncRNA 495810 and PKM2 were greater than 0.5 (RF=0.65, SVM=0.65).
The data suggested that lncRNA 495810 may bind to PKM2 (Table SIV), which was also verified by
RNA pull-down assay in HCT116 and HT-29 cells (Fig. 4F). In addition, a potential lncRNA
495810-binding region of PKM2 (401–452 nt) was predicted using
catRAPI (Fig. 4G). To confirm the
impact of lncRNA 495810 on PKM2, the expression of PKM2 was
detected in CRC cells after depletion or overexpression of lncRNA
495810. The results identified that silencing lncRNA 495810
expression declined, whereas ectopic lncRNA 495810 expression
enhanced the protein level of PKM2 (Fig. 4H and I).
LncRNA 495810 enhances PKM2 protein
stability via the ubiquitin-proteasome pathway
Increasing evidence has demonstrated that lncRNAs
regulate protein expression by post-translational modification
(17,23–25).
Its regulation mode of 495810 was then explored. The results showed
that silencing lncRNA 495810 expression shortened, whereas ectopic
lncRNA 495810 expression prolonged the half-life of PKM2 protein
(Fig. 5A and B). In addition,
suppression of proteasome activity by MG132, an inhibitor of
proteasome, prevented the lncRNA-induced PKM2 expression (Fig. 5C and D), suggesting that PKM2
protein degradation was mediated by the ubiquitin-proteasome
pathway.
Discussion
Patients with advanced CRC have a poor prognosis.
Reliable prediction of recurrence risk would be of great benefit in
improving outcomes for patients with colon cancer. However, the
molecular mechanisms underlying its development and recurrence
remain unclear, limiting the development of effective molecular
biomarkers (26,27). Emerging evidence has revealed that
dysregulation of lncRNAs plays a crucial role in cancer development
and progression (28,29). In the present study, it was
identified that a novel glycolysis-associated lncRNA 495810 was
highly expressed in CRC cells and tissues. Moreover, lncRNA 495810
is a new candidate oncogene in the CRC. Clinical analysis revealed
that high expression of lncRNA 495810 was associated with poor
survival, and was correlated with an advanced clinical grade and
stage. Thus, lncRNA 495810 could be used to evaluate prognosis in
colon cancer. The COX regression analysis indicated that the
expression of lncRNA 495810 was a prognostic factor for CRC and may
be a biomarker of CRC. More notably, functional experiments
presented that lncRNA 495810 significantly promoted proliferation
and inhibited apoptosis in CRC cells. Therefore, it was indicated
that lncRNA 495810 is a new candidate oncogene and promotes the
occurrence of CRC. However, the potential molecular mechanism of
lncRNA 495810 on CRC remains to be further investigated.
The involvement of lncRNAs in key oncogenic
signaling pathways is an important piece of evidence demonstrating
the functional role of lncRNAs in cancer (17,30).
The present study demonstrated that the target proteins of lncRNA
495810, which were identified by RNA pull-down combined with mass
spectrometry, were enriched in glycolysis pathway. It is well known
that increased aerobic glycolysis is one of the hallmarks of cancer
cells (31,32). Aerobic glycolysis not only provides
ATP as an important energy source for cell growth, but also
provides raw materials for the synthesis of various biological
macromolecules in tumor cells (33). Aberrantly expressed lncRNA in
cancer cells is associated with aerobic glycolysis. Further study
found that silencing lncRNA 495810 expression decreased, whereas
overexpression of lncRNA 495810 increased the glycolysis-related
indicators including glucose consumption, pyruvate acid production
and ATP production. Collectively, the data suggested that 495810
could promote aerobic glycolysis and thereby provide energy for
colon cancer proliferation.
Growing evidence suggests that lncRNAs are involved
in cellular behavior, including metabolism, inflammation, cell
differentiation and tumorigenesis, by interacting with specific
proteins (34). The present
findings indicated that PKM2, which positively correlated with
lncRNA 495810, was identified as a key downstream of lncRNA 495810.
Pyruvate kinase, which is a rate-limiting enzyme, catalyzes
phosphoenolpyruvate to produce pyruvate in the last step of aerobic
glycolysis. Pyruvate kinase isozyme M2 (PKM2) has been reported to
be enriched in cancer cells and plays a key role in metabolic
reprogramming and carcinogenesis (35–37).
The ubiquitin-proteasome system is the main pathway of
intracellular protein degradation (38) and is involved in the degradation of
PKM2 proteins (39). In the
present study, it was revealed that lncRNA 495810 may directly bind
to PKM2 to protect it from ubiquitin-mediated degradation.
The expression levels of LncRNA 495810 are elevated
in CRC cells and its tissues. However, the molecular mechanism of
its abnormal expression in colon cancer remains unclear. Previous
studies have shown that the regulation of key transcription factors
can lead to abnormal expression of lncRNA in cancer cells.
Chaudhary et al (40)
reported that p53 as a transcription factor regulates the
expression of lncRNA PINCR and promotes the development of CRC;
transcription factor YY1 activates lncRNA-PVT1 and modulates lung
cancer progression (41);
transcription factors p53 and TP63 bind to enhancers in the
promoter region of lincRNA00475 (linc-475) and lincRNA01503
respectively, resulting in the high expression of related lncRNA in
tumor cells (42,43). Therefore, it was hypothesized that
transcription factors play a key role in the aberrant expression of
lncRNA 495810. The aforementioned findings will be further
validated in a follow-up study.
Given the clinical, biochemical and functional
significance of 495810 in CRC, the first evidence was provided of a
novel lncRNA 495810 in colorectal carcinogenesis as well as the
glycolytic pathway, which may consist a novel biomarker and
therapeutic target for CRC.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the National Natural Science
Foundation of China (grant nos. 32072220 and 82072718) and the
Project of the Central Government Guiding Local Science and
Technology (grant nos. YDZX20201400001436 and YDZJSX2022A005).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
ZL and HW confirm the authenticity of all the raw
data. KC conducted investigation, developed methodology and wrote
the original draft. HW conducted investigation, wrote, reviewed and
edited the manuscript, and provided resources. LZ performed
software analysis and data validation. HL and GI wrote, reviewed
and edited the manuscript, and performed software analysis. ZL
conceptualized and supervised the study, wrote, reviewed and edited
the manuscript, and acquired funding. All authors read and approved
the final manuscript.
Ethics approval and consent to
participate
The present study was approved (approval no.
SXULL2020014; approval date: 2020.04.08) by the Clinical Research
Ethics Committees of the participating institutions (Taiyuan,
China). Informed consent was provided by all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD, Fuchs HE and Jemal
A: Cancer statistics, 2021. CA Cancer J Clin. 71:7–33. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Seymour MT, Thompson LC, Wasan HS,
Middleton G, Brewster AE, Shepherd SF, O'Mahony MS, Maughan TS,
Parmar M, Langley RE, et al: Chemotherapy options in elderly and
frail patients with metastatic colorectal cancer (MRC FOCUS2): An
open-label, randomised factorial trial. Lancet. 377:1749–1759.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhang Y, Chen Z and Li J: The current
status of treatment for colorectal cancer in China: A systematic
review. Medicine (Baltimore). 96:e82422017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Carethers JM and Jung BH: Genetics and
genetic biomarkers in sporadic colorectal cancer. Gastroenterology.
149:1177–1190. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
de Oliveira JC, Oliveira LC, Mathias C,
Pedroso GA, Lemos DS, Salviano-Silva A, Jucoski TS, Lobo-Alves SC,
Zambalde EP, Cipolla GA and Gradia DF: Long non-coding RNAs in
cancer: Another layer of complexity. J Gene Med.
21:e30652019.PubMed/NCBI
|
|
6
|
Godinho M, Meijer D, Setyono-Han B,
Dorssers LC and van Agthoven T: Characterization of BCAR4, a novel
oncogene causing endocrine resistance in human breast cancer cells.
J Cell Physiol. 226:1741–1749. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gupta RA, Shah N, Wang KC, Kim J, Horlings
HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al: Long
non-coding RNA HOTAIR reprograms chromatin state to promote cancer
metastasis. Nature. 464:1071–1076. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Mourtada-Maarabouni M, Pickard MR, Hedge
VL, Farzaneh F and Williams GT: GAS5, a non-protein-coding RNA,
controls apoptosis and is downregulated in breast cancer. Oncogene.
28:195–208. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zou A, Liu R and Wu X: Long non-coding RNA
MALAT1 is up-regulated in ovarian cancer tissue and promotes
SK-OV-3 cell proliferation and invasion. Neoplasma. 63:865–872.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Piao HY, Guo S, Wang Y and Zhang J: Long
noncoding RNA NALT1-induced gastric cancer invasion and metastasis
via NOTCH signaling pathway. World J Gastroenterol. 25:6508–6526.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bian Z, Zhang J, Li M, Feng Y, Wang X,
Zhang J, Yao S, Jin G, Du J, Han W, et al: LncRNA-FEZF1-AS1
promotes tumor proliferation and metastasis in colorectal cancer by
regulating PKM2 signaling. Clin Cancer Res. 24:4808–4819. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Song J, Shu H, Zhang L and Xiong J: Long
noncoding RNA GAS5 inhibits angiogenesis and metastasis of
colorectal cancer through the Wnt/β-catenin signaling pathway. J
Cell Biochem. 120:6937–6951. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gu W, Rauschecker AM, Hsu E, Zorn KC, Sucu
Y, Federman S, Gopez A, Arevalo S, Sample HA, Talevich E, et al:
Detection of neoplasms by metagenomic next-generation sequencing of
cerebrospinal fluid. JAMA Neurol. 78:1355–1366. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hosios AM, Fiske BP, Gui DY and Heiden MG:
Lack of evidence for PKM2 protein kinase activity. Mol Cell.
59:850–857. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gatenby RA and Gillies RJ: Why do cancers
have high aerobic glycolysis? Nat Rev Cancer. 4:891–899. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhang Y, Huang W, Yuan Y, Li J, Wu J, Yu
J, He Y, Wei Z and Zhang C: Long non-coding RNA H19 promotes
colorectal cancer metastasis via binding to hnRNPA2B1. J Exp Clin
Cancer Res. 39:1412020. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tang J, Yan T, Bao Y, Shen C, Yu C, Zhu X,
Tian X, Guo F, Liang Q, Liu Q, et al: LncRNA GLCC1 promotes
colorectal carcinogenesis and glucose metabolism by stabilizing
c-Myc. Nat Commun. 10:34992019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hong J, Guo F, Lu SY, Shen C, Ma D, Zhang
X, Xie Y, Yan T, Yu T, Sun T, et al: nucleatum targets lncRNA
ENO1-IT1 to promote glycolysis and oncogenesis in colorectal
cancer. Gut. 70:2123–2137. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chen J, Yu Y, Li H, Hu Q, Chen X, He Y,
Xue C, Ren F, Ren Z, Li J, et al: Long non-coding RNA PVT1 promotes
tumor progression by regulating the miR-143/HK2 axis in gallbladder
cancer. Mol Cancer. 18:332019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhao Y, Liu Y, Lin L, Huang Q, He W, Zhang
S, Dong S, Wen Z, Rao J, Liao W and Shi M: The lncRNA MACC1-AS1
promotes gastric cancer cell metabolic plasticity via AMPK/Lin28
mediated mRNA stability of MACC1. Mol Cancer. 17:692018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Cui K, Wu H, Fan J, Zhang L, Li H, Guo H,
Yang R and Li Z: The mixture of ferulic acid and P-Coumaric acid
suppresses colorectal cancer through lncRNA 495810/PKM2 mediated
aerobic glycolysis. Int J Mol Sci. 23:121062022. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Xu L, Huan L, Guo T, Wu Y, Liu Y, Wang Q,
Huang S, Xu Y, Liang L and He X: LncRNA SNHG11 facilitates tumor
metastasis by interacting with and stabilizing HIF-1α. Oncogene.
39:7005–7018. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Tan YT, Lin JF, Li T, Li JJ, Xu RH and Ju
HQ: LncRNA-mediated posttranslational modifications and
reprogramming of energy metabolism in cancer. Cancer Commun (Lond).
41:109–120. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ni W, Yao S, Zhou Y, Liu Y, Huang P, Zhou
A, Liu J, Che L and Li J: Long noncoding RNA GAS5 inhibits
progression of colorectal cancer by interacting with and triggering
YAP phosphorylation and degradation and is negatively regulated by
the m(6)A reader YTHDF3. Mol Cancer. 18:1432019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Malki A, ElRuz RA, Gupta I, Allouch A,
Vranic S and Al Moustafa AE: Molecular mechanisms of colon cancer
progression and metastasis: Recent insights and advancements. Int J
Mol Sci. 22:1302020. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Biller LH and Schrag D: Diagnosis and
treatment of metastatic colorectal cancer: A review. Jama.
325:669–685. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Tay Y, Rinn J and Pandolfi PP: The
multilayered complexity of ceRNA crosstalk and competition. Nature.
505:344–352. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang KC and Chang HY: Molecular mechanisms
of long noncoding RNAs. Mol Cell. 43:904–914. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Bassett AR, Akhtar A, Barlow DP, Bird AP,
Brockdorff N, Duboule D, Ephrussi A, Ferguson-Smith AC, Gingeras
TR, Haerty W, et al: Considerations when investigating lncRNA
function in vivo. ELife. 3:e030582014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ganapathy-Kanniappan S and Geschwind JF:
Tumor glycolysis as a target for cancer therapy: Progress and
prospects. Mol Cancer. 12:1522013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Li T, Han J, Jia L, Hu X, Chen L and Wang
Y: PKM2 coordinates glycolysis with mitochondrial fusion and
oxidative phosphorylation. Protein Cell. 10:583–594. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Feng J, Li J, Wu L, Yu Q, Ji J, Wu J, Dai
W and Guo C: Emerging roles and the regulation of aerobic
glycolysis in hepatocellular carcinoma. J Exp Clin Cancer Res.
39:1262020. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hua Q, Mi B and Huang G: The emerging
co-regulatory role of long noncoding RNAs in epithelial-mesenchymal
transition and the Warburg effect in aggressive tumors. Crit Rev
Oncol Hematol. 126:112–120. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Hua Q, Mi B, Xu F, Wen J, Zhao L, Liu J
and Huang G: Hypoxia-induced lncRNA-AC020978 promotes proliferation
and glycolytic metabolism of non-small cell lung cancer by
regulating PKM2/HIF-1α axis. Theranostics. 10:4762–4778. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zahra K, Dey T, Mishra SP and Pandey U:
Pyruvate kinase M2 and cancer: The role of PKM2 in promoting
tumorigenesis. Front Oncol. 10:1592020. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhang Z, Deng X, Liu Y, Liu Y, Sun L and
Chen F: PKM2, function and expression and regulation. Cell Biosci.
9:522019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Pohl C and Dikic I: Cellular quality
control by the ubiquitin-proteasome system and autophagy. Science.
366:818–822. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Yu S, Zang W, Qiu Y, Liao L and Zheng X:
Deubiquitinase OTUB2 exacerbates the progression of colorectal
cancer by promoting PKM2 activity and glycolysis. Oncogene.
41:46–56. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Chaudhary R, Gryder B, Woods WS,
Subramanian M, Jones MF, Li XL, Jenkins LM, Shabalina SA, Mo M,
Dasso M, et al: Prosurvival long noncoding RNA PINCR regulates a
subset of p53 targets in human colorectal cancer cells by binding
to Matrin 3. ELife. 6:e232442017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Huang T, Wang G, Yang L, Peng B, Wen Y,
Ding G and Wang Z: Transcription factor YY1 modulates lung cancer
progression by activating lncRNA-PVT1. DNA Cell Biol. 36:947–958.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Xie JJ, Jiang YY, Jiang Y, Li CQ, Lim MC,
An O, Mayakonda A, Ding LW, Long L, Sun C, et al:
Super-enhancer-driven long non-coding RNA LINC01503, regulated by
TP63, is over-expressed and oncogenic in squamous cell carcinoma.
Gastroenterology. 154:2137–2151. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Melo CA, Léveillé N, Rooijers K, Wijchers
PJ, Geeven G, Tal A, Melo SA, de Laat W and Agami R: A p53-bound
enhancer region controls a long intergenic noncoding RNA required
for p53 stress response. Oncogene. 35:4399–4406. 2016. View Article : Google Scholar : PubMed/NCBI
|