1. Introduction
MicroRNAs (miRNAs/miRs) are a type of naturally
occurring non-coding single-stranded RNA that range between 19 and
22 nucleotides in length. They are involved in the
post-transcriptional regulation of gene by effectively recognizing
the target genes' 3'-untranslated region (UTR) and cleaving mRNA
molecules or partially performing complementary binding (1). By contrast, in certain
circumstances, miRNAs also bind to the 5'-UTR or open reading frame
of the target genes, resulting in translation activation (2). In addition, their generation
proceeds through a series of complex processes from the nucleus to
the cytoplasm (Fig. 1). An
estimated 30% of human protein-coding genes are thought to be
regulated by miRNAs (3).
Furthermore, miRNAs are found on tumor-related chromosomes or
fragile chromosomal sites, suggesting that miRNAs are often
involved in the formation and progression of malignant carcinomas
(4,5). In addition, individual miRNAs are
engaged in more than one paramount physiological or pathological
cellular process, such as cell proliferation (6), cell cycle (7), apoptosis (8), metastasis (9) and angiogenesis (10).
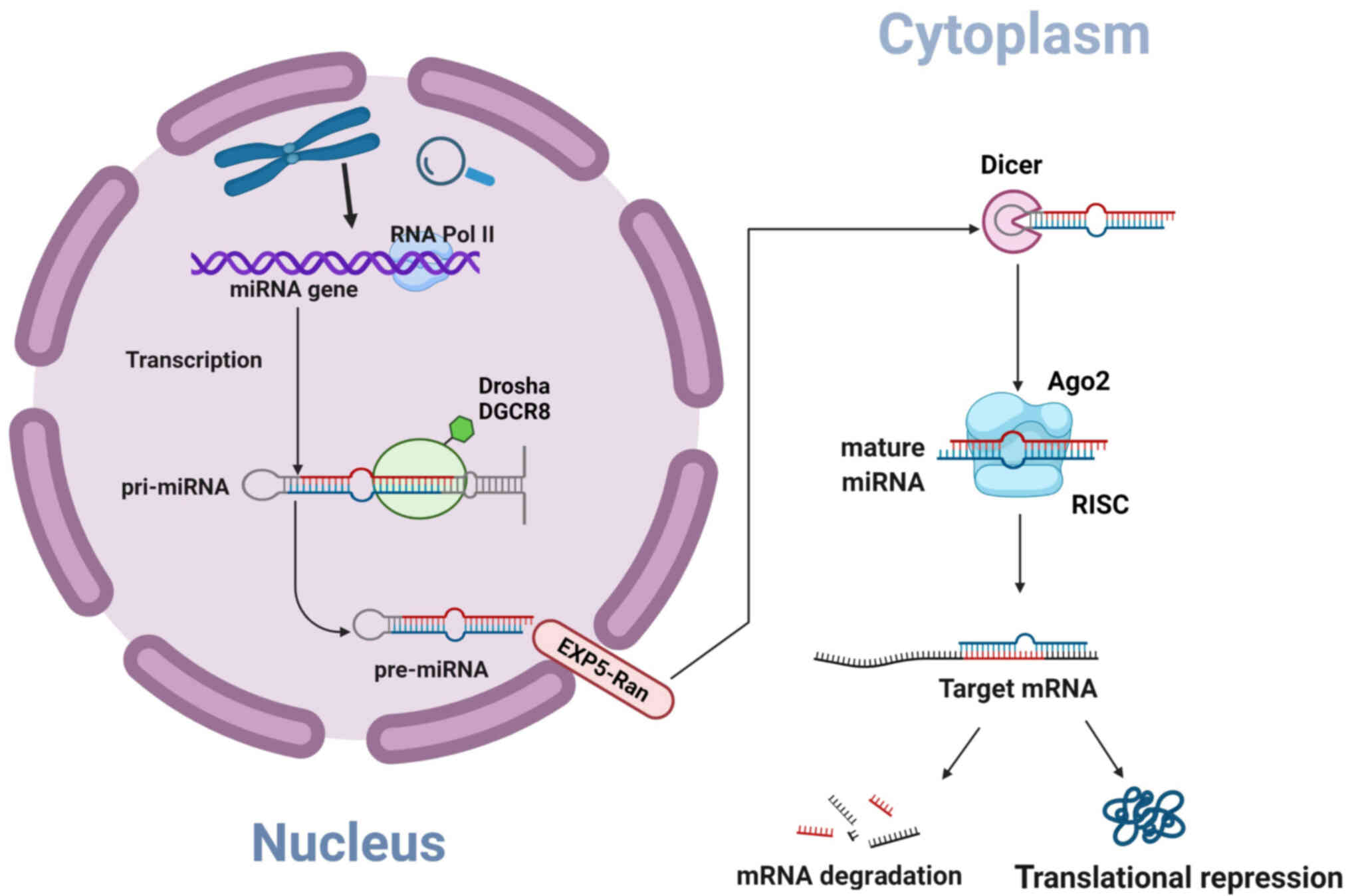 | Figure 1MiRNA biosynthesis and mechanisms. In
eukaryotic cells, miRNAs first transcribe a long pri-miRNA within
the nucleus and then are processed by Drosha and DGCR8 into a
hairpin pre-miRNA containing 60-70 nucleotides, which are
transported out of the nucleus with the help of EXP5-Ran, and cut
into mature miRNA by Dicer in the cytoplasm, It is then integrated
into the RISC to regulate gene expression based on complete or
incomplete pairing with mRNA. Created with Biorender software
(http://biorender.com). MiRNA, microRNA; Pol,
polymerase; Drosha, Drosha ribonuclease III; DGCR8, DiGeorge
syndrome chromosomal region 8; Dicer, Dicer ribonuclease III; Ago2,
Argonaute 2; RISC, RNA-induced silencing complex. |
Among the numerous miRNAs, miR-409-3p has attracted
a huge amount of attention. Initially found in embryonic stem cells
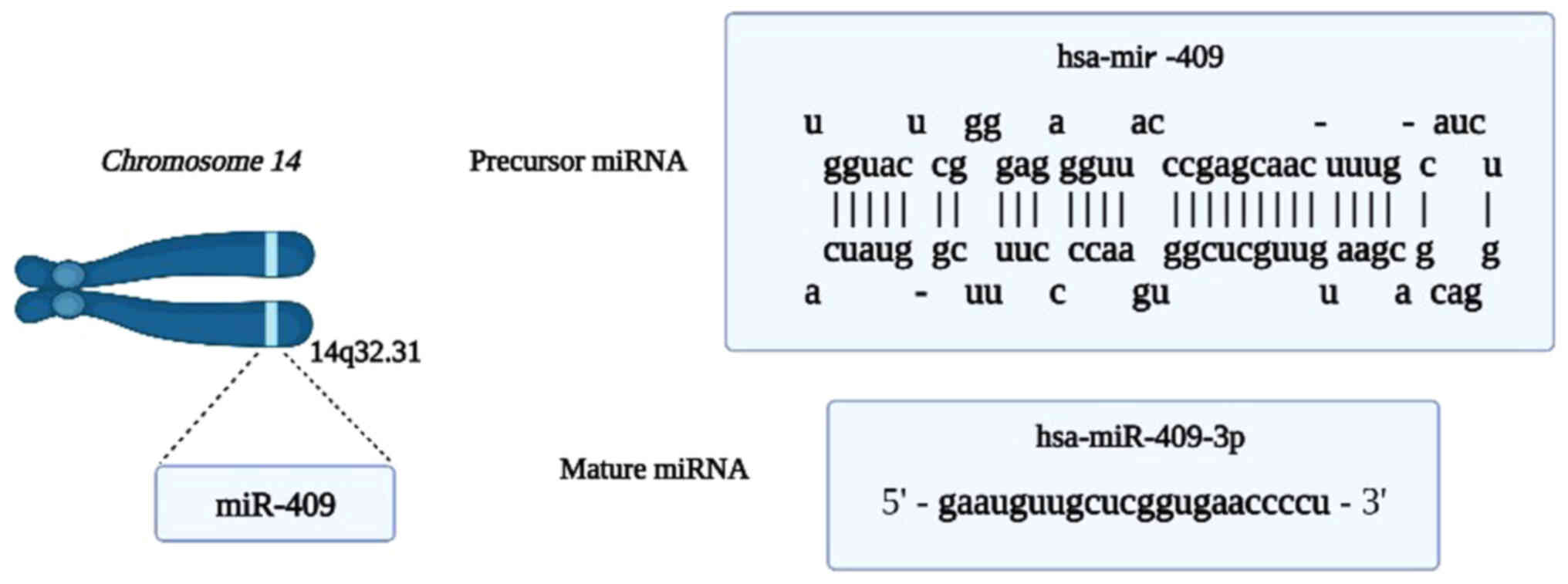
(11), miR-409 (NCBI gene ID:
574413) consists of 79 bases located on chromosome 14q32.31.
MiR-409 is being reprocessed to produce mature miR-409-3p (Fig. 2). Furthermore, increasing data
point to the broad expression of miR-409-3p in malignancies,
particularly non-small cell lung cancer (NSCLC) (12-21), breast cancer (BC) (22-30), cervical cancer (CC) (31-35), osteosarcoma (36-39), gastric cancer (GC) (40-44), colorectal cancer (CRC) (45-52), ovarian cancer (OC) (53-56), renal cell carcinoma (ccRCC)
(57,58), bladder cancer (BCa) (59-61), papillary thyroid cancer (PTC)
(62), pancreatic cancer (PC) and
biliary tract cancer (BTC) (63),
diffuse large B-cell lymphoma (DLBCL) (64), acute myeloid leukemia (AML)
(65,66), oligodendroglioma (67), melanoma (68), nasopharyngeal carcinoma (NPC)
(69), tongue squamous cell
carcinoma (TSCC) (70), prostate
cancer (PCa) (71-76), meningioma (77), intracranial artery tumors
(78), glioma (79,80), fibrosarcoma (81), glioblastoma (82) and hepatocellular carcinoma (HCC)
(83,84). Specifically, it has been shown
that miR-409-3p exerts anti- or pro-tumorigenic roles in cancer
progression by targeting downstream mRNAs or modulated by the
upstream regulators of long non-coding RNAs (lncRNAs) or circular
RNAs (circRNAs).
The present review outlines the expression, target
genes and functional mechanisms of miR-409-3p in various
malignancies, offering recommendations for future research and
clinical applications regarding miR-409-3p.
2. Aberrant expression of miR-409-3p in
cancers
Growing evidence has shown that multiple cancer
types have abnormal miR-409-3p expression, with both upregulation
and downregulation observed, as summarized in Table I. MiR-409-3p not only exists in
tissues and cells but also stably exists in circulating body
fluids. For the most part, miR-409-3p acts as a tumor suppressor
and was reported to be downregulated in a minimum of 13 cancers,
including NSCLC (12,14-17,19-21), BC (24,26,27,29,30), CC (31-35), osteosarcoma (36-39), GC (40-44), CRC (45,46,48-51), OC (53-55), ccRCC (57,58), BCa (59,61), PTC (62), AML (65,66), oligodendrogliomas (67), melanoma (68), TSCC (70), glioma (79), fibrosarcoma (81) and HCC (83,84). However, miR-409-3p also serves as
an oncogene to promote tumor progression. MiR-409-3p was reported
to be upregulated in carcinomas such as PC and BTC (63), meningioma (77), intracranial artery tumors
(78) and glioblastoma (82). However, as shown in Table I, there are inconsistent results
of miR-409-3p expression in NSCLC (13,18), BC (22,23,25,28), CRC (47,49), OC (56), DLBCL (64) and PC (71,73,74,76).
 | Table IExpression of miR-409-3p in cancers
based on published literature. |
Table I
Expression of miR-409-3p in cancers
based on published literature.
| Authors, year | Type of cancer | Normal cells | Cancer cells | Cells expression
(tumor/normal) | Tissues | Tissues expression
(tumor/normal) | Plasma/ serum | Plasma/serum
expression (tumor/normal) | (Refs.) |
|---|
| Wan et al,
2014 | Lung cancer | HBE | A549, SPC-A1,
PC9 | Downregulation | 34 pairs of NSCLC
tissues and matched adjacent normal tissues | Downregulation | - | - | (12) |
| Zhou et al,
2017 | | - | - | - | 19 pairs of NSCLC
tissues and adjacent normal tissues | Upregulation | Plasma | Upregulation | (13) |
| Song et al,
2018 | | - | A549, H460 | Downregulation | 85 pairs of NSCLC
tissues and matched adjacent normal tissues | Downregulation | - | - | (14) |
| Qu et al,
2018 | | HBE | A549, NCI-H1299,
NCI-H1650, SPC-A1, PC-9 | Downregulation | - | - | - | - | (15) |
| Yin et al,
2020 | | BEAS-2B | H1299, A549, H460,
PC-9 | Downregulation | 66 pairs of NSCLC
tissues and the adjacent normal tissues | Downregulation | - | - | (16) |
| Liu et al,
2020a | | HBE | A549, PC-9,
NCI-H1299, NCI, H460, NCI-H1650, NCI-H520 | Downregulation | 18 pairs of NSCLC
tumor and adjacent normal tissue samples | Downregulation | - | - | (17) |
| Wang et al,
2020a | | - | - | - | - | - | Serum | Downregulation | (18) |
| Wang et al,
2020b | | BEAS-2B | A549, SK-MES-1,
H1703, H460, H522 | Downregulation | 61 pairs of NSCLC
tissues and adjacent normal tissues | Downregulation | - | - | (19) |
| Liu et al,
2022 | | HBE | A549, H1650, H520,
H460, H1299, PC9 | Downregulation | 48 pairs of
cancerous and paracancerous tissues | Downregulation | - | - | (20) |
| Yang et al,
2022b | | MRC-5 | A549, H2170 | Downregulation | 67 pairs of NSCLC
tissues and adjacent normal tissues | Downregulation | - | - | (21) |
| Cuk et al,
2013b | BC | - | - | - | - | - | Plasma | Upregulation | (22) |
| Cuk et al,
2013a | | - | - | - | 24 primary BC
surgery tissue samples and 8 benign breast biopsies | Downregulation | Plasma | Upregulation | (23) |
| Li et al,
2013 | | - | - | - | 21 pairs of DCIS
and the corresponding normal tissues | Downregulation | - | - | (24) |
| Shen et al,
2014 | | - | - | - | - | - | Plasma | Upregulation | (25) |
| Zhang et al,
2016 | | HBL-100 | MCF-7, T47D,
MDA-MB-468, MDAMB-231 | Downregulation | 30 pairs of human
BC tissues and adjacent non-tumor samples | Downregulation | - | - | (26) |
| Ma et al,
2016 | | HBL-100,
MCF-10A | MCF-7, BT-474,
MDA-MB-231, SK-BR3 | Downregulation | 40 pairs of tumor
tissues and transitional tissues normal tissues | Downregulation | - | - | (27) |
| Venkatadri et
al, 2016 | | - | MCF-7 | Upregulation | - | - | - | - | (28) |
| Su et al,
2021 | | MCF-10A | MCF-7, SKBR-3 | Downregulation | 44 pairs BC tissue
and adjacent normal tissue samples | Downregulation | - | - | (29) |
| Yang et al,
2022a | | MCF10A | MCF7,
MDA-MB-231 | Downregulation | 64 pairs BC tissue
(including 30 sensitive and 30 resistant to tamoxifen) and 64
normal tissue samples | Downregulation | - | - | (30) |
| Shukla et
al, 2019 | CC | NCE | SiHa, Hela,
CaSki | Downregulation | - | - | Serum | Downregulation | (31) |
| Sommerova et
al, 2019 | | | HeLa, CaSki,
C4-I | Downregulation | Precancerous
cervical lesions | Downregulation | - | - | (32) |
| Cui et al,
2020 | | ECT1/E6E | HeLa, Caski, C33A,
Siha | Downregulation | 55 pairs of CC
tissues and adjacent non-tumor tissues | Downregulation | - | - | (33) |
| Zhou et al,
2021 | | HcerEpic | SiHa, SW756, CaSki,
C-33A | Downregulation | GEO datasets
(GSE102686) and 47 pairs of CC tissues and marched normal
tissues | Downregulation | - | - | (34) |
| Wu et al,
2021 | | HcerEpic | CaSki, HeLa | Downregulation | - | - | - | - | (35) |
| Wu et al,
2016 | Osteosa- rcoma | hFOB | U2OS, MG-63,
SAOS-2 | Downregulation | 58 pairs of
osteosarcoma tissue specimens and adjacent non-tumorous tissue
specimens | Downregulation | - | - | (36) |
| Zhang et al,
2017 | | NHOst, hFOB 1
19 | MG63, SaOS-2, U2OS,
G292 | Downregulation | 36 pairs of human
osteosarcoma tissues and normal bone tissues | Downregulation | - | - | (37) |
| Wu et al,
2019 | | hFOB 1 19 | HOS (GDC76), MG63
(GDC074) | Downregulation | 49 pairs of
osteosarcoma tumor and adjacent non-tumor tissues | Downregulation | - | - | (38) |
| Long et al,
2020 | | hFOB 1 19 | SJSA1, U2OS | Downregulation | 30 pairs of
osteosarcoma and adjacent tissues | Downregulation | - | - | (39) |
| Li et al,
2012 | GC | GES-1 | MKN45, MKN28,
SGC-7901, NCI-N87, AGS | Downregulation | 67 pairs of primary
GC tissues and its matched non-tumor tissues | Downregulation | - | - | (40) |
| Zheng et al,
2012 | | GES-1 | SGC-7901, HGC-27,
AGS, MGC-803, NCI-N87 | Downregulation | 90 paired of GC and
their corresponding non-tumorous tissues | Downregulation | - | - | (41) |
| Yu et al,
2021 | | GES-1 | XGC-1, MKN45 | Downregulation | 30 pairs of GC
tissues and paracancerous normal tissues | Downregulation | - | - | (42) |
| Feng et al,
2021 | | GES-1 | MKN45, BGC823,
MGC803, HGC27, SGC7901 | Downregulation | 94 pairs of GC
tissues and adjacent non-tumorous tissues | Downregulation | - | - | (43) |
| Liu et al,
2015 | CRC | - | SW480, SW1116 | Downregulation | 45 pairs of primary
CRC and their corresponding adjacent non-tumor tissues | Downregulation | - | - | (45) |
| Bai et al,
2015 | | - | HCT116, RKO, DLD1,
SW480 | Downregulation | 82 pairs of CRC
samples and corresponding non-tumorous tissues | Downregulation | - | - | (46) |
| Wang et al,
2015 | | - | - | - | - | - | Plasma | Upregulation | (47) |
| Tan et al,
2016 | | FHC, CCD-18Co | LoVo, HCT 116,
DLD-1, SW480, HT-29, RKO | Downregulation | 20 human CRC tissue
samples and 10 human normal tissues | Downregulation | - | - | (48) |
| López-Rosas et
al, 2018 | | - | SW-480/
trophozoites, Caco2/ trophozoites | Downregulation | - | - | - | - | (49) |
| Han et al,
2020 | | - | HCT-116/ | Downregulation | - | - | - | - | (50) |
| Chen et al,
2022 | | | L-OHP SW480,
HCT116 | - | 45 cases of CRC and
adjacent non-tumorous tissues | Downregulation | | | (51) |
| Zhang et al,
2018a | | - | - | - | - | - | Serum | Ratio of
miR-130a-3p/ miR-409-3p and miR-148a-3p/ miR-409-3p
upregulation | (52) |
| Gharpure et
al, 2018 | OC | IO180 | HeyA8, HeyA8 MDR,
A2780, A2780 CP20 SKOV3ip1, Ovcar5, Ovcar3, Ovca432, IGROV, IGROV
CP20, Ovcar8 | Downregulation | GEO datasets
(GSE15190) | Downregulation | - | - | (53) |
| Cheng et al,
2018 | | SV40, iMOSEC | OV-1063, CoC1,
CaoV-3, OVCAR3, and SKOV3 | Downregulation | 13 OC tumor samples
and 12 tumor-adjacent normal tissue samples | Downregulation | - | - | (54) |
| Zhang et al,
2018b | | IO180 | KOV3_ip1, A2780_PAR
and HEYA8 | Downregulation | - | - | - | - | (55) |
| Li et al,
2020b | | OSE80 | SKOV3, Vcar | - | 6 OC tissues | Upregulation | - | - | (56) |
| Zhu et al,
2016 | ccRCC | - | - | - | 13 cases of
localized renal cell carcinoma and 15 cases of metastatic renal
cell carcinoma | Downregulation | - | - | (57) |
| Wang et al,
2019 | | - | A-498, 769-P | Downregulation | 56 pairs of ccRCC
tissues and tumor- adjacent normal tissue samples | Downregulation | - | - | (58) |
| Xu et al,
2013 | BCa | SV-HUC-1 | T24, 5637, J82, UM
UC3 | Downregulation | 10 pairs of human
BCa tissues and adjacent normal mucosal tissues | Downregulation | - | - | (59) |
| Lian et al,
2018 | | - | - | - | - | - | Serum | Downregulation | (61) |
| Zhao et al,
2018 | PTC | Nthy-ori3-1 cell
l | B-CPAP, TPC-1,
GLAG-66 | Downregulation | 20 pairs of primary
PTC tissues and adjacent non-tumor | Downregulation | - | - | (62) |
| Kim et al,
2019 | | - | - | - | - | - | Serum | Upregulation | (63) |
| Leivonen et
al, 2017 | DLBCL | SV_Co10 | SU-DHL-4 | Upregulation | Discovery cohort: 7
pairs of DLBCL primary and relapse validation cohort: 13 pairs of
DLBCL primary and relapse | Down in the relapse
sample | - | - | (64) |
| Li et al,
2020a | AML | - | - | - | 73 patients with
AML and 15 healthy controls | Downregulation | - | - | (65) |
| Xie et al,
2023 | AML | HS-5 | NB4, THP-1 | Downregulation | - | - | - | - | (66) |
| Kumar et al,
2018 | Oligodendr-
ogliomas | - | HS683 | - | TCGA: 153 Oligoden-
drogliomassamples and 5 controls | Downregulation | - | - | (67) |
| Venza et al,
2015 | Melanoma | NHEM | G361, GR-M,
OCM-1 | Downregulation | - | - | - | - | (68) |
| Chen and Dai,
2018 | TSCC | HOK | Tca8113, SCC9,
SCC25, Ca127 | Downregulation | 68 patients (38
males and 30 females) | Downregulation | - | - | (70) |
| Josson et
al, 2014 | PCa | - | ARCaP, E
ARCaPM, LNCaPNeo, LNCaPRANKL | Upregulation in
ARCaPM and LNCaPNeo | Benign prostatic
hyperplasia (n=14), Gleason 6 (n=26), and Gleason ≥7 (n=35) | Upregulation in
Gleason ≥7 compared with Gleason 6; upregulation in Gleason 6 and
Gleason ≥7 compared with benign prostatic hyperplasia | - | - | (71) |
| Josson et
al, 2015 | | - | [SON, SOC,
HS-27aRWV, HS-27aC4-2, MG-63RWV,
MG-63LNCaP, MG-63C4-2 | Upregulation in the
cancer- associated stromal cell (SOC, HS-27aC4-2, MG-63LNCaP,
MG-63C4-2) | Gleason 6, (n=25)
and Gleason score ≥7 (Gleason 7, 8, 10, n=30) | Upregulation in
Gleason ≥7 | - | - | (73) |
| Yu et al,
2018 | | - | - - | - - | - PCa set (n=32)
and benign prostatic hyperplasia set (n=43) | -
Downregulation | Serum exosomes
- | Upregulation before
carbon ion radiotherapy - | (74) |
| Nguyen et
al, 2013 | | - | - | - | 42 primary tumors
of low-risk, localized PCa and 28 normal prostate tissue | Downregulation in
the low-risk, localized PCa | Serum | Downregulation in
castration resistant PCa compared with low-risk | (76) |
| Zhi et al,
2016 | Meningioma | - | - | - | - | - | Serum | Upregulation in
pre-operative | (77) |
| Ding et al,
2021 | Intracranial artery
tumors | - | Human brain
vascular smooth muscle cells exposed to
H2O2 | Upregulation | - | - | - | - | (78) |
| Cao et al,
2017 | Glioma | HAs | 172, SHG44, U251,
U87 | Downregulation | Glioma tissue
(n=20) and normal brain tissue (n=8) | Downregulation | - | - | (79) |
| Ma et al,
2022 | | NHA | U-87 MG, U-138 MG,
U-118 MG, T98-G, LN-229, LN-18 | Downregulation | 47 pairs of primary
glioma tissues and adjacent non-tumor | Downregulation | - | - | (80) |
| Weng et al,
2012 | Fibrosarcoma | - | HT1080, Cos-7 | Downregulation | - | - | - | - | (81) |
| Khalil et
al, 2016 | Glioblastoma | - | 98G, U251 and
U373 | Upregulation | Glioblastoma
tissues (n=26) | Upregulation | - | - | (82) |
| Chang et al,
2023 | HCC | - | Huh-7 | Downregulation | 45 pairs of liver
HCC tissue and adjacent normal tissue; tumor (n=375) and normal
tissues (n=50) based on TCGA and Genotype-Tissue Expression
database | Downregulation | - | - | (83) |
| Li et al,
2023 | | LX2 | Hep3B, Huh7 | Downregulation | 5 liver transplant
donors, 5 patients with liver cirrhosis, and 5 patients with
HCC | Downregulation | - | - | (84) |
To sum up, it was found that miR-409-3p expression
levels in tissues, cells and circulating fluids, such as plasma or
serum, were inconsistent. While the exact regulatory mechanism is
currently elusive, the reasons for the discrepancy found by certain
scholars are as follows. First, various types of cancer cell are
capable of selectively releasing specific miRNAs (85). Theoretically, the selective miRNAs
released into the circulating fluids may lead to an increase in
plasma or serum and a decrease in the level of tumor tissues/cells
from which they are derived. In addition, miRNAs can be secreted
into the extracellular space and packaged into all types of
membrane-bound vesicles, including exosomes, microvesicles and
apoptotic bodies (86), and
influence the biological function of the recipient cells, thereby
protecting these specific miRNAs from RNase-mediated degradation
and upregulation in circulating fluids (87-89). Secondly, it has been hypothesized
that miRNAs in the circulation may mainly represent by-products of
dead cells (90,91). Furthermore, in consideration of
the stromal compartment and tumor microenvironment, circulating
miRNAs in the plasma/serum of cancer patients may also come from
these parts (92). It was
inferred that this may be due to differences in sample size, study
cohort, batch effect, sensitivity and specificity of the assay and
clinical factors such as gender, age and TNM stage. The
contradictory findings on miR-409-3p in certain cancers require
further exploration.
3. Clinical applications
MiR-409-3p as a biomarker in cancers
A huge effort has gone into finding acceptable
miRNAs and it has been indicated that miRNAs are promising
biomarkers for the diagnosis and monitoring of several
malignancies. Circulating miR-409-3p was found to be dysregulated
in various types of cancers (Table
II). Zhou et al (13)
examined six miRNAs, including miR-409-3p, in the plasma, which may
contribute to the detection of LUAD to a certain extent. Similarly,
Wang et al (18) revealed
that miR-409-3p had high sensitivity and specificity in
differentiating lung adenocarcinomas (LUADs) from healthy
individuals and was able to detect early-stage LUAD in patients. In
addition, it was able to diagnose stage I or II BC when compared to
healthy controls (22). In
patients with BC, miR-409-3p was shown to have the highest
sensitivity and specificity for distinguishing patients with BC
from healthy subjects (23,25). Besides, when combined with
miR-32-5p, the sensitivity and specificity of the detection of CC
in tissue and serum samples were the highest (31). The combined panel of ratios of
five miRNAs possessed a diagnostic capability to make a distinction
between CRC and colorectal adenoma (52). Furthermore, Wang et al
(47) used a panel of miR-409-3p,
miR-7 and miR-93, which had great diagnostic accuracy in
distinguishing CRC from a healthy group. In PC and BTC, Kim et
al (63) demonstrated that
miR-409-3p was also downregulated by sequencing serum miRNAs. In
AML, Li et al (65)
discovered that miR-409-3p had the best diagnostic value, with a
sensitivity of 93.3% and a specificity of 87.9%. In addition,
miR-409-3p could be combined with other miRNAs to create a
signature model for the diagnosis of NPC (69). Most strikingly, when comparing the
serum concentrations of eight preoperative and postoperative
patients with carbon ion radiotherapy (CIRT)-localized PCa, Yu
et al (74) found that the
expression level of miR-409-3p was higher than that of patients
with CIRT following the surgical removal of primary tumors, and the
patients with higher preoperative miR-409-3p levels had a better
response to CIRT. The same result was found in the prediction of
meningioma (68).
 | Table IIDiagnostic value of miR-409-3p in
cancers. |
Table II
Diagnostic value of miR-409-3p in
cancers.
| Authors, year | Type of cancer | AUC | Sensitivity | Specificity | 95% CI | Combined diagnostic
molecular signature | (Refs.) |
|---|
| Zhou et al,
2017 | Lung cancer | 0.61 | - | - | 0.53-0.69 |
miR-19b-3p+miR-21-5p+miR-221-3p+miR-409-3p+miR-425-5p+miR-584-5p | (13) |
| Wang et al,
2020a | | 0.76 | 57.32% | 86.67% | 0.68-0.82 |
miR-409-3p+miR-142-5p+miR-223-3p+miR-146a-5p | (18) |
| Cuk et al,
2013b | BC | - | - | - | - |
miR-127-3p+miR-148b+miR-376a+ miR-376c+
miR-409-3p+miR-652+miR-801 | (22) |
| Cuk et al,
2013a | | 0.66 | - | - | 0.59-0.74 |
miR-148b+miR-409-3p+miR-801 | (23) |
| Shen et al,
2014 | | 0.78 | - | - | - |
miR-148b+miR-409-3p+miR-801 | (25) |
| Shukla et
al, 2019 | CC | - | - | - | - |
miR-32-5p+miR-409-3p | (31) |
| Wang et al,
2015 | CRC | - | - | - | - |
miR-409-3p+miR-7+miR-93 | (47) |
| Zhang et al,
2018a | | - | - | - | - |
let-7b/miR-367-3p+miR-130a-3p/miR-409-3p+miR-148-3p/miR-27b+
miR-148a-3p/miR-409-3p+miR-21-5p/miR-367-3p | (52) |
| Li et al,
2020a | AML | 0.93 | 93.30% | 87.90% | 0.86-1.00 | - | (65) |
| Jiang et al,
2021 | NPC | | | | |
miR-134-5p+miR-205-5p+miR-409-3p+miR-484+miR-486-3p+miR-486-5p+
miR-92b-3p | (69) |
| Fredsøe et
al, 2020 | PCa | - | - | - | - |
miR-375*+miR-33a-5p+miR-16-5p*+miR-409-3p | (75) |
| Zhi et al,
2016 | Meningioma | - | - | - | - |
miR-106a-5p+miR-219-5p+miR-375+miR-409-3p+miR-197+miR-224 | (77) |
In general, the aforementioned results proved that
miR-409-3p may serve as a promising biomarker for the diagnosis and
therapeutic monitoring of cancers.
Prognostic value of miR-409-3p in
cancers
MiR-409-3p has been discovered to be a prognostic
marker in a variety of malignancies. The varied expression of
miR-409-3p in various neoplasms may indicate that miR-409-3p
expression and survival have different relationships.
In NSCLC, miR-409-3p expression was an independent
prognostic marker. In general, patients with low miR-409-3p
expression exhibit poor prognosis. Patients with LUAD with low
miR-409-3p expression display a lower overall survival (12,14,16,20), recurrence-free survival (12) and disease-free survival (14,20) than patients with high miR-409-3p
expression. Furthermore, a low median survival time was associated
with pTNM (III+IV) and lymph node metastasis. Clinically, reduced
miR-409-3p expression was found to be substantially associated with
poor tumor differentiation, tumor size, advanced pTNM stage, lymph
node metastasis, pleural invasion and smoking (12,14,20). In BC, the level of tissue
miR-409-3p was negatively associated with TNM stage, lymph node
metastasis, pathological differentiation, tumor size and Ki-67
status (27). In osteosarcoma,
miR-409-3p expression was associated with advanced clinical stage
and distant metastasis (38). In
addition, miR-409-3p was usually decreased in GC. Among patients
with GC, changes in miR-409-3p expression were associated with
local invasion, TNM stage and tumor size (40-42). Studies investigating miRNA
profiles in CRC showed that downregulation of miR-409-3p was
associated with tumor size, local invasion and metastasis (45,46). In addition, in OC, lower
miR-409-3p levels were closely associated with the International
Federation of Gynecology and Obstetrics stage (55). Furthermore, miR-409-3p, when
combined with other miRNAs, was negatively associated with age and
the age at which non-muscle-invasive BCa develops (61). In patients with AML, a low level
of miR-409-3p was associated with poorer event-free survival and
white blood cell count (65).
Another study assessing oligodendrogliomas confirmed that
miR-409-3p was inversely associated with progression-free survival
(67). With regard to TSCC,
downregulation of miR-409-3p was associated with TNM stage and
lymph node metastasis (70). On
the contrary, a previous study has shown that in other neoplasms,
including PCa, patients with high miR-409-3p expression had a
poorer prognosis than those with low miR-409-3p expression. More
importantly, a higher Gleason score was positively associated with
the expression of miR-409-3p (71). In addition, the high recurrence
rate of meningioma was substantially connected with miR-409-3p
expression, indicating that miR-409-3p may have predictive utility
for patients with meningioma following tumor removal (77).
In summary, these findings suggested that miR-409-3p
was significantly linked to the survival of cancer patients.
Therefore, miR-409-3p may function as an independent prognostic
molecular biomarker in malignancies.
4. Biological roles of miR-409-3p in cancers
in vitro
MiR-409-3p plays a role in the initiation and
progression of cancer, and its abnormal expression is involved in
several biological processes (Table
III). The specific mechanisms are described below.
 | Table IIITargets and functions of miR-409-3p
in human cancers. |
Table III
Targets and functions of miR-409-3p
in human cancers.
| Authors, year | Tumor type | Upstream genes | Downstream
genes | Proliferation and
apoptosis (used cells) | Invasion and
migration (used cells) | Other functions
(used cells) | (Refs.) |
|---|
| Wan et al,
2014 | Lung cancer | - | MET | ↑miR-409-3p:
Proliferation↓, Colony formation↓, Apoptosis↑; ΔmiR-409-3p:
Proliferation↑, (A549, SPC-1) | ↑miR-409-3p:
Invasion and Migration↓; Δ miR-409-3p: Invasion and Migration↑
(A549, SPC-1) | - | (12) |
| Song et al,
2018 | | - | SPIN1 | ↑miR-409-3p:
Proliferation↓, Colony formation↓, Apoptosis↑; ΔmiR-409-3p:
Proliferation↑ (A549) | ↑miR-409-3p:
Invasion and Migration↓; ΔmiR-409-3p: Invasion and Migration↑
(A549) | - | (14) |
| Yin et al,
2020 | | DUXAP8 | HK2, LDHA | ΔmiR-409-3p: Cell
viability↑, Colony formation↑ (A549, H1299) | ΔmiR-409-3p:
Migration↑ (A549, H1299) | ΔmiR-409-3p:
Glycolysis ↑ (A549, H1299) | (16) |
| Liu et al,
2020a | | - | SOD1 | ↑miR-409-3p:
Proliferation↓, Colony formation↓, Apoptosis↑ (H1299) | - | - | (17) |
| Wang et al,
2020b | | PSMA3-AS1 | SPIN1 | ΔmiR-409-3p:
Proliferation↑, Apoptosis↓ (A549, H460) | ΔmiR-409-3p:
Invasion and Migration↑ (A549, H460) | - | (19) |
| Liu et al,
2022 | | CBR3-AS1 | SOD1 | ↑miR-409-3p:
Proliferation↓ (H1650) | ↑miR-409-3p:
Invasion and Migration↓ (H1650) | ΔmiR-409-3p:
Radiosensitivity ↓ (H520) | (20) |
| Yang et al,
2022b | |
Hsa_circ_0079530 | AQP4 | ↑miR-409-3p:
Proliferation↓; ΔmiR-409-3p: Proliferation↑ (H1270, A549) | ↑miR-409-3p:
Invasion and Migration↓, EMT↓; ΔmiR-409-3p:Invasion and Migration↑
(H1270, A549) | ↑miR-409-3p:
Radiosensitivity ↑; ΔmiR-409-3p: Radiosensitivity ↓ (H1270,
A549) | (21) |
| Zhang et al,
2016 | BC | - | AKT1 | ↑miR-409-3p:
Proliferation↓, Colony formation↓, Apoptosis↑ (MDA-MB-231,
MDA-MB-468); ΔmiR-409-3p: Proliferation↑ (T47D) | ↑miR-409-3p:
Invasion and Migration↓ (MDA-MB-231, MDA-MB-468); ΔmiR-409-3p:
Invasion and Migration ↑ (T47D) | - | (26) |
| Ma et al,
2016 | | - | ZEB1 | ↑miR-409-3p:
Proliferation↓, Colony formation↓ (MDA-MB-231) | ↑miR-409-3p:
Invasion and Migration↓ (MDA-MB-231) | - | (27) |
| Su et al,
2021 | | CircCNOT2 | TWIST1 | ↑miR-409-3p:
Proliferation↓, Apoptosis↑ (MCF-7) | ↑miR-409-3p:
Invasion and Migration↓ (MCF-7) | ↑miR-409-3p: EMT↓
(MCF-7) | (29) |
| Yang et al,
2022a | | CircTRIM28 | HMGA2 | ΔmiR-409-3p:
Proliferation↑, Colony formation↑, Apoptosis↓ (MCF7/R,
MDA-MB-231/R) | ΔmiR-409-3p:
Invasion and Migration ↑ (MCF7/R, MDA-MB-231/R) | ΔmiR-409-3p:
Tamoxifen sensitivity↓ (MCF7/R, MDA-MB-231/R) | (30) |
| Shukla et
al, 2019 | CC | - | MTF2 | ↑miR-409-3p:
Proliferation↓ (SiHa) | - | - | (31) |
| Sommerova et
al, 2019 | | - | HPV16/ 18 E6 | ↑miR-409-3p:
Proliferation↓; ΔmiR-409-3p: Proliferation↑ (C-4I, CaSki) | ↑miR-409-3p:
Migration↓; ΔmiR-409-3p: Migration↑, (CaSki) | - | (32) |
| Cui et al,
2020 | | Circ_0000745 | ATF1 | ↑miR-409-3p: Colony
formation↓ (CaSki, Siha) | ↑miR-409-3p:
Invasion and Migration↓ (CaSki, Siha) | ↑miR-409-3p:
Glycolysis↓ (CaSki, Siha) | (33) |
| Zhou et al,
2021 | | CircFAT1 | CDK8 | ΔmiR-409-3p:
Proliferation↑, Apoptosis↓ (CaSki, C-33A) | ΔmiR-409-3p:
Invasion and Migration ↑ (CaSki, C-33A) | - | (34) |
| Wu et al,
2016 | Osteosarcoma | - | CTNND1 | - | ↑miR-409-3p:
Invasion and Migration↓ (U2OS, SAOS-2) | - | (36) |
| Zhang et al,
2017 | | - | ELF2 | ↑miR-409-3p:
Proliferation↓, Cell cycle↓, Apoptosis↑: ΔmiR-409-3p:
Proliferation↑, Cell cycle↑, Apoptosis↓ (MG63, SaOS-2) | - | - | (37) |
| Wu et al,
2019 | | - | ZEB1 | ↑miR-409-3p:
Proliferation↓ (MG63, HOS) | ↑miR-409-3p:
Invasion↓ (MG63, HOS) | - | (38) |
| Long et al,
2020 | | Circ0000285 | IGFBP3 | ↑miR-409-3p:
Proliferation↓, Colony formation↓, Apoptosis↑; ΔmiR-409-3p:
Proliferation↑, Colony formation↑, Apoptosis↓ (SJSA1, U2OS) | ↑miR-409-3p:
Invasion and Migration↓; ΔmiR-409-3p: Invasion and Migration↑
(SJSA1, U2OS) | - | (39) |
| Zhang et al,
2021 | | CircATRNL1 | LDHA | - | - | ↑miR-409-3p:
Glycolysis↓ (Saos-2); ΔmiR-409-3p: Glycolysis↑ (MG63) | (112) |
| Li et al,
2012 | GC | - | PHF10 | ↑miR-409-3p:
Proliferation↓, Colony formation↓, Cell cycle↓, Apoptosis↑
(SGC-7901) | - | - | (40) |
| Zheng et al,
2012 | | - | RDX | - | ↑miR-409-3p:
Invasion and Migration↓ (SGC-7901, HGC-27, MKN45) | - | (41) |
| Yu et al,
2021 | | CircNEK9 | MAP7 | ↑miR-409-3p:
Proliferation↓, Colony formation↓, Cell cycle↓; ΔmiR-409-3p:
Proliferation↑, Colony formation↑, Cell cycle↑ (XGC-1, MKN45) | ↑miR-409-3p:
Invasion and Migration↓; ΔmiR-409-3p: Invasion and Migration↑
(XGC-1, MKN45) | - | (42) |
| Wang et al,
2020c | | Circ0001023 | PHF10 | ↑miR-409-3p:
Proliferation↓, Colony formation↓, Apoptosis↑ (AGS); ΔmiR-409-3p:
Proliferation↑, Colony formation↑, Apoptosis↓ (MKN-28,
SGC-7901) | ↑miR-409-3p:
Invasion and Migration↓ (AGS); ΔmiR-409-3p: Invasion and Migration↑
(MKN-28, SGC-7901) | - | (44) |
| Liu et al,
2015 | CRC | - | NLK | ↑miR-409-3p:
Proliferation↓, Colony formation↓, Apoptosis↑ (SW480, SW1116) | ↑miR-409-3p:
Invasion and Migration↓ (SW480, SW1116) | - | (45) |
| Bai et al,
2015 | | - | GAB1 | - | ↑miR-409-3p:
Invasion and Migration↓; ΔmiR-409-3p: Invasion and Migration↑
(HCT116, RKO) | - | (46) |
| Tan et al,
2016 | | - | beclin-1 | ↑miR-409-3p:
Proliferation↓, Colony formation↓, Apoptosis↑ (LoVo Oxa R) | - | ↑miR-409-3p:
Chemoresistance↓, autophagic activity↓ (LoVo Oxa R) | (48) |
| Han et al,
2020 | | - | ERCC1 | ↑miR-409-3p: Cell
cycle↓, Apoptosis↑; ΔmiR-409-3p: Cell cycle↑, Apoptosis↓
(HCT-116/L-OHP) | ↑miR-409-3p:
Invasion and Migration↓: ΔmiR-409-3p: Invasion and Migration↑
(HCT-116/L-OHP) | ↑miR-409-3p:
Chemoresistance↓; ΔmiR-409-3p: Chemoresistance↑
(HCT-116/L-OHP) | (50) |
| Chen et al,
2022 | | LINC00630 | HK2 | ↑miR-409-3p:
Proliferation↓, Apoptosis↑ (SW480, HCT116) | - | ↑miR-409-3p:
Glycolysis↓ (SW480, HCT116) | (51) |
| Gharpure et
al, 2018 | OC | - | FABP4 | - | ↑miR-409-3p:
Invasion and Migration↓; ΔmiR-409-3p: Invasion and Migration↑
(HeyA8 MDR, Ovcar5) | - | (53) |
| Cheng et al,
2018 | | - | FIP200 | ↑miR-409-3p:
Proliferation↓, Colony formation↓, Apoptosis↑ (OV-1063,
OV-1063R) | - | ↑miR-409-3p:
Chemoresistance↓; Autophagic activity↓ (OV-1063 R) | (54) |
| Li et al,
2020b | | - | RAB10 | ↑miR-409-3p:
Proliferation↓, Colony formation↓, Apoptosis↑; ΔmiR-409-3p:
Proliferation↑, Colony formation↑, Apoptosis↓ (SKOV3) | Δ miR-409-3p:
Migration↑ (SKOV3) | - | (56) |
| Wang et al,
2019 | ccRCC | - | PDK1 | - | - | ↑miR-409-3p:
Glycolysis↓ (A-498,769-P) | (58) |
| Xu et al,
2013 | BCa | - | MET | - | ↑miR-409-3p:
Invasion and Migration↓ (T24, 5637) | - | (59) |
| Xu et al,
2016 | | - | MET | ↑miR-409-3p: Cell
motility↓ (T24, UM-UC-3) | - | - | (60) |
| Zhao et al,
2018 | PTC | - | CCDN2 | ↑miR-409-3p:
Proliferation↓, Colony formation↓, Cell cycle↓ (TPC-1,
GLAG-66) | - | - | (62) |
| Leivonen et
al, 2017 | DLBCL | - | - | ↑miR-409-3p: Cell
viability↓ (SU-DHL-4, SV_Co10) | - | ↑miR-409-3p:
Chemoresistance↓ (SU-DHL-4, SV_Co10) | (64) |
| Xie et al,
2023 | AML | - | RAB10 | ↑miR-409-3p:
Proliferation↓, Apoptosis ↑ (THP-1) | - | - | (66) |
| Chen and Dai,
2018 | TSCC | - | RDX | ↑miR-409-3p:
Proliferation↓; ΔmiR-409-3p: Proliferation↑ (Tca8113) | ↑miR-409-3p:
Invasion and Migration↓; ΔmiR-409-3p: Invasion and Migration↑
(Tca8113) | - | (70) |
| Josson et
al, 2014 | PCa | - | RSU1 | ΔmiR-409-3p:
Apoptosis↑ (ARCaPM) | ↑miR-409-3p:
Invasion and Migration↑ (ARCaPE-409), EMT↑
(ARCaPE, LNCaP) | - | (71) |
| Gururajan et
al, 2014 | | - | - | - | ↑miR-409-3p: EMT↑;
ΔmiR-409-3p: EMT↓ (ARCaPM,ARCaPE) | - | (72) |
| Josson et
al, 2015 | | - | RSU1 | ↑miR-409-3p:
Proliferation↑ (ARCaPE, SON-409 CM, SON-C CM) | ↑miR-409-3p: EMT↑
(ARCaPE, SON-409 CM, C4-2B PCa) | - | (73) |
| Ding et al,
2021 | Intracranial artery
tumors | Circ_DOCK1 | MCL1 | ↑miR-409-3p:
Proliferation↓, PCNA↓, Apoptosis↑; ΔmiR-409-3p: proliferation↑,
PCNA↑, Apoptosis ↓ (HBVSMCs) | - | - | (78) |
| Cao et al,
2017 | Glioma | - | HMGN5 | ↑miR-409-3p:
Proliferation↓, Colony formation↓, Cell cycle↓ (U251); ΔmiR-409-3p:
Proliferation↑, Colony formation↑, Cell cycle↑ (U87) | ↑miR-409-3p:
Invasion↓ (U251); ΔmiR-409-3p: Invasion↑ (U87) | - | (79) |
| Ma et al,
2022 | | circATRNL1 | PDK1 | ↑miR-409-3p: Cell
cycle↓, Proliferation↓; ΔmiR-409-3p: Cell cycle↑, Proliferation↑
(LN-229 and T98-G) | - | - | (80) |
| Weng et al,
2012 | Fibrosarcoma | - | ANG | ↑miR-409-3p:
Proliferation↓ (HUVEC, HT1080) | - | ↑miR-409-3p:
Vascularization↓ (HUVEC, HT1080) | (81) |
| Chang et al,
2023 | HCC | - | BRF2 | - | ↑miR-409-3p:
Invision and Migration↓; ΔmiR-409-3p: Invision and Migration↑
(Huh-7) | - | (83) |
| Li et al,
2023 | | LINC00886 | RAB10 | ↑miR-409-3p:
Proliferation↓, Apoptosis ↑; ΔmiR-409-3p: Proliferation↑,
Apoptosis↓ (Hep3B, Huh7) | ↑miR-409-3p:
Invision and Migration↓; ΔmiR-409-3p: Invision and Migration↑
(Hep3B, Huh7) | - | (84) |
Growth and apoptosis
Several diseases, particularly cancer, are caused by
a disruption in the balance between cell growth and death. Targeted
genes of miR-409-3p are closely associated with the development and
apoptotic processes of tumor cells. In most cases, miR-409-3p acts
as a tumor suppressor. A tyrosine kinase receptor called MET
proto-oncogene, receptor tyrosine kinase (MET) triggers the
mitogenic signaling pathway, which leads to the development of
tumors and malignant transformation (93,94).
In LUAD, miR-409-3p suppressed the proliferation of
A549 and SPC-1 NSCLC cells and promoted caspase-3-dependent
apoptosis by directly targeting MET (12). Subsequently, Song et al
(14) reported that miR-409
upregulation clearly inhibited proliferation by inhibiting spindlin
1 (SPIN1), a component of the SPIN/SSTY gene family. A study
by Qu et al (15) unveiled
that lncRNA zinc finger E-box binding homeobox 1 antisense 1
(ZEB1-AS1) functioned as a competing endogenous RNA (ceRNA) to
facilitate NSCLC tumorigenesis by regulating the
ZEB1-AS1/miR-409-3p/ZEB1 signaling pathway. Mounting
evidence suggests that the ZEB1-AS1 signaling pathway may aggravate
NSCLC cell proliferation and reduce apoptosis (15). Furthermore, Yin et al
(16) revealed that by regulating
the miR-409-3p/hexokinase 2 (HK2) and lactate dehydrogenase
A (LDHA) axis, double homeobox A pseudogene 8 (DUXAP8)
promoted the viability of A549 and H1299 NSCLC cells. Liu et
al (17) also demonstrated
that loss of miR-409-3p inhibited the expression of superoxide
dismutase 1 (SOD1) and its oncogenic activity. Recently, a
publication described carbonyl reductase 3 (CBR3)-AS1 as a ceRNA
interacting with miR-409-3p in NSCLC. Furthermore, NSCLC
development and miR-409-3p suppression were facilitated by the
overexpression of CBR3-AS1. In addition, the
CBR3-AS1/miR-409-3p/SOD1 axis could promote the
proliferation of H1650 NSCLC cells (20). In addition to CBR3-AS1,
circ_0079530 also had an oncogenic role in NSCLC through the
inhibition of miR-409-3p. Yang et al (21) revealed that the
circ_0079530/miR-409-3p/aquaporin 4 (AQP4) axis promoted the
proliferation and invasion of H1270 and A549 NSCLC cells.
According to Zhang et al (26), overexpression of miR-409-3p, which
has a crucial role in cell development, significantly hampered the
proliferation and colony formation of MDA-MB-231 and MDA-MB-468 BC
cells and accelerated apoptosis by targeting AKT serine/threonine
kinase 1 (AKT1), which was essential for cell growth
(95). By contrast,
anti-miR-409-3p led to the inhibition of the proliferation of T47D
BC cells. In addition, despite the fact that upregulation of
miR-409-3p did not affect AKT1 mRNA, the protein level
decreased, indicating that the regulation of AKT1 by miR-409-3p was
post-transcriptional. Ma et al (27) also concluded that increased
miR-409-3p expression impeded cell proliferation and regulated the
process of BC by targeting the important nuclear transcription
factor ZEB1. Furthermore, miR-409-3p was predicted to mainly
target caspases, BCL2 and cell cycle cyclins involved in anti-tumor
drug-induced apoptosis (27).
However, the finding was not demonstrated by a corresponding
functional analysis in animals. Recently, by targeting miR-409-3p,
circRNA tripartite motif-containing 28 (circTRIM28) was reported to
upregulate oncogenic high mobility group AT-hook 2 (HMGA2)
in BC. Regulation of the miR-409-3p/HMGA2 axis also
accounted for the influence of circTRIM28 on cell proliferation and
apoptosis (30).
The direct binding between miR-409-3p and HPV16/18
E6 mRNA was identified in cervical dysplasia tissues. Despite the
regulation of miR-409-3p levels having no effect on colony
development, miR-409-3p overexpression also attenuated the
proliferation of CaSki, C-4I and Hela cells. Vice versa, miR-409-3p
knockdown promoted the proliferation of CaSki and C-4I CC cells,
but there was no change in HeLa CC cells (32). Cui et al (33) revealed that miR-409-3p was
involved in CC progression through the ceRNA mechanism. On the one
hand, circ0000745 regulated activating transcription factor 1
(ATF1) expression to regulate CC progression by sponging
miR-409-3p. Of note, the proliferation of cells was negatively
related to the expression level of miR-409-3p. Co-transfection of
miR-409-3p with circ0000745 alleviated the promoting effects of
overexpression of circ0000745 on CC cell proliferation (33). On the other hand, circRNA FAT
atypical cadherin 1 (circFAT1) also had a crucial role in CC by
acting as a sponge for miR-409-3p. Increased circFAT1 promoted CC
progression, including enhancing proliferation and repressing
apoptosis via the miR-409-3p/cyclin-dependent kinase 8
(CDK8) axis (34). In
addition, Wu et al (35)
reported that miR-409-3p, targeted by circEPSTI1, regulated the
expression of solute carrier family 7 membrane 11 (SLC7A11)
expression. The proliferation of Hela and CaSki cells in CC was
influenced by the circEPSTI1-miR-409-3p-SLC7A11 axis.
With regards to osteosarcoma, Zhang et al
(37) revealed that E74-like
factor 2 (ELF2), an ETS family transcription factor, served
as an oncogene and played prominent roles in cell proliferation,
differentiation and apoptosis. Therefore, the ectopic expression of
miR-409-3p clearly hampered cell proliferation, induced G0/G1
arrest and promoted apoptosis in osteosarcoma cells. Shortly after,
Wu et al (38) found that
ZEB1 was also regulated by miR-409-3p in HOS and MG63
osteosarcoma cells, thus inhibiting cell proliferation.
Furthermore, Long et al (39) discovered that circ0000285, which
is increased in osteosarcoma, acted as a ceRNA of miR-409-3p to
promote insulin-like growth factor binding protein 3
(IGFBP3). More importantly, miR-409-3p overexpression
clearly reduced cell viability and colony numbers of osteosarcoma
cells, and enhanced cell apoptosis, which was reversed by
IGFBP3. In conclusion, circ0000285 could promote the
proliferation and suppress the apoptosis of osteosarcoma via the
miR-409-3p/IGFBP3 axis. It was also the first ceRNA
mechanism to be explored in osteosarcoma.
MiR-409-3p was also mentioned in connection with the
cellular biological processes in GC. The 2020 study by Wang et
al (44) demonstrated that,
compared with adjacent tissue, circ0001023 expression was
conspicuously upregulated. Mechanistically, miR-409-3p was
positively correlated with PHD finger protein 10 (PHF10),
which was identified as a direct target of circ0001023 and
negatively modulated by circ0001023 via the ceRNA network.
Circ0001023 silencing could inhibit the proliferation and trigger
the apoptosis of GC cells, while miR-409-3p inhibitors could
partially inhibit apoptosis and promote the proliferation of GC
cells. Similarly, Li et al (40) also found that miR-409-3p
overexpression reduced the level of PHF10, thus markedly
limiting the proliferation and colony numbers of tumor cells and
slowing the transition from the G1 to the S phase of the cell
cycle. CircRNA NIMA-related kinase 9, which is enhanced in GC
cells, could promote the proliferation of GC cells by targeting the
miR-409-3p/microtubule associated protein 7 (MAP7) axis.
MiR-409-3p has been shown to bind to MAP7 mRNA directly,
causing cell cycle arrest in the G0/G1 phase and limiting GC cell
growth and colony formation (42). Furthermore, consistent with
previous findings, the expression of miR-409-3p had been confirmed
to decrease, and high levels of miR-409-3p were found to be able to
inhibit proliferation and trigger apoptosis. Surprisingly, when GC
cells were treated with precision hyperthermia, miR-409-3p could
positively regulate the targeting gene Kruppel-like factor 17
(KLF17) and induce GC cell apoptosis (43).
Liu et al (45) showed that miR-409-3p
overexpression in CRC targeted nemo-like kinase (NLK), which
is implicated in the formation, progression and signaling pathways
of cancer. In addition, in cells resistant to oxaliplatin, a
platinum-based chemotherapeutic drug, such as oxaliplatin-resistant
LoVo (Oxa-R) and HCT-116/L-OHP cells, miR-409-3p was found to
regulate cell proliferation, the cell cycle and apoptosis by
targeting beclin-1 and ERCC excision repair 1
(ERCC1), respectively (48,50). This phenomenon was also observed
in drug-resistant OV-1063 cells by inhibiting FIP200
(54). In CRC, LINC00630
functioned as an endogenous sponge for miR-409-3p and relieved the
inhibition of miR-409-3p on HK2. By repressing miR-409-3p,
LINC00630 upregulated HK2 and promoted cell proliferation,
and inhibited apoptosis in the CRC cell lines SW480 and HCT116
(51).
In addition, another group provided evidence that
miR-409-3p had effects on OC cells, including proliferation, colony
formation and apoptosis, by targeting RAB10, member RAS oncogene
family (RAB10), whereas its downregulation inhibited the
apoptosis and promoted the proliferation as well as colony
formation (56).
In BCa, Xu et al (60) also observed that MET could
regulate 14q32.2 miRNA clusters, including miR-409-3p, and
miR-409-3p could directly target MET. In addition,
miR-409-3p participated in the miR-433-mediated inhibition of cell
motility.
Of note, Zhao et al (62) reported that miR-409-3p
upregulation in PTC weakened PTC cell proliferation and induced
cell cycle arrest in G0/G1 phase by modulating cyclin D2. In DLBCL,
miR-409-3p overexpression significantly inhibited the proliferation
of SU-DHL-4 compared with SV_Co10 (64). In AML, Xie et al (66) revealed that miR-409-3p negatively
regulated the expression of RAB10, sequentially inhibiting
proliferation and promoting apoptosis. In TSCC, Chen and Dai
(70) concluded that miR-409-3p
negatively regulated the expression of radixin (RDX),
sequentially inhibiting proliferation in vitro. Furthermore,
miR-409-3p could act as an oncogene. In PCa, Josson et al
(71,73) noted that through the negative
regulation of Ras suppressor protein 1, miR-409-3p enhanced cell
proliferation and inhibited cell apoptosis. Simultaneously,
circular dedicator of cytokinesis 1, which promoted proliferation
and inhibited apoptosis in human brain vascular smooth muscle cells
through the miR-409-3p/myeloid cell leukemia sequence 1
(MCL1) axis, sponged miR-409-3p to boost MCL1
expression, which helped inhibit certain targets in the treatment
of intracranial artery tumors (78). Of note, a study by Cao et
al (79) found that high
mobility group nucleosome binding domain 5 (HMGN5) acted as
an oncogene in glioma. By suppressing HMGN5, miR-409-3p
suppressed glioma cell proliferation and colony formation and
blocked glioma cells in the G0/G1 phase. Angiogenin (ANG), a
gene involved in cell proliferation, has also been studied in the
context of fibrosarcoma. HT1080 cell transfection with a miR-409-3p
mimic could suppress cell proliferation by targeting ANG
(81). In general, miR-409-3p has
been strongly connected with cell growth and apoptosis in various
cancers. In HCC, Li et al (84) found that silencing LINC00886
upregulated the expression of miR-409-3p, weakened the
proliferation of HCC cells (Hep3B, Huh7) and promoted apoptosis.
Conversely, it promoted the proliferation of Hep3B as well as Huh7
proliferation and inhibited apoptosis.
Migration and invasion
The process through which independent cells travel
from the main tumor to distal organs through blood arteries or
lymphatic vessels is referred to as metastasis and invasiveness of
malignant tumors (96).
Metastasis, the most prominent feature of malignant tumors, is
responsible for the majority of cancer-related fatalities and has
become a major impediment to cancer treatment. MiR-409-3p
influences tumor migration and invasion by altering target genes to
further regulate the progression of cancer (Fig. 3).
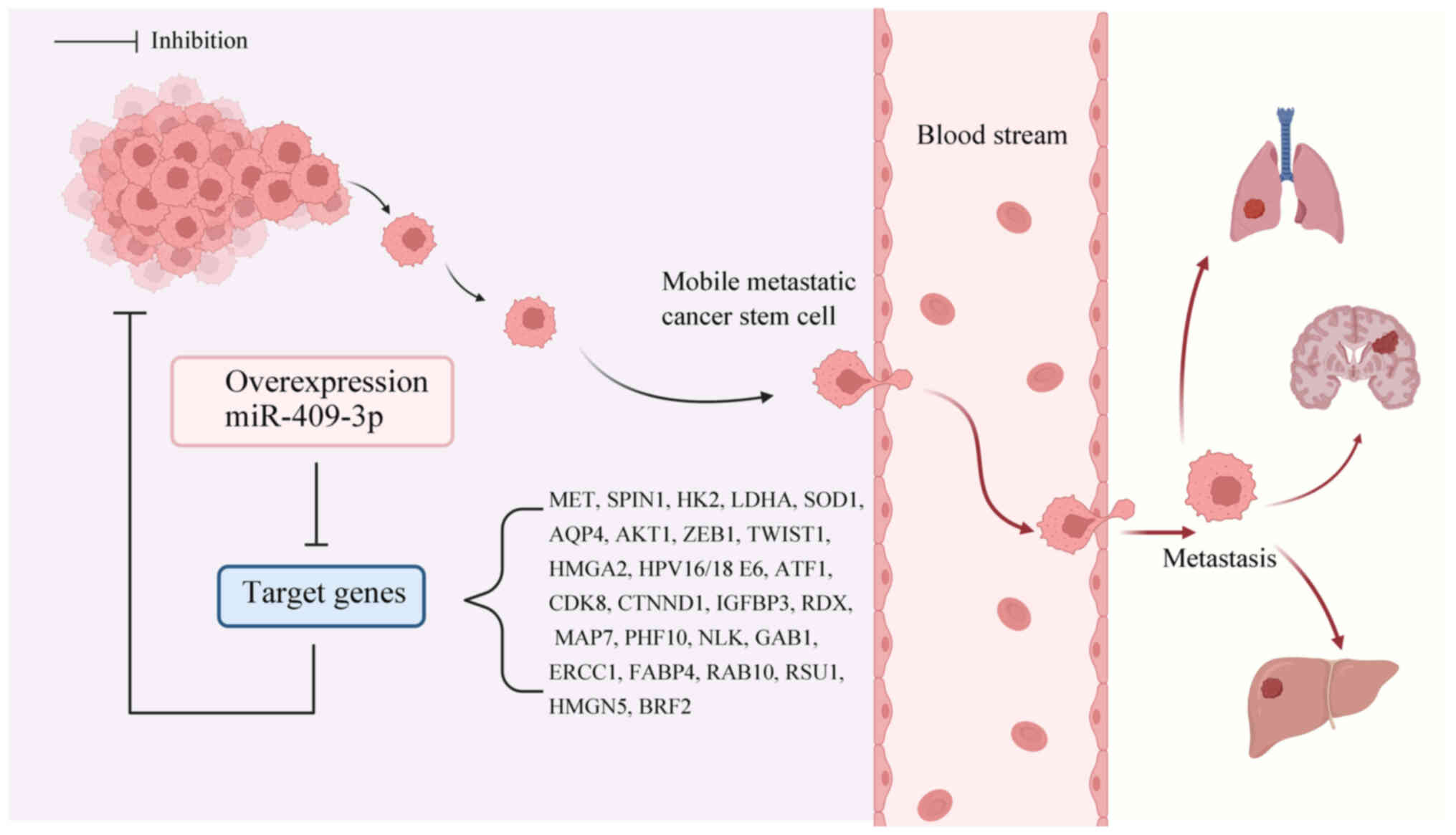 | Figure 3Effect of miR-409-3p target genes on
migration and invasion of cancer cells. Overexpression of
miR-409-3p inhibits downstream target genes, such as ZEB1, MET and
CDK8, and further regulates cell invasion and metastasis. Created
with BioRender software (http://biorender.com). MiR, microRNA; MET, MET
proto-oncogene, receptor tyrosine kinase; SPIN1, spindlin1; HK2,
hexokinase 2; LDHA, lactate dehydrogenase A; SOD1, superoxide
dismutase 1; AQP4, aquaporin 4; AKT1, AKT serine/threonine kinase
1; ZEB1, zinc finger E-box binding homeobox 1; TWIST1, Twist family
bHLH transcription factor; HMGA2, high mobility group AT-hook 2;
ATF1, activating transcription factor 1; CDK8, cyclin dependent
kinase 8; CTNND1, catenin-δ1; IGFBP3, insulin like growth factor
binding protein 3; RDX, radixin; MAP7, microtubule associated
protein 7; PHF10, PHD finger protein 10; NLK, nemo like kinase;
GAB1, GRB2 associated binding protein 1; ERCC1, ERCC excision
repair 1; FABP4, fatty acid binding protein 4; RAB10, RAB10, member
RAS oncogene family; RSU1, Ras suppressor protein 1; HMGN5, high
mobility group nucleosome binding domain 5; BRF2, BRF2 RNA
polymerase III transcription initiation factor subunit. |
A consensus has been reached that
epithelial-mesenchymal transition (EMT) plays a significant role in
cancer metastasis (97), which is
characterized by decreased E-cadherin expression and increased
N-cadherin/vimentin expression (98,99) and allows the cells to acquire the
properties of migration and invasion. Twist family bHLH
transcription factor 1 could induce EMT and participate in cancer
metastasis (100). A Transwell
assay performed by Su et al (29) found that miR-409-3p overexpression
could limit the EMT in MCF7 BC cells. KLF17 is a member of a
conserved family of transcription factors that regulates cell
migration and invasion. A certain range of heat treatments
accelerated the expression of miR-409-3p in SGC-7901 and BGC-823 GC
cells and indirectly upregulated KLF17, further inhibiting
the migration, invasion and the EMT pathway (43). In addition, Josson et al
(71,73) found an elevated level of
miR-409-3p/5p in PCa, suggesting that it may promote tumorigenesis,
EMT and bone metastasis of PCa. In addition, increased miR-409-3p
has also been observed in PCa cells and tissues in bone
metastasis.
Matrix metalloproteinases (MMPs), a type of
proteolytic enzyme that break down the structural components of
extracellular matrix and contributes to the loss of intercellular
connection, promote cell migration and invasion, in addition to the
EMT process.
In NSCLC, miR-409-3p mimics were able to suppress
the protein levels of phosphorylated AKT (p-AKT), ultimately
resulting in the decrease of the expression of MMP2 and MMP9, which
in turn reduced the migration and invasion of A549 and SPC-1 NSCLC
cells (12). In addition, a study
by Song et al (14) found
that SPIN1 is associated with cancer metastasis, as a direct
miR-409-3p target in A549 NSCLC cells. In line with this,
miR-409-3p overexpression evidently inhibited A549 NSCLC cell
invasion and migration, but this inhibition could be reversed by
anti-miR-409-3p. In addition, Yin et al (16) reported that the inhibition of
miR-409-3p in NSCLC rescued the number of migrated H1299 and A549
NSCLC cells transfected with small interfering RNAs targeting
DUXAP8. Wang et al (19)
verified the above mechanism in another NSCLC cell line, H1299.
They used miR-409-3p inhibitor and negative control, leading to the
weakening of the inhibitory effect on SPIN1, which then
promoted H1299 NSCLC cell metastasis and invasion. Liu et al
(20) showed that miR-409-3p
mimics also mitigated the effects of CBR3-AS1 on NSCLC cell
invasion and migration. A ceRNA named circ_0079530 was
overexpressed in NSCLC cells and sequestered miR-409-3p. Increased
rates of cell migration and invasion are carried on by low levels
of the miRNA, on the contrary, the increase of migration and
invasion rate (21).
By targeting AKT1, Zhang et al
(26) found that miR-409-3p
overexpression also hindered BC cell motility and invasion. Ma
et al (27) also found
that a low abundance of miR-409-3p and high expression of
ZEB1 in BC may be responsible for the high metastasis rate
of BC. Yang et al (30)
reported that circRNA tripartite motif-containing 28 (circTRIM28)
contributed to the migration and invasion of BC cells by
downregulating miR-409-3p.
An array of studies observed that when using
particular inhibitors and mimics of miR-409-3p in SiHa CC cell
lines, miR-409-3p displayed a pattern of overexpressed and
downregulated HPV16/18 E6, boosting or inhibiting migration,
respectively (32). Despite the
inhibition of miR-409-3p on cell migration and invasion reported by
Cui et al (33),
miR-409-3p may suppress ATF1 expression, and thus negatively
regulate the migration and invasion of CaSki and Siha CC cells. The
tumor suppressor role of miR-409-3p lies at least in part in the
negative regulation of CDK8. miR-409-3p/CDK8
modulated CaSki and C-33A CC cell invasion and migration behaviors
(34).
In osteosarcoma, Wu et al (36) found that functionally decreased
miR-409-3p markedly increased the migration and invasion of
osteosarcoma cell lines (U2OS and SAOS-2) by directly targeting
catenin-δ1, further promoting the metastasis of osteosarcoma. In a
different study, the miR-409-3p-mediated inhibition of the invasion
of another two osteosarcoma cell lines (MG63 and HOS) was examined
and ZEB1 was identified as a new target gene (38). Another publication reported that
overexpression of miR-409-3p clearly reduced SJSA1 as well as U2OS
cell invasion and migration in osteosarcoma, whereas its
downregulation increased migration and invasion rates (39).
In GC, one group indicated that miR-409-3p
functioned as a tumor suppressor by validating its targeting of
PHF10, inhibiting AGS GC cell invasion and migration, and
stimulating invasion and migration of MKN-28 and SGC-7901 GC cells
(44). In addition, the
constitutive expression of miR-409-3p in three GC cell lines,
including SGC-7901, HGC-27 and MKN45, reduced their migration and
invasion rates (41).
Furthermore, Yu et al (42) also found that this miRNA could
downregulate MMP2 and MMP9, while MAP7 was
another gene tightly associated with EMT and could partly rescue
its expression, thereby boosting the invasion and migration of
XGC-1 and MKN45 cells in GC. In addition, a similar regulation
mechanism pattern was also observed by Xu et al (59) in T24 and 5637 BCa cells in that
miR-409-3p may regulate cell migration and invasion in part by
indirectly controlling MMP2 and MMP9.
In CRC, Liu et al (45) showed that high expression of
miR-409-3p induced SW480 and SW1116 CRC cell migration and invasion
by repressing the expression of NLK. Furthermore, Bai et
al (46) detected that
miR-409-3p was a metastasis-specific miRNA in CRC. MiR-409-3p
directly linked to cell invasion and migration in CRC through
targeting GRB2-associated binding protein 1 (46). Another study found that miR-409-3p
has an inhibitory role in CRC, where it noticeably reduced HCT-116
and HCT-116/L-OHP migration and invasion, which occurred by
suppressing ERCC1 under certain experimental conditions
(50).
Simultaneously, Chen and Dai (70) also reported that miR-409-3p
strongly modulated the development of TSCC by targeting RDX,
thus inhibiting or stimulating the migration and invasion of
Tca8113 TSCC cells.
In OC, miR-409-3p reversely regulated fatty acid
binding protein 4 (FABP4) in OC cell lines (HeyA8 MDR and
Ovcar5). In addition, hypoxia and hypoxia-inducible factor-1α
(HIF-1α) could reduce miR-409-3p, which may eliminate its
inhibitory effect on FABP4, leading to increased
FABP4 levels to further promote the metastasis of OC cells
(53). MiR-409-3p suppressed cell
migration of SKOV3 OV cells (56), whose mediated target RAB10,
a type of small GTPases belonging to the Ras protein family
(101). The MET oncogene
is a tyrosine kinase with a well-defined receptor, which is
generally upregulated in various cancers. Tumor invasion and
metastasis are aided by MET activation caused by aberrant
paracrine stimulation of hepatocyte growth factor (102). In NSCLC, miR-409-3p inhibited
the invasion and migration of A549 and SPC-1 NSCLC cells by
negatively regulating MET, thereby exerting a
tumor-suppressive effect (12).
Similarly, following the transfection of miR-409-3p into T24 and
5637 BCa cell lines, the expression of MET, which could
induce migration and invasion, was directly reduced (59). In addition, the downregulation of
HMGN5 also accounted for the anti- and pro-metastatic action
of miR-409-3p in U251 and U87 glioma cells, respectively. In
addition, the downregulation of HMGN5 also accounted for the
anti-metastatic action of miR-409-3p in glioblastoma (79). In HCC, Chang et al
(83) observed that the relative
migration and invasion rate of Huh-7 cells treated with miR-409-3p
mimics was significantly decreased by regulating BRF2 RNA
polymerase III transcription initiation factor subunit
(BRF2). After silencing LINC00886, the expression level of
miR-409-3p was upregulated, which weakened the invasion and
migration of HCC cells (Hep3B, Huh7). Conversely, overexpression of
LINC00886 has the opposite effect (84).
Autophagy, chemoresistance and
radioresistance
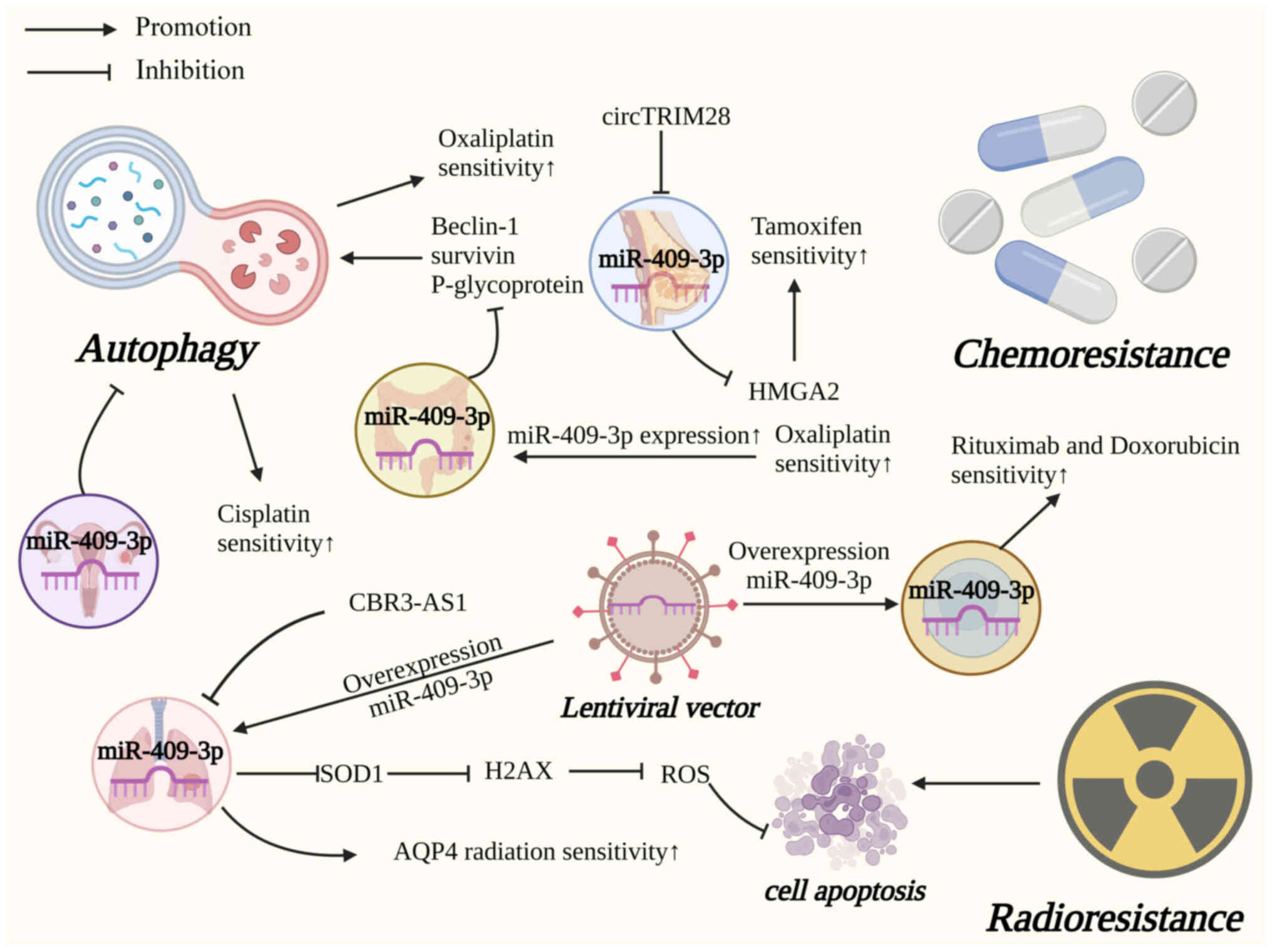
MiR-409-3p regulates numerous genes involved in
autophagy, chemoresistance and radioresistance (Fig. 4).
Generally speaking, autophagy is a normal
physiological process within cells, which involves loading damaged
proteins and organelles from the cytoplasm into autophagosomes, and
then fusing with lysosomes for degradation and reuse (103). Phosphatidylinositol 3-kinase
class III and beclin-1 are involved in autophagosome formation
(104). In a previous study,
when the ratio of light chain (LC)3I and LC3II decreased, it was
used as one of the criteria for autophagy. Autophagy is thought to
be a primary mechanism of cancer cell chemoresistance (105,106). Recent studies have discovered
that abnormal miRNA expression has a role in the molecular
mechanisms of chemoresistance. The regulatory function of
miR-409-3p on cancer cells' chemoresistance has been discussed in
previous studies. In BC, circTRIM28 knockdown improved tamoxifen
sensitivity through the miR-409-3p/HMGA2 axis (30). Furthermore, miR-409-3p, as a
promising therapeutic target in CRC, has been shown to improve CRC
cell susceptibility to chemotherapeutic drugs such as oxaliplatin
(48,50). Of note, Tan et al (48) found that miR-409-3p expression was
clearly upregulated in CRC cells with sensitivity to oxaliplatin,
and promoted the chemosensitivity of LoVo Oxa-R CRC cells by
inhibiting beclin-1-mediated autophagy. Most importantly, the
autophagy-related protein LCII was higher and lower expressed in
LoVo Oxa-R and LoVo CRC cells, respectively. Furthermore, Han et
al (50) discovered that
curcumin treatment increased the expression of miR-409-3p in
HCT-116/L-OHP CRC cells in a concentration-dependent manner,
increasing the sensitivity of CRC cells to chemotherapeutic drugs
through the partial inhibition of survivin, P-glycoprotein and
multidrug resistance-related protein. Of note, Cheng et al
(54) found that miR-409-3p
acting as an anti-cancer factor enhanced the sensitivity of OC
cells to cisplatin treatment by inhibiting FIP200-mediated
autophagy. Furthermore, in contrast to OC cells responsive to
cisplatin treatment, cells resistant to cisplatin treatment had a
lower miR-409-3p expression and significantly higher autophagic
activity. It was indicated that miR-409-3p could also be a
therapeutic target in OC resistance. In addition, DLBCL is a
heterogeneous disease and the most common lymphoid malignancy.
Using lentiviral vectors, Leivonen et al (64) found that the functional
overexpression of miR-409-3p improved the sensitivity of DLBCL
cells to chemotherapeutic agents such as rituximab and doxorubicin.
These findings suggested that miR-409-3p can enhance the effects of
chemotherapeutic agents in several types of cancers and act as a
chemosensitizer. Of note, despite abundant evidence pinpointing
that chemoresistance is caused by autophagy, two research teams
found the opposite results: The first was that L-6-hydroxymethyl-c
hiro-inositol 2[R]-2-O-methyl-3-O-octadecyl carbonate, an AKT
inhibitor, could accelerate radiosensitivity by guiding autophagy
(107), and the other was that
rapamycin, an inhibitor of mTOR, radiosensitized HCC827 NSCLC cells
by inhibiting the action of phosphatase and tensin homolog
(108), a tumor suppressor, to
positively regulate autophagy (109). Hence, based on the relationships
between autophagy and treatment resistance, whether these
contrasting findings reflect the differences between different
treatment regimens such as chemotherapy or radiotherapy and
different types of cancer is worthy of further exploration.
Another popular cancer treatment is radiotherapy.
In NSCLC, Liu et al (20)
found that CBR3-AS1/miR-409-3p/SOD1 signaling reduced
reactive oxygen species levels by reducing H2AX foci, which in turn
decreased apoptosis following ionizing radiation. In addition,
H1270 and A549 NSCLC cells with high expression of miR-409-3p
exhibited enhanced sensitivity to irradiation by direct targeting
of AQP4 (21).
Cell glycolysis
To meet their energy needs for proliferation and
metastasis, cancer cells mostly rely on glycolysis. Aerobic
glycolysis is an abnormal process of energy metabolism in cancer
(110). Rate-limiting enzymes
HK2, LDHA and GLUT1 are the key components of the glycolysis
pathway. In NSCLC, DUXAP8 served as an miR-409-3p sponge to promote
HK2 and LDHA expression. By regulating the
miR-409-3p/HK2/LDHA axis, DUXAP8 promoted glycolysis
(16). In addition, Chen et
al (51) demonstrated that
LINC00630 controlled glycolysis primarily by targeting the
miR-409-3p/HK2 axis, which may explain the development of
CRC and offer a possible target for its treatment. Likewise,
ATF1, an miR-409-3p target, conspicuously reduced the
extracellular acidification rate, glucose uptake and lactic acid
generation in CC (33). A study
by Wang et al (58)
indicated that miR-409-3p served as a suppressor in glycolysis. In
the A-498 and 769-P ccRCC cell lines, HIF-1α, an important molecule
for cancer cells to respond to a hypoxic microenvironment (111), could induce the decrease of
miR-409-3p expression and weaken the inhibitory effect of
miR-409-3p on 3-phosphoinositide dependent kinase 1, a molecule
required for metabolic activation. In osteosarcoma, miR-409-3p
decreased LDH2 expression, which was associated with a
decrease in glucose absorption, lactate generation and
extracellular acidification rate (112).
Angiogenesis
Angiogenesis refers to the process of producing new
blood vessels from pre-existing posterior venules of capillaries
and has a critical role in embryonic development, wound healing and
inflammation (113). In
addition, the growth and metastasis of tumors largely depend on
angiogenesis (114). MiR-409-3p
appears to have a significant role in tumor angiogenesis, according
to new research. The capacity of HT1080 fibrosarcoma cells to form
tubular structures on Matrigel® was significantly
diminished when miR-409-3p was overexpressed compared with control
cells, according to in vitro investigations. However, this
inhibition could be reversed by ANG overexpression (81).
5. Biological roles of miR-409-3p in cancer
in vivo
MiR-409-3p has been studied in various cancers and
it has been found to have a conspicuous role in arresting
tumorigenesis or promoting tumor progression (Table IV). A large number of studies
using xenograft models have indicated a tumor suppressor role for
miR-409-3p, since its upregulation led to inhibitory effects on
tumor growth and distant metastasis. By contrast, miR-409-3p could
act as an oncogene to significantly promote tumorigenesis in the
xenograft models of PCa (71,73). Most importantly, in the xenograft
model of CRC, overexpression of miR-409-3p in LoVo Oxa-R CRC cells
injected into nude mice restricted tumor growth and improved the
sensitivity to chemotherapeutic drugs (48). Xie et al (66) found that overexpression of
miR-409-3p could reduce the tumor volume and weight in SCID mice.
In addition, Chen and Dai (70)
discovered that the lymphatic microvessel density of TSCC nude mice
transfected with miR-409-3p was considerably reduced when compared
to the control group. At the same time, in vivo experiments
showed that the miR-409-3p overexpression group had fewer tubular
structures, and miR-409-3p promoted the development of tumors by
inhibiting cell proliferation and tumor vascularization (81). In HCC, Chang et al
(83) discovered that decreased
miR-409-3p led to the development of lung and liver metastases.
 | Table IVOutline of studies on the function of
miR-409-3p in animal models. |
Table IV
Outline of studies on the function of
miR-409-3p in animal models.
| Authors, year | Tumor type | Animal models | Results | (Refs.) |
|---|
| Song et al,
2018 | Lung cancer | 6-week-old BALB/c
mice | ↑miR-409-3p: Tumor
growth↓, lung burden↓, photonic radiance intensity of the
lungs↓ | (14) |
| Zhang et al,
2016 | BC | 4-5-week-old nude
mice | ↑miR-409-3p: Tumor
growth↓, average tumor volume and weight↓, Ki-67 antigen
staining↓ | (26) |
| Ma et al,
2016 | | 5-week-old male
athymic nude mice | ↑miR-409-3p: Tumor
size↓, tumor weight↓ | (27) |
| Zhang et al,
2017 | Osteosarcoma | 6-8-week-old male
nude BALB/c mice | ↑miR-409-3p: Tumor
volume↓, tumor weight↓ | (37) |
| Li et al,
2012 | GC | 4-week-old male
nude mice | ↑miR-409-3p: Tumor
growth↓, tumor weight↓, Ki-67 antigen staining↓, apoptosis↑ | (40) |
| Zheng et al,
2012 | | week-old
BALB/c-nu/nu | ↑miR-409-3p:
Pulmonary metastasis assays: Number and size of metastatic
nodules↓; Peritoneal dissemination↓ | (41) |
| Liu et al,
2015 | CRC | 4-week-old male
BALB/C nude mice | ↑miR-409-3p:
Peritoneal nodules↓, metastasis↓ | (45) |
| Bai et al,
2015 | | 4-5-week-old female
BALB/c-nu/nu mice | ↑miR-409-3p:
Pulmonary metastatic nodules↓, Ki-67 antigen staining↓ | (46) |
| Tan et al,
2016 | | 6-week-old female
BALB/c nude mice | ↑miR-409-3p:
Chemosensitivity↓, autophagic activity↓ | (48) |
| Gharpure et
al, 2018 | OC | Female athymic nude
mice | ↑miR-409-3p: Tumor
weight↓, number of tumor nodules↓, metastasis↓ | (53) |
| Cheng et al,
2018 | | 5-6-week-old female
BALB/c nude mice | ↑miR-409-3p: Tumor
volume↓ | (54) |
| Xie et al,
2023 | AML | 4-week-old male
SCID-Beige mice | ↑miR-409-3p: Tumor
volume↓, tumor weight↓ | (66) |
| Chen and Dai,
2018 | TSCC | 4-6-week-old female
BALB/C mice | ↑miR-409-3p: Tumor
volume↓, tumor weight↓, lymphatic metastasis↓, lymphatic
microvessel density ↓ | (70) |
| Josson et
al, 2014 | PCa | 4-week-old male
nude mice | ↑miR-409-3p: Tumor
size↑, Ki-67 antigen staining↑, EMT↑ | (71) |
| Josson et
al, 2015 | | Athymic mice | ↑miR-409-3p: Tumor
incidence↑, tumor size↑, Ki-67 antigen staining↑, EMT↑ | (73) |
| Weng et al,
2012 | Fibrosarcoma | 4-6-week-old female
BALB/c nude mice | ↑miR-409-3p: Tumor
growth↓, vascularization↓, metastasis↓, tumor sizes↓, Ki-67 antigen
staining↓ | (81) |
| Chang et al,
2023 | HCC | 4-5-week-old female
nude mice | ↑miR-409-3p: Lung
and liver metastases↑ | (83) |
6. MiR-409-3p-related signaling
pathways
Of note, by influencing target genes or being
controlled by upstream genes, miR-409-3p can alter the activities
of numerous signaling pathways. The most significant signaling
pathways influenced by miR-409-3p that contribute to tumorigenesis
include the Wnt/β-catenin pathway, PI3K/AKT/mTORC1 pathway and the
MAPK pathway (Fig. 5).
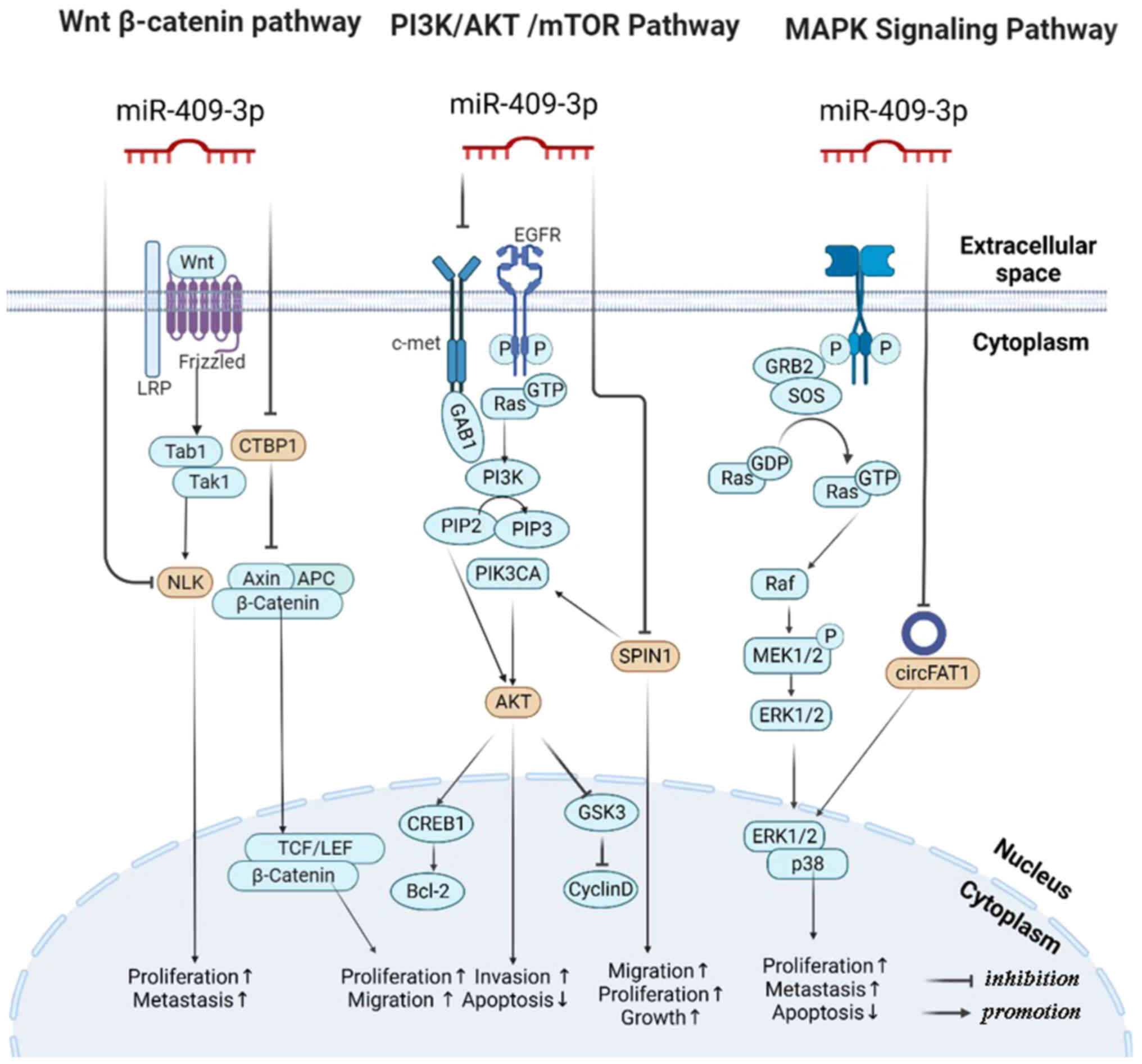 | Figure 5MiR-409-3p related signaling pathways
in cancer. MiR-409-3p can influence cancer development and regulate
the biological processes of cells by participating in the
Wnt/β-Catenin, PIK3/AKT/mTOR, MAPK signaling pathways. Created with
Biorender software (http://biorender.com). MiR, microRNA; LRP,
low-density lipoprotein receptor-related protein; Tab1, TGF-β
activated kinase 1/MAP3K7 binding protein 1; CTBP1, C-terminal
binding protein 1; Tak1, TGF-β-activated kinase 1; NLK, nemo-like
kinase; APC, adenomatous polyposis coli; TCF, T-cell factor; LEF,
lymphoid enhancer-binding factor; EGFR, epidermal growth factor
receptor; MET, MET proto-oncogene, receptor tyrosine kinase; GTP,
guanosine triphosphate; GAB, growth factor receptor-bound protein
2-associated binder; PIP2, phosphatidylinositol 4,5-bisphosphate;
PIP3, phosphatidylinositol 3,4,5-trisphosphate; PIK3CA,
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit α;
AKT1, AKT serine/threonine kinase 1; GSK3, glycogen synthase kinase
3; CREB1, cAMP responsive element binding protein 1; Bcl-2, B-cell
lymphoma 2; GRB2, growth factor receptor-bound protein 2; SOS, Son
of sevenless; GDP, guanosine diphosphate; Ras, Rat sarcoma; Raf,
rapidly accelerated fibrosarcoma; MEK1/2, mitogen-activated protein
kinase kinase 1/2; circFAT1, circular RNA FAT atypical cadherin 1;
ERK1/2, extracellular signal-regulated kinase 1/2. |
Wnt/β-Catenin signaling pathway
The incidence and progression of malignancies are
linked to the Wnt/β-Catenin signaling pathway, which enhances tumor
stem cell proliferation, survival and differentiation (115). NLK is an evolutionarily
conserved MAPK, highly expressed in nerve tissues (116,117). When NLK phosphorylation is
activated, it can phosphorylate substrates that participate in the
Wnt/β-Catenin signaling pathway (118). Hence, in CRC, miR-409-3p
downregulation could boost NLK expression and increase the
expression of the Wnt/β-Catenin signaling pathway, regulating CRC
cell proliferation and metastasis (45). In addition, using functional
enrichment analysis, Zhang et al (55) demonstrated that the targeted genes
of miR-409-3p are implicated in the pathway, and the negative
regulatory association between miR-409-3p and C-terminal binding
protein 1 (CTBP1) was verified using OC data in The Cancer
Genome Atlas (TCGA). A study by Deng et al (119) indicated that CTBP1
activates the expression gene of Wnt genes, thereby regulating the
signaling pathway. Chang et al (83) found that miR-409-3p regulated
BRF2, which enhanced invasion and metastasis in HCC via the
Wnt/β-catenin signaling pathway.
PI3K/AKT signaling pathway
According to a vast number of studies, miR-409-3p
is found in most oncogenic signaling pathways. One of the many
signaling pathways involved in cell growth and survival is the
PI3K/AKT/mTOR signaling pathway. MET is a tyrosine kinase protein
in response to a hepatocyte growth factor that plays an essential
part in cancer progression, including in morphogenesis,
mitogenesis, metastasis, proliferation and survival (120). In vitro experiments
confirmed that miR-409-3p has MET as its direct target; it
inactivated AKT signaling and achieved the effect of inhibiting
cancer proliferation, invasion and migration, as well as promoting
apoptosis in NSCLC (12). Another
group also found that, by inhibiting the expression of
SPIN1, miR-409-3p could inhibit components of PI3K/AKT,
including the expression of BCL2, cAMP responsive element binding
protein 1, p-AKT and cyclin D, thereby blocking the progression of
NSCLC (14). Besides, as
previously mentioned, AKT1 is a crucial downstream target kinase in
the PI3K signaling cascade (121). In GC, miR-409-3p/AKT1
axis could influence GC cell lines, including MDA-ZMB-231,
MDA-MB-468 and T47D cell proliferation and metastasis in
vivo and vitro (26).
In addition, in DLBCL, Leivonen et al (64) demonstrated that PIK3R1 was a
targeted gene of miR-409-3p in SU-DHL-4 DLBCL cells by using
lentiviral vectors. Most importantly, the PI3K/AKT pathway has been
found to be involved in the progression of DLBCL, which exerts an
obviously significant contribution to cellular processes and is
expected to become a promising therapeutic target. Besides, in PCa,
Yu et al (74) indicated
that PCa cells subjected to CIRT delivered exosomal miR-409-3p,
inhibiting AKT in the PI3K/AKT pathways in recipient cells, which
is the main signaling pathway linked to apoptosis induction and
proliferation inhibition, and which may be involved in the
mechanism of action of CIRT.
MAPK signaling pathway
MAPK cascade pathways are highly conserved and
modulate cell proliferation, differentiation and migration by
phosphorylating specific target protein substrates. MAPK consists
of the following subfamilies, including extracellular
signal-regulated kinases, c-Jun N-terminal kinase, p38 protein
kinases and extracellular-signal-regulated kinase 5 (122). Furthermore, circFAT1 promoted
cancer progression in the C-33A and CaSki CC cell lines,
stimulating proliferation and metastasis, as well as inhibiting
their apoptosis by activating the ERK1/2 and p38 MAPK pathway
(34). Of note, in malignant
hematological diseases such as DLBCL, miR-409-3p has been indicated
to suppress MAPK1 mRNA and ERK1/2 protein levels and improve the
chemosensitivity of cells in vitro, indicating that the MAPK
pathway is involved in the progression of DLBCL (64).
7. Discussion
The development of genomics, proteomics,
high-throughput sequencing and array technology has resulted in the
introduction of miRNA gene profiles of various malignancies, which
are currently available in the respective databases, such as TCGA
and Gene Expression Omnibus. The present review outlined that
miR-409-3p, an important molecule in current research, has been
proven to exert a role as a tumor suppressor or as an oncogenic
miRNA in several malignancies. Molecular studies to analyze cancer
development dependent on the levels of miRNAs have become a notable
aspect of genetic research. Of note, miR-409-3p exhibits
considerable promise for use as a diagnostic and therapeutic target
for cancer based on the potent biological roles it possesses.
In the fight against cancer, miR-409-3p deserves to
be a focus of research, since it has a critical role in the
emergence of numerous cancers. Due to its stability in circulating
body fluids, such as serum or plasma, miR-409-3p can also be used
for cancer diagnosis and prognosis. It is frequently present at
different concentrations in fluids from cancer patients compared to
those from healthy individuals. These levels can even vary
depending on the type of cancer, indicating the potential use of
miR-409-3p as a noninvasive biomarker in the future. In addition,
throughout the history of cancer treatment, resistance has become
the most influential factor in the treatment effect. Interestingly,
other experiments have shown that miR-409-3p could be used in
combination therapy to fight different cancers. Of note, enhancing
the expression of miR-409-3p can increase sensitivity to
chemotherapy and radiotherapy of patients, significantly extend
patient survival and hopefully brighten the outlook for patients.
More importantly, changes in miR-409-3p expression levels could be
used to evaluate the effectiveness of surgical treatment. The
changes in miR-409-3p expression provide a reliable way to assess
effectiveness.
However, at present, the roles of miR-409-3p in
different cancer types are only now beginning to be investigated,
with studies focusing solely on how miR-409-3p affects the apparent
biological function of cancer. More in-depth mechanisms need to be
further clarified. In the future, the biological processes through
which miR-409-3p participates in tumorigenesis and development
should be further explored. At present, the tumor microenvironment,
ferroptosis, pyroptosis, necroptosis, cell senescence, mitophagy,
fatty acid and amino acid metabolism and cancer stemness are
research hot-spots. The association between miR-409-3p and these
biological processes has remained largely elusive. In addition,
research on the molecular mechanisms of miR-409-3p is mostly
confined to its differential expression in cancer and tissues
adjacent to cancer. It is necessary to further study the mechanisms
of miR-409-3p affecting tumor growth at the transcriptional and
post-transcriptional levels to analyze the complex interaction
network of miR-409-3p in pan-cancer panels. At this stage, miRNAs
may combine with different target genes or corresponding upstream
or downstream genes to form a network of mutual regulation to
participate in the development of tumors, which is a widely
recognized molecular mechanism. miR-409-3p has been proved to
target MET, c-myc, CyclinD1, MMP2, ZEB1
and ELF2, etc. In addition, these target genes also have
prominent molecular regulatory functions, such as cell
proliferation, apoptosis, migration, invasion, autophagy,
resistance, angiogenesis and glycolysis. It is worth noting that
few studies have been conducted on miR-409-3p in malignancies, such
as hematological malignancies, melanoma and liver cancer, and the
upstream signaling of miR-409-3p, such as lncRNAs or circRNAs.
Studying the ceRNA mechanism in which miR-409-3p is widely involved
can provide new ideas for disease discovery, diagnosis and
treatment. In addition, current research shows that miR-409-3p is
dysregulated in 23 types of cancer and brings into play the role of
oncogenes or tumor suppressor genes in tumorigenesis. However,
miR-409-3p has tumor specificity and its specific mechanism remains
to be elucidated. In addition, in order to better study the
functions and mechanisms of miRNAs, online tools such as miRDB
(http://www.mirdb.org/) (123) and TargetScan (http://www.targetscan.org/) (124) may be used for miRNA target
prediction and functional annotations. These websites have the
function of predicting target genes. According to the prediction
results, validation at the cellular and animal levels will greatly
accelerate and boost the progress of miRNA research.
MiR-409-3p has been identified as a strong
candidate molecule in cancer diagnosis and treatment. Increasing
evidence shows that the combination of miR-409-3p with other
biomarkers can improve the sensitivity and specificity of
diagnosis. However, there is currently no mature miRNA detection
technology in clinical practice, so miRNA as a biomarker has not
been used in clinical practice. In addition, it is positive that,
based on the rapidly developing targeted delivery strategy,
treatment schemes involving miR-409-3p regulatory target tissues
are expected to be applied in medical practice. However, due to the
limitations of medical equipment and technology, how to better
design targeted drugs for miR-409-3p remains elusive and there is
still a long way to go to better deliver drugs to the designated
sites. Although miR-409-3p-targeted therapy is a potentially
effective therapeutic approach, the molecular mechanisms underlying
the changes in miR-409-3p expression in various malignancies remain
unclear, significantly diminishing the utility of miR-409-3p in
clinical treatment. More research should be conducted in the
future.
In summary, compared to previous studies, the
present article is a comprehensive review that provides a detailed
summary of the expression, function and clinical application
prospects of miR-409-3p in various malignant tumors. It provided a
more comprehensive perspective and demonstrated the importance of
miR-409-3p in the field of cancer. In addition to emphasizing the
importance of miR-409-3p in cancer, its potential as a biomarker
and therapeutic target was also highlighted.
In addition, the current review not only summarized
existing research results but also provided specific suggestions
for future research directions. It emphasized the relationship
between miR-409-3p and the tumor microenvironment, cell death
mechanisms and other areas that have not been fully explored in
current research. Proposing these future research directions
provides guidance for further development in this field.
It is expected that the present review on
miR-409-3p will prompt further research to fully understand the
basic biological mechanisms of miR-409-3p, as well as its potential
as a tool for clinical application in cancer management and therapy
in the future.
Availability of data and materials
Not applicable.
Authors' contributions
WJX and ZCW wrote major parts of the manuscript and
prepared the figures and tables. XW and JKW revised the manuscript.
HZG oversaw the process and wrote the manuscript. ZCW, XW and JKW
conceptualized the study and oversaw the process. Data
authentication is not applicable. All authors have read and
approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Abbreviations:
|
miRNA/miR
|
microRNA
|
|
UTR
|
untranslated region
|
|
NSCLC
|
non-small cell lung cancer
|
|
BC
|
breast cancer
|
|
CC
|
cervical cancer
|
|
GC
|
gastric cancer
|
|
CRC
|
colorectal cancer
|
|
OC
|
ovarian cancer
|
|
ccRCC
|
renal cell carcinoma
|
|
BCa
|
bladder cancer
|
|
PTC
|
papillary thyroid cancer
|
|
PC
|
pancreatic cancer
|
|
BTC
|
biliary tract cancer
|
|
DLBCL
|
diffuse large B-cell lymphoma
|
|
AML
|
acute myeloid leukemia
|
|
NPC
|
nasopharyngeal carcinoma
|
|
TSCC
|
tongue squamous cell carcinoma
|
|
PCa
|
prostate cancer
|
|
HCC
|
hepatocellular carcinoma
|
|
lncRNA
|
lncRNA
|
|
circRNA
|
circular RNA
|
|
LUAD
|
lung adenocarcinoma
|
|
CIRT
|
carbon ion radiotherapy
|
|
MET
|
MET proto-oncogene, receptor tyrosine
kinase
|
|
SPIN1
|
spindlin1
|
|
ZB1-AS1
|
zinc finger E-box binding homeobox 1
antisense 1
|
|
ceRNA
|
competing endogenous RNA
|
|
HK2
|
hexokinase 2
|
|
LDHA
|
lactate dehydrogenase A
|
|
DUXAP8
|
double homeobox A pseudogene 8
|
|
SOD1
|
superoxide dismutase 1
|
|
CBR3-AS1
|
CBR3 antisense RNA 1
|
|
AQP4
|
aquaporin4
|
|
AKT1
|
AKT serine/threonine kinase 1
|
|
circTRIM28
|
circular RNA tripartite
motif-containing 28
|
|
HMGA2
|
high mobility group AT-hook 2
|
|
ATF1
|
activating transcription factor 1
|
|
circFAT1
|
circular RNA FAT atypical cadherin
1
|
|
CDK8
|
cyclin-dependent kinase 8
|
|
SLC7A11
|
solute carrier family 7 membrane
11
|
|
ELF2
|
E74-like factor 2
|
|
IGFBP3
|
insulin-like growth factor binding
protein 3
|
|
PHF10
|
PHD finger protein 10
|
|
circNEK9
|
circRNA NIMA-related kinase 9
|
|
MAP7
|
microtubule associated protein 7
|
|
KLF17
|
Kruppel-like factor 17
|
|
NLK
|
nemo-like kinase
|
|
Oxa-R
|
oxaliplatin resistance
|
|
ERCC1
|
ERCC excision repair 1
|
|
RAB10
|
RAB10, member RAS oncogene family
|
|
CCDN2
|
cyclin D2
|
|
RDX
|
radixin
|
|
RSU1
|
Ras suppressor protein 1
|
|
DOCK1
|
dedicator of cytokinesis 1
|
|
HBVSMCs
|
human brain vascular smooth muscle
cells
|
|
MCL1
|
myeloid cell leukemia sequence 1
|
|
HMGN5
|
high mobility group nucleosome
binding domain 5
|
|
ANG
|
Angiogenin
|
|
EMT
|
epithelial-mesenchymal transition
|
|
TWIST1
|
Twist family bHLH transcription
factor
|
|
MMP
|
matrix metalloproteinase
|
|
p-AKT
|
phosphorylated AKT
|
|
CTNND1
|
catenin-δ1
|
|
GAB1
|
GRB2 associated binding protein 1
|
|
circTRIM28
|
circRNA tripartite motif-containing
28
|
|
FABP4
|
fatty acid binding protein 4
|
|
HIF-1α
|
hypoxia-inducible factor-1α
|
|
BRF2
|
BRF2 RNA polymerase III transcription
initiation factor subunit
|
|
ROS
|
reactive oxygen species
|
|
PDK1
|
3-phosphoinositide dependent kinase
1
|
|
CTBP1
|
C-terminal binding protein 1
|
|
TCGA
|
The Cancer Genome Atlas
|
|
GEO
|
Gene Expression Omnibus
|
Acknowledgements
Not applicable.
Funding
This work was supported by the Natural Science Foundation of
Shandong Province, China (grant no. ZR2020MH311). The funding body
neither had any involvement in the study design, nor in the
collection, analysis and interpretation of data, in the writing of
the report, or in the decision to submit the article for
publication.
References
|
1
|
Lee YS and Dutta A: MicroRNAs in cancer.
Annu Rev Pathol. 4:199–227. 2009. View Article : Google Scholar :
|
|
2
|
Oliveto S, Mancino M, Manfrini N and Biffo
S: Role of microRNAs in translation regulation and cancer. World J
Biol Chem. 8:45–56. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Cortez MA, Bueso-Ramos C, Ferdin J,
Lopez-Berestein G, Sood AK and Calin GA: MicroRNAs in body
fluids-the mix of hormones and biomarkers. Nat Rev Clin Oncol.
8:467–477. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Macfarlane LA and Murphy PR: MicroRNA:
Biogenesis function and role in cancer. Curr Genomics. 11:537–561.
2010. View Article : Google Scholar
|
|
6
|
Croce CM and Calin GA: miRNAs, cancer, and
stem cell division. Cell. 122:6–7. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bueno MJ and Malumbres M: MicroRNAs and
the cell cycle. Biochim Biophys Acta. 1812:592–601. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Su Z, Yang Z, Xu Y, Chen Y and Yu Q:
MicroRNAs in apoptosis, autophagy and necroptosis. Oncotarget.
6:8474–8490. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ma L: MicroRNA and metastasis. Adv Cancer
Res. 132:165–207. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Tiwari A, Mukherjee B and Dixit M:
MicroRNA key to angiogenesis regulation: MiRNA biology and therapy.
Curr Cancer Drug Targets. 18:266–277. 2018. View Article : Google Scholar
|
|
11
|
Altuvia Y, Landgraf P, Lithwick G, Elefant
N, Pfeffer S, Aravin A, Brownstein MJ, Tuschl T and Margalit H:
Clustering and conservation patterns of human microRNAs. Nucleic
Acids Res. 33:2697–2706. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wan L, Zhu L, Xu J, Lu B, Yang Y, Liu F
and Wang Z: MicroRNA-409-3p functions as a tumor suppressor in
human lung adenocarcinoma by targeting c-Met. Cell Physiol Biochem.
34:1273–1290. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhou X, Wen W, Shan X, Zhu W, Xu J, Guo R,
Cheng W, Wang F, Qi LW, Chen Y, et al: A six-microRNA panel in
plasma was identified as a potential biomarker for lung
adenocarcinoma diagnosis. Oncotarget. 8:6513–6525. 2017. View Article : Google Scholar :
|
|
14
|
Song Q, Ji Q, Xiao J, Li F, Wang L, Chen
Y, Xu Y and Jiao S: miR-409 inhibits human non-small-cell lung
cancer progression by directly targeting SPIN1. Mol Ther Nucleic
Acids. 13:154–163. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Qu R, Chen X and Zhang C: LncRNA
ZEB1-AS1/miR-409-3p/ ZEB1 feedback loop is involved in the
progression of non-small cell lung cancer. Biochem Biophys Res
Commun. 507:450–456. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yin D, Hua L, Wang J, Liu Y and Li X: Long
non-coding RNA DUXAP8 facilitates cell viability, migration, and
glycolysis in non-small-cell lung cancer via regulating HK2 and
LDHA by inhibition of miR-409-3p. Onco Targets Ther. 13:7111–7123.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liu S, Li B, Xu J, Hu S, Zhan N, Wang H,
Gao C, Li J and Xu X: SOD1 promotes cell proliferation and
metastasis in non-small cell lung cancer via an
miR-409-3p/SOD1/SETDB1 epigenetic regulatory feedforward loop.
Front Cell Dev Biol. 8:2132020. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang J, Zhang C, Peng X, Liu K, Zhao L,
Chen X, Yu H and Lai Y: A combination of four serum miRNAs for
screening of lung adenocarcinoma. Hum Cell. 33:830–838. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang L, Wu L and Pang J: Long noncoding
RNA PSMA3-AS1 functions as a microRNA-409-3p sponge to promote the
progression of non-small cell lung carcinoma by targeting spindlin
1. Oncol Rep. 44:1550–1560. 2020.PubMed/NCBI
|
|
20
|
Liu S, Zhan N, Gao C, Xu P, Wang H, Wang
S, Piao S and Jing S: Long noncoding RNA CBR3-AS1 mediates
tumorigenesis and radiosensitivity of non-small cell lung cancer
through redox and DNA repair by CBR3-AS1/miR-409-3p/SOD1 axis.
Cancer Lett. 526:1–11. 2022. View Article : Google Scholar
|
|
21
|
Yang X, Li M, Zhao Y, Tan X, Su J and
Zhong X: Hsa_circ_0079530/AQP4 axis is related to non-small cell
lung cancer development and radiosensitivity. Ann Thorac Cardiovasc
Surg. 28:307–319. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cuk K, Zucknick M, Madhavan D, Schott S,
Golatta M, Heil J, Marmé F, Turchinovich A, Sinn P, Sohn C, et al:
Plasma microRNA panel for minimally invasive detection of breast
cancer. PLoS One. 8:e767292013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Cuk K, Zucknick M, Heil J, Madhavan D,
Schott S, Turchinovich A, Arlt D, Rath M, Sohn C, Benner A, et al:
Circulating microRNAs in plasma as early detection markers for
breast cancer. Int J Cancer. 132:1602–1612. 2013. View Article : Google Scholar
|
|
24
|
Li S, Meng H, Zhou F, Zhai L, Zhang L, Gu
F, Fan Y, Lang R, Fu L, Gu L and Qi L: MicroRNA-132 is frequently
down-regulated in ductal carcinoma in situ (DCIS) of breast and
acts as a tumor suppressor by inhibiting cell proliferation. Pathol
Res Pract. 209:179–183. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Shen J, Hu Q, Schrauder M, Yan L, Wang D,
Medico L, Guo Y, Yao S, Zhu Q, Liu B, et al: Circulating miR-148b
and miR-133a as biomarkers for breast cancer detection. Oncotarget.
5:5284–5294. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhang G, Liu Z, Xu H and Yang Q:
miR-409-3p suppresses breast cancer cell growth and invasion by
targeting Akt1. Biochem Biophys Res Commun. 469:189–195. 2016.
View Article : Google Scholar
|
|
27
|
Ma Z, Li Y, Xu J, Ren Q, Yao J and Tian X:
MicroRNA-409-3p regulates cell invasion and metastasis by targeting
ZEB1 in breast cancer. IUBMB Life. 68:394–402. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Venkatadri R, Muni T, Iyer AKV, Yakisich
JS and Azad N: Role of apoptosis-related miRNAs in
resveratrol-induced breast cancer cell death. Cell Death Dis.
7:e21042016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Su Q, Shen H, Gu B and Zhu N: Circular RNA
CNOT2 knockdown regulates twist family BHLH transcription factor
via targeting microRNA 409-3p to prevent breast cancer invasion,
migration and epithelial-mesenchymal transition. Bioengineered.
12:9058–9069. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yang S, Zou C, Li Y, Yang X, Liu W, Zhang
G and Lu N: Knockdown circTRIM28 enhances tamoxifen sensitivity via
the miR-409-3p/HMGA2 axis in breast cancer. Reprod Biol Endocrinol.
20:1462022. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Shukla V, Varghese VK, Kabekkodu SP,
Mallya S, Chakrabarty S, Jayaram P, Pandey D, Banerjee S, Sharan K
and Satyamoorthy K: Enumeration of deregulated miRNAs in liquid and
tissue biopsies of cervical cancer. Gynecol Oncol. 155:135–143.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Sommerova L, Anton M, Bouchalova P,
Jasickova H, Rak V, Jandakova E, Selingerova I, Bartosik M,
Vojtesek B and Hrstka R: The role of miR-409-3p in regulation of
HPV16/18-E6 mRNA in human cervical high-grade squamous
intraepithelial lesions. Antiviral Res. 163:185–192. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Cui X, Chen J, Zheng Y and Shen H:
Circ_0000745 promotes the progression of cervical cancer by
regulating miR-409-3p/ATF1 axis. Cancer Biother Radiopharm.
37:766–778. 2022.
|
|
34
|
Zhou B, Li T, Xie R, Zhou J, Liu J, Luo Y
and Zhang X: CircFAT1 facilitates cervical cancer malignant
progression by regulating ERK1/2 and p38 MAPK pathway through
miR-409-3p/CDK8 axis. Drug Dev Res. 82:1131–1143. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wu P, Li C, Ye DM, Yu K, Li Y, Tang H, Xu
G, Yi S and Zhang Z: Circular RNA circEPSTI1 accelerates cervical
cancer progression via miR-375/409-3P/515-5p-SLC7A11 axis. Aging
(Albany NY). 13:4663–4673. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wu S, Du X, Wu M, Du H, Shi X and Zhang T:
MicroRNA-409-3p inhibits osteosarcoma cell migration and invasion
by targeting catenin-δ1. Gene. 584:83–89. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhang J, Hou W, Jia J, Zhao Y and Zhao B:
MiR-409-3p regulates cell proliferation and tumor growth by
targeting E74-like factor 2 in osteosarcoma. FEBS Open Bio.
7:348–357. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wu L, Zhang Y, Huang Z, Gu H, Zhou K, Yin
X and Xu J: MiR-409-3p inhibits cell proliferation and invasion of
osteosarcoma by targeting zinc-finger E-box-binding homeobox-1.
Front Pharmacol. 10:1372019. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Long Z, Gong F, Li Y, Fan Z and Li J:
Circ_0000285 regulates proliferation, migration, invasion and
apoptosis of osteosarcoma by miR-409-3p/IGFBP3 axis. Cancer Cell
Int. 20:4812020. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Li C, Nie H, Wang M, Su L, Li J, Yu B, Wei
M, Ju J, Yu Y, Yan M, et al: MicroRNA-409-3p regulates cell
proliferation and apoptosis by targeting PHF10 in gastric cancer.
Cancer Lett. 320:189–197. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zheng B, Liang L, Huang S, Zha R, Liu L,
Jia D, Tian Q, Wang Q, Wang C, Long Z, et al: MicroRNA-409
suppresses tumour cell invasion and metastasis by directly
targeting radixin in gastric cancers. Oncogene. 31:4509–4516. 2012.
View Article : Google Scholar
|
|
42
|
Yu L, Xie J, Liu X, Yu Y and Wang S:
Plasma exosomal CircNEK9 accelerates the progression of gastric
cancer via miR-409-3p/MAP7 axis. Dig Dis Sci. 66:4274–4289. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Feng J, Li K, Liu G, Feng Y, Shi H and
Zhang X: Precision hyperthermia-induced miRNA-409-3p upregulation
inhibits migration, invasion, and EMT of gastric cancer cells by
targeting KLF17. Biochem Biophys Res Commun. 549:113–119. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Wang Y, Zhang J, Chen X and Gao L:
Circ_0001023 promotes proliferation and metastasis of gastric
cancer cells through miR-409-3p/PHF10 axis. Onco Targets Ther.
13:4533–4544. 2020. View Article : Google Scholar :
|
|
45
|
Liu M, Xu A, Yuan X, Zhang Q, Fang T, Wang
W and Li C: Downregulation of microRNA-409-3p promotes
aggressiveness and metastasis in colorectal cancer: An indication
for personalized medicine. J Transl Med. 13:1952015. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Bai R, Weng C, Dong H, Li S, Chen G and Xu
Z: MicroRNA-409-3p suppresses colorectal cancer invasion and
metastasis partly by targeting GAB1 expression. Int J Cancer.
137:2310–2322. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Wang S, Xiang J, Li Z, Lu S, Hu J, Gao X,
Yu L, Wang L, Wang J, Wu Y, et al: A plasma microRNA panel for
early detection of colorectal cancer. Int J Cancer. 136:152–161.
2015. View Article : Google Scholar
|
|
48
|
Tan S, Shi H, Ba M, Lin S, Tang H, Zeng X
and Zhang X: miR-409-3p sensitizes colon cancer cells to
oxaliplatin by inhibiting Beclin-1-mediated autophagy. Int J Mol
Med. 37:1030–1038. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
López-Rosas I, López-Camarillo C,
Salinas-Vera YM, Hernández-de la Cruz ON, Palma-Flores C,
Chávez-Munguía B, Resendis-Antonio O, Guillen N, Pérez-Plasencia C,
Álvarez-Sánchez ME, et al: Entamoeba histolytica up-regulates
MicroRNA-643 to promote apoptosis by targeting XIAP in human
epithelial colon cells. Front Cell Infect Microbiol. 8:4372019.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Han W, Yin H, Ma H, Wang Y, Kong D and Fan
Z: Curcumin regulates ERCC1 expression and enhances oxaliplatin
sensitivity in resistant colorectal cancer cells through its
effects on miR-409-3p. Evid Based Complement Alternat Med.
2020:83945742020. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Chen J, Wang R, Lu E, Song S and Zhu Y:
LINC00630 as a miR-409-3p sponge promotes apoptosis and glycolysis
of colon carcinoma cells via regulating HK2. Am J Transl Res.
14:863–875. 2022.PubMed/NCBI
|
|
52
|
Zhang J, Raju GS, Chang DW, Lin SH, Chen Z
and Wu X: Global and targeted circulating microRNA profiling of
colorectal adenoma and colorectal cancer. Cancer. 124:785–796.
2018. View Article : Google Scholar
|
|
53
|
Gharpure KM, Pradeep S, Sans M, Rupaimoole
R, Ivan C, Wu SY, Bayraktar E, Nagaraja AS, Mangala LS, Zhang X, et
al: FABP4 as a key determinant of metastatic potential of ovarian
cancer. Nat Commun. 9:29232018. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Cheng Y, Ban R, Liu W, Wang H, Li S, Yue
Z, Zhu G, Zhuan Y and Wang C: MiRNA-409-3p enhances
cisplatin-sensitivity of ovarian cancer cells by blocking the
autophagy mediated by Fip200. Oncol Res. Jan 2–2018.Epub ahead of
print. View Article : Google Scholar
|
|
55
|
Zhang S, Zhang X, Fu X, Li W, Xing S and
Yang Y: Identification of common differentially-expressed miRNAs in
ovarian cancer cells and their exosomes compared with normal
ovarian surface epithelial cell cells. Oncol Lett. 16:2391–2401.
2018.PubMed/NCBI
|
|
56
|
Li Y, Chen L, Zhang B, Ohno Y and Hu H:
miR-409-3p inhibits the proliferation and migration of human
ovarian cancer cells by targeting Rab10. Cell Mol Biol
(Noisy-le-grand). 66:197–201. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Zhu J, Ma X, Zhang Y, Ni D, Ai Q, Li H and
Zhang X: Establishment of a miRNA-mRNA regulatory network in
metastatic renal cell carcinoma and screening of potential
therapeutic targets. Tumour Biol. Nov 2–2016.Epub ahead of print.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Wang Y, He Y, Bai H, Dang Y, Gao J and Lv
P: Phosphoinositide-dependent kinase 1-associated glycolysis is
regulated by miR-409-3p in clear cell renal cell carcinoma. J Cell
Biochem. 120:126–134. 2019. View Article : Google Scholar
|
|
59
|
Xu X, Chen H, Lin Y, Hu Z, Mao Y, Wu J, Xu
X, Zhu Y, Li S, Zheng X and Xie L: MicroRNA-409-3p inhibits
migration and invasion of bladder cancer cells via targeting c-Met.
Mol Cells. 36:62–68. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Xu X, Zhu Y, Liang Z, Li S, Xu X, Wang X,
Wu J, Hu Z, Meng S, Liu B, et al: c-Met and CREB1 are involved in
miR-433-mediated inhibition of the epithelial-mesenchymal
transition in bladder cancer by regulating Akt/GSK-3β/Snail
signaling. Cell Death Dis. 7:e20882016. View Article : Google Scholar
|
|
61
|
Lian J, Lin SH, Ye Y, Chang DW, Huang M,
Dinney CP and Wu X: Serum microRNAs as predictors of risk for
non-muscle invasive bladder cancer. Oncotarget. 9:14895–14908.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Zhao Z, Yang F, Liu Y, Fu K and Jing S:
MicroRNA-409-3p suppresses cell proliferation and cell cycle
progression by targeting cyclin D2 in papillary thyroid carcinoma.
Oncol Lett. 16:5237–5242. 2018.PubMed/NCBI
|
|
63
|
Kim K, Yoo D, Lee HS, Lee KJ, Park SB, Kim
C, Jo JH, Jung DE and Song SY: Identification of potential
biomarkers for diagnosis of pancreatic and biliary tract cancers by
sequencing of serum microRNAs. BMC Med Genomics. 12:622019.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Leivonen SK, Icay K, Jäntti K, Siren I,
Liu C, Alkodsi A, Cervera A, Ludvigsen M, Hamilton-Dutoit SJ,
d'Amore F, et al: MicroRNAs regulate key cell survival pathways and
mediate chemosensitivity during progression of diffuse large B-cell
lymphoma. Blood Cancer J. 7:6542017. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Li M, Cui X and Guan H: The expression and
clinical significance of MicroRNA-409-3p in acute myeloid leukemia.
Clin Lab. 66:2020. View Article : Google Scholar
|
|
66
|
Xie W, Wang Z, Guo X and Guan H:
MiR-409-3p regulates the proliferation and apoptosis of THP-1
through targeting Rab10. Leuk Res. 132:1073502023. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Kumar A, Nayak S, Pathak P, Purkait S,
Malgulawar PB, Sharma MC, Suri V, Mukhopadhyay A, Suri A and Sarkar
C: Identification of miR-379/miR-656 (C14MC) cluster downregulation
and associated epigenetic and transcription regulatory mechanism in
oligodendrogliomas. J Neurooncol. 139:23–31. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Venza I, Visalli M, Beninati C, Benfatto
S, Teti D and Venza M: IL-10Rα expression is post-transcriptionally
regulated by miR-15a, miR-185, and miR-211 in melanoma. BMC Med
Genomics. 8:812015. View Article : Google Scholar
|
|
69
|
Jiang L, Zhang Y, Li B, Kang M, Yang Z,
Lin C, Hu K, Wei Z, Xu M, Mi J, et al: miRNAs derived from
circulating small extracellular vesicles as diagnostic biomarkers
for nasopharyngeal carcinoma. Cancer Sci. 112:2393–2404. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Chen H and Dai J: miR-409-3p suppresses
the proliferation, invasion and migration of tongue squamous cell
carcinoma via targeting RDX. Oncol Lett. 16:543–551.
2018.PubMed/NCBI
|
|
71
|
Josson S, Gururajan M, Hu P, Shao C, Chu
GY, Zhau HE, Liu C, Lao K, Lu CL, Lu YT, et al: miR-409-3p/-5p
promotes tumorigenesis, epithelial-to-mesenchymal transition, and
bone metastasis of human prostate cancer. Clin Cancer Res.
20:4636–4646. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Gururajan M, Josson S, Chu GC, Lu CL, Lu
YT, Haga CL, Zhau HE, Liu C, Lichterman J, Duan P, et al: miR-154*
and miR-379 in the DLK1-DIO3 microRNA mega-cluster regulate
epithelial to mesenchymal transition and bone metastasis of
prostate cancer. Clin Cancer Res. 20:6559–6569. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Josson S, Gururajan M, Sung SY, Hu P, Shao
C, Zhau HE, Liu C, Lichterman J, Duan P, Li Q, et al: Stromal
fibroblast-derived miR-409 promotes epithelial-to-mesenchymal
transition and prostate tumorigenesis. Oncogene. 34:2690–2699.
2015. View Article : Google Scholar
|
|
74
|
Yu Q, Li P, Weng M, Wu S, Zhang Y, Chen X,
Zhang Q, Shen G, Ding X and Fu S: Nano-vesicles are a potential
tool to monitor therapeutic efficacy of carbon ion radiotherapy in
prostate cancer. J Biomed Nanotechnol. 14:168–178. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Fredsøe J, Rasmussen AKI, Mouritzen P,
Bjerre MT, Østergren P, Fode M, Borre M and Sørensen KD: Profiling
of circulating microRNAs in prostate cancer reveals diagnostic
biomarker potential. Diagnostics (Basel). 10:1882020. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Nguyen HCN, Xie W, Yang M, Hsieh CL,
Drouin S, Lee GS and Kantoff PW: Expression differences of
circulating microRNAs in metastatic castration resistant prostate
cancer and low-risk, localized prostate cancer. Prostate.
73:346–354. 2013. View Article : Google Scholar
|
|
77
|
Zhi F, Shao N, Li B, Xue L, Deng D, Xu Y,
Lan Q, Peng Y and Yang Y: A serum 6-miRNA panel as a novel
non-invasive biomarker for meningioma. Sci Rep. 6:320672016.
View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Ding X, Wang X, Han L, Zhao Z, Jia S and
Tuo Y: CircRNA DOCK1 regulates miR-409-3p/MCL1 axis to modulate
proliferation and apoptosis of human brain vascular smooth muscle
cells. Front Cell Dev Biol. 9:6556282021. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Cao Y, Zhang L, Wei M, Jiang X and Jia D:
MicroRNA-409-3p represses glioma cell invasion and proliferation by
targeting high-mobility group nucleosome-binding domain 5. Oncol
Res. 25:1097–1107. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Ma Z, Chen Z, Zhou Y, Li Y, Li S, Wang H
and Feng J: Hsa_circ_0000418 promotes the progression of glioma by
regulating microRNA-409-3p/pyruvate dehydrogenase kinase 1 axis.
Bioengineered. 13:7541–7552. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Weng C, Dong H, Chen G, Zhai Y, Bai R, Hu
H, Lu L and Xu Z: miR-409-3p inhibits HT1080 cell proliferation,
vascularization and metastasis by targeting angiogenin. Cancer
Lett. 323:171–179. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Khalil S, Fabbri E, Santangelo A, Bezzerri
V, Cantù C, Di Gennaro G, Finotti A, Ghimenton C, Eccher A,
Dechecchi M, et al: miRNA array screening reveals cooperative
MGMT-regulation between miR-181d-5p and miR-409-3p in glioblastoma.
Oncotarget. 7:28195–28206. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Chang JH, Xu BW, Shen D, Zhao W, Wang Y,
Liu JL, Meng GX, Li GZ and Zhang ZL: BRF2 is mediated by
microRNA-409-3p and promotes invasion and metastasis of HCC through
the Wnt/β-catenin pathway. Cancer Cell Int. 23:462023. View Article : Google Scholar
|
|
84
|
Li L, Ai R, Yuan X, Dong S, Zhao D, Sun X,
Miao T, Guan W, Guo P, Yu S and Nan Y: LINC00886 facilitates
hepatocellular carcinoma tumorigenesis by sequestering
microRNA-409-3p and microRNA-214-5p. J Hepatocell Carcinoma.
10:863–881. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Pigati L, Yaddanapudi SCS, Iyengar R, Kim
DJ, Hearn SA, Danforth D, Hastings ML and Duelli DM: Selective
release of microRNA species from normal and malignant mammary
epithelial cells. PLoS One. 5:e135152010. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Witwer KW, Buzás EI, Bemis LT, Bora A,
Lässer C, Lötvall J, Nolte-'t Hoen EN, Piper MG, Sivaraman S, Skog
J, et al: Standardization of sample collection, isolation and
analysis methods in extracellular vesicle research. J Extracell
Vesicles. 2:2013. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Valadi H, Ekström K, Bossios A, Sjöstrand
M, Lee JJ and Lötvall JO: Exosome-mediated transfer of mRNAs and
microRNAs is a novel mechanism of genetic exchange between cells.
Nat Cell Biol. 9:654–659. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Gallo A, Tandon M, Alevizos I and Illei
GG: The majority of microRNAs detectable in serum and saliva is
concentrated in exosomes. PLoS One. 7:e306792012. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Mathivanan S, Ji H and Simpson RJ:
Exosomes: Extracellular organelles important in intercellular
communication. J Proteomics. 73:1907–1920. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Arroyo JD, Chevillet JR, Kroh EM, Ruf IK,
Pritchard CC, Gibson DF, Mitchell PS, Bennett CF,
Pogosova-Agadjanyan EL, Stirewalt DL, et al: Argonaute2 complexes
carry a population of circulating microRNAs independent of vesicles
in human plasma. Proc Natl Acad Sci USA. 108:5003–5008. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Turchinovich A, Weiz L, Langheinz A and
Burwinkel B: Characterization of extracellular circulating
microRNA. Nucleic Acids Res. 39:7223–7233. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Finak G, Bertos N, Pepin F, Sadekova S,
Souleimanova M, Zhao H, Chen H, Omeroglu G, Meterissian S, Omeroglu
A, et al: Stromal gene expression predicts clinical outcome in
breast cancer. Nat Med. 14:518–527. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Goetsch L, Caussanel V and Corvaia N:
Biological significance and targeting of c-Met tyrosine kinase
receptor in cancer. Front Biosci (Landmark Ed). 18:454–473. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Cecchi F, Rabe DC and Bottaro DP:
Targeting the HGF/Met signalling pathway in cancer. Eur J Cancer.
46:1260–1270. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Dienstmann R, Rodon J, Serra V and
Tabernero J: Picking the point of inhibition: A comparative review
of PI3K/AKT/mTOR pathway inhibitors. Mol Cancer Ther. 13:1021–1031.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Friedl P and Wolf K: Tumour-cell invasion
and migration: Diversity and escape mechanisms. Nat Rev Cancer.
3:362–374. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Brabletz T, Kalluri R, Nieto MA and
Weinberg RA: EMT in cancer. Nat Rev Cancer. 18:128–134. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Singh M, Yelle N, Venugopal C and Singh
SK: EMT: Mechanisms and therapeutic implications. Pharmacol Ther.
182:80–94. 2018. View Article : Google Scholar
|
|
100
|
Liu T, Zhao X, Zheng X, Zheng Y, Dong X,
Zhao N, Liao S and Sun B: The EMT transcription factor, Twist1, as
a novel therapeutic target for pulmonary sarcomatoid carcinomas.
Int J Oncol. 56:750–760. 2020.PubMed/NCBI
|
|
101
|
Stenmark H: Rab GTPases as coordinators of
vesicle traffic. Nat Rev Mol Cell Biol. 10:513–525. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Cheng HL, Trink B, Tzai TS, Liu HS, Chan
SH, Ho CL, Sidransky D and Chow NH: Overexpression of c-met as a
prognostic indicator for transitional cell carcinoma of the urinary
bladder: A comparison with p53 nuclear accumulation. J Clin Oncol.
20:1544–1550. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Tan Q, Joshua AM, Wang M, Bristow RG,
Wouters BG, Allen CJ and Tannock IF: Correction to: Up-regulation
of autophagy is a mechanism of resistance to chemotherapy and can
be inhibited by pantoprazole to increase drug sensitivity. Cancer
Chemother Pharmacol. Jan 19–2024.Epub ahead of print. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Helgason GV, Holyoake TL and Ryan KM: Role
of autophagy in cancer prevention, development and therapy. Essays
Biochem. 55:133–151. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Zhou Y, Sun K, Ma Y, Yang H, Zhang Y, Kong
X and Wei L: Autophagy inhibits chemotherapy-induced apoptosis
through downregulating Bad and Bim in hepatocellular carcinoma
cells. Sci Rep. 4:53822014. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Sui X, Chen R, Wang Z, Huang Z, Kong N,
Zhang M, Han W, Lou F, Yang J, Zhang Q, et al: Autophagy and
chemotherapy resistance: A promising therapeutic target for cancer
treatment. Cell Death Dis. 4:e8382013. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Fujiwara K, Iwado E, Mills GB, Sawaya R,
Kondo S and Kondo Y: Akt inhibitor shows anticancer and
radiosensitizing effects in malignant glioma cells by inducing
autophagy. Int J Oncol. 31:753–760. 2007.PubMed/NCBI
|
|
108
|
Kim EJ, Jeong JH, Bae S, Kang S, Kim CH
and Lim YB: mTOR inhibitors radiosensitize PTEN-deficient
non-small-cell lung cancer cells harboring an EGFR activating
mutation by inducing autophagy. J Cell Biochem. 114:1248–1256.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Arico S, Petiot A, Bauvy C, Dubbelhuis PF,
Meijer AJ, Codogno P and Ogier-Denis E: The tumor suppressor PTEN
positively regulates macroautophagy by inhibiting the
phosphatidylinositol 3-kinase/protein kinase B pathway. J Biol
Chem. 276:35243–35246. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Liberti MV and Locasale JW: The Warburg
effect: How does it benefit cancer cells? Trends Biochem Sci.
41:211–218. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Orang AV, Petersen J, McKinnon RA and
Michael MZ: Micromanaging aerobic respiration and glycolysis in
cancer cells. Mol Metab. 23:98–126. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Zhang Q, Wang L, Cao L and Wei T: Novel
circular RNA circATRNL1 accelerates the osteosarcoma aerobic
glycolysis through targeting miR-409-3p/LDHA. Bioengineered.
12:9965–9975. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Chang SH and Hla T: Gene regulation by RNA
binding proteins and microRNAs in angiogenesis. Trends Mol Med.
17:650–658. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Folkman J, Watson K, Ingber D and Hanahan
D: Induction of angiogenesis during the transition from hyperplasia
to neoplasia. Nature. 339:58–61. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Zhang Y and Wang X: Targeting the
Wnt/β-catenin signaling pathway in cancer. J Hematol Oncol.
13:1652020. View Article : Google Scholar
|
|
116
|
Huang Y, Yang Y, He Y and Li J: The
emerging role of Nemo-like kinase (NLK) in the regulation of
cancers. Tumour Biol. 36:9147–9152. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Masoumi KC, Daams R, Sime W, Siino V, Ke
H, Levander F and Massoumi R: NLK-mediated phosphorylation of HDAC1
negatively regulates Wnt signaling. Mol Biol Cell. 28:346–355.
2017. View Article : Google Scholar :
|
|
118
|
Smit L, Baas A, Kuipers J, Korswagen H,
van de Wetering M and Clevers H: Wnt activates the Tak1/Nemo-like
kinase pathway. J Biol Chem. 279:17232–17240. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Deng Y, Deng H, Liu J, Han G, Malkoski S,
Liu B, Zhao R, Wang XJ and Zhang Q: Transcriptional down-regulation
of Brca1 and E-cadherin by CtBP1 in breast cancer. Mol Carcinog.
51:500–507. 2012. View Article : Google Scholar
|
|
120
|
Ma PC, Maulik G, Christensen J and Salgia
R: c-Met: Structure, functions and potential for therapeutic
inhibition. Cancer Metastasis Rev. 22:309–325. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Revathidevi S and Munirajan AK: Akt in
cancer: Mediator and more. Semin Cancer Biol. 59:80–91. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Johnson GL and Lapadat R:
Mitogen-activated protein kinase pathways mediated by ERK, JNK, and
p38 protein kinases. Science. 298:1911–1912. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Chen Y and Wang X: miRDB: An online
database for prediction of functional microRNA targets. Nucleic
Acids Res. 48(D1): D127–D131. 2020. View Article : Google Scholar :
|
|
124
|
McGeary SE, Lin KS, Shi CY, Pham TM,
Bisaria N, Kelley GM and Bartel DP: The biochemical basis of
microRNA targeting efficacy. Science. 366:eaav17412019. View Article : Google Scholar : PubMed/NCBI
|