Introduction
Tuberculosis (TB) caused by Mycobacterium
tuberculosis (MTB) has the highest mortality rate worldwide of
any infectious disease and has been declared a global health
emergency by the World Health Organization (1,2). The
attenuated M. bovis Bacillus Calmette Guerin (BCG) is the
only available vaccine against TB. However, BCG exhibits varying
efficacy (0-80%) against adult pulmonary TB (3). Emergence of drug-resistant isolates
of MTB highlight the continued necessity for the discovery and
development of drugs active against the bacterium (4–8).
Aptamers have high affinity and specificity for
their targets and have been developed for use as oligonucleotide
analogs of antibodies. These molecules exhibit several advantages
over antibodies and antibiotics. Aptamers are smaller than
antibodies and therefore exhibit improved cell penetration, blood
clearance and chemical modification. They are also nonimmunogenic
and readily synthesized and therefore do not induce an immune host
response, which may cause harmful side-effects (9,10).
Single-stranded DNA (ssDNA) aptamers exhibit more variable
structures, longer relative temperature stability and shelf-life
than antibodies and therefore demonstrate significant potential for
in vivo use as therapeutics (11,12).
In previous years the Systematic Evolution of
Ligands by Exponential Enrichment (SELEX) technique has become
increasingly important for the study of protein function, as well
as in drug discovery and identification of antagonists against a
number of functional proteins (13,14).
However, little is known regarding the in vitro SELEX
selection strategy utilized for the generation of inhibitors
against whole bacteria. Our previous study extracted special
aptamers (NK2) and aptamer pools (10th pool) from a whole bacterial
SELEX strategy (15) and
demonstrated that aptamers inhibit MBT H37Rv invasion of
macrophages in vitro(16).
In the present study, we further evaluated the function of the 10th
pool and NK2 against MBT H37Rv in a mouse model. This investigation
may aid the development of a new antitubercular agent based on an
aptamer species.
Materials and methods
Bacterial strain and animals
MTB H37Rv (strain ATCC 93009) was purchased from the
Beijing Biological Product Institute (Beijing, China). Bacteria
were maintained on Lowenstein-Jensen (L-J) medium and harvested in
log phase growth. Prior to use, bacilli were washed in 0.05%
Tween-80 saline and triturated to uniformity. C57BL/6 mice (from
the Experimental Building of the Animal Laboratory Center, Wuhan
University, Wuhan, China) of either sex were used at 5–6 weeks of
age. Bacterial cultures and animal tests were performed in the
Animal Biosafety Level 3 Laboratory (ABSL-III) of Wuhan University
School of Medicine. The research procedures and the animal
protocols in this study were approved by the HPKLTM Ethics &
Animal Use Committee (approval ID: HPKLTME201005).
Fluorescence microscopy and phagocytic
index
C57BL/6 mouse peritoneal macrophages were collected
and cultured in RPMI-1640 medium with 10% FBS and penicillin and
streptomycin. Following 48 h, cell number was estimated using
trypan blue staining. To analyze the effect of aptamers on H37Rv
invasion of macrophages, 108 cfu H37Rv bacteria was
incubated with aptamers (pretreated at 85°C for 15 min and then
incubated on ice for 3 min) at 37°C for 15 min (control group was
untreated with aptamer). Following this, centrifugation at 12,000
rpm for 5 min was performed and the supernatant was discarded.
Mouse peritoneal macrophages (106) were mixed with
108 cfu H37Rv in 2 ml medium. Phagocytosis was allowed
to occur for 1 h during mixing at 37°C in 6-well costar plates. A
pretreated (polylysine, 0.1 mg/ml) coverslip was applied to each
analytic well. The coverslip was dislodged and fixed by frigorific
acetone and then stained with auramine O. The coverslip was then
observed under a fluorescence microscope (CW4000, Leica, Wetzlar,
Germany). Counts of 200 cells from each coverslip were performed
and the phagocytic index was calculated using the formula:
phagocytic index = (total MTB phagocytosed by
macrophage)/(macrophage number of phagocytosed MTB).
Flow cytometry analysis of the effect of
aptamers on the invasion of H37Rv to CD4+ and
CD8+ T cells
H37Rv (108 cfu; prestained with Rhodamine
B) was incubated for 1 h with 106 peripheral blood
mononuclear cells (PBMCs) collected from the tail vein of C57BL/6
mice. Experimental groups were pretreated with the 10th pool or NK2
as previously described (15).
Following this, the cells were washed twice with PBS and incubated
with 5 μl mouse anti-CD3-PE-Cy5, anti-CD4-FITC (Caltag
Laboratories, Buckingham, UK) or isotype control antibodies, at the
concentration recommended by the manufacturer’s instructions, for
30 min at 4°C. The cells were then fixed with 70% ethanol and
analyzed using a fluorescence-activated cell sorter (FACS; Epics
Altra II, Beckman Coulter, Miami, FL, USA).
Effect of aptamers on the intracellular
expression of IFN-γ of CD4+ and CD8+ T cells
in H37Rv-infected splenocytes
Murine splenocytes (2×106)were incubated
with 108 cfu H37Rv or pretreated with the 10th pool and
NK2 for 1 h. Following this, 1 μg/ml monensin (eBioscience, San
Diego, CA, USA) was added and incubated at 37°C for 2 h in a 5%
(v/v) CO2 atmosphere. Cells were washed twice with PBS,
resuspended in 100 μl cold PBS and incubated with 5 μl mouse
anti-CD3-PE-Cy5 and anti-CD4-FITC antibodies (Caltag Laboratories)
at 4°C for 30 min. The cells were fixed using 70% ethanol and
incubated with 5 μl PE-conjugated mouse anti-IFN-γ (Caltag
Laboratories) at 37°C for 30 min in the dark. Stained cells were
washed with PBS and intracellular cytokine expression of IFN-γ in
CD3+ CD4+ and CD3+ CD8+
T cells was analyzed by FACS.
Challenge infection and analysis of
survival rate
C57BL/6 female mice (18–22 g) were used in the
study. H37Rv bacteria used for in vivo experiments underwent
several passages in C57BL/6 mice to enhance virulence (17). Mice were injected intravenously
with 107 cfu H37Rv/mouse in 0.4 ml saline. Equal MTB was
pretreated with 8 μg of the 10th pool or NK2 aptamers, the
supernatant was then discarded, and pellets were centrifuged prior
to injection with 0.4 ml saline. Mice were fed with standard
pelleted food and water for 20 days and each group was composed of
8 mice. Mortality was monitored daily. Survival analysis was
analyzed by Kaplan-Meier (SPSS 13.0, Chicago, IL, USA).
Histopathology and acid-fast stain
Lungs or spleens from mice sacrificed 20 days
post-infection were homogenized in saline and plated on L-J medium.
Following 4–6 weeks incubation at 37°C, the number of viable
organisms in the lungs or spleens was determined. Sections of the
lungs and spleens were soaked in 10% paraformaldehyde for a minimum
of 12 h and examined with hematoxylin and eosin stain and acid-fast
stain, respectively.
Statistical analysis
Data were presented as the mean ± SEM and were
analyzed using one-way analysis of variance followed by the
Student-Newman-Keuls post-hoc test. P<0.05 was considered to
indicate a statistically significant difference.
Results
Inhibition of MTB invasion of mouse
peritoneal macrophages
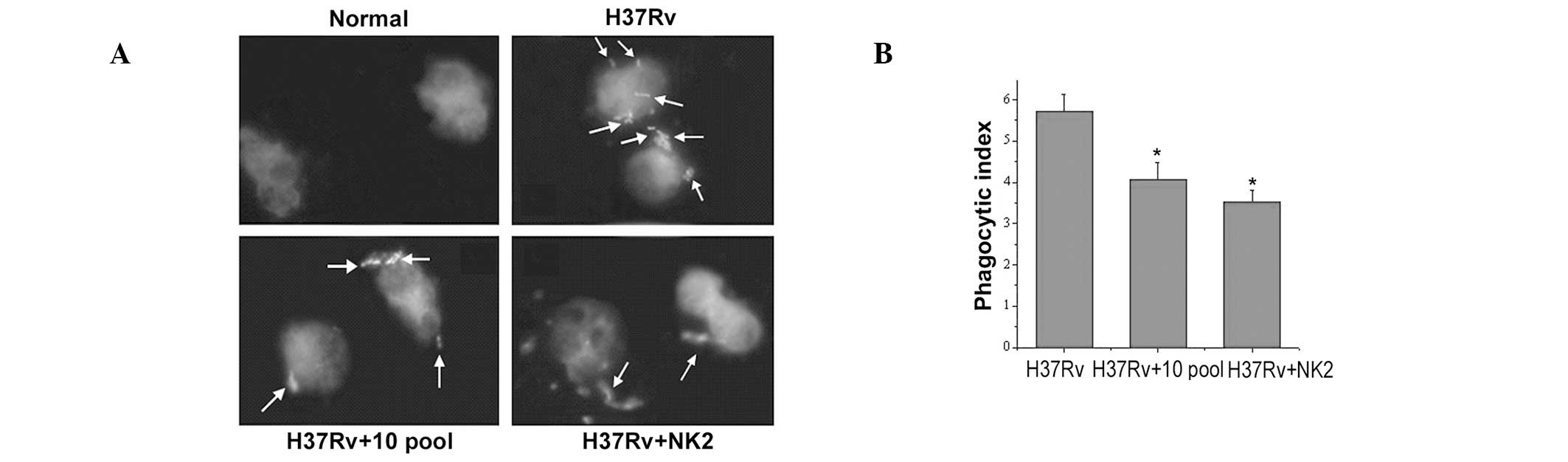
Compared with untreated, groups pretreated with
aptamers (NK2 or 10th pool) inhibited MTB invasion of mouse
peritoneal macrophages (Fig. 1A).
Similar results were obtained by comparing the phagocytic index
with fluorescence microscope observations (Fig. 1B). MTB invasion of mouse peritoneal
macrophages was higher in NK2 than the 10th pool.
Inhibition of H37Rv invasion to
CD3+ CD8+ T cells
H37Rv invasion to CD3+ CD8+ T
cells was inhibited by the aptamers. Invasion to CD3+
CD4+ T cells was not inhibited. H37Rv invasion to
CD3+ CD8+ T cells was found to be
significantly decreased from 50.4% (without aptamers) to 33.9, 28,
21 and 20% with NK2 aptamer, 10th, 7th and 3rd aptamer pool,
respectively (Fig. 2A). These data
indicate that aptamer pools protect CD8+ T cells against
H37Rv infection.
Aptamers increase intracellular IFN-γ
secretion of CD3+ CD4+ T cells
Intracellular IFN-γ levels in CD3+
CD4+ and CD3+ CD8+ T cells were
detected by flow cytometry. When PBMCs from mice were infected by
H37Rv in vitro, increased levels of intracellular IFN-γ were
observed in the presence of aptamers in CD3+
CD4+ T cells, but not in CD3+/CD8+
T cells. Intracellular IFN-γ levels in CD3+
CD4+ T cells were increased from 35% (without aptamers)
to 48.1, 50.1, 46.2, 53.6 and 54.9% with 3rd, 7th, 10th and 12th
aptamer pools and NK2 aptamer, respectivly. The NK2 aptamer had the
strongest stimulatory effect on intracellular IFN-γ levels in
CD3+ CD4+ T cells (Fig. 2B). These data indicate that
aptamers, particularly the NK2 aptamer, stimulate IFN-γ production
and decrease the infection efficiency of MTB.
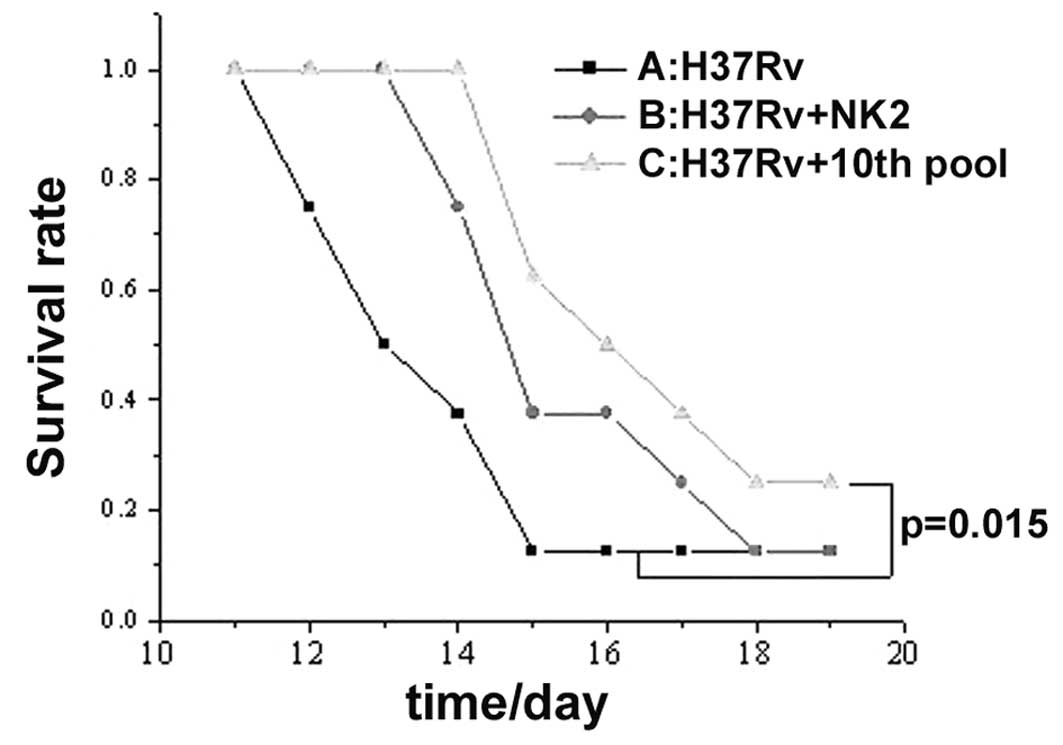
Treatment of H37Rv in vivo
Effect of 10th round aptamer pool or NK2 aptamer on
acute tuberculosis in mice was examined. In the control group,
H37Rv injection for 13 days, resulted in 50% mortality. Survival
rate of the mice was prolonged for 2 days with a single injection
of aptamer NK2 and 3 days with a single injection of aptamer pool
(10th round selection; P<0.05; Fig.
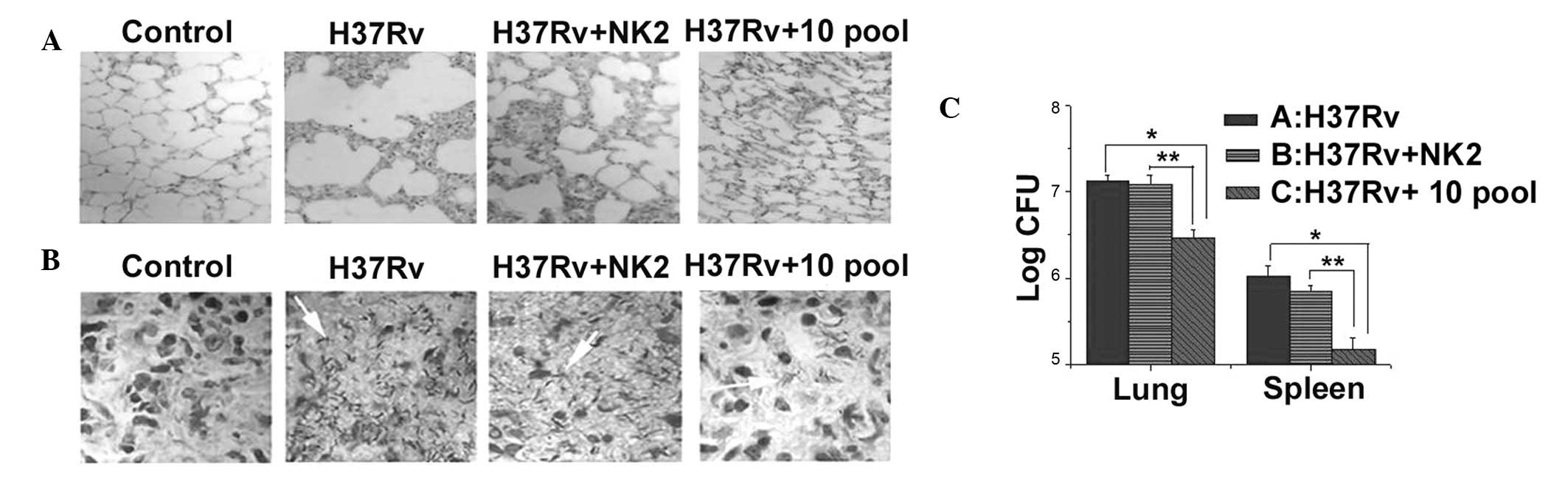
3). Histopathological examination of lungs from mice injected
with H37Rv revealed a marked acute inflammatory reaction compared
with normal mice. NK2- or aptamer pool treated-mice demonstrated
decreased pulmonary alveoli fusion and swelling and more prominent
air spaces, similar to lungs of normal mice (Fig. 4A). Furthermore, compared with
normal mice the density of acid fast bacillus in the lung was
higher in control mice than mice that had received a single
injection of aptamer NK2 or 10th pool aptamers (Fig. 4B). Number of H37Rv colonies was
higher in control mice than in the 10th aptamer pool or NK2 groups
(Fig. 4C). The present study
revealed that, in vivo, the 10th pool of aptamers and NK2
had an active therapeutic effect on H37Rv infection and the 10th
pool of aptamers was more effective than NK2.
Discussion
The use of aptamers as therapeutic drugs has been
reported in numerous research fields, including HIV (18) and cancer (19). Aptamers may inhibit MTB infection
through blockage of virulence components or epitopes of H37Rv.
In vitro, inhibition of H37Rv macrophage infection by NK2
aptamers was higher when compared with the 10th aptamer pool
(Fig. 1), however, the 10th
aptamer pool prolonged the survival rate of mice, enhancing
clearance of the bacterium in vivo to a more significant
degree (Figs. 3 and 4). This may be due to the complex surface
of MTB and the degradation of DNA aptamers by nuclease enzymes
(20). The 10th aptamer pool
contained various non-special aptamers that may aid resistance to
nucleases and function in synergy with the NK2 aptamer. Thus, the
10th aptamer pool was more effective than the NK2 aptamer in
vivo. Previously, modified aptamers with longer half-lives were
developed to resist nuclease digestion. However, disadvantages have
been identified, including an increased rate of integration into
the chromosomal DNA of host T cells. There are multiple binding
sites and antigen epitopes on the surface of H37Rv and synergism of
several aptamers is required to inhibit H37Rv infection. In the
present study, we noted that inhibition of H37Rv invasion to
CD8+ T cells decreased as the screening process
progressed (Fig. 2A). This
observation may be due to inhibitory aptamers being missed through
SELEX. This may be explained by important components in the reverse
target (BCG) which stimulate the cytotoxic T-cell effect.
Currently, BCG is the only vaccine against tuberculosis, however,
its immune protective effects are not always effective. A possible
explanation for reduced efficacy may be due to the effect of
aptamers on CD4+ and CD8+ T cells (Fig. 2).
The present study demonstrates that the aptamer pool
and NK2 aptamer exhibit protection against tuberculosis, however,
their mechanisms of action are different. NK2 revealed improved
inhibition of H37Rv invasion to mice peritoneal macrophages and
stimulation of CD4+ T-cell INF-γ secretion compared with
the 10th pool. However, survival rate and histological analysis
revealed that the 10th pool has a better therapeutic effect
compared with NK2. The results demonstrated we should not only pay
attention to those aptamers in the majority (including NK2), but
also investigate the role of early deserted aptamers. The present
study demonstrates the limitations of ssDNA aptamers, highlighting
a number of factors which must be considered to avoid available
aptamer loss.
Acknowledgements
This study was supported by Wuhan Health Bureau
Funded Projects (WH12A03) and the Hainan Natural Science Fund (nos.
808162 and 812199).
References
|
1
|
Raviglione MC, Dye C, Schmidt S and Kochi
A: Assessment of worldwide tuberculosis control. WHO global
surveillance and monitoring project. Lancet. 350:624–649. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Dye C, Scheele S, Dolin P, Pathania V and
Raviglione MC: Consensus statement. Global burden of tuberculosis:
estimated incidence, prevalence and mortality by country WHO Global
Surveillance and Monitoring Project. JAMA. 282:677–686. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Fine PE: Variation in protection by BCG:
implications of and for heterologous immunity. Lancet.
346:1339–1345. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Pablos-Mendez A, Raviglione MC, Laszlo A,
Binkin N, Rieder HL, Bustreo F, Cohn DL, Lambregts-van Weezenbeek
CS, Kim SJ, Chaulet P and Nunn P: Global surveillance for
antituberculosis-drug resistance 1994–1997. World Health
Organization-International Union against tuberculosis and lung
disease working group on anti-tuberculosis drug resistance
surveillance. N Engl J Med. 338:1641–1649. 1998.
|
|
5
|
Ahmed N and Hasnain SE: Genomics of
Mycobacterium tuberculosis: old threats and new trends.
Indian J Med Res. 120:207–212. 2004.
|
|
6
|
Baptista IMFD, Oelemann MC, Opromolla DVA
and Suffys PA: Drug resistance and genotypes of strains of
Mycobacterium tuberculosis isolated from human
immunodeficiency virus-infected and non-infected tuberculosis
patients in Bauru, São Paulo, Brazil. Mem Inst Oswaldo Cruz.
97:1147–1152. 2002.PubMed/NCBI
|
|
7
|
Pereira M, Tripathy S, Inamdar V, Ramesh
K, Bhavsar M, Date A, Iyyer R, Acchammachary A, Mehendale S and
Risbud A: Drug resistance pattern of Mycobacterium
tuberculosis in seropositive and seronegative HIV-TB patients
in Pune, India. Indian J Med Res. 121:235–239. 2005.PubMed/NCBI
|
|
8
|
Yew WW and Leung CC: Update in
tuberculosis 2007. Am J Respir Crit Care Med. 177:479–485. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jayasena SD: Aptamers: An emerging class
of molecules that rival antibodies in diagnostics. Clin Chem.
45:1628–1650. 1999.PubMed/NCBI
|
|
10
|
Guthrie JW, Hamula CLA, Zhang H and Le XC:
Assays for cytokines using aptamers. Methods. 38:324–330. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ulrich H, Alves MJM and Colli W: RNA and
DNA aptamers as potential tools to prevent cell adhesion in
disease. Braz J Med Bio. 34:295–300. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Marimuthu C, Tang TH, Tominaga J, Tan SC
and Gopinath SC: Single-stranded DNA (ssDNA) production in DNA
aptamer generation. Analyst. 137:1307–1315. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Shimada T, Fujita N, Maeda M and Ishihama
A: Systematic search for the Cra-binding promoters using genomic
SELEX system. Genes Cells. 10:907–918. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Jensen KB, Atkinson BL, Willis MC, Koch TD
and Gold L: Using in vitro selection to direct the covalent
attachment of human immunodeficiency virus type 1 Rev protein to
high-affinity RNA ligands. Proc Natl Acad Sci USA. 92:12220–12224.
1995. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chen F, Zhou J, Luo F, Mohammed AB and
Zhang ZL: Aptamer from whole-bacterium SELEX as new therapeutic
reagent against virulent Mycobacterium tuberculosis. Biochem
Biophys Res Commun. 357:743–748. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chen F, Zhang XL, Zhou J, Liu S and Liu J:
Aptamer inhibits Mycobacterium tuberculosis (H37Rv) invasion
of macrophage. Mol Biol Rep. 39:2157–2162. 2012.PubMed/NCBI
|
|
17
|
Jagannath C, Emanuele MH and Hunter RL:
Activity of poloxamer CRL-1072 against drug-sensitive and resistant
strains of Mycobacterium tuberculosis in macrophages and in
mice. Intern J Anti Agent. 15:55–63. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Jing N and Hogan ME: Structure-activity of
tetrad-forming oligonucleotides as a potent anti-HIV therapeutic
drug. J Biol Chem. 273:34992–34999. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Blank M, Weinschenk T, Priemer M and
Schluesener H: Systematic evolution of a DNA aptamer binding to the
rat brain tumor microvessels: selective targeting of endothelial
regulatory protein pigpen. J Biol Chem. 276:16464–16468. 2001.
View Article : Google Scholar
|
|
20
|
Cerchia L, Ducongé F, Pestourie C, Boulay
J, Aissouni Y, Gombert K, Tavitian B, de Franciscis V and Libri D:
Neutralizing aptamers from whole-cell SELEX inhibit the RET
receptor tyrosine kinase. PLoS Biol. 3:e1232005. View Article : Google Scholar : PubMed/NCBI
|