Introduction
Tyrosine-phosphorylated proteins govern a host of
cell functions, such as growth, division, adhesion and motility
(1). These proteins are critical
regulators of signaling in the majority of eukaryotic cellular
pathways, and deregulated phosphorylation is involved in an array
of diseases (2), including cancer
(3).
Given their importance, a number of techniques have
been used to study tyrosine-phosphorylated proteins, such as
MS-based phosphoproteomic methods (4), redox-based probes (5), an Src homology 2 (SH2) profiling
method based on far-western blot analysis (6), and the use of Grb2-SH2 domain binding
proteins with SILAC (7).
The Nck adaptor protein consists of three SH3
domains followed by a C-terminal SH2 domain, and is capable of
binding to numerous receptor tyrosine kinases via its SH2 domain
(8). Dierck et al have
developed an alternative phosphoproteomic method (termed SH2
profiling) to explore phosphotyrosine signaling in cancer cells
(9), and have demonstrated that it
is an ideal method to detect phosphotyrosine, due to its being
highly sensitive and throughput.
However, these aforementioned studies were performed
in cell lines. Therefore, the state of tyrosine-phosphorylated
proteins in tumor tissues remains unknown. Our previous study
successfully combined the GST-Nck1-SH2 pull-down with
two-dimensional electrophoresis (2-DE) to detect
tyrosine-phosphorylated proteins in liver tissues (10).
Although the combination of GST-Nck1-SH2 pull-down
and 2-DE was able to detect tyrosine-phosphorylated proteins, it
continues to involve numerous challenges. The greatest of those
challenges was how to harvest samples from GST-Nck1-SH2 pull-down
that are also compatible with downstream 2-DE. The efficiency of
the GST-Nck1-SH2 pull-down method requires improvement in order
that it yields sufficient tyrosine-phosphorylated proteins for
downstream 2-DE. In addition, the samples obtained though the
GST-Nck1-SH2 pull-down method include different types of
interferential material, such as fragments of GST beads and iron,
which are likely to affect the success of isoelectric focusing
(IEF). Therefore, there is a requirement to deplete the
interferential materials, while retaining the activity of the
tyrosine-phosphorylated proteins. At present, to the best of our
knowledge, few effective and detailed methods have been devised to
obtain tyrosine-phosphorylated proteins from tissues by the use of
the GST-Nck1-SH2 pull-down method. The present study set out to
explore the effective techniques of GST-Nck1-SH2 pull-down, and to
search for detailed methods regarding sample preparation for
downstream 2-DE.
Materials and methods
Hepatocellular carcinoma (HCC) patient
samples
HCC tissues were collected from 21 HCC patients who
underwent hepatectomy at the First Affiliated Hospital of Sun-Yat
Sen University (Guangzhou, China). None of these patients had
received preoperative chemotherapy or radiotherapy. Normal liver
tissues were obtained from 8 patients diagnosed with liver
hemangioma or cholelithiasis. Specimens were obtained with written
informed consent from all patients. The study was conducted with
prior approval from the Committees for Ethical Review of Research
involving Human Subjects of the First Affiliated Hospital of
Sun-Yat Sen University.
Plasmid constructs and transfection
The full-length dermcidin cDNA was amplified and
cloned into the pReciever M06 expression vector (FulenGen Co.,
Ltd., Guangzhou, China). The GST-tagged SH2 domain of Nck was
generated by PCR amplification of the human Nck template, and then
ligated into the pGEX-4T-3 expression vector.
GST fusion protein purification
Escherichia coli (BL21) was transformed with
pGEX-4T-3 or pGEXNck-SH2 incubated with 0.2 mM
isopropyl-β-D-1-thiogalactopyranoside (IPTG) for 4 h. The GST
fusion proteins were purified from bacterial lysates with
GSH-Sepharose 4B beads, according to the manufacturer’s
instructions (Amersham Biosciences Corp., Picataway, NJ, USA).
Tissue/cell lysates were prepared and spun at 15,000 × g for 15
min.
GST pull-down
The liver proteins were incubated with
GST-Nck1-SH2-conjugated sepharose beads for 2 h at 4°C. Following
incubation, the supernatant was removed and the beads were washed
with a Tris-sucrose solution (10 mM Tris-HCl; 150 mM NaCl; 1%
Triton X-100, pH 7.5) to remove any non-specific or non-covalently
bound proteins. The fusion proteins were eluted with 2 ml 2-D lysis
buffer (10 mM reduced glutathione in 50 mM Tris-HCl, pH 8.0) and
desalted by ultrafiltration.
2-D clean-up
2-D clean-up was performed according to the
manufacturer’ instructions (Amersham Biosciences Corp.).
Ultrafiltration
Samples were loaded into centrifuge tubes with a
10-kDa membrane, in order to concentrate the proteins and remove
small interference molecules. Samples were centrifuged at 5,000 ×
g, at 4°C for 1 h.
2-DE and image analysis
Protein samples (250 μg) were diluted to 450 μl with
a rehydration solution (7 M urea, 4% CHAPS, 0.5% IPG ampholyte, 65
mM DTE, 2 M thiourea, and 0.0002% bromophenol blue), and then
loaded onto IPG gel strips (pH 3.0–10.0 linear, 24 cm long;
Amersham Biosciences Corp.). The first dimensional separation was
performed using the IPGphor system (Amersham Biosciences Corp.) at
18°C with 8,000 V, for a total of 90 k VHS. Following IEF, the IPG
strips were subjected to reduction with 2% DTE in equilibration
solution (50 mM Tris-HCl, pH 8.8; 6 M urea; 2% SDS; 30% glycerol),
followed by alkylation with 2.5% iodoacetamide in the same buffer.
The gels were stained with Silver Staining kit (Amersham
Biosciences Corp.) according to the manufacturer’s instructions.
The developed gels were scanned as 2-DE images using an image
scanner, and then analyzed using ImageMaster software (Amersham
Biosciences Corp.).
In-gel digestion and protein
identification
2-DE gels of interest were washed in
water/acetonitrile (ACN; 1:1) and then dehydrated in ACN. The gel
pieces were air-dried and rehydrated in 20 μl of 10 mM DTT and 0.1
M NH4HCO3. Reduction of disulfide bonds was
performed at 56°C for 45 min. The supernatant was discarded and
cysteine residues were modified to S-carboxyamidomethylcysteine in
55 mM iodoacetamide and 0.1 M NH4HCO3. After
washing with 0.1 M NH4HCO3/ACN (1:1) for 15
min, followed by ACN, the gel pieces were air-dried, rehydrated in
chilled 50 mM NH4HCO3 and 12.5 ng/μl trypsin,
and then incubated at 37°C overnight. The supernatant was collected
and peptides were extracted twice from the gel with 50 mM
NH4HCO3/ACN (1:1) followed by 5% formic
acid/ACN (1:1). The combined extracts were evaporated to dryness in
a vacuum centrifuge. Prior to mass spectrometric analysis, peptides
were re-dissolved in 10 μl of 0.1% formic acid. Online peptide
separation was performed after trapping each sample on a 180 μm ×
20 mm Symmetry® C18 Nano Acquity™ UPLC™ column with 1%
ACN and 0.1% formic acid at a 15 ml/min flow rate; following
separation on a 75 μm × 250 mm BEH130 column (Nano Aquity™ UPLC™)
with a 50-min gradient from 5 to 95% ACN and 0.1% formic acid, at a
flow rate of 300 nl/min. A tapered fused silica was used as an
emitter. Mass analyses were performed with a quadrupole
time-of-flight mass spectrometer (QTOF, Waters Corp., Milford, MA,
USA). The mass spectrometer was operated in a data-dependent mode
to automatically switch between MS and MS/MS acquisition. Survey MS
spectra (m/z 400–1800) were acquired in the QTOF, and the four most
intense ions in each survey scan were fragmented and analyzed.
Proteins were identified by automated database searching (Spectrum
Mill; Agilent Technologies UK Ltd., Wokingham, UK) and MASCOT
(Matrix Science, London, UK), of all MS and MS/MS spectra using the
IPI Human, Swiss-Prot and NCBinr databases. Raw data files were
converted to .pkl files by the Protein Lynx Global Server (PLGS;
Waters Corp.). Search parameters were set as follows: MS accuracy,
0.15 Da; MS/MS accuracy, 0.15 Da; two missed cleavage allowed.
Variable carbamidomethyl modification of cystine and variable
oxidation of methionine, and all entries of the databases were
searched.
Results
Induction and purification of
GST-Nck1-SH2
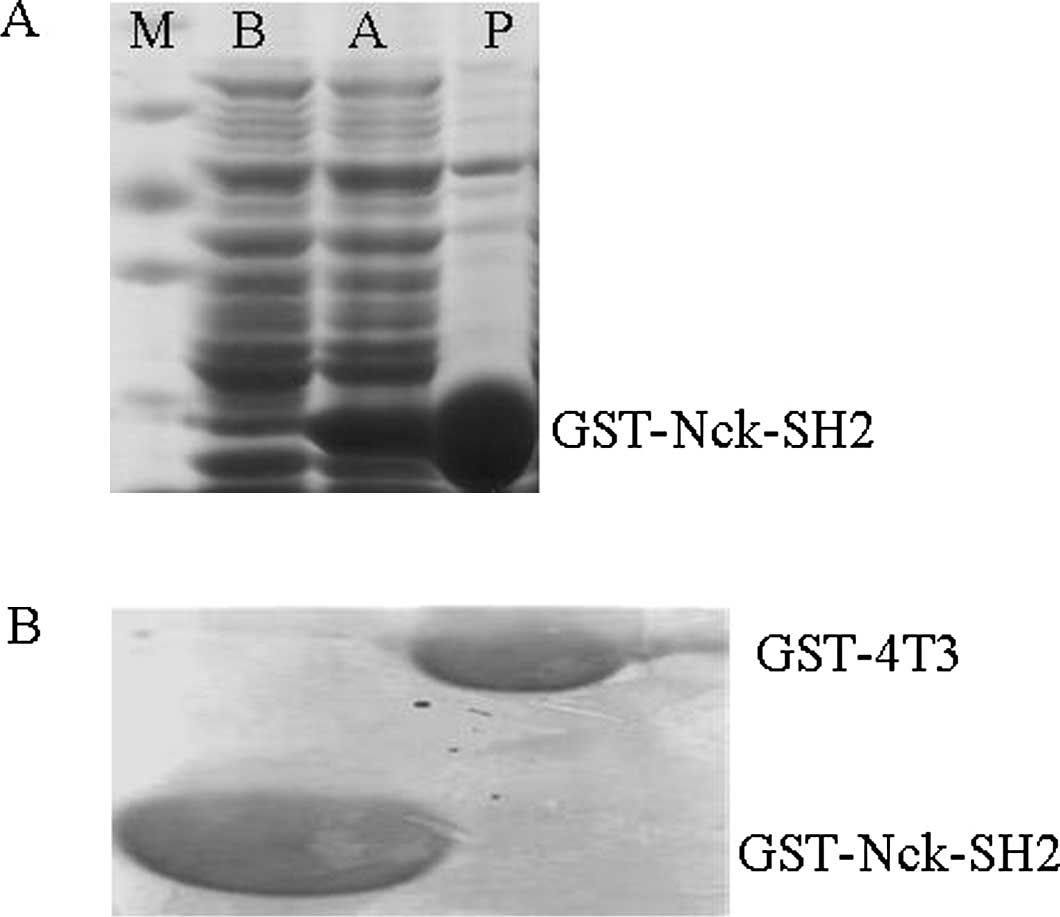
The first key step of our study was to induce and
purify the GST-Nck1-SH2 fusion proteins. As shown in Fig. 1A, following induction with
isopropyl-β-D-1-thiogalactopyranoside (IPTG), the level of
GST-Nck1-SH2 fusion proteins notably increased. To confirm the
specificity of tyrosine-phosphorylated proteins which are the
‘prey’ proteins that are pulled down by by GST-Nck1-SH2, we
performed negative control experiments with GST-4T3, under the same
conditions. The comparison between GST-4T-3 and GST-Nck1-SH2 is
shown in Fig. 1B. These results
demonstrated that the induction and purification of GST-Nck1-SH2
fusion proteins were successful.
Comparison of different strategies for
capturing tyrosine-phosphorylated proteins by the GST-Nck1-SH2
pull-down method
In order to yield sufficient tyrosine-phosphorylated
proteins by GST-Nck1-SH2 pull-down, we explored different pull-down
strategies to detect the ‘prey’ proteins in liver tissues (Table I). According to the general
guidelines of GST pull-down in cells, we employed 100 μl
GST-Nck1-SH2 fusion proteins to pull down the ‘prey’
tyrosine-phosphorylated proteins among 1 mg liver proteins in
method A. However, in method B, we optimized the quantity of
GST-Nck1-SH2 fusion protein to 200 μl; while in methods C and D, 10
and 20 mg of liver protein was added, respectively. Protein yields
pulled down by methods A, B, C and D were 100±5, 123±6, 1202±28 and
1301±32 μg, respectively. As demonstrated in Fig. 2A, method C (100 μl fusion
protein/10 mg liver protein) and method D (100 μl fusion protein/20
mg liver protein) were able to markedly increase the protein
accounts, while methods A and B yielded insufficient quantities of
protein for downstream tests.
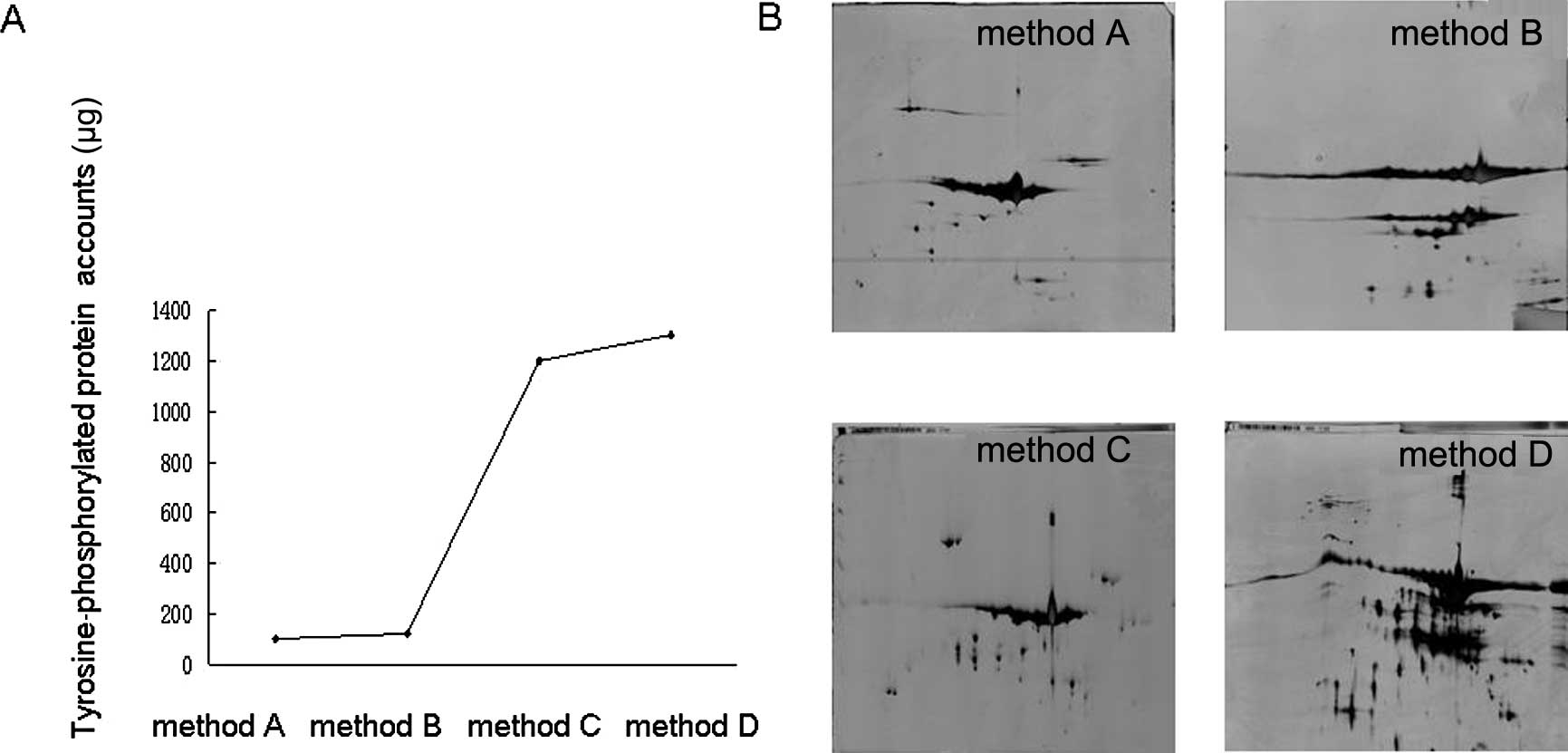 | Figure 2Different strategies employed to yield
tyrosine-phosphorylated proteins. (A) Comparison of different
strategies to yield tyrosine-phosphorylated proteins. Protein
yields obtained by methods A, B, C and D were 100±5, 123±6, 1202±28
and 1301±32 μg, respectively. (B) Two-dimensional electrophoresis
(2-DE) was used to confirm the effect of the above strategies.
Protein spots (50±5, 70±15, 200±38 and 238±43) were detected on the
2-DE gels of methods A, B, C and D, respectively. |
 | Table IApproximate yield using different
methods. |
Table I
Approximate yield using different
methods.
| Method | GST-Nck1-SH2 fusion
protein (μl) | Liver protein
(mg) | Yield (μg) | Protein spots |
|---|
| A | 100 | 1 | 100±5 | 50±5 |
| B | 200 | 1 | 123±6 | 70±15 |
| C | 100 | 10 | 1202±28 | 200±38 |
| D | 100 | 20 | 1301+32 | 238±43 |
We set out to further investigate the effect of the
previous strategies; samples pulled down by the aforementioned
methods were loaded to 2-DE gels. As shown in Fig. 2B, protein spots of method A, B, C
and D were 50±5, 70±15, 200±38 and 238±43, respectively. The 2-D
gel results for methods C and D contained more protein spots than
methods A and B; however, the 2-D gel for method D exhibited
horizontal streaks and blurry protein spots. Additionally, the 2-D
gel for method B exhibited only large GST beads. Overall, method C
was selected for further study.
Comparison of different types of lysis
buffer for dissolving proteins
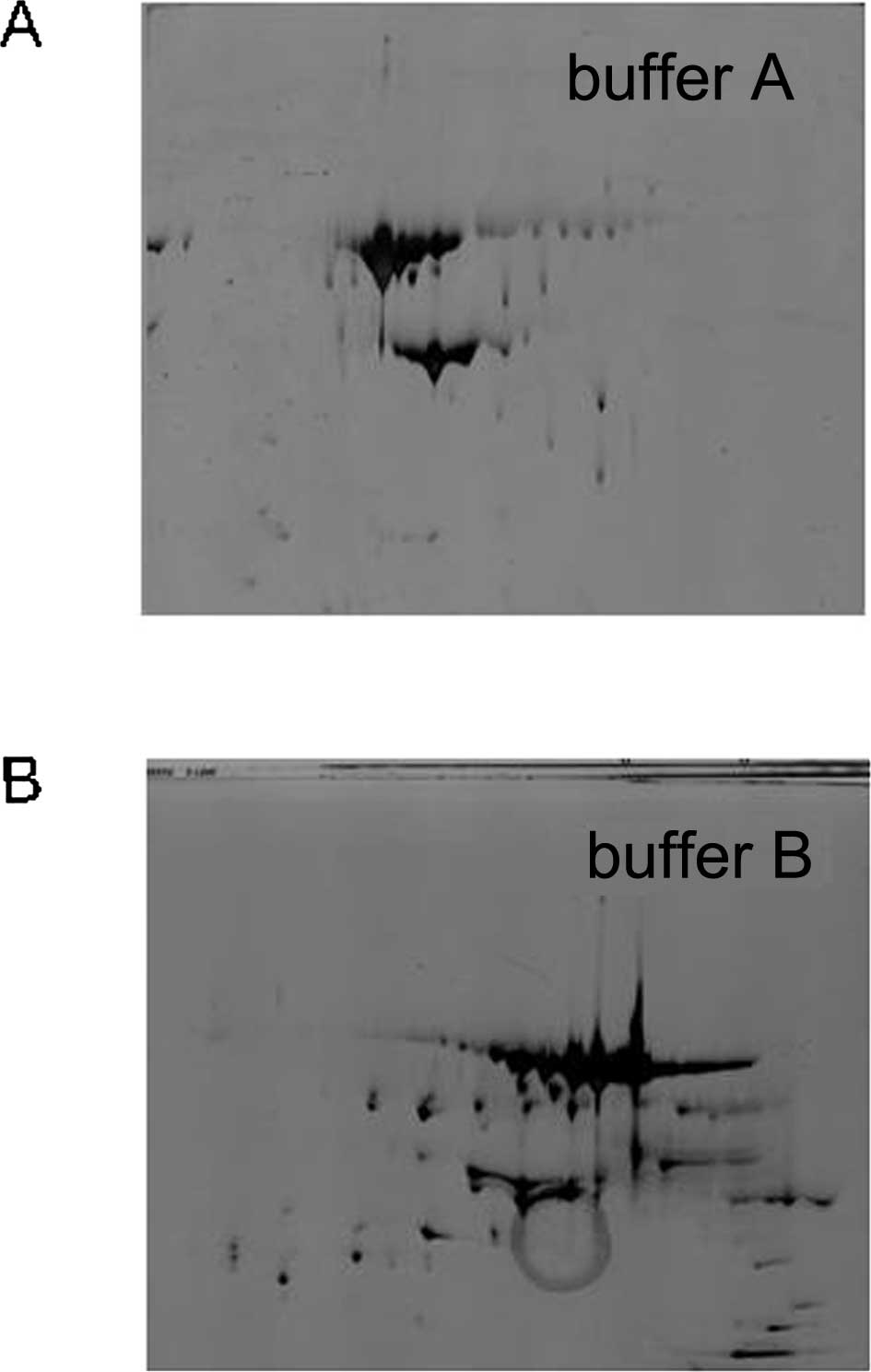
Commonly, there are two types of lysis buffer for
dissolving proteins. The standard cocktail contains 8 M urea
(chaotropic), 4% CHAPS (detergent) and 50 mM DTT (lysis buffer A);
while the other buffer contains 7 M urea and 2 M thiourea (lysis
buffer B) in place of 8 M urea. The latter buffer type is capable
of increasing the solubility of certain proteins and producing more
spots, which is consistent with our results: Buffer A, 89±40
protein spots; buffer B, 200±38 protein spots (Fig. 3A and B). Overall, buffer B was used
for further sample preparation.
Comparison of different methods for
depleting the interferential materials
The resulting samples from GST-Nck1-SH2 pull-down
include different types of material that affect the success of IEF.
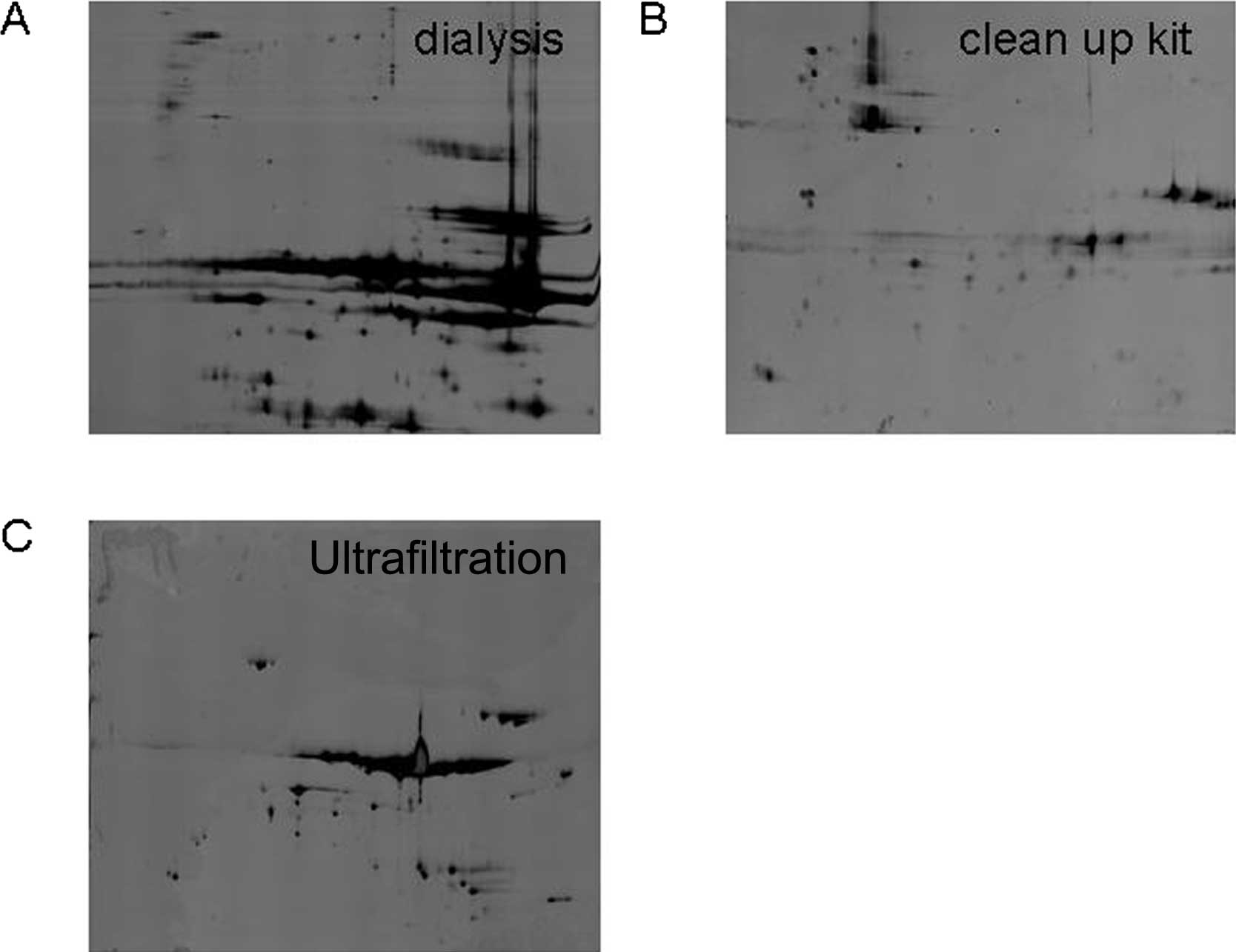
Therefore, we selected three types of typical methods for deleting
the interferential materials to explore. These methods included a
mini dialysis kit (8 kDa, Amersham Biosciences Corp.), a 2-D
clean-up kit (Amersham Biosciences Corp.) and ultrafiltration (10
kDa, Millipore, Biosciences, NJ, USA). To delete the interferential
materials, the samples must be loaded onto 2-DE gels. As shown in
Fig. 4A, the 2-D gel for the mini
dialysis kit was of poor quality, with horizontal streaks and
blurry protein spots, suggesting that IEF had failed due to an
incomplete removal of salts and GST fragments by the kit. However,
the 2-D clean-up kit removed the fragments of the GST beads
effectively, while also cleaning the majority of the
tyrosine-phosphorylated proteins (Fig.
4B); only a few protein spots remained following depletion. As
shown in Fig. 4C, 210±18 spots
were detected after ultrafiltration depletion. Overall,
ultrafiltration was chosen to be further tested.
Negative control of the GST-Nck1-SH2
pull-down procedure
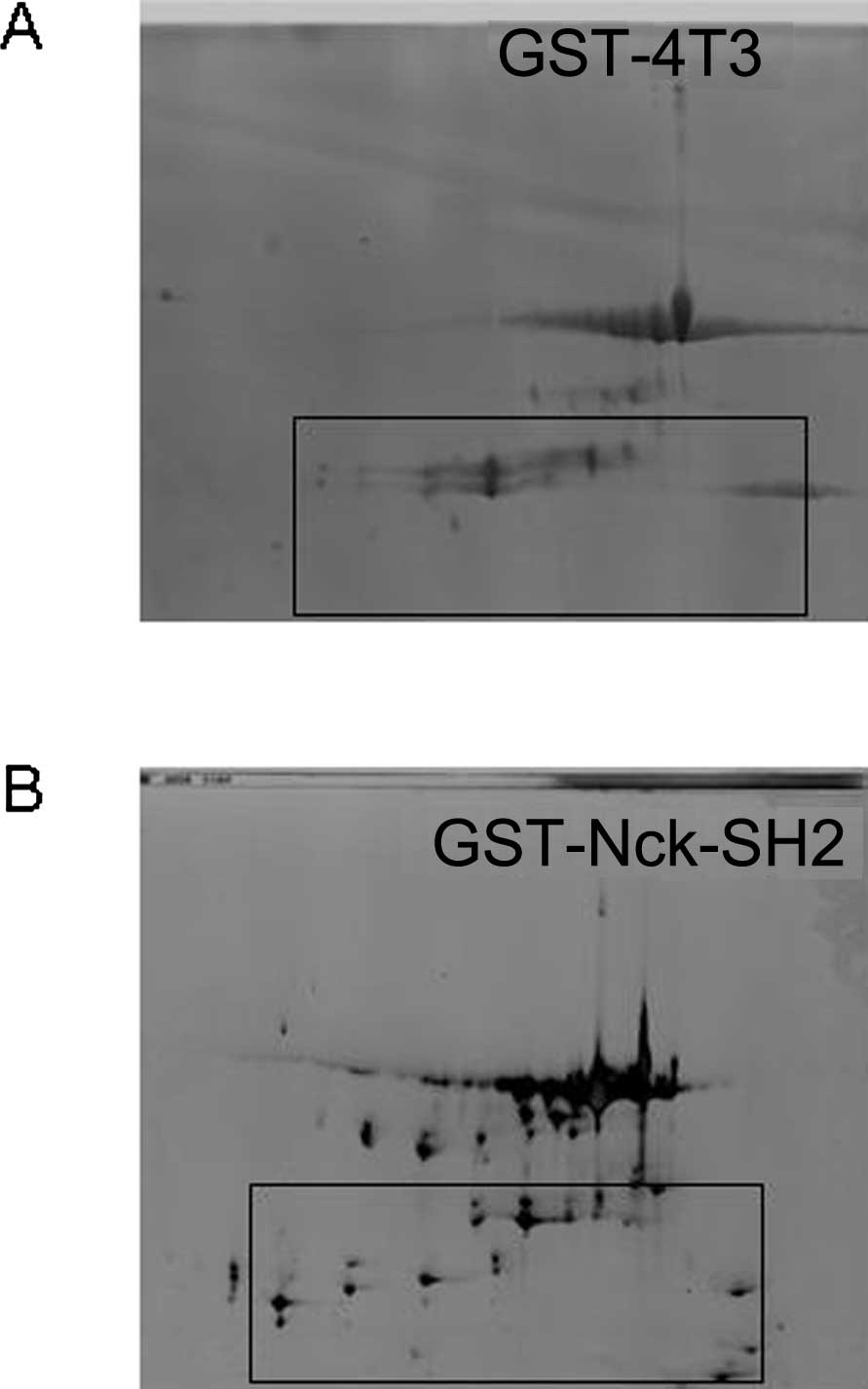
An appropriate control experiment should be
conducted during the GST-Nck1-SH2 pull-down procedure. In the
present study, we performed a negative control experiment by the
use of GST-4T3. The proteins pulled down by GST-Nck1-SH2 fusion or
GST-4T3 underwent removal of the interferential materials by
ultrafiltration, and were then loaded onto 2-DE gels under the same
conditions. As shown in Fig. 5,
the 2-D gel for GST-Nck1-SH2 separated 200±38 protein spots while
that of GST-4T3 possessed GST fragments and few protein spots.
Proteins obtained using our method
interacted with Nck in a tyrosine phosphorylation-dependent
manner
To demonstrate whether the proteins that were pulled
down by our method interacted with Nck in a tyrosine
phosphorylation-dependent manner, we firstly identified two
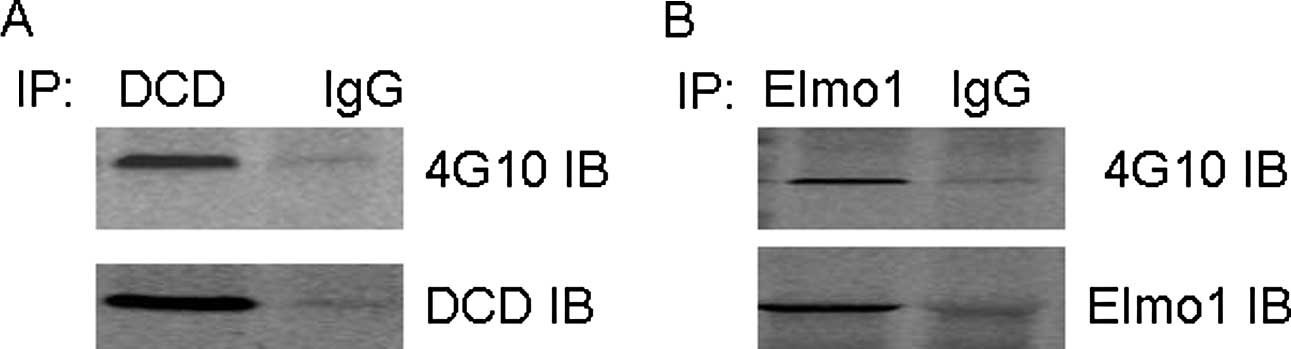
proteins by MALDI-TOF/TOF MS [Fig.
5B; lane 1, dermcidin (DCD); lane 2, engulfment and cell
motility proteins (Elmo1)]. These results were published in our
previous study (10).
Co-immunoprecipitation (co-IP) experiments were used
to examine the correlation between endogenous Elmo1/DCD and Nck in
SK-HEP-1 cells. As demonstrated in Fig. 6A, DCD was detected by anti-DCD
antibody in the anti-Nck immunoprecipitate, but not in the
precipitate obtained by IgG. In Fig.
6B, Elmo1 was detected by anti-Elmo1 antibody in the anti-Nck
immunoprecipitate, but not in the precipitate obtained by IgG.
Subsequent tests used to demonstrate that the proteins interact
with Nck in a tyrosine phosphorylation-dependent manner have been
published in our previous study (10).
Discussion
Given the importance of signaling mediated by
tyrosine-phosphorylated proteins, there is significant interest in
strategies to define or profile the global state of tyrosine
phosphorylation in the cell (11).
A limited number of studies have focused on tyrosine-phosphorylated
proteins in tumor tissues thus far, although these regulate many
important cancer-related activities, including cell proliferation,
survival, invasion/metastasis and angiogenesis (12). Therefore, profiling the global
state of tyrosine-phosphorylated proteins in a tumor is likely to
provide a wealth of information that may be used to classify tumors
for prognosis and prediction (13). To detect the state of
tyrosine-phosphorylated proteins in tumor tissues, we selected
liver tissue from HCC patients for further study. HCC is one of the
most common and aggressive human malignancies (14), and is also regulated by
tyrosine-phosphorylated proteins.
Machida et al demonstrated that SH2 binding
methods may serve as a valuable complement in large-scale proteomic
analyses (6). GST pull-down is an
important tool for the validation of suspected protein-protein
interactions, or for identifying novel protein interactions. 2-DE
is one of the most commonly used methods in proteome analysis and
classified tumors (15). Given
this background, we employed GST-Nck1-SH2 pull-down to detect
tyrosine-phosphorylated proteins in liver tissues, and then
combined this with 2-DE to separate the proteins.
Although 2-DE is a powerful way to separate proteins
for proteomics analysis, it presents a challenge for sample
preparation (16). Based on the
present study, the major barriers include the quantity of protein,
depletion of interferential materials and certainty that the
proteins obtained by GST-Nck1-SH2 pull-down are
tyrosine-phosphorylated proteins.
Protein amounts
Yielding sufficient proteins is the initial step
required for 2-DE, as the protein concentration of the loading
sample for silver-stained 2-DE gels should not be <0.5 μg/μl
(17). To capture sufficient
tyrosine-phosphorylated proteins by GST-Nck1-SH2 pull-down, we
explored four different methods. The first method involved
following the guidelines for GST pull-down in cells. However, our
results demonstrated that this method was not suitable for tissue
samples as it only yielded a limited number of
tyrosine-phosphorylated proteins. Subsequently, we optimized the
quantity of GST-Nck1-SH2 particles or liver proteins in methods B,
C and D, respectively. The quantity of tyrosine-phosphorylated
protein in methods C and D markedly increased. We further confirmed
the effect of the four different strategies by 2-DE. Overall, our
results demonstrated that protein accounts obtained by GST
pull-down were dependent on the ratio of GST-Nck1-SH2 fusion
proteins to liver proteins. Only with an appropriate ratio (method
C) are ideal accounts obtained, employing a greater quantity of
liver proteins (method D) or fusion proteins (method B) did not
lead to a greater amount of tyrosine-phosphorylated proteins being
obtained. By contrast, using a large excess of particles or liver
proteins would result in non-specific interactions between the
proteins and particles.
Protein denaturization is another key step of 2-DE
that is often achieved by the addition of chaotropic agents, such
as urea and thiourea. Variations in the components and
concentration of chaotropic agents markedly affect protein amounts
and patterns. Thiourea is known to be able to increase the
solubility of certain proteins and produce more protein spots,
which is consistent with the present results.
Depletion efficiency and compatibility
for downstream 2-DE
2-DE is often limited by the presence of non-protein
impurities in the samples. Excess salts originate from sample
preparation and may render the solution too conductive for
effective IEF. The samples resulting from GST-Nck1-SH2 pull-down
included iron in the wash buffer and GST particle fragments, all of
which negatively impact IEF. Therefore, depletion of such
interferential material is an important step to insure successful
IEF. To ensure samples pulled down by GST-Nck1-SH2 may be used for
downstream proteomics studies, we selected three common methods,
acetone, a 2-D clean-up kit and ultrafiltration, to deplete the
aforementioned materials, and then loaded the samples onto 2-DE
gels.
To evaluate the depletion method, the depletion
efficiency and the protein yield post-depletion were assessed.
Dialysis is a simple and straightforward technique to de-salting
with a dialysis membrane. The capped tube with the sample is
inverted in a stirred beaker containing the solution against salts
and molecules smaller than the molecular weight cut-off of the
dialysis membrane exchange. However, our results demonstrated that
dialysis could not be used for present study. A possible reason for
this is that the samples resulting from GST-Nck1-SH2 pull-down
included the majority of the GST bead fragments, which would block
the dialysis membrane and result in failure of dialysis.
The 2-D clean-up kit is the classical depletion kit
and it may be used to prepare proteins from sources that are
diluted, and that contain high levels of salt and other interfering
substances. However, our study demonstrated that this method was
not suitable for tyrosine-phosphorylated proteins as a limited
number of protein spots remained following depletion. We suggest
that the reason for this finding is that the 2-D clean-up kit
applies chemicals to the precipitant proteins, and the
tyrosine-phosphorylated proteins require a tender method in order
to retain their activity, thus any type of chemical modification
would result in a loss of activity.
Ultrafiltration is a mild method for desalting and
removal of materials of low molecular weight by centrifugalization
that does not require a phase change. Thus, it is able to maintain
the activity of tyrosine-phosphorylated proteins effectively.
Overall, we suggest that ultrafiltration is a more appropriate
method compared with the other methods.
Confirmation that the proteins obtained
by GST-Nck1-SH2 pull-down are tyrosine-phosphorylated proteins
The disadvantage of GST-4T3 pull-down is the
interferential interaction by any non-specific or non-covalently
bound proteins. To eliminate false positives resulting from
non-specific interactions, we performed a negative control
experiment using GST-4T3 during the pull-down procedure, and
subsequently loaded the samples pulled down by both GST-4T3 and
GST-Nck1-SH2 onto 2-DE gels. Our results demonstrated that the 2-D
gel for GST-Nck1-SH2 harvested more protein spots than that of
GST-4T3.
We further identified DCD and Elmo1 by MALDI-TOF/TOF
MS on the 2-D gel and demonstrated that the proteins pulled down by
our method, which interacted with Nck in a tyrosine
phosphorylation-dependent manner, were either DCD or Elmo1. As the
SH2 domain is a small, modular protein domain that binds
specifically to tyrosine-phosphorylated peptide ligands, we suggest
that the present strategy is effective for identifying novel SH2
domains associated with phosphorylated proteins in tumor
tissues.
In summary, we have optimized the GST-Nck1-SH2
pull-down procedure to obtain tyrosine-phosphorylated proteins in
tumor tissues, and the sample preparation for downstream 2-DE.
Moreover, the successful identification of protein spots by
MALDI-TOF/TOF MS and the proteins pulled down by our method, which
interacted with Nck in a tyrosine phosphorylation-dependent manner,
demonstrated that GST-Nck1-SH2 pull-down combined with 2-DE is an
effective molecular diagnostic approach to identifying novel SH2
domains associated with phosphorylated proteins in tumor tissue,
thus facilitating future research.
Acknowledgements
This study was supported by a grant (No. 81070554)
from the National Science Foundation of China.
References
|
1
|
Barr AJ: Protein tyrosine phosphatases as
drug targets: strategies and challenges of inhibitor development.
Future Med Chem. 2:1563–1576. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Soderblom EJ, Philipp M, Thompson JW,
Caron MG and Moseley MA: Quantitative label-free phosphoproteomics
strategy for multifaceted experimental designs. Anal Chem.
83:3758–3764. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Morishita A, Gong J, Nomura T, Yoshida H,
Izuishi K, Suzuki Y, Kushida Y, Haba R, D’Armiento J and Masaki T:
The use of protein array to identify targetable receptor tyrosine
kinases for treatment of human colon cancer. Int J Oncol.
37:829–835. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Alcolea MP, Kleiner O and Cutillas PR:
Increased confidence in large-scale phosphoproteomics data by
complementary mass spectrometric techniques and matching of
phosphopeptide data sets. J Proteome Res. 8:3808–3815. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kuban-Jankowska A, Gorska M, Debicki A,
Popowska U, Knap N and Wozniak M: Protein tyrosine phosphatases -
endogenous markers of oxidative stress. Postepy Biochem.
56:269–273. 2010.(In Polish).
|
|
6
|
Machida K, Mayer BJ and Nollau P:
Profiling the global tyrosine phosphorylation state. Mol Cell
Proteomics. 2:215–233. 2003.
|
|
7
|
Blagoev B, Kratchmarova I, Ong SE, Nielsen
M, Foster LJ and Mann M: A proteomics strategy to elucidate
functional protein-protein interactions applied to EGF signaling.
Nat Biotechnol. 21:315–318. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bladt F, Aippersbach E, Gelkop S, Strasser
GA, Nash P, Tafuri A, Gertler FB and Pawson T: The murine Nck
SH2/SH3 adaptors are important for the development of
mesoderm-derived embryonic structures and for regulating the
cellular actin network. Mol Cell Biol. 23:4586–4597. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Dierck K, Machida K, Voigt A, Thimm J,
Horstmann M, Fiedler W, Mayer BJ and Nollau P: Quantitative
multiplexed profiling of cellular signaling networks using
phosphotyrosine-specific DNA-tagged SH2 domains. Nat Methods.
3:737–744. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Shen SL, Qiu FH, Dayarathna TK, Wu J,
Kuang M, Li SS, Peng BG and Nie J: Identification of Dermcidin as a
novel binding protein of Nck1 and characterization of its role in
promoting cell migration. Biochim Biophys Acta. 1812:703–710. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Machida K, Thompson CM, Dierck K, et al:
High-throughput phosphotyrosine profiling using SH2 domains. Mol
Cell. 26:899–915. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Blume-Jensen P and Hunter T: Oncogenic
kinase signalling. Nature. 411:355–365. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Machida K, Eschrich S, Li J, Bai Y, Koomen
J, Mayer BJ and Haura EB: Characterizing tyrosine phosphorylation
signaling in lung cancer using SH2 profiling. PLoS One.
5:e134702010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kudo M and Okanoue T; Japan Society of
Hepatology. Management of hepatocellular carcinoma in Japan:
consensus-based clinical practice manual proposed by the Japan
Society of Hepatology. Oncology. 72(Suppl 1): 2–15. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kikuchi T and Carbone DP: Proteomics
analysis in lung cancer: challenges and opportunities. Respirology.
12:22–28. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wu Y, Zhou J, Zhang X, Zheng X, Jiang X,
Shi L, Yin W and Wang J: Optimized sample preparation for
two-dimensional gel electrophoresis of soluble proteins from
chicken bursa of Fabricius. Proteome Sci. 7:382009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liu B, Qiu FH, Voss C, Xu Y, Zhao MZ, Wu
YX, Nie J and Wang ZL: Evaluation of three high abundance protein
depletion kits for umbilical cord serum proteomics. Proteome Sci.
9:242011. View Article : Google Scholar : PubMed/NCBI
|