Introduction
Gastric cancer remains a leading cause of mortality
in certain countries. Although the occurrence rate of gastric
cancer has been on the decrease worldwide, certain Asian countries
(including Korea and Japan), and certain European and South
American countries still exhibit a high incidence rate (1). As is the case with other types of
cancer, early-stage gastric cancer may be treated by surgery.
However, a large number of patients are already in advanced stages
at the time of diagnosis, and thus have a worse prognosis. Although
numerous chemotherapeutic agents have been used to treat gastric
cancer patients, one of the predominant obstacles in treating these
patients is chemoresistance. Cisplatin is the most frequently used
drug for the treatment of patients with advanced gastric cancer.
However, gastric cancer cells may be inherently insensitive to
cisplatin and acquire resistance against cisplatin during treatment
(2). Acquired drug resistance is
suggested to be multifactorial, involving host factors and numerous
genetic and molecular events (3).
Studies have demonstrated that resistance may be related to
decreased drug accumulation, alteration of intracellular drug
distribution, changes in drug-target interaction, cell-cycle
deregulation, increased damaged-DNA repair and reduced apoptotic
response (4–12). Therefore, it is important to
determine the mechanism of resistance to cisplatin.
Annealing control primer-based-reverse
transcriptase-polymerase chain reaction (ACP RT-PCR) technology
regulated by an annealing control primer has been used to identify
differentially expressed genes in several tissues including cancer
tissues (13–15). This method specifically targets
sequence hybridization to the template via a polydeoxyinosine (poly
dI) linker. The ACP-based PCR system may be facilitated to identify
differentially expressed genes (DEGs) from samples exhibiting low
mRNA levels possibly without generating false positives. Therefore,
using this technique, it was possible to search for related genes
that may be responsible for cisplatin resistance.
This study aimed to determine the correlation
between gene expression changes and cisplatin-resistance in gastric
cancer cells. In our previous study, a cisplatin-resistant gastric
cancer cell line (YCC-3/R) was established by chronic exposure of a
human gastric carcinoma cell line (YCC-3) to cisplatin, and it was
observed that p27 expression was upregulated in the YCC-3/R cells
(16). In order to identify other
genes that may be responsible for resistance to cisplatin, in the
present study, DEGs between YCC-3 and YCC-3/R cells were
investigated using ACP-based RT-PCR technology. It was demonstrated
that interferon-induced transmembrane protein 1 (9-27) and
interferon α-inducible protein 27 (IFI-27) genes were highly
expressed in YCC-3/R cells. Subsequent studies suggested that the
Brahma-related gene 1 (BRG1)-associated expression of 9-27
and IFI-27 genes may be involved in resistance to cisplatin
in YCC-3/R cells.
Materials and methods
Cell lines and culture conditions
YCC-3 (donated by Professor Hyun-Chul Chung, Yonsei
Cancer Center, Yonsei University, Seoul) a human gastric cancer
cell line, was cultured with Dulbecco’s modified Eagle’s medium
(DMEM) (Invitrogen Life Technologies, Carlsbad, CA, USA)
supplemented with 10% heat-inactivated fetal bovine serum (Hyclone
Laborotories Inc., Logan, UT, USA), 3.7 mg/ml sodium bicarbonate, 2
mM L-glutamine, 25 mM HEPES and 1% penicillin/streptomycin. The
culture was maintained under a humidified 5% CO2
atmosphere at 37°C. The culture was passaged twice or three times a
week. YCC-3/R was established by chronic exposure to equal
concentrations (0.5 μg/ml) of cisplatin for 6 months as described
previously (16).
ACP RT-PCR analysis
Total RNA extracted from YCC-3 and YCC-3/R cells was
used for the synthesis of first-strand cDNA by reverse
transcriptase. Reverse transcription was performed for 1.5 h at
42°C in a final reaction volume of 20 μl containing 3 μg of the
purified total RNAs, 4 μl 5X reaction buffer (Promega, Madison, WI,
USA), 5 μl dNTPs (each 2 mM), 2 μl 10 μM dT-ACP1
[5′-CGTGAATGCTGCGACTACGATIIIIIT(18)-3′, Seegene Inc., Seoul, Korea], 0.5
μl RNasin® RNase Inhibitor (40 U/μl; Promega) and 1 μl
Moloney murine leukemia virus reverse transcriptase (200 U/μl;
Promega). First-strand cDNA was diluted by the addition of 80 μl
ultra-purified water for the GeneFishing™ PCR (Seegene Inc., Seoul,
Korea), and stored at −20°C until use. Differentially expressed
genes were screened by the ACP-based PCR method using the
GeneFishing DEG kits (Seegene Inc.) (13). Briefly, second-strand cDNA
synthesis was conducted at 50°C during one cycle of first-stage PCR
in a final reaction volume of 20 μl containing 3–5 μl (~50 ng)
diluted first-strand cDNA, 1 μl dT-ACP2 (10 μM), 1 μl 10 μM
arbitrary ACP, and 10 μl 2X Master Mix (Seegene Inc.). The PCR
protocol for second-strand synthesis was one cycle at 94°C for 1
min, followed by 50°C for 3 min, and 72°C for 1 min. When the
second-strand DNA synthesis was completed, the second-stage PCR
amplification protocol was 40 cycles of 94°C for 40 sec, followed
by 65°C for 40 sec, 72°C for 40 sec and a 5 min final extension at
72°C. The amplified PCR products were separated on 2% agarose gel
stained with ethidium bromide. The differentially expressed bands
were re-amplified and extracted from the gel using the
GENCLEAN® II kit (Q-BIO Gene, Carlsbad, CA, USA) and
directly sequenced with ABI PRISM® 3100-Avant
Genetic Analyzer (Applied Biosystems, Foster City, CA, USA). The
identity of each product was confirmed by sequence homology
analysis using the Basic Local Alignment Search Tool (BLAST)
(17).
Western blot analysis
Cell lysates were boiled in Laemmli sample buffer
for 5 min at 95°C and 30 μg protein lysates from each sample were
electrophoresed on a 12% sodium dodecyl sulphate-polyacrylamide gel
and then transferred to a polyvinylidine fluoride membrane
(Bio-Rad, Hercules, CA, USA) and electrophoresed at a constant 40 V
for 90 min. Western blot analysis was conducted as described
previously (18). The membrane was
pre-blocked for 1 h at room temperature (RT) in Tris-buffered
saline Tween-20 (TBST) containing 5% skimmed milk. The blot was
incubated for 3 h at RT with anti-BRG1 antibody (Amersham
Biosciences, Seoul, Korea) and β-actin (Abcam, Cambridge, UK) in
TBST. The blot was then washed three times for 10 min with TBST.
The membrane was incubated for 1 h with peroxidase-linked
species-specific whole antibodies. Protein was detected using an
enhanced chemiluminescence western blot analysis system (Amersham
Biosciences).
Results
To screen DEGs between YCC-3 and YCC-3/R cells,
mRNAs from the two types of cells were extracted and subjected to
ACP RT-PCR analysis using 20 arbitrary ACPs provided by Seegene
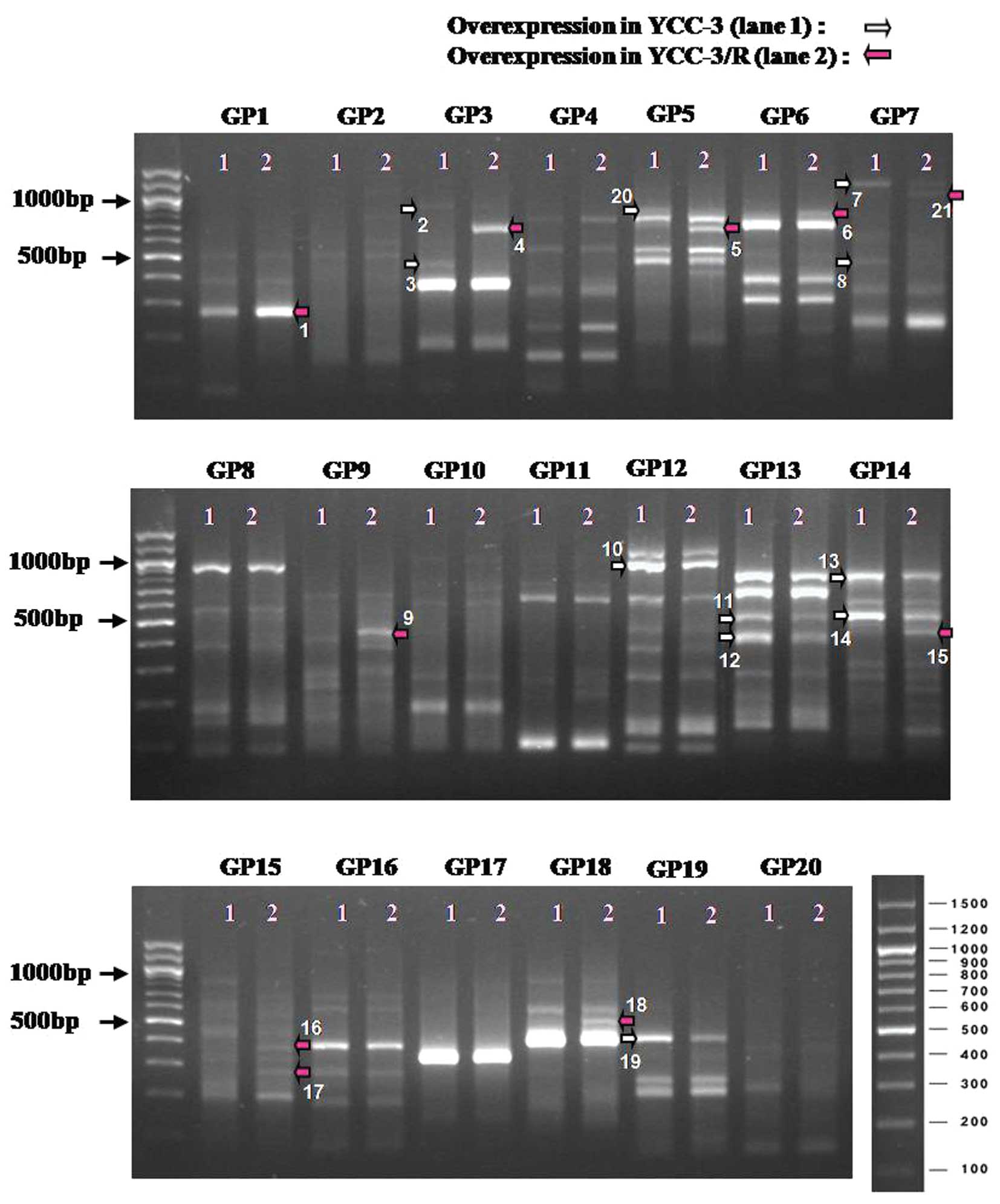
Inc. (13–15). As a result, 21 bands appeared to be
differentially expressed between YCC-3 and YCC-3/R cells, and 11
bands appeared to be upregulated in YCC-3 cells (Fig. 1, white arrows), while 10 bands
appeared to be upregulated in YCC-3/R cells (Fig. 1, red arrows). DNA bands were
excised and sequenced for further analysis. However, only 11 DNA
fragments including DEG1, 4, 5, 7, 9, 14, 15, 18, 19, 20 and 21
were successfully sequenced (Table
I). A sequence homology search using BLAST revealed that the
bands were IFI-27, 9-27, transmembrane emp24 protein
transport domain containing 3, deltex 4 homolog, unknown cDNA
clone, mitochondrial DNA, Mongolian 259 mitochondrial control
region, and ribosomal protein. Seven DEGs were upregulated in
YCC-3/R cells while 4 DEGs were downregulated. Notably, three out
of seven upregulated DEGs in YCC-3/R were interferon α-inducible
gene 9-27 and 1 DEG was another interferon α-inducible gene
IFI-27.
 | Table IList of genes differentially expressed
in YCC-3/R. |
Table I
List of genes differentially expressed
in YCC-3/R.
| DEG no. | GeneBank accession
no. | Sequence homology
search |
|---|
| DEG1a | BC15492 | Homo sapiens
interferon, α-inducible protein 27 (IFI-27) |
| DEG4a | BC000897 | Homo sapiens
interferon induced transmembrane protein 1 (9-27)
(IFITM1) |
| DEG5a | BC000897 | Homo sapiens
interferon induced transmembrane protein 1 (9-27)
(IFITM1) |
| DEG7 | BC000027 | Homo sapiens
transmembrane emp24 protein transport domain containing 3 |
| DEG9a | BC000897 | Homo sapiens
interferon induced transmembrane protein 1 (9-27)
(IFITM1) |
| DEG14 | BC015592 | Homo sapiens
deltex 4 homolog (Drosophilla) |
| DEG15a | AV73350 (EST) | Homo sapiens
cDNA clone |
| AP008920 | Homo sapiens
mitochondrial DNA, complete genome |
| DEG18a | AY244097 | Homo sapiens
isolate Mongolian 259 mitochondrial control region |
| DEG19 | BC070194 | Homo sapiens
ribosomal protein, large, P0 |
| DEG20 | AL110297 | Homo sapiens
mRNA |
| DEG21a | BC012355 | Homo sapiens
solute carrier family 6 (neurotransmitter transporter,
creatine) |
Interferon α-inducible genes 9-27 and
IFI-27 were investigated as a previous study suggested that
BRG1, a key component of the mammalian ATP-dependent
chromatin-remodeling SW12-SNF2 complex, selectively activates a
subset of interferon-α-inducible genes including 9-27 and
IFI-27(19). Therefore,
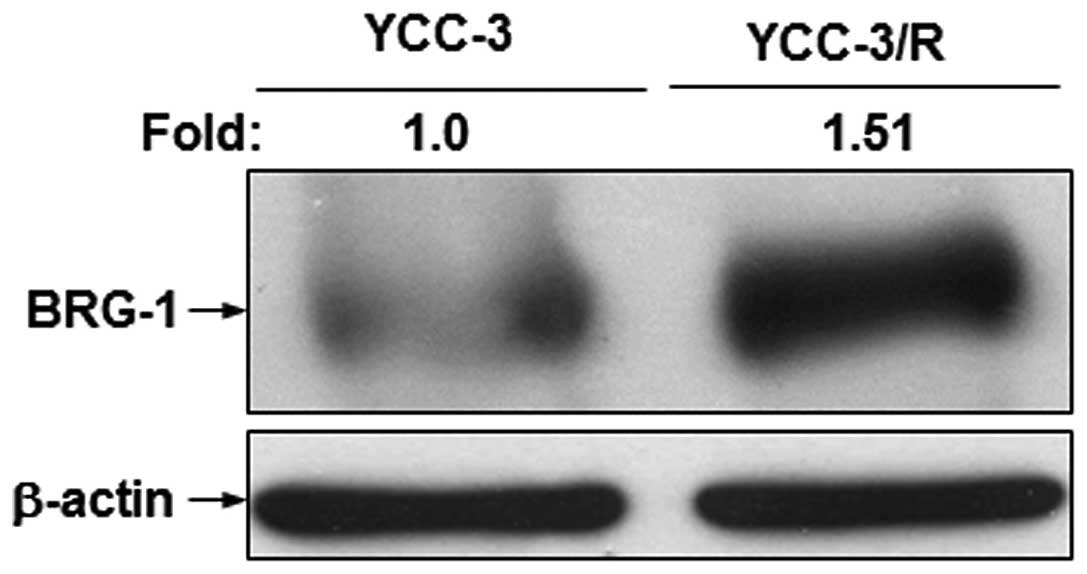
BRG1 expression was investigated by western blot analysis to
identify whether BRG1 expression is associated with the increased
expression of 9-27 and IFI-27 (Fig. 2). As expected, the BRG1 protein was
upregulated in YCC-3/R cells compared with that in the YCC-3 cells,
suggesting that the BRG1-associated expression of 9-27 and
IFI-27 is involved in acquired cisplatin resistance of
gastric cancer cells.
Discussion
In our previous study, a cisplatin-resistant gastric
cancer cell line (YCC-3/R) was established by the chronic exposure
of a human gastric carcinoma cell line (YCC-3) to cisplatin
(16). To screen genes that may be
related to cisplatin resistance in YCC-3/R, ACP RT-PCR technology
was used as conventional display methods are labor intensive and
lead to a high degree of false positivity. The basis of ACP
technology is the unique tripartite structure of a specific
oligonucleotide primer (ACP), which contains distinct 3′- and
5′-end regions separated by a regulator and the interactions of
each portion of this primer during two-stage PCR (13–15).
ACP RT-PCR technology yielded 11 novel DEGs and among these
IFI-27 and 9-27 were particularly noteworthy
(Fig. 1). IFI-27 and
9-27 are known as interferon-inducible proteins that are
involved in the control of cell growth. The 9-27 gene is
suspected to encode a protein that associates with other membrane
proteins, forming a multimeric complex involved in the transduction
of anti-proliferative and homotypic adhesion signals (20). In addition, 9-27 was shown
to be correlated with susceptibility to natural killer cells and
their invasiveness in gastric cancer cells (21). Luker et al(22) also showed that a
paclitaxel-resistant cell line overexpresses IFN-inducible p27
(IFI-27), and confers resistance to paclitaxel in breast
cancer cells. A previous study suggested that 9-27 arrests
cell cycle progression in the G1 phase (23). In a previous study, p27-dependent
cell cycle arrest was demonstrated to be involved in acquired
cisplatin resistance of YCC-3/R cells (16). Therefore, it may be speculated that
the induction of 9-27 and IFI-27 contributes to
acquired cisplatin resistance of YCC-3/R cells.
BRG1 is a predominant component of the mammalian
ATP-dependent chromatin-remodeling SW12/SNF2 complex and interacts
with signal transducer and activator of transcription (STAT2).
Notably, BRG1 overexpression is associated with advanced-stage
human gastric carcinomas (24).
Subsequent studies demonstrated that BRG1 selectively activated a
subset of interferon-α-inducible genes including 9-27 and
IFI-27(19). A recent study
also demonstrated that the downregulation of BRG1 and brahma (Brm)
results in enhanced cell sensitivity to cisplatin (23). According to these studies,
incomplete repair of cisplatin intrastrand adducts, interstrand
crosslinks and interstrand crosslinks-induced DNA double-strand
breaks are observed in BRG1- and Brm-depleted cells. BRG1 and Brm
deficiency also results in impaired chromatin relaxation, altered
check point activation and enhanced apoptosis (25). Thus, BRG1 and Brm may modulate
cisplatin cytotoxicity via chromatin remodeling and facilitate the
DNA repair factors following DNA damage recognition and sensitivity
to UV radiation (25). In
conclusion, an increased expression of BRG1 and BRG1-inducible
genes, 9-27 and IFI-27, may contribute to acquired
resistance to cisplatin in gastric cancer cells.
Acknowledgements
The authors would like to thank Professor Hyun-Chul
Chung (Yonsei Cancer Center, Yonsei University, Seoul) for donating
the YCC-3 cell line. This study was funded by the Samsung Research
Fund, Sungkyunkwan University, 2006, and the Inje University
Research Grant in 2005.
References
|
1
|
Kelley JR and Duggan JM: Gastric cancer
epidemiology and risk factors. J Clin Epidemiol. 56:1–9. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Banerjee D, Mayer-Kuckuk P, Capiaux G,
Budak-Alpdogan T, Gorlick R and Bertino JR: Novel aspects of
resistance to drugs targeted to dihydrofolate reductase and
thymidylate synthase. Biochim Biophys Acta. 1587:164–173. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Gottesman MM: Mechanisms of cancer drug
resistance. Annu Rev Med. 53:615–627. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Larsen AK, Escargueil AE and Skladanowski
A: Resistance mechanisms associated with altered intracellular
distribution of anticancer agents. Pharmacol Ther. 85:217–229.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Fuertes MA, Alonso C and Pérez JM:
Biochemical modulation of Cisplatin mechanisms of action:
enhancement of antitumor activity and circumvention of drug
resistance. Chem Rev. 103:645–662. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Konishi H, Usui T, Sawada H, Uchino H and
Kidani Y: Effects of anticancer platinum compounds on
Escherichia coli strains with normal and defective DNA
repair capacity. Gann. 72:627–630. 1981.PubMed/NCBI
|
|
7
|
Beck DJ, Popoff S, Sancar A and Rupp WD:
Reactions of the UVRABC excision nuclease with DNA damaged by
diamminedichloroplatinum(II). Nucleic Acids Res. 13:7395–7412.
1985. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Fram RJ, Cusick PS and Marinus MG: Studies
on mutagenesis and repair induced by platinum analogs. Mutat Res.
173:13–18. 1986. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kartalou M and Essigmann JM: Mechanisms of
resistance to cisplatin. Mutat Res. 478:23–43. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Brown R, Hirst GL, Gallagher WM, et al:
hMLH1 expression and cellular responses of ovarian tumour cells to
treatment with cytotoxic anticancer agents. Oncogene. 15:45–52.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Fink D, Nebel S, Aebi S, et al: The role
of DNA mismatch repair in platinum drug resistance. Cancer Res.
56:4881–4886. 1996.PubMed/NCBI
|
|
12
|
Vaisman A, Varchenko M, Umar A, et al: The
role of hMLH1, hMSH3, and hMSH6 defects in cisplatin and
oxaliplatin resistance: correlation with replicative bypass of
platinum-DNA adducts. Cancer Res. 58:3579–3585. 1998.PubMed/NCBI
|
|
13
|
Kim YJ, Kwak CI, Gu YY, Hwang IT and Chun
JY: Annealing control primer system for identification of
differentially expressed genes on agarose gels. Biotechniques.
36:424–426. 2004.PubMed/NCBI
|
|
14
|
Hwang KC, Cui XS, Park SP, et al:
Identification of differentially regulated genes in bovine
blastocysts using an annealing control primer system. Mol Reprod
Dev. 69:43–51. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lee KY, Huang SM, Li S and Kim JM:
Identification of differentially expressed genes in papillary
thyroid cancers. Yonsei Med J. 50:60–67. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Le TV, Seo Y, Ryu CJ, Lee HR and Park HJ:
Increased expression of p27 is associated with the cisplatin
resistance in gastric cancer cell line YCC-3. Arch Pharm Res.
33:1127–1132. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Altschul SF, Gish W, Miller W, Myers EW
and Lipman DJ: Basic local alignment search tool. J Mol Biol.
215:403–410. 1990. View Article : Google Scholar
|
|
18
|
Yoon IS, Chung JH, Hahm SH, et al:
Ribosomal protein S3 is phosphorylated by Cdk1/cdc2 during G2/M
phase. BMB Rep. 44:529–534. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Huang M, Qian F, Hu Y, Ang C, Li Z and Wen
Z: Chromatin-remodelling factor BRG1 selectively activates a subset
of interferon-alpha-inducible genes. Nat Cell Biol. 4:774–781.
2002. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Deblandre GA, Marinx OP, Evans SS, et al:
Expression cloning of an interferon-inducible 17-kDa membrane
protein implicated in the control of cell growth. J Biol Chem.
270:23860–23866. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kang HC, Kim IJ, Park JH, et al:
Identification of genes with differential expression in acquired
drug-resistant gastric cancer cells using high-density
oligonucleotide microarrays. Clin Cancer Res. 10:272–284. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Luker KE, Pica CM, Schreiber RD and
Piwnica-Worms D: Overexpression of IRF9 confers resistance to
antimicrotubule agents in breast cancer cells. Cancer Res.
61:6540–6547. 2001.PubMed/NCBI
|
|
23
|
Yang G, Xu Y, Chen X and Hu G: IFITM1
plays an essential role in the antiproliferative action of
interferon-gamma. Oncogene. 26:594–603. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sentani K, Oue N, Kondo H, et al:
Increased expression but not genetic alteration of BRG1, a
component of the SWI/SNF complex, is associated with the advanced
stage of human gastric carcinomas. Pathobiology. 69:315–320. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kothandapani A, Gopalakrishnan K, Kahali
B, Reisman D and Patrick SM: Downregulation of SWI/SNF chromatin
remodeling factor subunits modulates cisplatin cytotoxicity. Exp
Cell Res. 318:1973–1986. 2012. View Article : Google Scholar : PubMed/NCBI
|