Introduction
Natriuretic peptides (NPs) are a family of
polypeptide hormones that regulate blood pressure and fluid balance
by directly affecting the kidney and the systemic vasculature.
Since dendroaspis NP (DNP) has been isolated recently from the
venom of snakes, the NP family now contains four members: Atrial NP
(ANP), brain NP (BNP), C-type NP (CNP) and DNP (1–5).
While ANP is secreted from atrial myocytes in response to an
increased intravascular volume, BNP is synthesized mainly in the
ventricular myocardium and is released into the blood (6). The effects of ANP are largely on the
blood vessels and kidneys, where it leads to natriuresis, diuresis
and a decrease in intravascular volume and blood pressure (7). The actions of BNP are similar to
those of ANP, and these two peptides are hypothesized to be
circulating hormones. By contrast, CNP is likely to be an autocrine
or paracrine mediator, as it is only detected at extremely low
levels in the plasma (8). In fact,
CNP has lower hypotensive and natriuretic properties compared with
ANP and BNP (9). However, contrary
to our understanding of ANP, BNP and CNP, the role of DNP in
mammals remains unclear.
Our recent study demonstrated that the concentration
of DNP was relatively high in rabbit plasma, while the tissue
contents of DNP in various organs, including the liver, spleen,
intestine, atrium, ventricle, septum, kidney, cerebrum and
cerebellum, were quite low (10).
The major sites for DNP synthesis and secretion have not been
identified, although the concentration of DNP in the plasma was
markedly higher compared with that of ANP in the rabbit. Notably,
the high plasma concentrations of DNP were identified to be
correlated with the biological half-life of DNP in the circulation
of the rabbit (10). Since the
stability of DNP in plasma is a significant factor with regard to
its half-life and renal functions, the current experiments were
designed to confirm the stability of DNP versus ANP, BNP and CNP in
rat plasma, and to determine a physiological target site for the
degradation of DNP. In addition, the present study was undertaken
to investigate the metabolism of DNP and to determine whether
degradation of DNP stability may be mediated by specific
proteinases.
Materials and methods
Plasma and tissue preparation
A total of 30 male Sprague Dawley rats (Daehan
Biolink Co. Ltd., Chungbuk, South Korea) were used for the study.
All the experiments conformed to the guidelines of the National
Institutes of Health (Bethesda, MA, USA) and were performed with
the approval of the Institutional Animal Care and Use Committee of
Chonbuk National University (Jeonju, Korea). The rats were
euthanized under ketamine/xylazine (90 and 10 mg/kg, respectively)
anesthesia. For sampling of the plasma, the arterial blood was
collected into pre-chilled tubes containing
ethylenediaminetetraacetic acid (EDTA), 1 mg/ml whole blood,
phenylmethylsulfonyl fluoride (0.4%) and soybean trypsin inhibitor
(Nα-benzoyl-L-arginine ethyl ester, 50 U/ml). The
plasma samples were obtained following centrifugation at 10,000 × g
for 15 min at 4°C. The finely-diced pooled kidney, liver, spleen
and lung tissues from five rats were homogenized at 4°C in 30 mM
phosphate buffer (pH 7.4) and by three 30-sec bursts at 1,000 rpm
(Polytron homogenizer, Fisher Scientific, Waltham, MA, USA). The
homogenate was centrifuged at 1,000 × g for 10 min at 4°C and the
supernatant was collected.
Iodination of ANP, BNP, CNP and DNP
125I-ANP, -BNP, -CNP and -DNP were
prepared as described previously (11). Synthetic ANP, BNP, CNP and DNP (5
μg/5 μl 0.1 M acetic acid; Peninsula Laboratories, Belmont, CA,
USA) were introduced into vials containing 25 μl 0.5 M
phosphate-buffered saline (pH 7.4), followed by addition of 1 mCi
125I-Na (Amersham International, Buckinghamshire, UK).
Chloramine-T (10 μg/10 μl) was added to the reaction vials, which
were mixed gently. After 30 sec, bovine serum albumin (BSA; 60
mg/200 μl) was added. The reaction mixture was immediately applied
to a Sephadex G-25 column (1.0 × 24 cm; GE Healthcare Life Science,
Pittsburgh, PA, USA) and eluted with 0.1 N-acetic acid
containing 0.3% BSA, 0.3% lysozyme, 0.1% glycine and 200 KIU/ml of
aprotinin. Iodinated ANP, BNP, CNP and DNP were divided and stored
at −70°C until further use. The iodinated ANP, BNP, CNP and DNP
were purified by high-performance liquid chromatography (HPLC) on a
reversed-phase Bondapak Column (Waters Associates, Milford, MA,
USA) with a linear gradient (0 to 60% acetonitrile) elution
immediately prior to use (12,13).
Stability of 125I-labeled ANP,
BNP, CNP and DNP
The stability of the NPs was determined. In order to
compare the stability of ANP to DNP, 125I-labeled ANP,
BNP, CNP and DNP were incubated in rat plasma at 37°C for 1, 2 and
4 h. A reversed-phase HPLC C18 Bondapak column (4.5×2,500 mm;
Waters Associates) was used for analyzing the
125I-labeled ANP and DNP. In total, ~100 μl incubated
125I-labeled peptide solution in plasma from each
time-point (0, 1, 2 and 4 h after incubation) was loaded and then
eluted on a linear gradient of 0–60% acetonitrile with 0.1%
trifluoroacetic acid (40 min; flow rate, 1 ml/min). Degradation of
the 125I-labeled ANP, BNP, CNP and DNP in the plasma was
estimated by counting the radioactivity of the HPLC fractions.
Proteinase inhibitors
To investigate the main organ sites for DNP
degradation, 125I-labeled DNP was incubated at 37°C for
4 h in the rat kidney, spleen, liver and lung. The quantity of
protein in the tissue extracts from the spleen, lung and liver was
measured and used in equal quantities (5 μg/μl). In addition, to
further address the stability of DNP and determine which proteinase
degrades DNP, the following proteinase inhibitors were used: The
metalloproteinase, phenanthroline; the serine-cysteine proteinase,
leupeptin; the serine-proteinase, aprotinin; the acid-proteinase,
pepstatin; and the amino-proteinase, bacitracin.
Statistical analysis
The data are presented as the mean ± standard error
of the mean. Statistical comparisons were performed using an
unpaired Student’s t-test and one-way analysis of variance,
followed by the multiple comparison Bonferroni t-test. P<0.05
was considered to indicate a statistically significant
difference.
Results
HPLC profiles of 125I-labeled
ANP, BNP, CNP and DNP during incubation in rat plasma
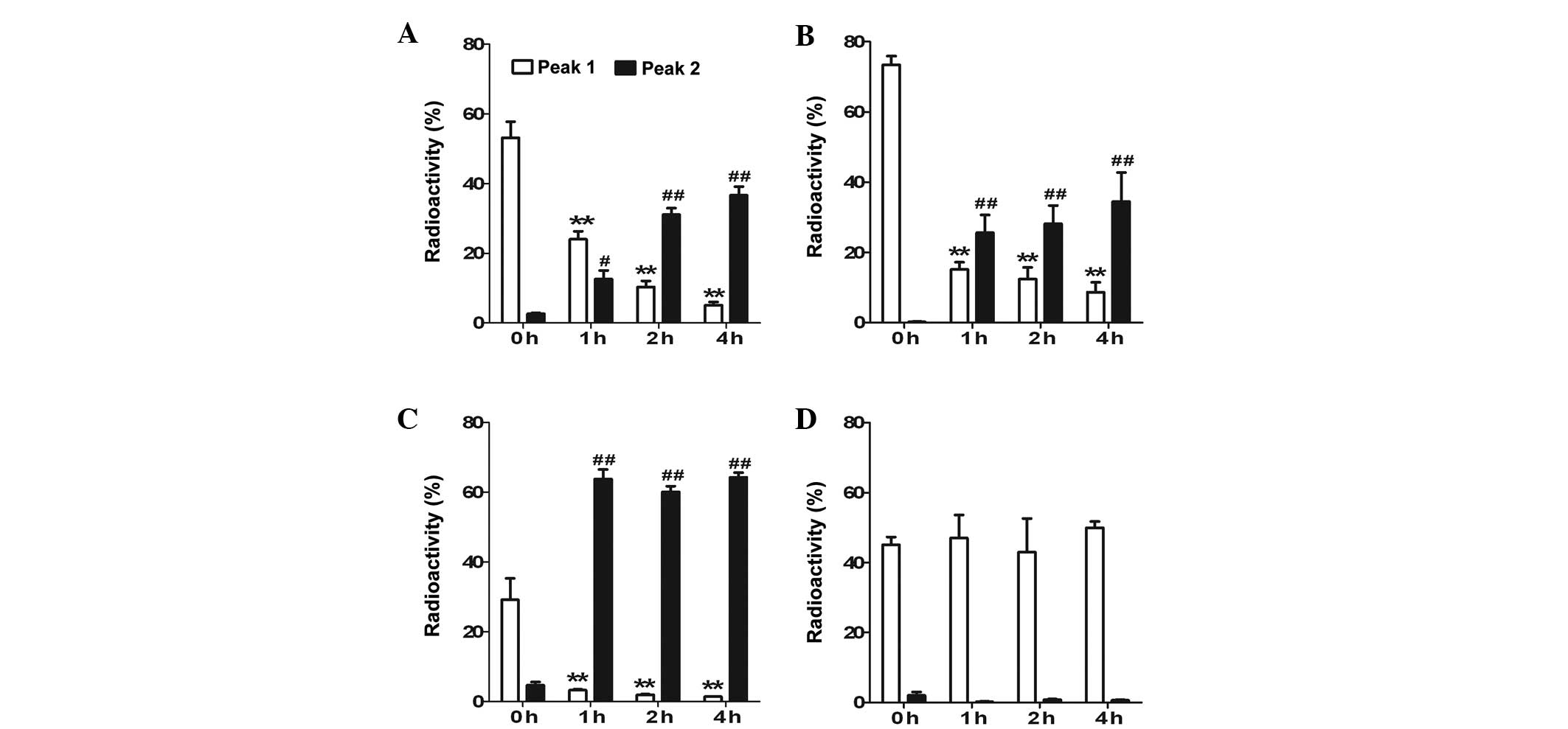
Radioactive values of 125I-labeled ANP,
BNP, CNP and DNP were determined. 125I-labeled ANP, BNP,
CNP and DNP each had one peak with a high level of radioactivity at
0 h of incubation of the rat plasma (Fig. 1, Aa, Ba, Ca and Da). Following a
1-h incubation in rat plasma, two peaks were observed with
125I-labeled ANP, BNP, CNP and DNP (Figs. 1 and 2; peak 1 is a biologically active form
and peak 2 is a degraded form of the peptide).
125I-labeled ANP, BNP and CNP demonstrated a
significantly increased degradation peak at 2 and 4 h following
incubation in the rat plasma (Figs.
1 and 2), while
125I-labeled DNP did not reveal any differences until 4
h of incubation in the rat plasma (Figs. 1 and 2). The relative stability of the peptides
in the rat plasma was DNP>>>ANP≥BNP>>CNP (Table I and Fig. 2; P<0.05). These results indicate
that DNP has the most stable structure amongst the various NPs in
rat plasma.
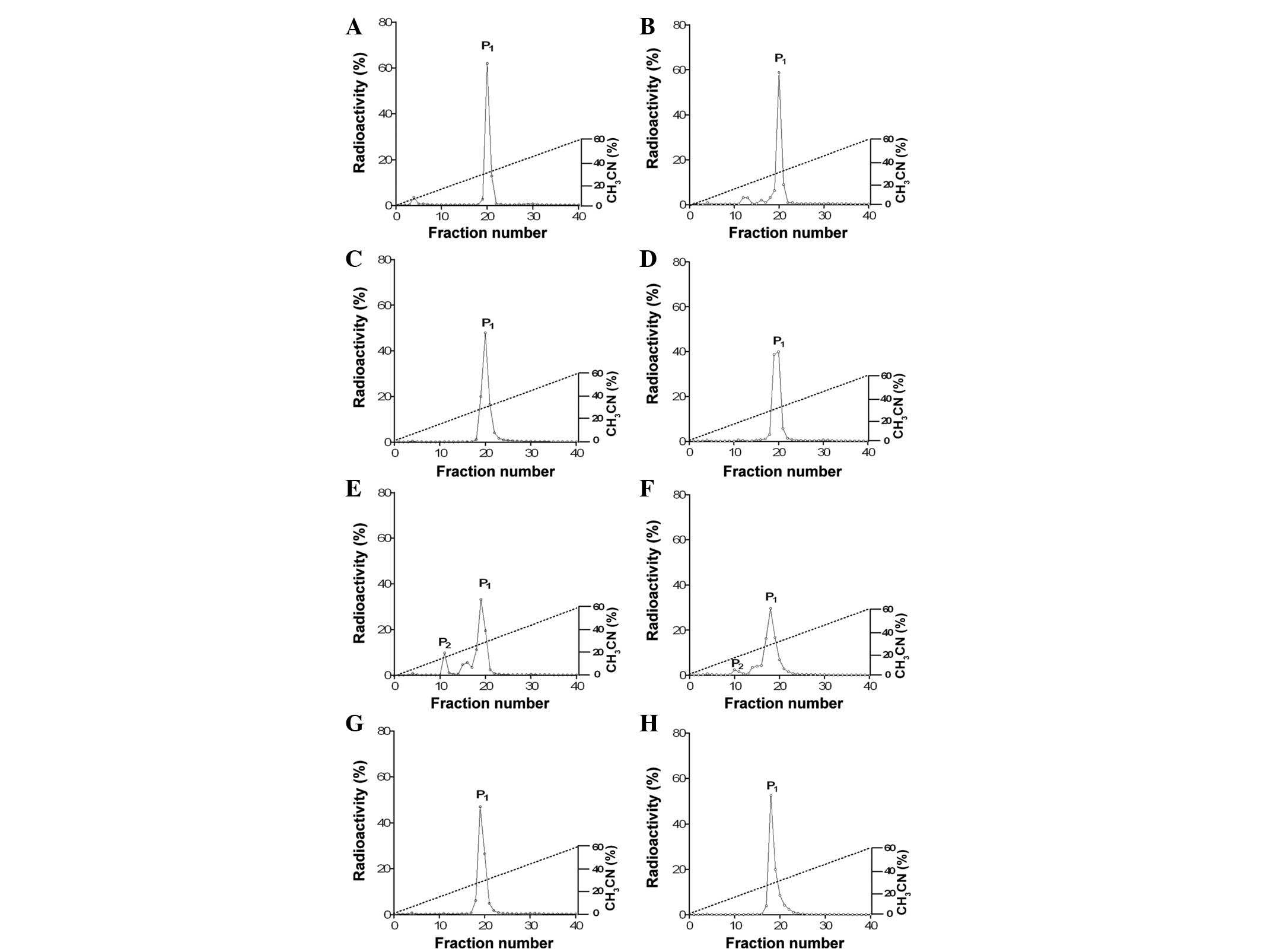
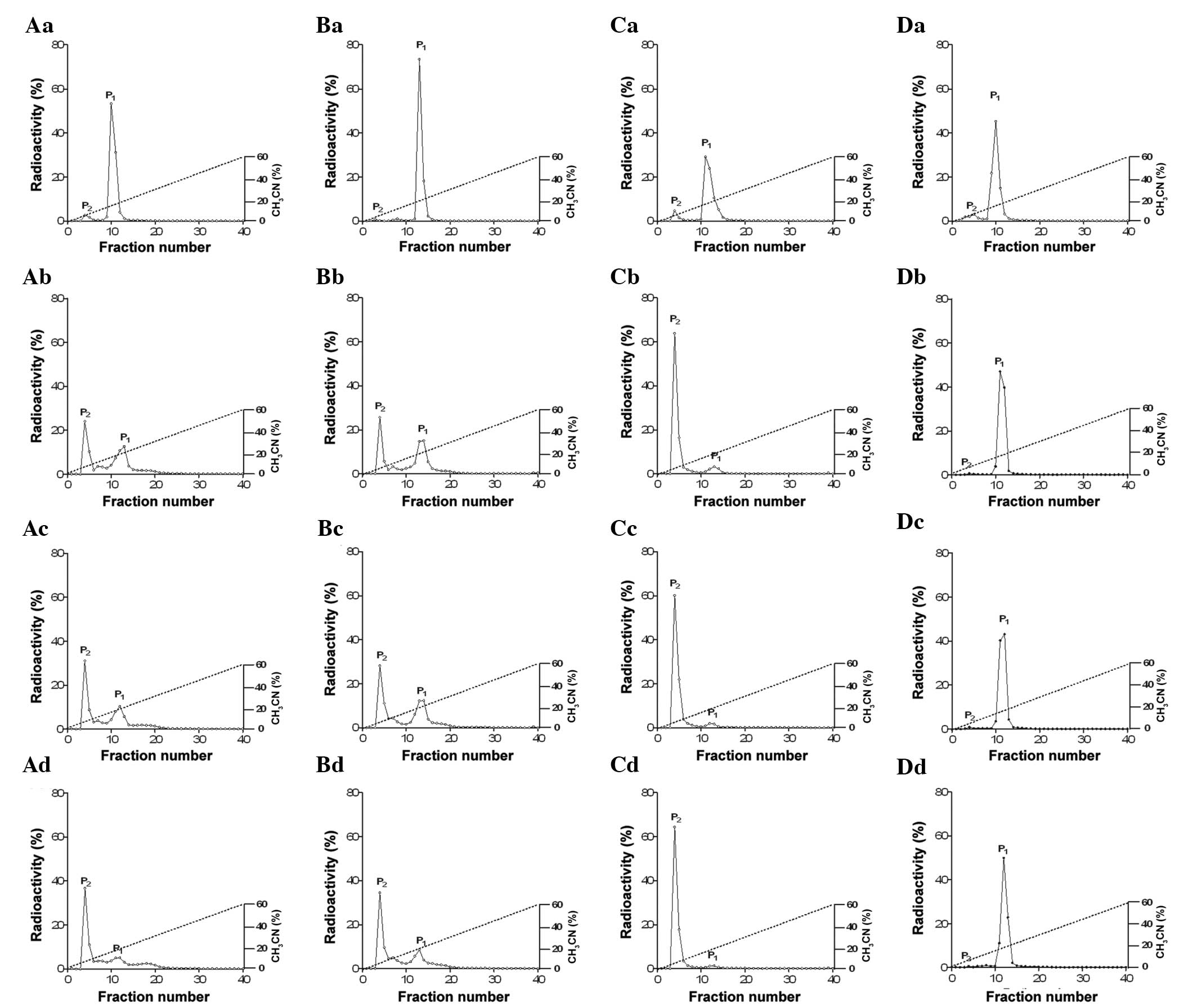 | Figure 1Comparison of the reversed-phase HPLC
profiles of (Aa) 125I-ANP, (Ba) 125I-BNP,
(Ca) 125I-CNP and (Da) 125I-DNP during
incubation in rat plasma at 37°C for 1 h (Ab, Bb, Cb and Db), 2 h
(Ac, Bc, Cc and Dc) and 4 h (Ad, Bd, Cd, and Dd). Peak 1 (P1) is a
biologically active form and peak 2 (P2) is a degraded form of the
peptide. All the experiments were performed more than five times.
In total, ~100 μl 125I-labeled peptide solution
incubated in plasma at each time point (0, 1, 2 and 4 h after
incubation) was loaded and then eluted on a linear gradient of
0–60% acetonitrile with 0.1% trifluoroacetic acid (40 min; flow
rate, 1 ml/min) using a reversed-phase HPLC C18 Bondapak column
(4.5×2,500 mm). The degradation of 125I-labeled ANP,
BNP, CNP and DNP in the plasma was estimated by counting the
radioactivity of the HPLC fractions and expressed with a percentage
ratio (%). ANP, atrial natriuretic peptide; BNP, brain natriuretic
peptide; CNP, C-type natriuretic peptide; DNP, dendroaspis
natriuretic peptide; HPLC, high-performance liquid
chromatography. |
 | Table ISummary of bioactive degradation of
125I-labeled natriuretic peptides in plasma with a
time-dependent manner.a |
Table I
Summary of bioactive degradation of
125I-labeled natriuretic peptides in plasma with a
time-dependent manner.a
| Peak | 0 h | 1 h | 2 h | 4 h |
|---|
| ANP peak 1 | 53.2±4.6 | 24.02±2.31b | 10.36±1.71b | 5.09±0.95b |
| ANP peak 2 | 2.63±0.2 | 12.6±2.48c | 31.09±1.86d | 36.66±2.49d |
| BNP peak 1 | 73.47±2.43 | 15.13±2.10b | 12.37±3.32b | 8.65±2.82b |
| BNP peak 2 | 0.25±0.15 | 25.67±5.03d | 28.15±5.22d | 34.50±8.25d |
| CNP peak 1 | 29.14±6.51 | 3.22±0.27b | 1.92±0.19b | 1.34±0.12b |
| CNP peak 2 | 4.7±0.9 | 63.76±2.79d | 60.12±1.61d | 64.29±1.36d |
| DNP peak 1 | 45.15±2.17 | 47.03±6.60 | 43.03±9.55 | 49.95±1.82 |
| DNP peak 2 | 2.03±0.97 | 0.21±0.16 | 0.86±0.14 | 0.66±0.19 |
Stability of 125I-labeled DNP
in tissue extracts from kidney, liver, lung, heart and spleen
Since DNP was observed to be the most stable NP in
plasma and may not be degraded rapidly by possible endogenous
plasma proteinases, the present study sought to determine whether
there is a physiological target site for the degradation of DNP in
various organs. The lung, liver, spleen, kidney and heart were
selected as target organs, as these organs play major roles in the
metabolism of hormones. Therefore, the stability of DNP was
determined by incubating 125I-labeled DNP in tissue
extracts from the lung, liver, spleen, renal medulla, renal cortex,
atrium and ventricle at 37°C for 1 h (Fig. 3). Following incubation, the
molecular profiles of 125I-labeled DNP were analyzed.
125I-labeled DNP was significantly degraded in the
tissue extracts from the renal cortex and medulla of the rats
(Fig. 3E and F), indicating that
the most favorable organ for the degradation of DNP may be the
kidney.
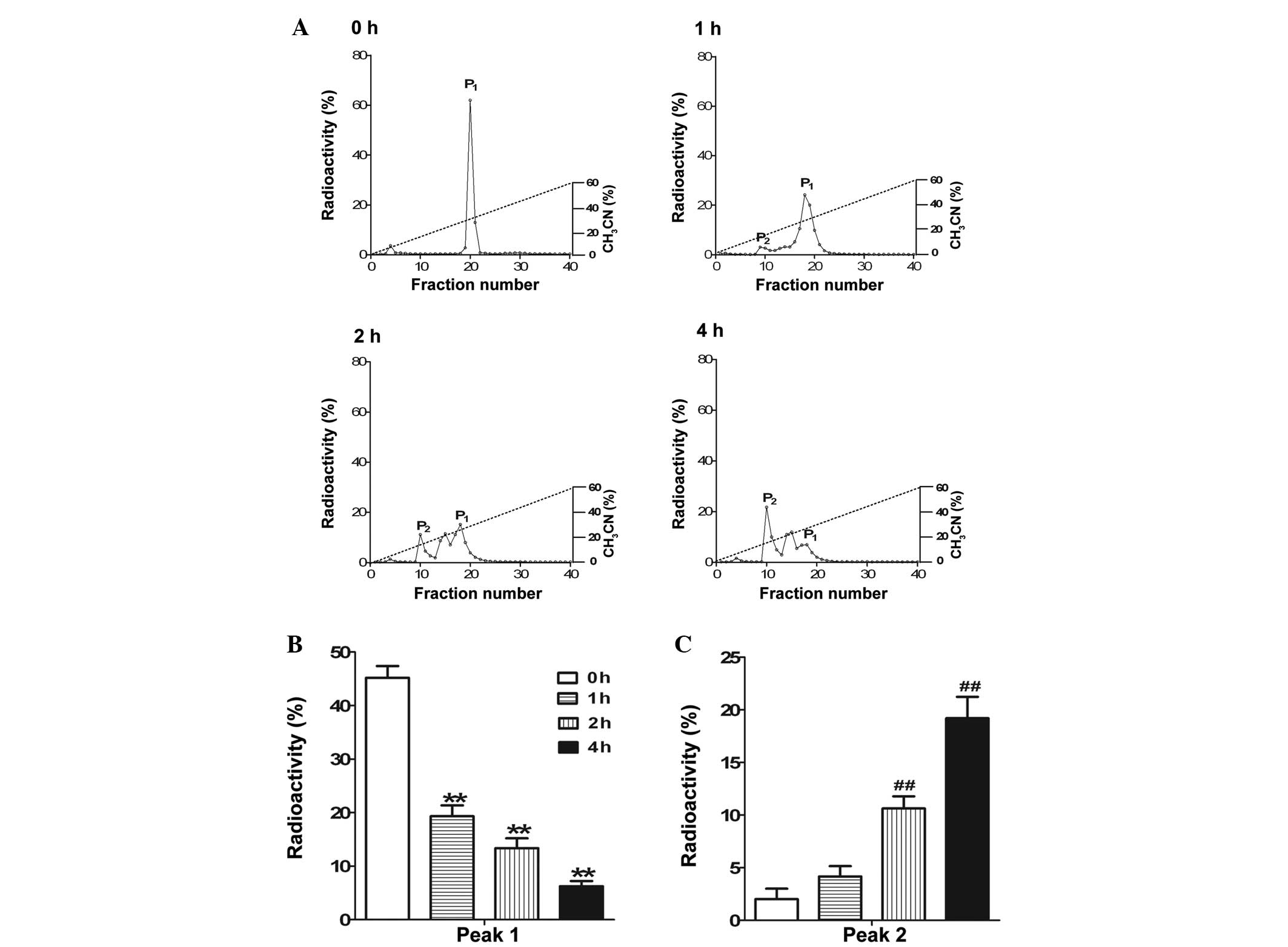
Degradation rate of
125I-labeled DNP during incubation in tissue extracts
from the kidney
125I-labeled DNP was incubated in tissue
extracts from the renal cortex at 37°C for 1, 2 and 4 h (Fig. 4A). 125I-labeled DNP was
significantly degraded in a time-dependent manner, as observed by
the shifting from peak 1 towards peak 2 (Fig. 4B and C).
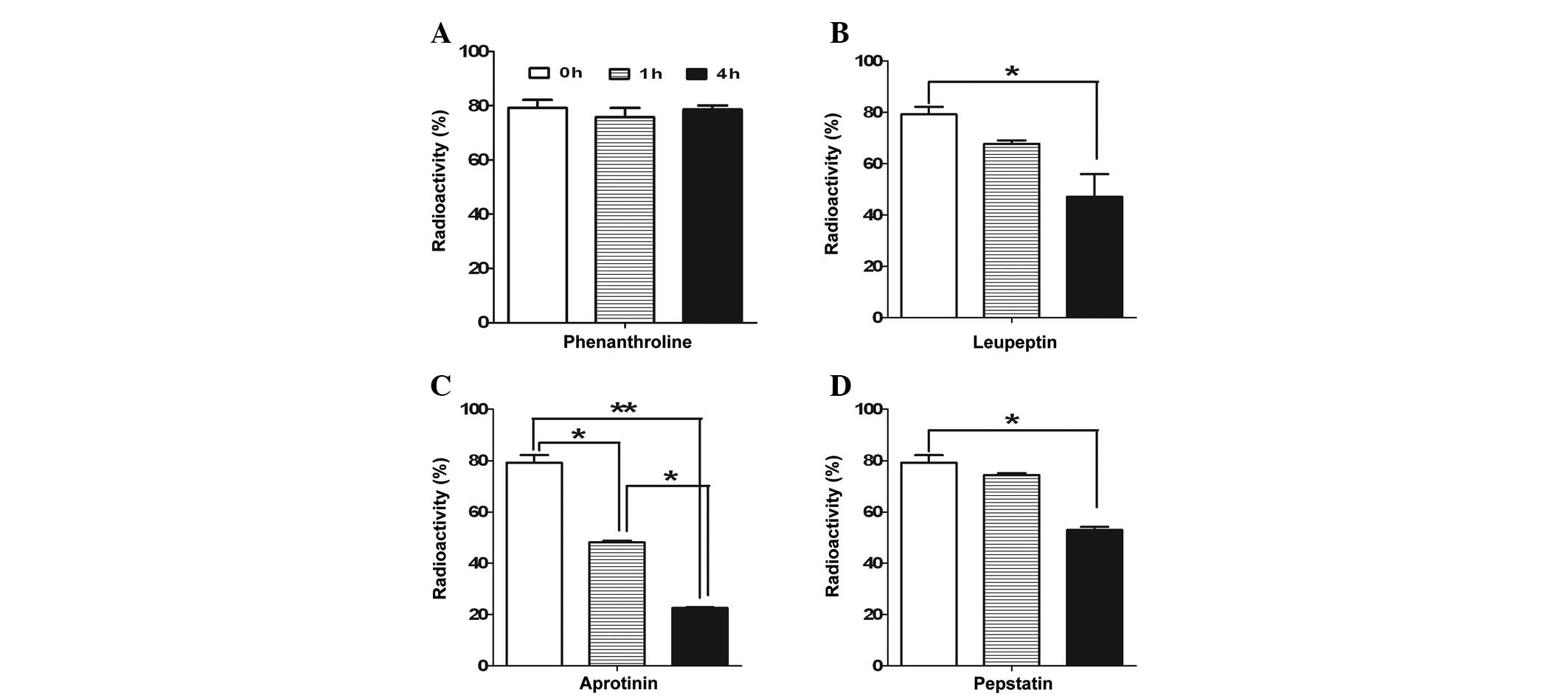
Effects of proteinase inhibitors on
125I-DNP stability
The mechanism by which DNP, mediated by other
proteinases, is degraded in the renal cortex was investigated.
Several types of proteinase inhibitors were used. As shown in
Fig. 5, DNP was resistant to
degradation by phenanthroline, a metalloproteinase inhibitor,
regardless of time. However, leupeptin, a serine-cysteine
proteinase inhibitor, significantly accelerated the degradation of
125I-labeled DNP following incubation for 4 h.
Aprotinin, a serine-proteinase inhibitor, also significantly
increased the degradation of 125I-labeled DNP from 1 h
of incubation in tissue extracts from the kidney cortex. Finally,
pepstatin, an acid-proteinase inhibitor, significantly increased
the degradation of the radioactivity of 125I-labeled
DNP. Therefore, these results indicate that the degradation of DNP
may be mediated by a metalloproteinase.
Discussion
The aim of the present study was to further analyze
the metabolic fate of DNP in rat plasma and to identify a
physiological target site for the degradation of DNP in the organs.
The results revealed that DNP is stable and may have a more
conservative molecular structure compared with any of the other NPs
against the endogenous peptidases in plasma. More significantly,
the organ that metabolizes DNP may be the kidney, and degradation
of DNP may be mediated by a metalloproteinase.
The present study was undertaken to compare NP
stability in plasma for the first time, and the results clearly
reveal evidence of the different stabilities of the NPs in rat
plasma. Time-dependent degradation was identified to occur in the
rank order of DNP>>>ANP≥BNP>>CNP in the incubated
rat plasma. DNP was less degraded than any other peptides, while
CNP was easily destroyed by theprotein enzyme. A noteworthy aspect
of the present study was the observation that
125I-labeled DNP was not degraded until undergoing a 4-h
incubation in plasma, while 125I-labeled CNP, BNP and
ANP were significantly degraded in a time-dependent manner. These
results are in agreement with earlier measurements from our
laboratory (Sung Zoo Kim, Department of Physiology, Chonbuk
National University, Jeonju, South Korea) demonstrating a
noticeably stronger stability of DNP compared with that of ANP in
rabbit plasma (14). EDTA was used
to inhibit coagulation when the blood was collected. Several
studies have revealed a correlation between EDTA and the stability
of NPs (15,16). Tan et al (15) demonstrated that no difference was
observed in ANP levels with EDTA and without EDTA in plasma
sampling conditions. In addition, degradation of
125I-ANP was shown to be antagonized by EDTA (16). In the present study, a stability
study of DNP was performed using no coagulator (serum), heparin or
EDTA when the blood was collected. However, there were no
significant differences in the DNP stabilities regardless of the
use of the anticoagulators (heparin and EDTA; data not shown).
Therefore, it was hypothesized that EDTA is not the main reason for
the varying degradation rates between ANP, BNP, CNP and DNP.
The metabolic fate of ANP in plasma is well known
(17), along with the fact that it
is degraded by a neutral endopeptidase (NEP), which is a
metallopeptidase (18). It has
been reported that the rank order of hydrolysis by NEP in NPs is
CNP>ANP>BNP, indicating that a longer length of the
C-terminus of the peptide results in a greater resistance to
hydrolysis by NEP (19). In the
present study, CNP, which has the shortest amino acid sequence (22
amino acids), was the most rapidly degraded, while the NP with the
longest amino acid sequence, DNP, was stable in the rat plasma.
Therefore, the length of the C-terminus appears to be a significant
factor affecting the stability of NPs. Perhaps the long half-life
of DNP in plasma may be due to the strong resistance of DNP to
degradation by an endopeptidase. Therefore, these results indicate
that DNP may have a different metabolic fate compared with other
NPs and that it may be applicable as a therapeutic agent for
cardiac diseases, including congestive heart failure or
hypertension. Thus, the present findings indicate that DNP is
extremely stable in plasma compared with other NPs. Nevertheless,
the reason for the strong stability of DNP in plasma is
unknown.
Unlike the other NPs, DNP did not easily degrade in
the plasma. In addition, the present study sought to determine the
physiological target sites for the clearance of DNP in organs. In
the present study, 125I-labeled DNP was rapidly degraded
in tissue extracts from the renal cortex and medulla during
incubation at 37°C for 1 h, while the tissue extracts from the
spleen, lung, heart and liver were relatively stable for the same
time. The kidney appears to be the major organ for the degradation
of DNP. It is noteworthy that the degradation of
125I-labeled DNP in the tissue extracts from the renal
cortex occurred in a time-dependent manner. ANP is degraded by the
renal brush border membrane of the proximal tubule, which is
extraordinarily rich in degradative enzymes (20–22).
The renal brush border is not the only site of ANP degradation by
NEP, although the kidney has the greatest capacity for peptide
degradation (23,24). NEP is also disseminated in the
thyroid, lung, brain, parts of the intestine, seminal vesicles,
prostate, vascular tissue and plasma (25,26).
BNP is hydrolyzed by NEP in renal microvilli (27,28).
Therefore, the findings of the present study are somewhat similar
to previous studies in that the kidney may be the likely organ
responsible for the degradation of DNP.
The effects of proteinase inhibitors were then
investigated on 125I-DNP stability in tissue extracts
from rat kidneys. Phenanthroline, a metalloproteinase inhibitor,
reduced the degradation of 125I-labeled DNP during
incubation for 1 h in tissue extracts from the renal cortex.
Furthermore, following incubation for 4 h, the degradation of
125I-labeled DNP remained attenuated by phenanthroline.
The present findings are consistent with previous studies (17,18)
and indicate the significance of NEP in the inactivation of DNP,
similar to ANP, revealing that NEP is involved in the inactivation
of BNP and ANP in the plasma. However, these observations are
fairly different from an earlier study, which revealed that DNP was
resistant to degradation by NEP (29). The discrepancy between the present
study and previous observations revealing different degradation
abilities of NEP for DNP is difficult to elucidate and further
experiments are likely to be required.
In conclusion, DNP was found to have the most stable
structure amongst the NPs, which may be responsible for its long
half-life. Furthermore, DNP was observed to be destroyed mainly in
the kidney by a metalloproteinase.
Acknowledgements
The authors would like to thank Professor Kyung Woo
Cho for the valuable critique and comments. This study was
supported by the Basic Science Research Program through the
National Research Foundation of Korea, funded by the Ministry of
Education, Science and Technology (grant nos. 2011-0014864 and
2012-0009322).
References
|
1
|
de Bold AJ: Atrial natriuretic factor: a
hormone produced by the heart. Science. 230:767–770.
1985.PubMed/NCBI
|
|
2
|
de Bold AJ: Atrial natriuretic factor: an
overview. Fed Proc. 45:2081–2085. 1986.
|
|
3
|
Sudoh T, Kangawa K, Minamino N and Matsuo
H: A new natriuretic peptide in porcine brain. Nature. 332:78–81.
1988. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sudoh T, Minamino N, Kangawa K and Matsuo
H: C-type natriuretic peptide (CNP): a new member of natriuretic
peptide family identified in porcine brain. Biochem Biophys Res
Commun. 168:863–870. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Schweitz H, Vigne P, Moinier D, Frelin C
and Lazdunski M: A new member of the natriuretic peptide family is
present in the venom of the green mamba (Dendroaspis
angusticeps). J Biol Chem. 267:13928–13932. 1992.PubMed/NCBI
|
|
6
|
Nakao K, Mukoyama M, Hosoda K, et al:
Biosynthesis, secretion, and receptor selectivity of human brain
natriuretic peptide. Can J Physiol Pharmacol. 69:1500–1506. 1991.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Stoupakis G and Klapholz M: Natriuretic
peptides: biochemistry, physiology, and therapeutic role in heart
failure. Heart Dis. 5:215–223. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Scotland RS, Ahluwalia A and Hobbs AJ:
C-type natriuretic peptide in vascular physiology and disease.
Pharmacol Ther. 105:85–93. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Januszewicz A: The natriuretic peptides in
hypertension. Curr Opin Cardiol. 10:495–500. 1995. View Article : Google Scholar
|
|
10
|
Kim SM, Kim YA, Kim SY, Kim SH, Cho KW and
Kim SZ: Presence of dendroaspis natriuretic peptide and its binding
to NPR-A receptor in rabbit kidney. Regul Pept. 167:42–49. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Cho KW, Kim SH, Kim CH and Seul KH:
Mechanical basis of ANP secretion in beating atria: atrial stroke
volume and ECF translocation. Am J Physiol. 268:R1129–R1136.
1995.PubMed/NCBI
|
|
12
|
Morris BJ: Specific radioactivity of
radioimmunoassay tracer determined by self-displacement: a
re-evaluation. Clin Chim Acta. 73:213–216. 1976. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Joseph LJ, Desai KB, Mehta MN and
Mathiyarasu R: Measurement of specific activity of radiolabelled
antigens by a simple radioimmunoassay technique. Int J Rad Appl
Instrum B. 15:589–590. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kim SM, Kim SY, Kim SH, Cho KW and Kim SZ:
Renal actions of dendroaspis natriuretic peptide in rabbits.
Peptides. 33:59–66. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tan AC, Rosmalen FM, Theelen BG,
Kloppenborg PW, Benraad HB and Benraad TJ: Atrial natriuretic
peptide - the influence of various physiological and sampling
conditions. Ann Clin Biochem. 24:500–507. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Rugg EL, Aiton JF and Cramb G: Degradation
of [125I]-atrial natriuretic peptide by a soluble metallopeptidase
isolated from rat ventricular myocytes. Biochem Biophys Res Commun.
152:294–300. 1988.
|
|
17
|
Olins GM, Spear KL, Siegel NR, Reinhard EJ
and Zurcher-Neely HA: Atrial peptide inactivation by rabbit-kidney
brush-border membranes. Eur J Biochem. 170:431–434. 1987.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lisy O, Jougasaki M, Schirger JA, Chen HH,
Barclay PT and Burnett JC Jr: Neutral endopeptidase inhibition
potentiates the natriuretic actions of adrenomedullin. Am J
Physiol. 275:F410–F414. 1998.PubMed/NCBI
|
|
19
|
Dussaule JC: Natriuretic factors of
cardiac origin: renal effects. Rev Prat. 43(20 Suppl): 13–17.
1993.(In French).
|
|
20
|
Stephenson SL and Kenny AJ: The hydrolysis
of alpha-human atrial natriuretic peptide by pig kidney microvillar
membranes is initiated by endopeptidase-24.11. Biochem J.
243:183–187. 1987.PubMed/NCBI
|
|
21
|
Stephenson SL and Kenny AJ: The metabolism
of neuropeptides. Hydrolysis of peptides by the
phosphoramidon-insensitive rat kidney enzyme ‘endopeptidase-2’ and
by rat microvillar membranes. Biochem J. 255:45–51. 1988.PubMed/NCBI
|
|
22
|
Johnson GR, Arik L and Foster CJ:
Metabolism of 125I-atrial natriuretic factor by vascular smooth
muscle cells. Evidence for a peptidase that specifically removes
the COOH-terminal tripeptide. J Biol Chem. 264:11637–11642.
1989.
|
|
23
|
Erdös EG and Skidgel RA: Neutral
endopeptidase 24.11 (enkephalinase) and related regulators of
peptide hormones. FASEB J. 3:145–151. 1989.
|
|
24
|
Erdös EG, Wagner B, Harbury CB, Painter
RG, Skidgel RA and Fa XG: Down-regulation and inactivation of
neutral endopeptidase 24.11 (enkephalinase) in human neutrophils. J
Biol Chem. 264:14519–14523. 1989.
|
|
25
|
Yandle TG, Brennan SO, Espiner EA,
Nicholls MG and Richards AM: Endopeptidase-24.11 in human plasma
degrades atrial natriuretic factor (ANF) to ANF(99-105/106-126).
Peptides. 10:891–894. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tamburini PP, Koehn JA, Gilligan JP, et
al: Rat vascular tissue contains a neutral endopeptidase capable of
degrading atrial natriuretic peptide. J Pharmacol Exp Ther.
251:956–961. 1989.PubMed/NCBI
|
|
27
|
Bourne A and Kenny AJ: The hydrolysis of
brain and atrial natriuretic peptides by porcine choroid plexus is
attributable to endopeptidase-24.11. Biochem J. 271:381–385.
1990.PubMed/NCBI
|
|
28
|
Vanneste Y, Pauwels S, Lambotte L and
Deschodt-Lanckman M: In vivo metabolism of brain natriuretic
peptide in the rat involves endopeptidase 24.11 and angiotensin
converting enzyme. Biochem Biophys Res Commun. 173:265–271. 1990.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Chen HH, Lainchbury JG and Burnett JC Jr:
Natriuretic peptide receptors and neutral endopeptidase in
mediating the renal actions of a new therapeutic synthetic
natriuretic peptide dendroaspis natriuretic peptide. J Am Coll
Cardiol. 40:1186–1191. 2002. View Article : Google Scholar
|