Introduction
Colorectal cancer (CRC) is the fourth most common
type of cancer and the third leading cause of cancer-related
mortalities in the western world (1). Thus, in recent years, an increasing
number of studies have focused on its mechanisms.
MicroRNAs (miRNAs) are endogenous small non-coding
RNAs that inhibit gene expression by binding complementary
sequences in the 3′-untranslated regions (3′-UTR) of the target
mRNAs (2,3). Mounting evidence has shown the
important role of miRNAs in regulating various functions, including
cell proliferation, apoptosis, differentiation and survival
(4). Over the past few decades, it
has become clear that miRNA is markedly altered in CRC and that
this aberrant miRNA expression is associated with diagnosis and
prognosis, as well as the therapeutic outcome of CRC (5–8).
miR-224 is located on the human X-chromosome and a
number of studies have demonstrated that miR-224 is upregulated in
hepatocellular (9,10), breast (11) and pancreatic cancers (12). More recently, the elevation of
miR-224 in hepatocellular carcinoma is through epigenetic
mechanisms (10), its
overexpression promotes cell proliferation, anti-apoptosis,
migration and invasion (9,13). miR-224 has been shown to be
involved in transforming growth factor (14) and raf kinase inhibitor protein
(RKIP) (11) pathway-mediated
tumor growth and metastasis.
Although miR-224 is underexpressed when colon cancer
cells are exposed to 5-fluorouracil (15) or in methotrexate-resistant colon
cancer cells (16), in the
majority of CRCs, miR-224 is upregulated. This has been confirmed
by miRNA microarray assay performed by Fu et al (17) and Wang et al (18). In the current study, miR-224 was
observed to be upregulated in 12 CRC tissues compared with
corresponding adjacent normal tissues. Overexpression of miR-224
may facilitate the proliferation of CRC cell lines. miR-224
promotes CRC cell line G1/S transition and this progress
may be mediated by the repression of cyclin-dependent kinase
inhibitors-P21 (CDKN1A), which was confirmed as a new target of
miR-224 in the study.
Materials and methods
Patient samples
A total of 12 matched CRC and their corresponding
normal mucosal tissues (>5 cm laterally from the edge of the
cancerous region), were collected from 12 patients undergoing tumor
resection in the First Affiliated Hospital of Liaoning Medical
University. All the samples were divided into smaller parts,
preserved in liquid nitrogen following retrieval and histologically
confirmed. The study was approved by the ethics committee of
Liaoning Medical University and written informed consent was
obtained from all patients.
Cell culture
The human CRC cell lines HCT-116 and SW-480, were
purchased from the Cell Bank of Shanghai (Shanghai, China) and were
cultured in RPMI-1640 medium supplemented with 10% fetal calf
serum, 100 U/ml penicillin and 100 U/ml streptomycin at 37°C in a
5% CO2 incubator.
RNA extraction and quantitative
polymerase chain reaction (qPCR)
Total RNA was isolated from patient samples or
cultured cells using TRIzol reagent (Invitrogen Life Technologies,
Carlsbad, CA, USA) and the miRNA was purified with the mirVana
miRNA Isolation kit (Ambion, Austin, TX, USA) according to the
manufacturer’s instructions. A stem-loop RT-PCR assay was performed
to detect the mature miRNA levels. The reverse transcription primer
for miR-224 was: 5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCAG
TTGAGAACGGAAC-3′ and U6 snRNA: 5′-AACGCTTCAC GAATTTGCGT-3′. The
cDNA was then amplified by SYBR® Premix Ex-Taq™ II
(Takara Biotechnology Inc., Dalian, China) using the primers:
miR224-forward, 5′-ACAC TCCAGCTGGGCAAGTCACTAGTGGT-3′ and reverse,
5′-TGGTGTCGTGGAGTCG-3′; U6-forward, 5′-CTCGCTT CGGCAGCACA-3′and
reverse, 5′-AACGCTTCACGAATTT GCGT-3′ and P21-forward,
5′-CGATGGAACTTCGACT TTGTCA-3′ and reverse, 5′-GCACAAGGGTACAAGACA
GTG-3′. PCR was performed using the following conditions: 95°C for
1 min, followed by 46 cycles of 95°C for 15 sec and 60°C for 40
sec.
MTT assay
Logarithmically growing HCT-116 and SW-480 cells
were seeded in 96-well plates (5×103 cells/100 μl
medium/well). The culture medium was replaced after 24 h with fresh
medium and the miR-224 mimics, miR-224 inhibitor and negative
control (100 nM) were transfected (GenePharma, Shanghai, China)
with Lipofetamine 2000 (Invitrogen Life Technologies). Cells were
incubated for 4 h in the presence of 20 μl MTT solution (5 g/l;
Sigma-Aldrich, St. Louis, MO, USA) and following 36 h transfection,
the supernatants were carefully discarded and DMSO (100 μl/well)
was added. The spectrophotometric absorbance of each sample was
measured at 490 nm. The experiments were repeated three times and
the average results were calculated.
Cell cycle analysis
HCT-116 and SW-480 cells were transfected with
miR-224 mimics, inhibitor or negative control miRNA. Following 36 h
culture, the cells were collected, washed with phosphate-buffered
saline and fixed with 70% ice-cold ethanol at 4°C overnight. The
fixed cells were washed twice and resuspended in 300 μl stain
buffer (50 mg/ml PI, 1 mg/ml Rnase A, 0.2% Tween-20) for 30 min at
37°C in the dark prior to flow cytometry (Beckman Coulter, Inc.,
Brea, CA, USA).
Target validation with luciferase
reporter
The reporter constructs containing 3′-UTR
P21WAF1/CIP1 were cloned into the pMIR-REPORT™ vector
(Ambion) using PCR-generated fragments. The mutant was constructed
with QuikChange XL Site Directed Mutagenesis kit (Stratagene, La
Jolla, CA, USA). The primers used were: P21-UTR-wt-forward,
5′-TCACGCGGT GAGCACAGCCTAGGGCTG and reverse, GTACTAGTGT
AAAGTCACTAAGAATCATTTATTGAGC; P21-UTR-mt-forward,
TCACGCGTGTGAGCACAGCCTAGGGCTG and P21-UTR-mt-reverse,
GTACTAGTGTAAACAGTGATAGAA TCATTTATTGAGC. HCT-116 and SW-480 cells
were seeded in a 24-well plate (1×105 cells/well) and
were co-transfected 24 h later with 400 ng of the reporter vector
and 10 ng pMIR-REPORT-β gal control plasmid, which was used to
normalize transfection efficiency and 100 nM miR-224 mimics,
inhibitor or negative control miRNA using Lipofectamine. The cells
were harvested for luciferase assays 36 h after transfection.
Reporter gene assays were performed in triplicate using Luciferase
Assays kits (Promega Corporation, Madison, WI, USA). The experiment
was repeated three times.
Western blot analysis
For western blot analysis, protein extracts were
prepared by resuspending cell pellets in 1% NP-40 (Sigma-Aldrich)
lysis buffer containing 50 mM Tris-HCl (pH 8.0), 150 mM NaCl, 1 mM
EDTA, 5 mM NaF, 2 mM PMSF, 1 mM Na-orthovanadate, and 10 μg/ml
leupeptin and aprotinin, respectively. The proteins were separated
by 10% SDS-PAGE and transferred to polyvinylidene difluoride
membranes (Amersham, Piscataway, NJ, USA ). The membranes were
blocked with 5% milk powder in TBS and 0.1% Tween-20 for 1 h and
then incubated overnight with a P21 antibody (Cell Signaling
Technology, Inc., Danvers, MA, USA; 1:1,000) and GAPDH antibody
(Kangchen Bio-tech, Inc., Shanghai, China; 1:10,000).
Statistical analysis
Data are expressed as mean ± SD or median values.
The expression of miR-224 in the CRC samples and adjacent non-tumor
tissues were compared using a paired t-test. Continuous variables
were compared by an independent two-sample t-test for two groups.
Statistical analysis was performed using IBM SPASS Statistics V17.0
(IBM Corporation, Armonk, NY, USA). P<0.05 was considered to
indicate a statistically significant difference.
Results
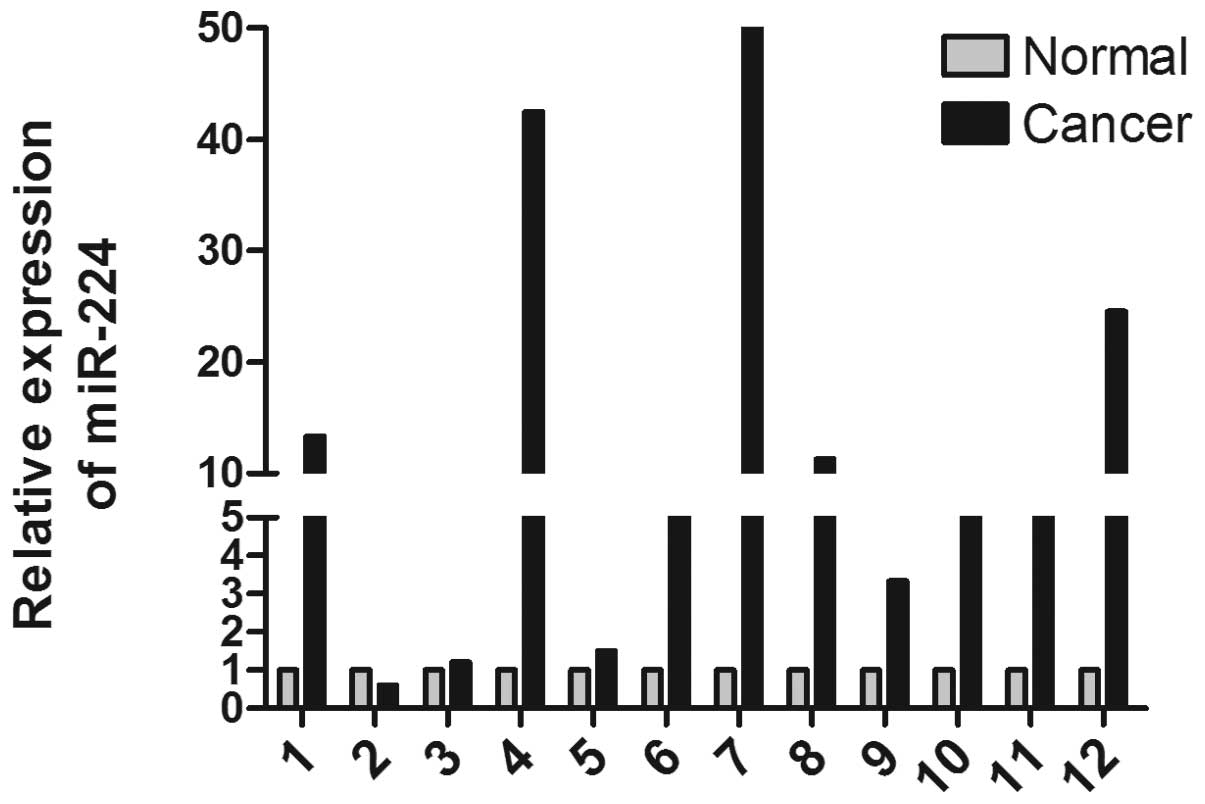
miR-224 is upregulated in CRC
miR-224 is an oncogene that is overexpressed in a
number of tumor tissues (11,12,19–21).
To detect the expression of miR-224 in CRC, 12 pairs of matched
human CRC tissues and the adjacent paracancerous tissue were
analyzed by qPCR (Fig. 1A). The
data indicate that miR-224 is markedly upregulated in CRC tissue
(paired t-test, P=0.0227).
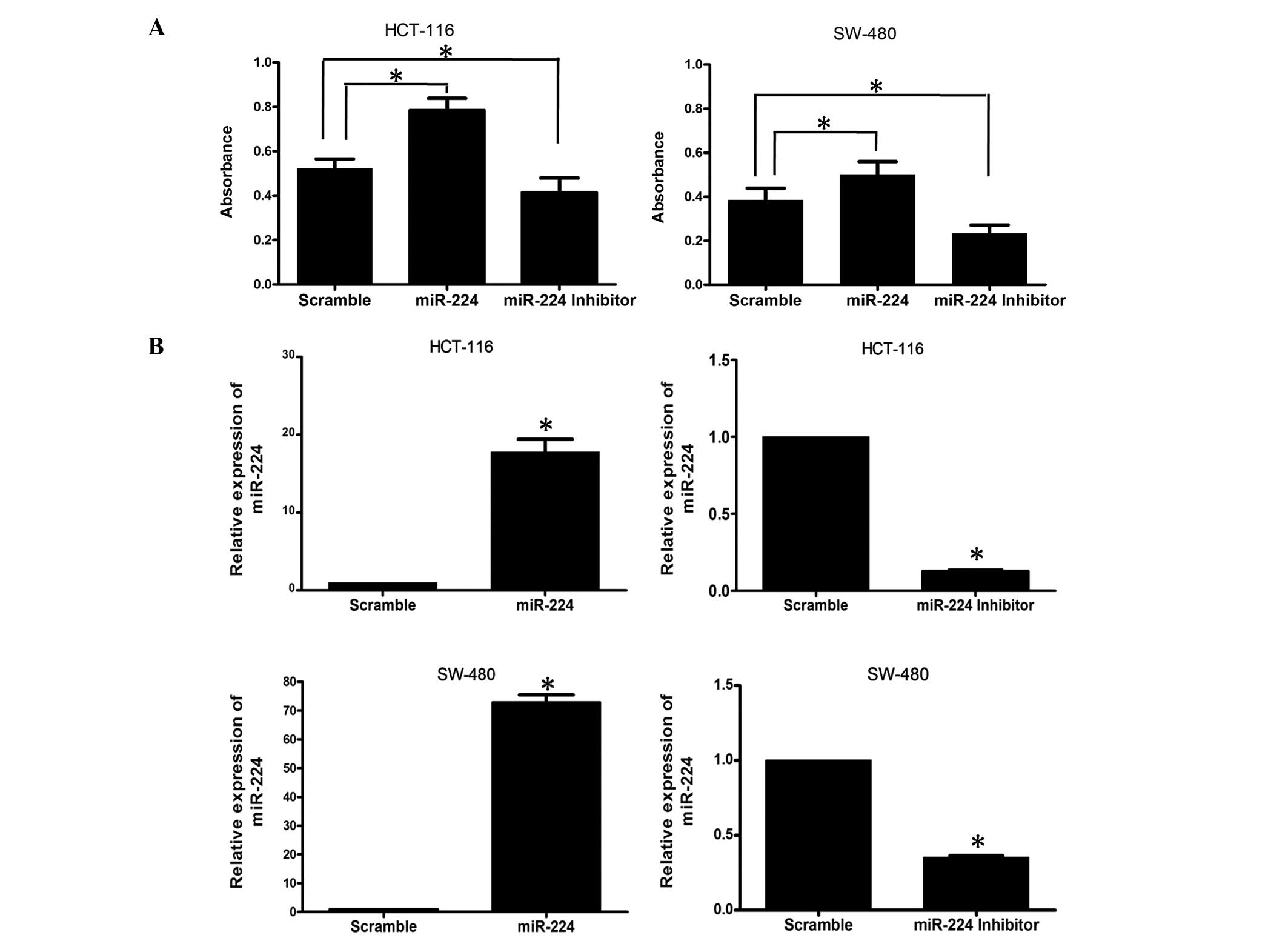
miR-224 overexpression promotes CRC cell
proliferation
To determine the impact of miR-224 on CRC cells,
HCT-116 and SW-480 cell lines were transfected with miR-224 mimics
and inhibitors and the proliferation of CRC cells was detected
using MTT. As shown in Fig. 2A,
the results of the MTT assay revealed that overexpression of
miR-224 significantly increased cell growth in the two CRC cell
lines. When miR-224 was inhibited by transfection with miR-224
inhibitors, this function was reversed. The transfection efficiency
was confirmed by qPCR (Fig.
2B).
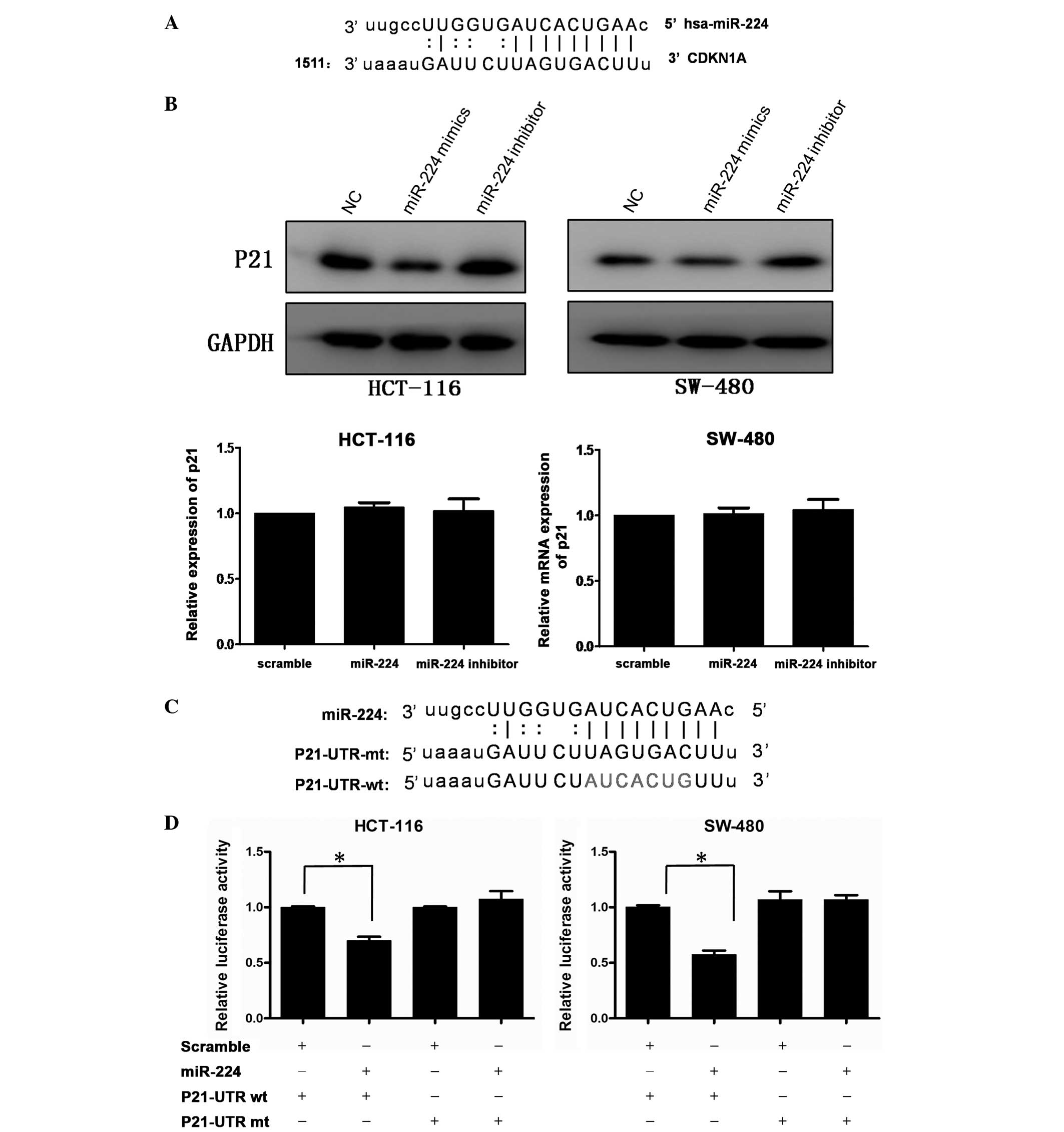
miR-224 directly targets
P21WAF1/CIP1 at the translational level
Considering the effect of miR-224 on CRC cell
growth, possible targets, which are associated with cell
proliferation were selected following bioinformatics analysis using
microRNA.org and targetscan. CDKN1A, also known as
P21WAF1/CIP1, was observed to have a potential miR-224
binding site (Fig. 3A). To
validate the miRNA-target interactions, the expression of P21 was
evaluated in HCT-116 and SW-480 cells transfected with miR-224
mimics, miR-224 inhibitor or negative control. As shown in Fig. 3B, the protein level of P21 was
downregulated following transfection with miR-224 and upregulated
when miR-224 was blocked, while P21 mRNA exhibited no significant
change. To confirm whether P21 is regulated by miR-224 through
direct binding to its 3′-UTR, a human P21 3′-UTR fragment
containing wild-type (wt-UTR) or mutant (mt-UTR) was cloned into
the pMIR-REPORT vector (Fig. 3C).
CRC cell lines were then co-transfected with wt or mt UTR vector.
As shown in Fig. 3D, the relative
luciferase activity of the reporter containing wt-UTR was
significantly suppressed following miR-224 transfection. However,
variations were not observed in the luciferase activity of cells
co-transfected with mt-UTR and miR-224. The results therefore
suggest that P21 is a target gene of miR-224.
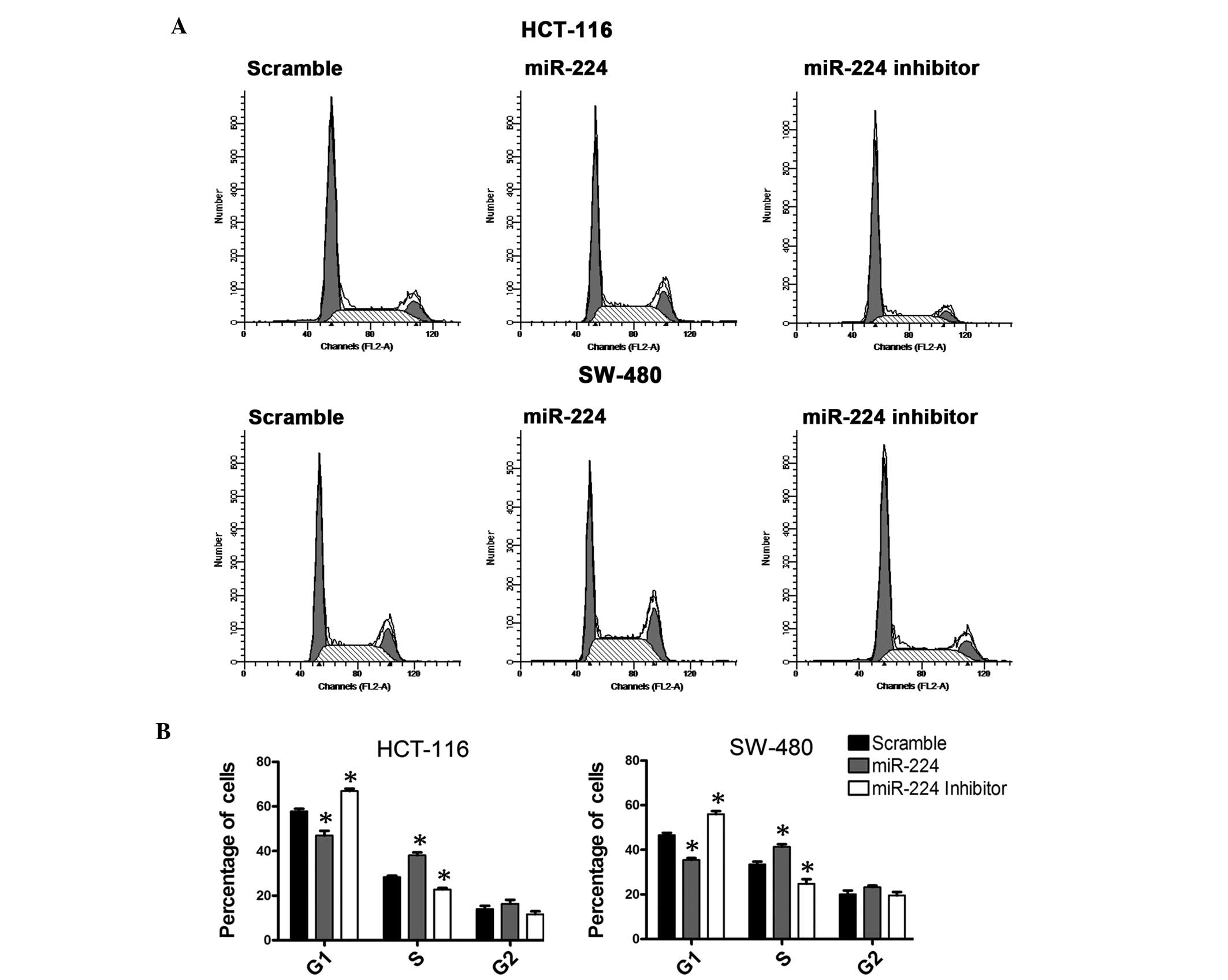
miR-224 regulates the cell cycle of CRC
cells
Due to the regulation of P21, the role of miR-224 in
the regulation of cell cycle in CRC cell lines was assessed. A flow
cytometric analysis was performed following transfection with
miR-224 mimics, inhibitors or negative control. The analysis
results revealed that CRC cells overexpressing miR-224 exhibited an
increase in the S-phase population and a decrease of G1
population, while the group treated with the miR-224 inhibitor
exhibited converse results compared with the negative control
(Fig. 4).
Discussion
Accumulating evidence has demonstrated an important
role of miRNAs in tumorigenesis and tumor progression, diagnosis
and treatment (22). Despite the
identification of a number of miRNA targets involved in human
tumors, the majority of mechanisms remain unclear. The expression
of miR-224 is abnormal in several types of cancer, and is
associated with histone acetylation (10) and inflammation (19). Although miR-224 represses
hepatocellular and breast cancer metastasis by targeting RKIP
(11,23), its mechanism on tumor growth
remains to be clarified.
Growth and proliferation of cancer cells is key to
tumor progression. miRNAs, including miR-21, miR-223 and miR-145
may regulate cancer cell proliferation by repressing the
translation of the targets (24–26).
The cell cycle consists of the G1, S, G2 and M
phases. Cyclins and cyclin-dependent kinases (CDKs) are the two
most important regulatory molecules in the cell cycle (27). Different cyclin-CDK complexes
control cell cycle progression by organized synthesis and
degradation. Cell cycle progression may be prevented by two
families of the inhibitor: CDK interacting protein/kinase
inhibitory protein (cip/kip) and the inhibitor of kinase
4/alternative reading frame. A previous study revealed that
dysregulation of the cell cycle leads to tumor formation (28).
P21 (CDKN1A), together with P27 (CDKN1B) and P57
(CDKN1C), are the members of cip/kip. The decreased expression of
P21 is associated with CRC proliferation and prognosis (29–31)
and is involved in numerous pathways. Furthermore, this function is
associated with the expression of the P53 status (32–34).
As a key molecule in the progression of the cell cycle, P21
negatively regulates cell cycle and transient expression in tumor
cells, resulting in the inhibition of cell proliferation (35,36).
However, despite its considerable role in tumor progression, when
present in low levels, P21 does not function efficiently.
The current data demonstrate that miR-224 regulates
CRC cell growth by targeting P21WAF1/CIP1. This
hypothesis is confirmed by western blotting and luciferase assays.
Overexpression of miR-224 decreased the P21 protein level and the
luciferase activity of P21 3′-UTR. The result was confirmed when
the miR-224-binding site was mutated and the luciferase activity
exhibited no specific change. The results of the study demonstrate
that a decrease of P21 in cancer may be caused by overexpression of
miR-224. This hypothesis was confirmed by the flow cytometric
analysis.
In conclusion, the results reveal the role of
miR-224 in CRC. The expression levels of miR-224 were significantly
upregulated in CRC tissue. It promotes cell proliferation and the
cell cycle phase transition from G1 to S by targeting
P21. Therefore, miR-224 may be a novel therapeutic target of
CRC.
Acknowledgements
This work was supported by the Natural Science
Foundation of Liaoning Province (no. 2013022068) and National
Natural Science Foundation of China (no. 81270696). The author
thanks Dr J Khan for generously providing the pMIR-REPORT
vector.
References
|
1
|
Ohtani H, Tamamori Y, Arimoto Y,
Nishiguchi Y, Maeda K and Hirakawa K: A meta-analysis of the short-
and long-term results of randomized controlled trials that compared
laparoscopy-assisted and conventional open surgery for colorectal
cancer. J Cancer. 2:425–434. 2011. View Article : Google Scholar
|
|
2
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lytle JR, Yario TA and Steitz JA: Target
mRNAs are repressed as efficiently by microRNA-binding sites in the
5′ UTR as in the 3′ UTR. Proc Natl Acad Sci USA. 104:9667–9672.
2007.PubMed/NCBI
|
|
4
|
Friedman RC, Farh KK, Burge CB and Bartel
DP: Most mammalian mRNAs are conserved targets of microRNAs. Genome
Res. 19:92–105. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yamashita S, Yamamoto H, Mimori K, et al:
MicroRNA-372 is associated with poor prognosis in colorectal
cancer. Oncology. 82:205–212. 2012.PubMed/NCBI
|
|
6
|
Rossi S, Di Narzo AF, Mestdagh P, et al:
microRNAs in colon cancer: a roadmap for discovery. FEBS Lett.
586:3000–3007. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Rawson JB and Bapat B: Epigenetic
biomarkers in colorectal cancer diagnostics. Expert Rev Mol Diagn.
12:499–509. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ahmed FE, Amed NC, Vos PW, et al:
Diagnostic microRNA markers to screen for sporadic human colon
cancer in blood. Cancer Genomics Proteomics. 9:179–192.
2012.PubMed/NCBI
|
|
9
|
Wang Y, Lee AT, Ma JZ, et al: Profiling
microRNA expression in hepatocellular carcinoma reveals
microRNA-224 up-regulation and apoptosis inhibitor-5 as a
microRNA-224-specific target. J Biol Chem. 283:13205–13215. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang Y, Toh HC, Chow P, et al:
MicroRNA-224 is up-regulated in hepatocellular carcinoma through
epigenetic mechanisms. FASEB J. 26:3032–3041. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Huang L, Dai T, Lin X, et al: MicroRNA-224
targets RKIP to control cell invasion and expression of metastasis
genes in human breast cancer cells. Biochem Biophys Res Commun.
425:127–133. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mees ST, Mardin WA, Sielker S, et al:
Involvement of CD40 targeting miR-224 and miR-486 on the
progression of pancreatic ductal adenocarcinomas. Ann Surg Oncol.
16:2339–2350. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang Y, Takahashi S, Tasaka A, Yoshima T,
Ochi H and Chayama K: Involvement of microRNA-224 in cell
proliferation, migration, invasion and anti-apoptosis in
hepatocellular carcinoma. J Gastroenterol Hepatol. 28:565–575.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yao G, Yin M, Lian J, et al: MicroRNA-224
is involved in transforming growth factor-beta-mediated mouse
granulosa cell proliferation and granulosa cell function by
targeting Smad4. Mol Endocrinol. 24:540–551. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Rossi L, Bonmassar E and Faraoni I:
Modification of miR gene expression pattern in human colon cancer
cells following exposure to 5-fluorouracil in vitro. Pharmacol Res.
56:248–253. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mencia N, Selga E, Noe V and Ciudad CJ:
Underexpression of miR-224 in methotrexate resistant human colon
cancer cells. Biochem Pharmacol. 82:1572–1582. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Fu J, Tang W, Du P, et al: Identifying
microRNA-mRNA regulatory network in colorectal cancer by a
combination of expression profile and bioinformatics analysis. BMC
Syst Biol. 6:682012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang YX, Zhang XY, Zhang BF, Yang CQ, Chen
XM and Gao HJ: Initial study of microRNA expression profiles of
colonic cancer without lymph node metastasis. J Dig Dis. 11:50–54.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Scisciani C, Vossio S, Guerrieri F, et al:
Transcriptional regulation of miR-224 upregulated in human HCCs by
NFκB inflammatory pathways. J Hepatol. 56:855–861. 2012.PubMed/NCBI
|
|
20
|
Wotschofsky Z, Busch J, Jung M, et al:
Diagnostic and prognostic potential of differentially expressed
miRNAs between metastatic and non-metastatic renal cell carcinoma
at the time of nephrectomy. Clin Chim Acta. 416:5–10. 2013.
View Article : Google Scholar
|
|
21
|
Liu ZR, Chen XW, Qiao XH, Wang R and Xiang
Z: Detection of miR-122a and miR-224 expression in hepatocellular
carcinoma by real-time fluorescence quantitative RT-PCR. Nan Fang
Yi Ke Da Xue Xue Bao. 29:751–753. 2009.(In Chinese).
|
|
22
|
Lu J, Getz G, Miska EA, et al: MicroRNA
expression profiles classify human cancers. Nature. 435:834–838.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Poma P, Labbozzetta M, Vivona N, Porcasi
R, D’Alessandro N and Notarbartolo M: Analysis of possible
mechanisms accounting for raf-1 kinase inhibitor protein
downregulation in hepatocellular carcinoma. OMICS. 16:579–588.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wu L, Li H, Jia CY, et al: MicroRNA-223
regulates FOXO1 expression and cell proliferation. FEBS Lett.
586:1038–1043. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang J, Xiao Z, Lai D, et al: miR-21,
miR-17 and miR-19a induced by phosphatase of regenerating liver-3
promote the proliferation and metastasis of colon cancer. Br J
Cancer. 107:352–359. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Xu Q, Liu LZ, Qian X, et al: MiR-145
directly targets p70S6K1 in cancer cells to inhibit tumor growth
and angiogenesis. Nucleic Acids Res. 40:761–774. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Nigg EA: Cyclin-dependent protein kinases:
key regulators of the eukaryotic cell cycle. Bioessays. 17:471–480.
1995. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zucca E and Nigg EA: Cell cycle regulation
and the function of cancer genes. Ann Oncol. 6:975–978.
1995.PubMed/NCBI
|
|
29
|
Wang L, Cao XX, Chen Q, Zhu TF, Zhu HG and
Zheng L: DIXDC1 targets p21 and cyclin D1 via PI3K pathway
activation to promote colon cancer cell proliferation. Cancer Sci.
100:1801–1808. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Mitomi H, Ohkura Y, Fukui N, et al:
P21WAF1/CIP1 expression in colorectal carcinomas is related to Kras
mutations and prognosis. Eur J Gastroenterol Hepatol. 19:883–889.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kim JA, Park KS, Kim HI, et al:
Troglitazone activates p21Cip/WAF1 through the ERK pathway in HCT15
human colorectal cancer cells. Cancer Lett. 179:185–195. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Pasz-Walczak G and Kordek R: Comparative
evaluation of the expression of cell cycle regulating proteins:
cyclin D1, P53 and P21 (WAF1) in colorectal cancer. Pol J Pathol.
51:63–69. 2000.PubMed/NCBI
|
|
33
|
Pasz-Walczak G, Kordek R and Faflik M: P21
(WAF1) expression in colorectal cancer: correlation with P53 and
cyclin D1 expression, clinicopathological parameters and prognosis.
Pathol Res Pract. 197:683–689. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Noske A, Lipka S, Budczies J, et al:
Combination of p53 expression and p21 loss has an independent
prognostic impact on sporadic colorectal cancer. Oncol Rep. 22:3–9.
2009.PubMed/NCBI
|
|
35
|
Park KS, Ahn Y, Kim JA, Yun MS, Seong BL
and Choi KY: Extracellular zinc stimulates ERK-dependent activation
of p21Cip/WAF1 and inhibits proliferation of colorectal
cancer cells. Br J Pharmacol. 137:597–607. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Al-Maghrabi J, Al-Ahwal M, Buhmeida A, et
al: Expression of cell cycle regulators p21 and p27 as predictors
of disease outcome in colorectal carcinoma. J Gastrointest Cancer.
43:279–287. 2011. View Article : Google Scholar : PubMed/NCBI
|