Introduction
Atopic dermatitis (AD), also known as atopic eczema
(AE), is a chronic relapsing inflammatory skin disease. It has been
classified into 3 sequential phases: infantile, childhood and
adult, each with characteristic physical findings (1). Approximately 70% of patients have a
family history of allergies (2).
The disease has an important impact not only on patients’ physical
and mental health, but also on the economy of society and affected
families (3). In recent years, the
incidence rate of AD has increased to almost 20% (4).
Various internal factors (i.e., gene mutations) and
external factors (i.e., infection and allergens) influence immune
cells and trigger abnormal immune response, which can eventually
lead to AD (5). Defects in the
innate immune system are also believed to contribute to
pathogenesis of AD (6,7). As the pathophysiologic complexity of
this disease is being elucidated, it becomes clear that the
development of AD is triggered by complex interactions between
genetic and environmental factors, along with interactions
occurring within a dense network of cytokines and chemokines;
however, a unifying concept for the pathogenesis of AD has not been
established (8). Basic therapeutic
treatment of AD includes optimal skin care, addressing skin barrier
defects, avoidance of specific and non-specific factors that
trigger AD, as well as local treatment, including wet-wrap or
antimicrobial therapy and administration of glucocorticosteroids
and other compounds (9). However,
these therapies have a relatively slow onset of action and cannot
systematically eradicate the symptoms interfering with daily life
of the patients. Therefore, understanding the immunological basis
of AD and developing effective immune modulators is expected to be
the most effective strategy for treatment of this disease (10).
Omics technologies including proteomics (11) and microarrays (12) are powerful tools to identify the
underlying molecular mechanisms of disease and potential
biomarkers. Park et al identified 31 atopic dermatitis-related
candidate proteins from a proteomic analysis (13). Microarray technology is also used
to analyze the differences between disease and healthy samples
(14). For instance, the study by
Nomura et al (15), used
DNA microarray analysis to compare the complex gene expression
pattern of skin lesions from AD and psoriasis.
In the present study, gene expression profiles of AD
skin were compared to those of healthy skin to identify
differentially expressed genes (DEGs). Gene network construction
combined with function and pathway enrichment analysis were
performed to investigate the functions and potential roles of the
identified DEGs in the pathogenesis of AD.
Materials and methods
Microarray data
The microarray dataset GSE6012 was downloaded from
the Gene Expression Omnibus database (GEO; http://www.ncbi.nlm.nih.gov/geo/) which contains data
from 10 human AD samples (aged 25–50 years) and 10 healthy skin
samples (aged 21–50 years). These data were generated from a
previous study (16), and had been
used for meta-analyses by other research groups (17). Briefly, the data were derived from
the profiling of patients diagnosed with AD based on the criteria
of Williams et al (18), which did
not undergo local or systemic treatment with glucocorticoids or
tacrolimus for the last 2 weeks prior to the study. The control
groups of the study comprised healthy, symptom-free individuals,
gender- and age-matched to the patient group, with normal levels of
IgE, and no history of AE or any other atopic or chronic disease.
Individuals in both the patient and the control group were of
Caucasian origin. Two punch biopsies (4 mm diameter) were collected
under local anaesthesia to obtain lesional skin samples from each
patient and skin samples from the matching location of each control
individual. Hybridizations were performed on the GPL96 Affymetrix
Human Genome U133A (HG-U133A) array. Annotation files for human
genes were also simultaneously gathered when the original
expression data were collected from the GEO database.
Data treatment and identification of
DEGs
The original CEL-format file was converted into an
expression profile format with the open-source package Affy
(19,20), available from the R project website
(http://www.r-project.org/). The data
were normalized with the median method. The expression profiles of
AD and healthy samples were then analyzed with the linear models
for microarray data (LIMMA) package available in Bioconductor and
implemented in R language (21).
Multiple testing correction was performed on the P-values using the
Benjamini-Hochberg (BH) method (22). False discovery rate (FDR) and fold
change (FC) were used as the criteria to identify DEGs, with
cut-offs being FDR <0.05 and |log fold change (FC)| >1.
Construction of interaction networks for
DEGs
In order to detect potential interactions among the
DEGs, the previously described software Osprey (23) was chosen to retrieve known
interaction relationships and construct interaction networks.
Osprey is a suitable tool for this analysis, since it integrates
information from the Biomolecular Interaction Network Database
(BIND) (24) and the Global
Resource Information Database (GRID) (25), while also including data on 50,000
interactions between proteins and nucleic acids. Osprey is used to
build color-coded graphical representations of the interaction
data. Networks built in Osprey can be saved as tab-delimited text
files for future manipulation, or exported as Joint Photographic
Experts Group (JPEG), portable network graphics (PNG), or scalable
vector graphic (SVG) files (23).
Network screening for high degree-node
DEGs
The degree of nodes represents the number of edges
(here, other genes) linked to a node (a given gene) (26). The number of gene interactions
correlates with the importance of the gene in the entire network
and this hypothesis has been experimentally validated in the yeast
Saccharomyces cerevisiae (27). In the present study, low-degree
nodes were gradually removed, which led to a final network
comprising DEGs with a degree of node >10.
Functional enrichment analysis
The Database for Annotation, Visualization, and
Integrated Discovery (DAVID) (28)
is widely used in functional enrichment analysis of DEGs. In this
study, DAVID was used to perform the functional enrichment analysis
for the genes comprised in the network of all DEGs and those
comprised in the network of DEGs with a high degree of node
(>10). The genes were mapped to Gene Ontology (GO) terms for
this purpose. The FDR cut-off associated with this analysis was set
at <0.05 in order to identify significantly enriched functional
terms within the networks.
Pathway enrichment analysis
The Web-based Gene Set Analysis Toolkit (WebGestalt)
(29,30) has become one of the most popular
software tools in pathway analysis. In this study, WebGestalt was
used to perform pathway enrichment analysis of the genes comprised
in the network of all DEGs and those comprised in the networks of
high degree-node DEGs. The genes were mapped to Kyoto Encyclopedia
of Genes and Genomes (KEGG) database-derived pathways for this
purpose. P<0.05 was set as the threshold for the identification
of significantly enriched pathways.
Results
Differentially expressed genes
Raw data were normalized prior to differential
expression analysis (Fig. 1A). The
comparison between the control and disease group using overall data
for all the samples is shown in Fig.
1B. The expression level of the disease group was significantly
lower (P=3.05E-5) compared to the control group. The genes meeting
the criteria for identification of DEGs (FDR <0.05, |logFC|
>1) were selected. A total of 337 differentially expressed genes
were identified, of which 185 were downregulated and 152 were
upregulated in AD (data not shown).
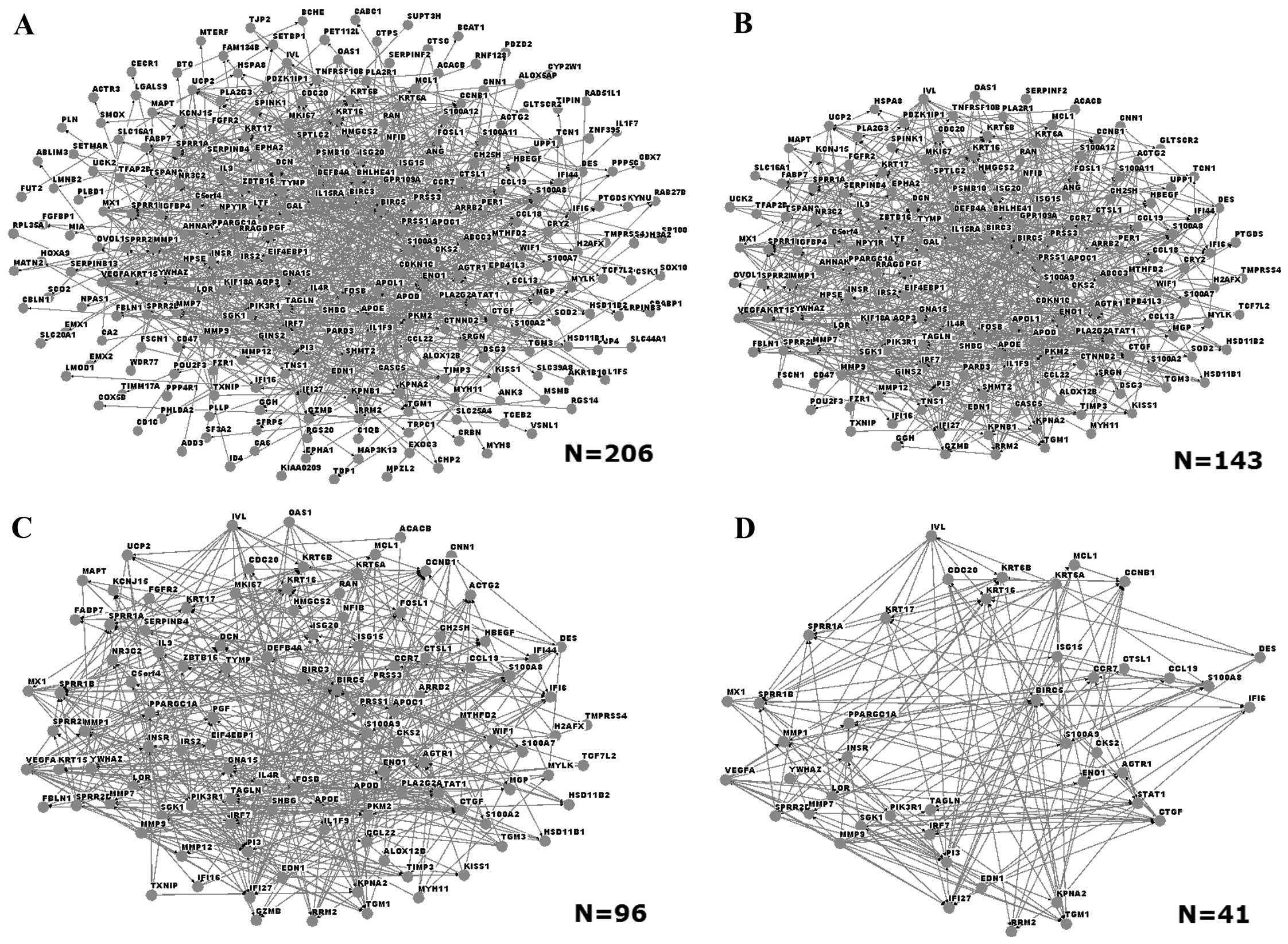
Interaction networks of DEGs
Interaction networks were built with Osprey software
in order to identify interactions among the DEGs. User-defined
interactions were added or subtracted from mouse-over pop-up
windows that link to the database (23). An initial network comprised a total
of 206 DEGs (Fig. 2A). Networks of
DEGs with different degrees of node were also built: Fig. 2B, C and D show the networks of DEGs
with degree of node of >3, >5 and >10, respectively. The
network that consisted of DEGs with degree of node >10 contained
41 genes, among which those coding for matrix metalloproteinase 1
(MMP1), MMP9, small proline-rich repeat protein 1A (SPRR1A) and
loricrin (LOR).
Functional enrichment analysis
Functional enrichment analysis using GO terms was
performed for the network of all DEGs and the network of DEGs with
degree of node >10, comprising 12 and 6 genes, respectively
(Table IA and B). Epidermis
development was the most significantly enriched functional term in
the two networks (FDR =5.01E-04 in the network of high degree-node
genes). This term was associated with a number of high node-degree
DEGs, coding for LOR, type I cytokeratin 17 (KRT17), connective
tissue growth factor (CTGF), SPRR2D, SPRR1A, SPRR1B,
transglutaminase 1 (TGM1) and involucrin (IVL).
 | Table IFunctional enrichment analysis of
Gene Ontology (GO) terms for (A) the network of all differentially
expressed genes (DEGs) and (B) the network comprising DEGs with
degree of node >10. |
Table I
Functional enrichment analysis of
Gene Ontology (GO) terms for (A) the network of all differentially
expressed genes (DEGs) and (B) the network comprising DEGs with
degree of node >10.
| A, Network of all
DEGs |
|---|
|
|---|
| Biological process,
GO term | No. of genes | P-value | FDR |
|---|
| 0008544: Epidermis
development | 15 | 2.95E-07 | 5.01E-04 |
| 0030216:
Keratinocyte differentiation | 9 | 3.60E-06 | 0.006099 |
| 0009913: Epidermal
cell differentiation | 9 | 6.98E-06 | 0.011834 |
| 0042493: Response
to drug | 14 | 1.07E-05 | 0.018058 |
| 0009991: Response
to extracellular stimulus | 14 | 1.30E-05 | 0.022048 |
| 0009725: Response
to hormone stimulus | 18 | 1.54E-05 | 0.026167 |
| 0009611: Response
to wounding | 22 | 1.64E-05 | 0.027821 |
| 0031667: Response
to nutrient levels | 13 | 2.08E-05 | 0.035279 |
| 0009615: Response
to virus | 10 | 2.15E-05 | 0.036422 |
| 0010033: Response
to organic substance | 26 | 2.41E-05 | 0.040820 |
| 0018149: Peptide
cross-linking | 6 | 2.62E-05 | 0.044354 |
| 0031424:
Keratinization | 7 | 2.72E-05 | 0.046187 |
|
| B, Network of DEGs
with degree of node >10 |
|
| Biological process,
GO term | No. of genes | P-value | FDR |
|
| 0008544: Epidermis
development | 9 | 4.32E-08 | 6.69E-05 |
| 0031424:
Keratinization | 6 | 1.36E-07 | 2.10E-04 |
| 0018149: Peptide
cross-linking | 5 | 8.42E-07 | 0.001306 |
| 0030216:
Keratinocyte differentiation | 6 | 1.20E-06 | 0.001861 |
| 0009913: Epidermal
cell differentiation | 6 | 1.86E-06 | 0.002877 |
| 0030855: Epithelial
cell differentiation | 7 | 2.40E-06 | 0.003715 |
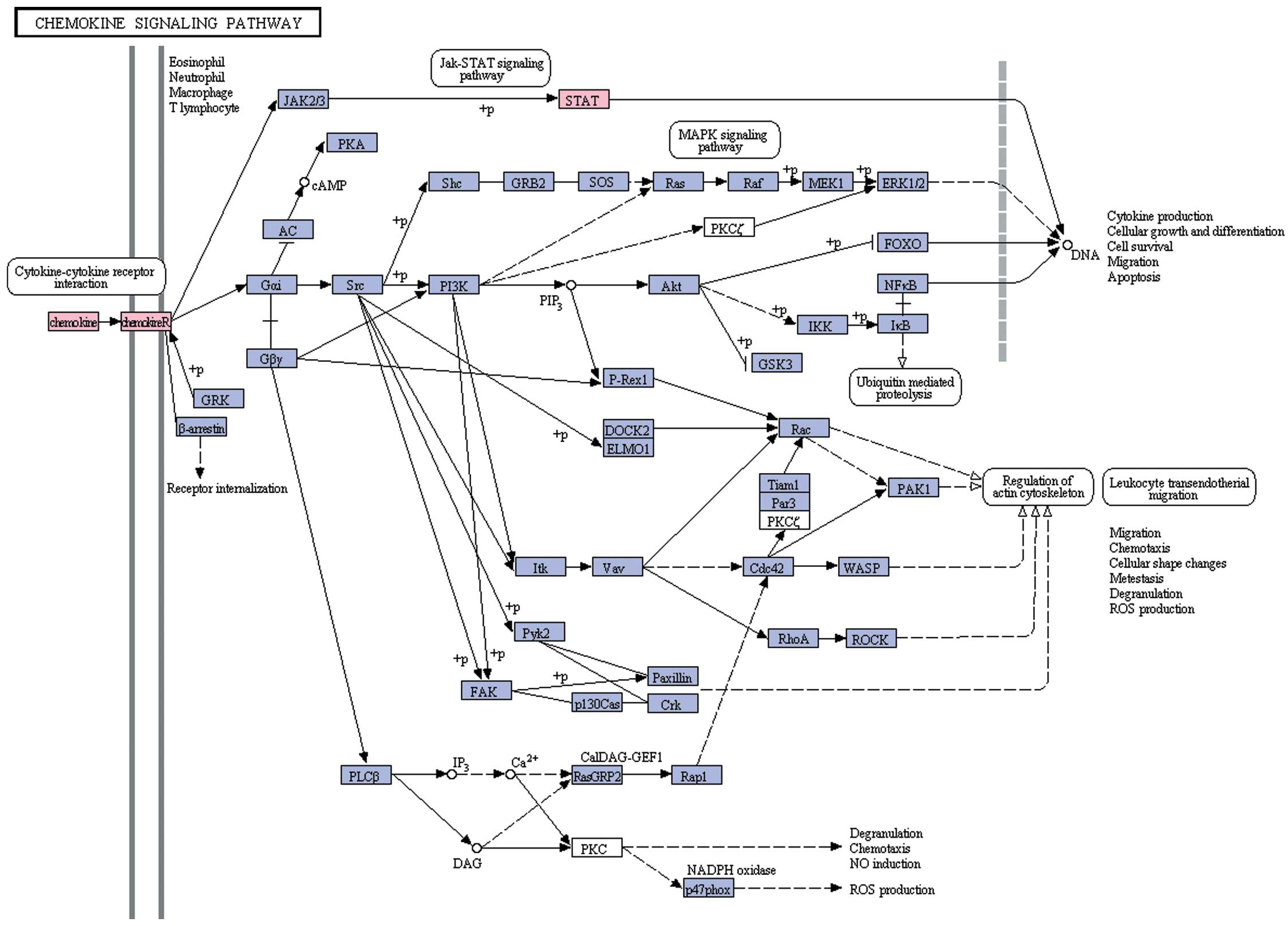
Pathway enrichment analysis
Genes from the network of all DEGs and the network
of DEGs with degree of node >10 were subjected to pathway
enrichment analysis using pathways derived from KEGG. According to
the cut off of P<0.05, only one pathway, the chemokine signaling
pathway, was enriched (Fig. 3)
(P=0.0490978 for the work of all the DEGs and P=0.0399795 for the
network of DEGs with degree of node >10). DEGs that belong to
the chemokine pathway include chemokine (C-C motif) receptor 7
(CCR7), chemokine (C-C motif) ligand 19 (CCL19), signal transducer
and activator of transcription 1 (STAT1) and
phosphoinositide-3-kinase regulatory subunit 1 (PIK3R1).
Discussion
Statistical comparison of gene expression profiles
between AD and healthy skin samples was carried out in the present
study to identify DEGs, followed by construction of DEGs networks,
functional and pathway enrichment analyses, aiming to investigate
the interactions and the functions of the identified DEGs. The
long-term goal of the study was to identify AD-related key
genes.
A range of genes related to epidermis development
were identified among the high degree-node DEGs, such as genes
coding for LOR, KRT17, KRT16, SPRR2D, SPRR1A, SPRR1B and IVL. LOR
and IVL are the main protein components of the cornified cell
envelope, found in terminally differentiated epidermal cells. Kim
et al showed that the two proteins were downregulated in
skin with AD compared to healthy skin (31). Those authors suggested that the two
proteins were inhibited by tyrosine hydroxylase 2 (Th2) cytokines
through STAT-6 (31), with a
deficiency in LOR protein expression leading to a reduction in the
expression of epidermal growth factor receptor (EGFR), e-cadherin,
and occludin (32). In addition,
Sevilla et al reported that mice deficient in IVL,
envoplakin and periplakin had a defective epidermal barrier
(33). Thus, it can be
hypothesized that LOR and IVL are important in maintaining the
status of the skin/epidermal barrier.
KRT17 is a type I intermediate filament chain,
expressed in nail beds, hair follicle, sebaceous glands and other
epidermal appendages. In our study, KRT17 was identified as part of
the network of DEGs with degree of node >10, and it was mapped
to the GO biological process ‘epidermis development’. In a previous
study, the expression of KRT17 in inflammatory skin diseases was
shown to be regulated by lymphocytes via cytokine production
(34). Other studies indicated
that keratine intermediate filament proteins acted as regulators of
inflammation and immunity in skin diseases (35), and that KRT17 promoted epithelial
cell proliferation (36). Findings
of a recent study suggested that the expression levels of KRT17 and
IVL were increased in canine atopic dermatitis (37). Therefore, KRT17 may be a useful
molecular target in AD treatment.
The genes coding for SPRRs locate near to the
LOR and IVL genes, within a cluster of 1.5 Mbp on the
chromosome 1q21, and most likely evolved from a common ancestor.
Moreover, SPRR genes are strongly induced during
differentiation of human epidermal keratinocytes (38). A study by Zimmermann et al
indicated that SPRR2 encoded an allergen and was induced by
IL-13 in experimental allergic responses that may be involved in
disease pathophysiology (39).
Notably, SPRR2A and SPRR2B proteins were considered to be involved
in epithelial differentiation, but not allergic disease (39), prior to this study. In the present
study, the genes coding for SPRR2D, SPRR1A and SPRR1B were found to
be differentially expressed in AD and mapped to the enriched
functional categories of epidermis development, keratinization,
peptide cross-linking and keratinocyte differentiation. Thus,
further studies are necessary to better understand the involvement
of this family of proteins in AD pathogenesis.
Pathway enrichment analysis revealed that AD-related
genes were significantly enriched for constituents of the chemokine
signaling pathway, and the relevant proteins extensively interacted
with other proteins, such as CCR7, CCL19, STAT1 and PIK3R1. The
chemokine receptor CCR7 can control the migration of memory T cells
to inflamed tissues, as well as stimulate dendritic cells
maturation (40). CCL19
specifically binds to CCR7, while the involvement of the CC
chemokine family proteins in AD has been reported in numerous
studies (41–43). STAT1 is also involved in the
chemokine signaling pathway: this protein is phosphorylated by
receptor-associated kinases and translocates to the cell nucleus
where it acts as a transcription activator. Heishi et al reported
that the STAT1 gene was differentially expressed in
peripheral blood mononuclear cells from AD patients compared to
controls and may thus be a useful marker for evaluating AD
(44). The gene can be activated
by IFNγ and is thus involved in immune-mediated inflammation.
STAT1 was also upregulated in lesional compared to
non-lesional AD (45). As for
PIK3R1, encoding the α regulatory subunit of
phosphoinositide 3-kinase (PI3K) (46), there are few reports concerning its
role in AD, limited to mouse models of atopic disorders (47,48).
In conclusion, the chemokine signaling pathway genes CCR7,
CCL19 and STAT1 may be key genes in AD, and
PIK3R1 may represent a new therapeutical target in AD.
In summary, we adopted a microarray approach to
identify key genes associated with AD. A number of potential AD
targets was identified, such as the DEGs coding for LOR, IVL, KRT17
and SPRRs, which are involved in epidermis development, as well as
the DEGs encoding CCR7, CCL19 and STAT1, proteins of the chemokine
signaling pathway. Among these DEGs, two represent potentially
novel therapeutical targets for AD, PIK3R1 and KRT17.
However, this study was limited by the small sample size, while we
did not experimentally confirm that the identified DEGs are
involved in AD. Future studies focusing on these genes may provide
important contributions to the diagnosis and treatment of AD.
References
|
1
|
Spergel JM and Paller AS: Atopic
dermatitis and the atopic march. J Allergy Clin Immunol.
112:S118–S127. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Leung DY: Atopic dermatitis: new insights
and opportunities for therapeutic intervention. J Allergy Clin
Immunol. 105:860–876. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Laughter D, Istvan JA, Tofte SJ and
Hanifin JM: The prevalence of atopic dermatitis in Oregon
schoolchildren. J Am Acad Dermatol. 43:649–655. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Johansson SG, Bieber T, Dahl R, et al:
Revised nomenclature for allergy for global use: Report of the
Nomenclature Review Committee of the World Allergy Organization,
October 2003. J Allergy Clin Immunol. 113:832–836. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Beltrani VS: Atopic dermatitis: an update.
J Allergy Clin Immunol. 104:S85–S86. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
McGirt LY and Beck LA: Innate immune
defects in atopic dermatitis. J Allergy Clin Immunol. 118:202–208.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
De Benedetto A, Agnihothri R, McGirt LY,
Bankova LG and Beck LA: Atopic dermatitis: a disease caused by
innate immune defects? J Invest Dermatol. 129:14–30.
2009.PubMed/NCBI
|
|
8
|
Novak N, Bieber T and Leung DY: Immune
mechanisms leading to atopic dermatitis. J Allergy Clin Immunol.
112:S128–S139. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Akdis CA, Akdis M, Bieber T, et al:
Diagnosis and treatment of atopic dermatitis in children and
adults: European Academy of Allergology and Clinical
Immunology/American Academy of Allergy, Asthma and
Immunology/PRACTALL Consensus Report. Allergy. 61:969–987. 2006.
View Article : Google Scholar
|
|
10
|
Leung DY: Atopic dermatitis: immunobiology
and treatment with immune modulators. Clin Exp Immunol. 107(Suppl
1): 25–30. 1997.PubMed/NCBI
|
|
11
|
Toda M and Ono SJ: Genomics and proteomics
of allergic disease. Immunology. 106:1–10. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Morar N, Willis-Owen SA, Moffatt MF and
Cookson WO: The genetics of atopic dermatitis. J Allergy Clin
Immunol. 118:24–34. 2006. View Article : Google Scholar
|
|
13
|
Park YD, Kim SY, Jang HS, et al: Towards a
proteomic analysis of atopic dermatitis: a
two-dimensional-polyacrylamide gel electrophoresis/mass
spectrometric analysis of cultured patient-derived fibroblasts.
Proteomics. 4:3446–3455. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Merryman-Simpson AE, Wood SH, Fretwell N,
et al: Gene (mRNA) expression in canine atopic dermatitis:
microarray analysis. Vet Dermatol. 19:59–66. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Nomura I, Gao B, Boguniewicz M, Darst MA,
Travers JB and Leung DY: Distinct patterns of gene expression in
the skin lesions of atopic dermatitis and psoriasis: a gene
microarray analysis. J Allergy Clin Immunol. 112:1195–1202. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Olsson M, Broberg A, Jernas M, et al:
Increased expression of aquaporin 3 in atopic eczema. Allergy.
61:1132–1137. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Mobini R, Andersson BA, Erjefalt J, et al:
A module-based analytical strategy to identify novel
disease-associated genes shows an inhibitory role for interleukin 7
receptor in allergic inflammation. BMC Syst Biol. 3:192009.
View Article : Google Scholar
|
|
18
|
Williams H, Jburney P, Hay R, et al: The
U.K. Working Party’s diagnostic criteria for atopic dermatitis. I
Derivation of a minimum set of discriminators for atopic
dermatitis. Br J Dermatol. 131:383–396. 1994.
|
|
19
|
Troyanskaya O, Cantor M, Sherlock G, et
al: Missing value estimation methods for DNA microarrays.
Bioinformatics. 17:520–525. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Fujita A, Sato JR, Rodrigues Lde O,
Ferreira CE and Sogayar MC: Evaluating different methods of
microarray data normalization. BMC Bioinformatics. 7:4692006.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Smyth GK: Limma: linear models for
microarray data. Bioinformatics and Computational Biology Solutions
Using R and Bioconductor. Gentleman R, Carey V, Huber W, Irizarry R
and Dudoit S: Springer; New York: pp. 397–420. 2005, View Article : Google Scholar
|
|
22
|
Dudoit S, Schaffer J and Boldrick J:
Multiple hypothesis testing in microarray experiments. Statist Sci.
18:71–103. 2003. View Article : Google Scholar
|
|
23
|
Breitkreutz BJ, Stark C and Tyers M:
Osprey: a network visualization system. Genome Biol. 4:R222003.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Willis RC and Hogue CW: Searching,
viewing, and visualizing data in the Biomolecular Interaction
Network Database (BIND). Curr Protoc Bioinformatics. Chapter 8(Unit
8.9)2006. View Article : Google Scholar
|
|
25
|
Breitkreutz BJ, Stark C and Tyers M: The
GRID: the general repository for interaction datasets. Genome Biol.
4:R232003. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Estrada E: Virtual identification of
essential proteins within the protein interaction network of yeast.
Proteomics. 6:35–40. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Jeong H, Mason SP, Barabasi AL and Oltvai
ZN: Lethality and centrality in protein networks. Nature.
411:41–42. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
28
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009.PubMed/NCBI
|
|
29
|
Zhang B, Kirov S and Snoddy J: WebGestalt:
an integrated system for exploring gene sets in various biological
contexts. Nucleic Acids Res. 33:W741–W748. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Duncan D, Prodduturi N and Zhang B:
WebGestalt2: an updated and expanded version of the Web-based Gene
Set Analysis Toolkit. BMC Bioinformatics. 11:1. 2010. View Article : Google Scholar
|
|
31
|
Kim BE, Leung DYM, Boguniewicz M and
Howell MD: Loricrin and involucrin expression is down-regulated by
Th2 cytokines through STAT-6. Clin Immunol. 126:332–337. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Nakai K, Yoneda K, Hosokawa Y, et al:
Reduced expression of epidermal growth factor receptor, E-cadherin,
and occludin in the skin of flaky tail mice is due to filaggrin and
loricrin deficiencies. Am J Pathol. 181:969–977. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Sevilla LM, Nachat R, Groot KR, et al:
Mice deficient in involucrin, envoplakin, and periplakin have a
defective epidermal barrier. J Cell Biol. 179:1599–1612. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Komine M, Freedberg IM and Blumenberg M:
Regulation of epidermal expression of keratin K17 in inflammatory
skin diseases. J Invest Dermatol. 107:569–575. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Hobbs RP, Lessard JC and Coulombe PA:
Keratin intermediate filament proteins - novel regulators of
inflammation and immunity in skin. J Cell Sci. 125:5257–5258. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Depianto D, Kerns ML, Dlugosz AA and
Coulombe PA: Keratin 17 promotes epithelial proliferation and tumor
growth by polarizing the immune response in skin. Nat Genet.
42:910–914. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
37
|
Theerawatanasirikul S, Sailasuta A,
Thanawongnuwech R and Suriyaphol G: Alterations of keratins,
involucrin and filaggrin gene expression in canine atopic
dermatitis. Res Vet Sci. 93:1287–1292. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Hohl D, de Viragh PA, Amiguet-Barras F,
Gibbs S, Backendorf C and Huber M: The small proline-rich proteins
constitute a multigene family of differentially regulated cornified
cell envelope precursor proteins. J Invest Dermatol. 104:902–909.
1995. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zimmermann N, Doepker MP, Witte DP, et al:
Expression and regulation of small proline-rich protein 2 in
allergic inflammation. Am J Respir Cell Mol Biol. 32:428–435. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Sallusto F: The role of chemokine
receptors in primary, effector and memory immune response. Exp
Dermatol. 11:476–478. 2002. View Article : Google Scholar
|
|
41
|
Kakinuma T, Saeki H, Tsunemi Y, et al:
Increased serum cutaneous T cell-attracting chemokine (CCL27)
levels in patients with atopic dermatitis and psoriasis vulgaris. J
Allergy Clin Immunol. 111:592–597. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Gunther C, Bello-Fernandez C, Kopp T, et
al: CCL18 is expressed in atopic dermatitis and mediates skin
homing of human memory T cells. J Immunol. 174:1723–1728. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Shimada Y, Takehara K and Sato S: Both Th2
and Th1 chemokines (TARC/CCL17, MDC/CCL22, and Mig/CXCL9) are
elevated in sera from patients with atopic dermatitis. J Dermatol
Sci. 34:201–208. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Heishi M, Kagaya S, Katsunuma T, et al:
High-density oligonucleotide array analysis of mRNA transcripts in
peripheral blood cells of severe atopic dermatitis patients. Int
Arch Allergy Immunol. 129:57–66. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Suarez-Farinas M, Tintle SJ, Shemer A, et
al: Nonlesional atopic dermatitis skin is characterized by broad
terminal differentiation defects and variable immune abnormalities.
J Allergy Clin Immunol. 127:954–964. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Parvaneh N, Casanova JL, Notarangelo LD
and Conley ME: Primary immunodeficiencies: a rapidly evolving
story. J Allergy Clin Immunol. 131:314–323. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Watanabe O, Tamari M, Natori K, et al:
Loci on murine chromosomes 7 and 13 that modify the phenotype of
the NOA mouse, an animal model of atopic dermatitis. J Hum Genet.
46:221–224. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Heinzmann A and Daser A: Mouse models for
the genetic dissection of atopy. Int Arch Allergy Immunol.
127:170–180. 2002. View Article : Google Scholar : PubMed/NCBI
|