Introduction
Tissue-specific stem cells are present in numerous
human tissues and have the potential to differentiate into one or
multiple mature cell types. Tissue-specific stem cells represent an
intermediate between pluripotent cells and fully committed mature
cells, and are excellent models for studying cellular
differentiation and tissue regeneration. Human bone marrow
mesenchymal stem cells (hBMMSCs) are derived from bone marrow
pluripotent stem cells and exhibit the homologous properties of
stem cells, including self-renewal and the capacity to develop into
multiple lineages (1–3). The isolation of hBMMSCs is simple due
to their plastic adherence and ability to expand in culture
(4–7). It has been reported that hBMMSCs have
considerable therapeutic potential in several malignancies,
including cardiovascular disease, cellular replacement therapy and
tissue engineering (8–11).
hBMMSCs express numerous surface markers, including
SH2, SH3, CD90, CD105 and CD106, but do not express hematopoietic
stem cell markers, such as CD14, CD34 and CD45. hBMMSCs are stable
and preserve their differentiation potential following long-term
ex vivo culture and cryopreservation. hBMMSCs have been
demonstrated to differentiate into myocytes, hepatocytes,
osteoblasts, chondrocytes, fibroblasts and adipocytes (12), but also differentiate into
hematopoietic cells and matrix cells, and under specific
conditions, form myotubes and tendon (13–15).
Furthermore, hBMMSC transplantation does not elicit immune
rejection. Therefore, they may become a new source for tissue
repair and regeneration. hBMMSCs are readily obtained from human
bone marrow and expanded ex vivo, and are then transplanted
back into patients following in vitro induction (16,17).
Therefore, hBMMSCs have potential clinical utility for the use of
stem cells for personalized medicine.
Even though hBMMSCs are efficiently recovered from
bone marrow aspirates of healthy individuals and from those of
patients suffering from severe diseases and injuries, they are an
uncommon cell type in the bone marrow, accounting for <0.1% of
nucleated cells in bone marrow aspirates. Therefore, expansion of
hBMMSCs to obtain a stable source of hBMMSCs without losing the
pluripotential of hBMMSCs is important. In the present study,
hBMMSCs were obtained from a human subject, grown to passage (P)65
and the morphological characteristics as well as pluripotent
differentiation potential of these hBMMSCs were investigated in
vitro and in vivo.
Materials and methods
Isolation and expansion of bone marrow
cells
The study was approved by the ethics committee of
Shanghai Chest Hospital, (Shanghai, China). Bone marrow cells were
isolated from the rib fragment from an adult surgical patient at
the Department of Thoracic Surgery, Shanghai Chest Hospital,
Shanghai Jiaotong University (Shanghai, China). Cells (1.5×108
) were plated in gelatin-coated T175 culture flasks and the
mesenchymal cell population was isolated based on plastic
adherence. The cells were cultured in Dulbecco’s modified Eagles’s
medium (DMEM; high glucose; Gibco BRL, Grand Island, NY, USA)
containing 10% fetal bovine serum (FBS), 50 μg/ml thymidine
phosphorylase (ECGF), 50 μg/ml heparin, penicillin 100 U/ml
and streptomycin 100 mg/ml (Invitrogen Life Technologies, Carlsbad,
CA, USA) at 37°C in a humidified atmosphere containing 5%
CO2 for 24 h. For expansion of the mesenchymal cell
population, the cells were incubated under standard culture
conditions. On day three, non-adherent cells were removed by
changing the medium. Thereafter, the medium was replaced twice a
week. Following reaching confluence, the cells were trypsinized
(0.25% v/v trypsin EDTA; Invitrogen Life Technologies),
re-suspended in culture media and seeded at 1×103
cells/cm2 [passage (P)1]. P10 to P57 cells were used in
the present study. Prior to administration, the cells were
resuspended in 250 μl serum-free DMEM. For the cellular
proliferation assays, hBMMSCs were grown in 24-well plates at a
density of 0.5×104 cells/ml and were counted at
different time intervals for plotting the growth curve and
calculating the proliferation index.
Transmission electron microscopy
For transmission electron microscopy, hBMMSCs
(2×105) were harvested and fixed with 2.5% electron
microscopy-grade glutaraldehyde, post-fixed in 1% osmium tetroxide
with 0.1% potassium ferricyanide, dehydrated in gradient ethanol
(30–90%) and embedded in Epon. Ultrathin sections (65 nm) were cut,
stained with 2% uranyl acetate and examined under a Hitachi H-600
transmission electron microscope (Hitachi, Tokyo, Japan).
Flow cytometric analysis
To verify the presentation of hBMSC markers,
phenotyping was routinely performed by flow cytometry on
culture-expanded hBMMSCs. Cells of P2 were collected from confluent
layers following incubation with 0.25% trypsin-EDTA for 5 min. The
single-cell suspensions were washed with phosphate-buffered saline
(PBS)/0.5% bovine serum albumin (BSA) prior to the staining. For
direct staining, the cells were centrifuged (250 × g, 5 min) and
re-suspended in cold PBS/0.5% BSA. A total of 1×106
cells were incubated for 15 min on ice in the dark in cold PBS/0.5%
BSA with titrated concentrations of R-phycoerythrin
(PE)-conjugated monoclonal mouse anti-human CD29, -CD31, -CD38,
-CD44, -CD59, -CD80, -CD86, -CD90, -CD105 and -CD106 antibodies or
monoclonal fluorescein isothiocyanate (FITC) conjugated mouse
anti-human CD9, -CD14 and -CD34 antibodies. The cells were then
washed twice by centrifugation (250 × g, 5 min) and re-suspended
with cold PBS/0.5% BSA prior to proceeding to flow cytometry. Dead
cells and debris were stained with propidium iodide (100 μg/ml) and
excluded from the measurements. Acquisitions were performed on a
FACS Calibur flow cytometer (Becton-Dickinson Bioscience, Franklin
Lakes, NJ, USA). Data analysis was conducted with FCS Express V2
software (version 2, De Novo Software, Los Angeles, CA, USA)
following gating for the designated population.
In vitro multipotent differentiation
assays
The multi-lineage differentiation potential of human
MSCs (n=3, P3) was analyzed by applying the standard procedures
used by Pittenger et al (12). For adipogenesis, 2×105
cells/cm2 were seeded. Five days after reaching
confluence, the hBMMSCs were treated for three days with induction
media consisting of α-MEM (Gibco Laboratories, Gaithersburg, MD,
USA) supplemented with 10% FBS, 1.0×10−7 M dexamethasone
(Sigma, St. Louis, MO, USA), 1.0×10−9 M insulin, 0.5 mM
1.0×10−7 M ascorbic acid and 7×10−3 M sodium
β-glycerophosphate for 18 days at 37°C in a humidified atmosphere
containing 5% CO2 and then for two days with maintenance
media consisting of α-MEM, FBS and 1.0×10−9 M insulin.
Sudan black B staining of the lipid droplets was performed as
previously described (12). Sudan
black B concentrations were calculated by comparison with a Sudan
black B dye standard curve and expressed as nmol/ml following
normalization against the total cellular protein, and is expressed
as nmol/mg protein. The above experiments were performed at least
three times independently and each experiment was conducted in
quadruplicate.
For osteogenesis, hBMMSCs were seeded at a density
of 2×105 cells/cm2 and grown under osteogenic
induction conditions in DMEM (Gibco Laboratories) supplemented with
1.0×10−7 M dexamethasone, 1.0×10−7 M ascorbic
acid and 7×10−3 M β-glycerophosphate sodium for seven
days at 37°C in a humidified atmosphere containing 5%
CO2. The cells were collected following incubation with
0.25% trypsin-EDTA for 5 min. Following centrifugation, the cells
were suspended in 100 ml normal saline and frozen and thawed three
times at −200°C. The suspension was centrifuged at 250 × g for 10
min and osteocalcin contents were determined using the enzyme
dynamics method.
In vivo multipotent differentiation
assays
For osteogenesis, five nude mice were inoculated
subcutaneously with P15 hBMMSCs (2.5×107) mixed with an
equal volume of Matrigel®, and osteogenesis was observed
as described previously (16).
Myogenesis was studied as described by Caterson et al
(16). For the controls, six nude
mice were injected in the hind leg with absolute alcohol (n=3) or
Dil-labeled hBMMSCs (2×106) (n=3). For the treatment
group, six nude mice were injected with absolute alcohol followed 2
h later by Dil-labeled hBMMSCs (2×106). The tissue
sections of leg muscles were prepared and incubated with the
monoclonal antibody against myoglobin at day three and with the
monoclonal antibody against MyoD1 at day 12 and were examined under
a fluorescent microscope (CK40/BK51, Olympus Corporation, Tokyo,
Japan).
Hematopoietic reconstitution
The nude mice (6 to 8-week old males), were obtained
from Shanghai Super-B&K laboratory animal Corp., Ltd (Shanghai,
China). The mice were handled and housed according to protocols
approved by the Shanghai Medical Experimental Animal Care
Commission. A total of 4 h after the nude mice were sublethally
(650 cGy) irradiated, each mouse was infused via the tail vein with
2×106 hBMMSCs. In addition, 0.5 μg granulocyte
colony-stimulating factor was administered daily for three days.
The control nude mice were infused with normal saline. The mice
were sacrificed at different time-points for analysis. Peripheral
blood samples were collected 48–144 h following infusion of hBMMSCs
for leukocyte count and smear. The cells were also removed from
bilateral femurs by flushing with PBS. The firmly adhered cells of
the endosteal region were recovered by digesting the crushed bone
with collagenase type IV (0.03%) and dispase (2 U/ml). The bone
marrow cells were pooled in a tube and the erythrocytes were lysed
by treatment with Grey’s solution. Mononuclear cells were collected
and prepared for smears. The smears were then fixed in acetone and
stained with PE-conjugated anti-human CD45 antibody and
FITC-conjugated anti-human CD34 antibody, examined under a
fluorescent microscope (Olympus Corporation) and the images were
captured accordingly.
RNA extraction and gene expression
profiling
Total cellular RNA was extracted from hBMMSCs of P7
and P65 using the TRIzol reagent as instructed by the manufacturer
(Invitrogen Life Technologies). RNA (1 μg; RIN value >6) was
used for generation of second-strand cDNA, and cRNA was amplified
with the Oligo dT primer, then biotinylated and fragmented with
One-Cycle Target Labeling and control reagents (Affymetrix, Inc.,
Santa Clara, CA, USA), followed by hybridization to U133 Plus 2.0
array overnight (17 h) according to the manufacturer’s
instructions. Finally, the hybridized DNA microarray was
fluorescence stained with GeneChipFluidics Station 450
(Affymetrics) and scanned with a GeneChip Scanner 3000
(Affymetrics). GCOS1.2 Affymetrix GeneChip Command Console (TGCC)
was employed to extract the data in the microarray followed by
analysis with the Robust Multi-Array Average (RMA) method. The
relative gene expression was calculated. A difference of >2 or
<0.5-fold and reliability (P<0.05) was considered to
indicate significant differences in gene expression.
Results
Morphological characteristics of cultured
hBMMSCs
The majority of hBMMSCs were elliptical following
one week of culture with abundant cytoplasms and prominent nuclei.
Light microscopy demonstrated that the cells resembled fibroblasts
in shape and were arranged in parallel or whorls (Fig. 1A–D). Electron microscopy of
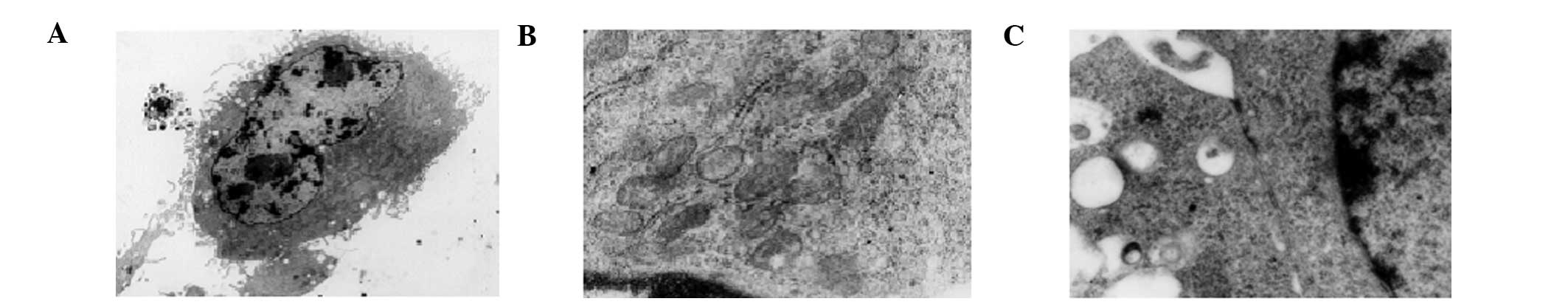
cultured hBMMSCs revealed that the nuclei were large and irregular
in shape with 2–3 nucleoli and scant nuceloplasm (Fig. 2A). The cytoplasms were rich in
mitochondria and rough endoplasmic reticula (Fig. 2B) with gap junctions (Fig. 2C). Transmission electron microscopy
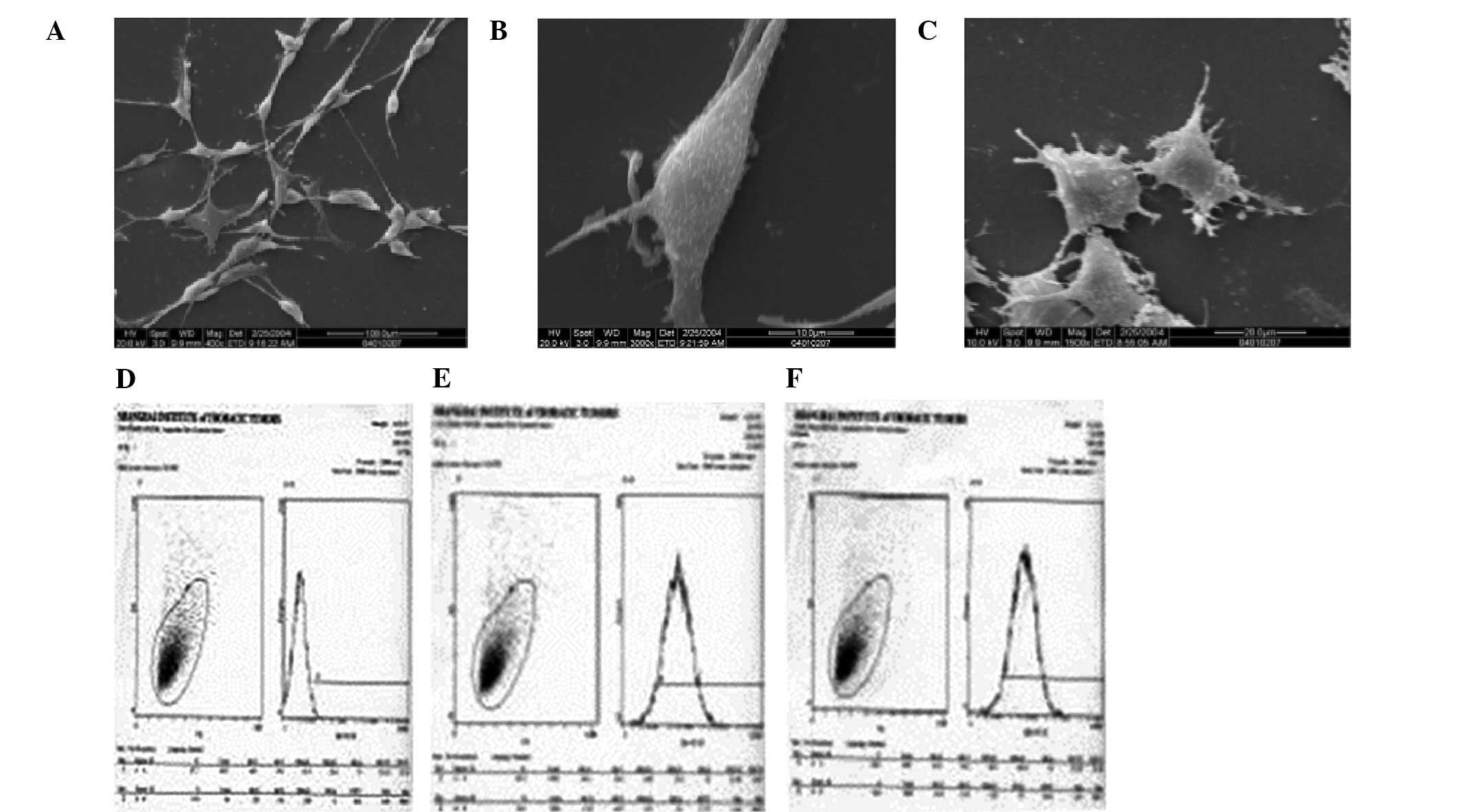
further demonstrated that the cultured hBMMSCs were elongated or
polygonal (Fig. 3A) with short
microvilli (Fig. 3B) and
projections (Fig. 3C). No apparent
changes in cell morphology were observed for cells up to P10. The
doubling time for P10, P20 and P40 hBMMSCs was 32, 36 and 32 h,
respectively.
Flow cytometric studies revealed that the positive
rate of CD31 was 31.1% in P4 hBMMSCs and 18.6% in P10 hBMMSCs.
CD105 and CD106 were expressed in 99 and 95% of P25 hBMMSCs,
respectively (Fig. 3D–F).
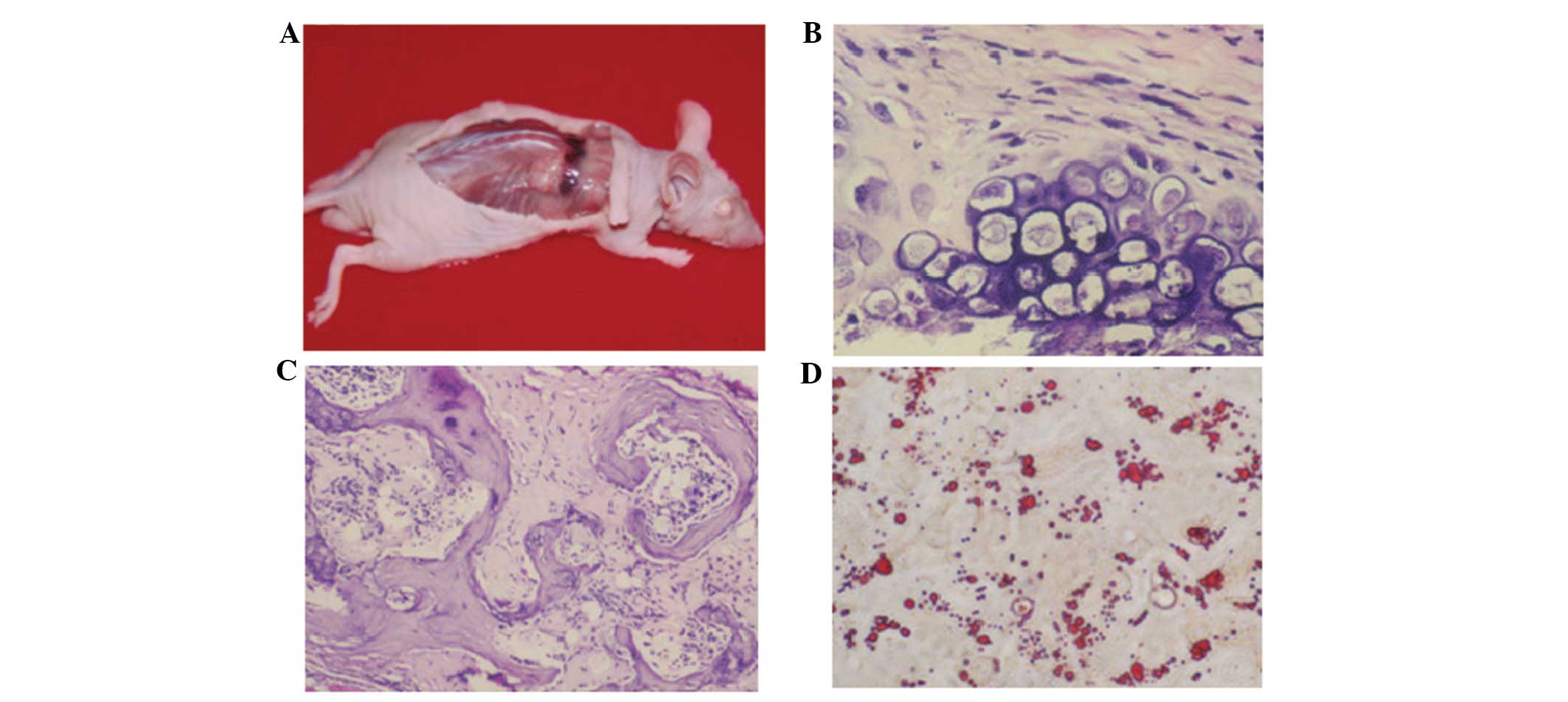
An in vivo osteogenesis test demonstrated
subcutaneous bony tissue formation on the back of the nude mice 35
days post-hBMMSCs inoculation (Fig.
4A). Histology analysis showed a predominant endochondral bone
formation process (Fig. 4B) in
in vivo bone tissue formation (Fig. 4C). In adipogenesis, lipid droplets
appeared at day 18 post induction of adipogenesis, which increased
in size and occupied the entire cell (Fig. 4D). In osteogenesis, it was
identified that the induced hBMMSCs were elongated, contained
numerous particles and were positive for osteocalcin (Fig. 4B). The osteocalcin content was
12±0.5 mg/ml for induced P10 hBMMSCs and 12±0.5 mg/ml for induced
P25 hBMMSCs, which was ~3× higher than that of the unstimulated
cells. Furthermore, no expression of osteocalcin was observed in
the induced P38 and P57 hBMMSCs.
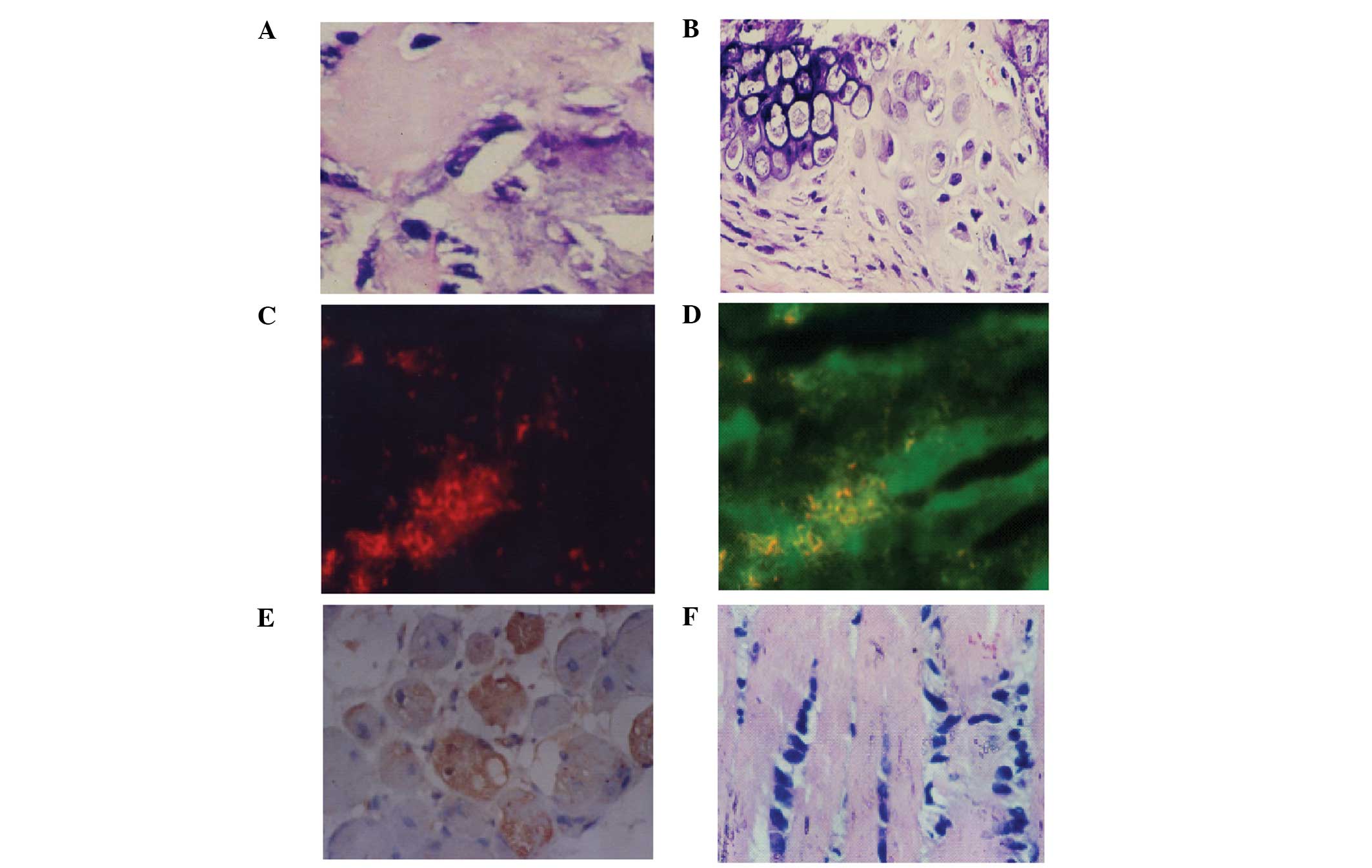
For osteogenesis, palpable masses were detected from
day 35 post inoculation of hBMMSCs, and bone tissue formation was
observed (Fig. 5A). Hematoxylin
and eosin (H&E) staining further revealed chondrocytes and the
formation of bone tissues (Fig.
5B). For myogenesis, at day six after subcutaneous inoculation,
hBMMSCs differentiated to myocytes and were positive for myoglobin
(Fig. 5C and D). At day 12,
H&E staining demonstrated striations in the cytoplasm of
differentiated hBMMSCs at day 12 post inoculation (Fig. 5E) and the cells were positive for
MyoD1 (Fig. 5F). The control nude
mice revealed no formation of muscle cells and were negative for
myoglobin and MyoD1.
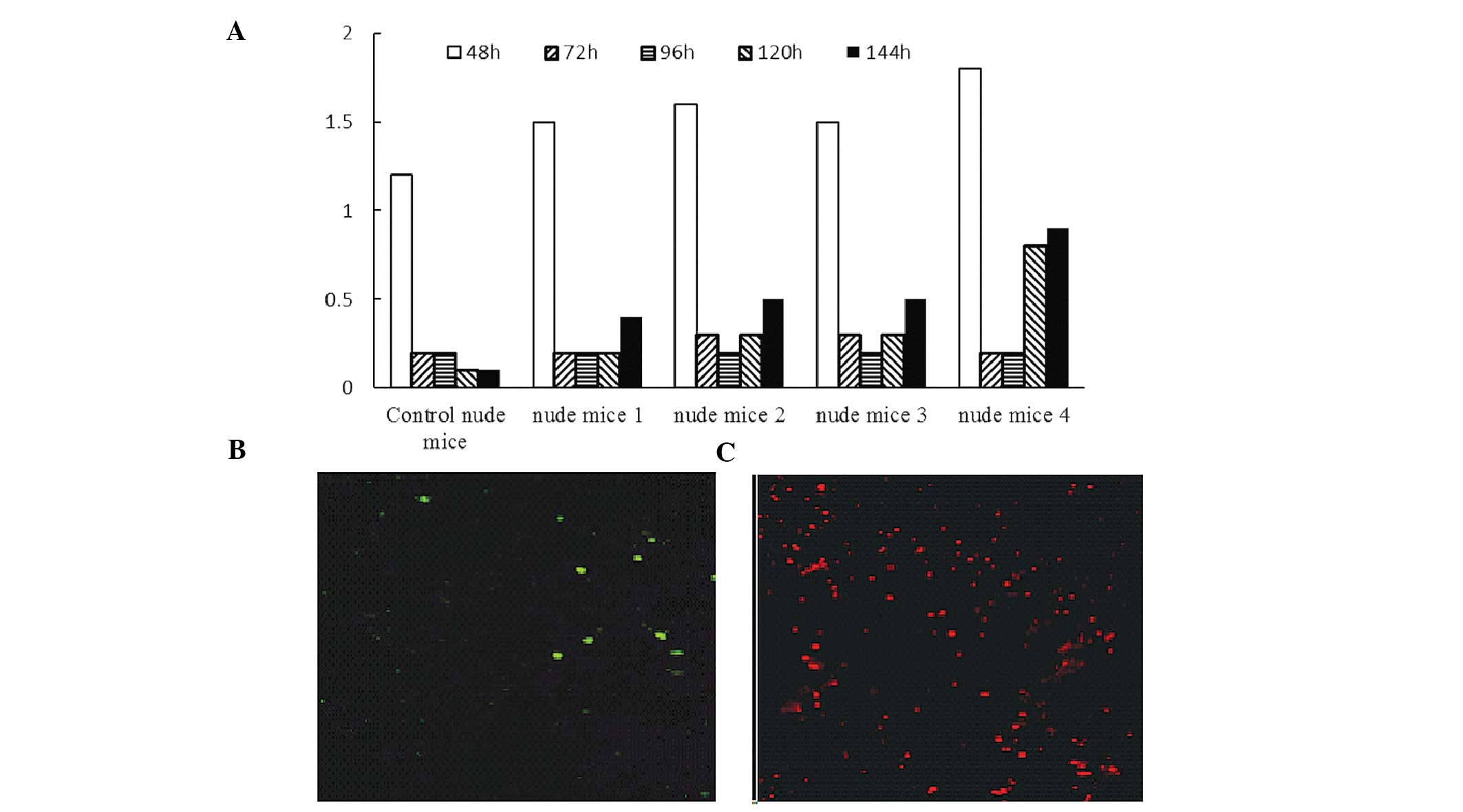
White blood cell counts in the peripheral blood of
control mice irradiated at sublethal doses gradually decreased and
did not recover to normal up to 144 h following irradiation
(Fig. 6A). In irradiated nude mice
reconstituted by hBMMSCs, the white blood cell count briefly
dropped following irradiation but gradually recovered. CD45- and
CD34-positive cells were not detected in the peripheral blood and
bone marrow of the control nude mice. In irradiated nude mice
reconstituted by hBMMSCs, CD45- and CD34-positive cells were
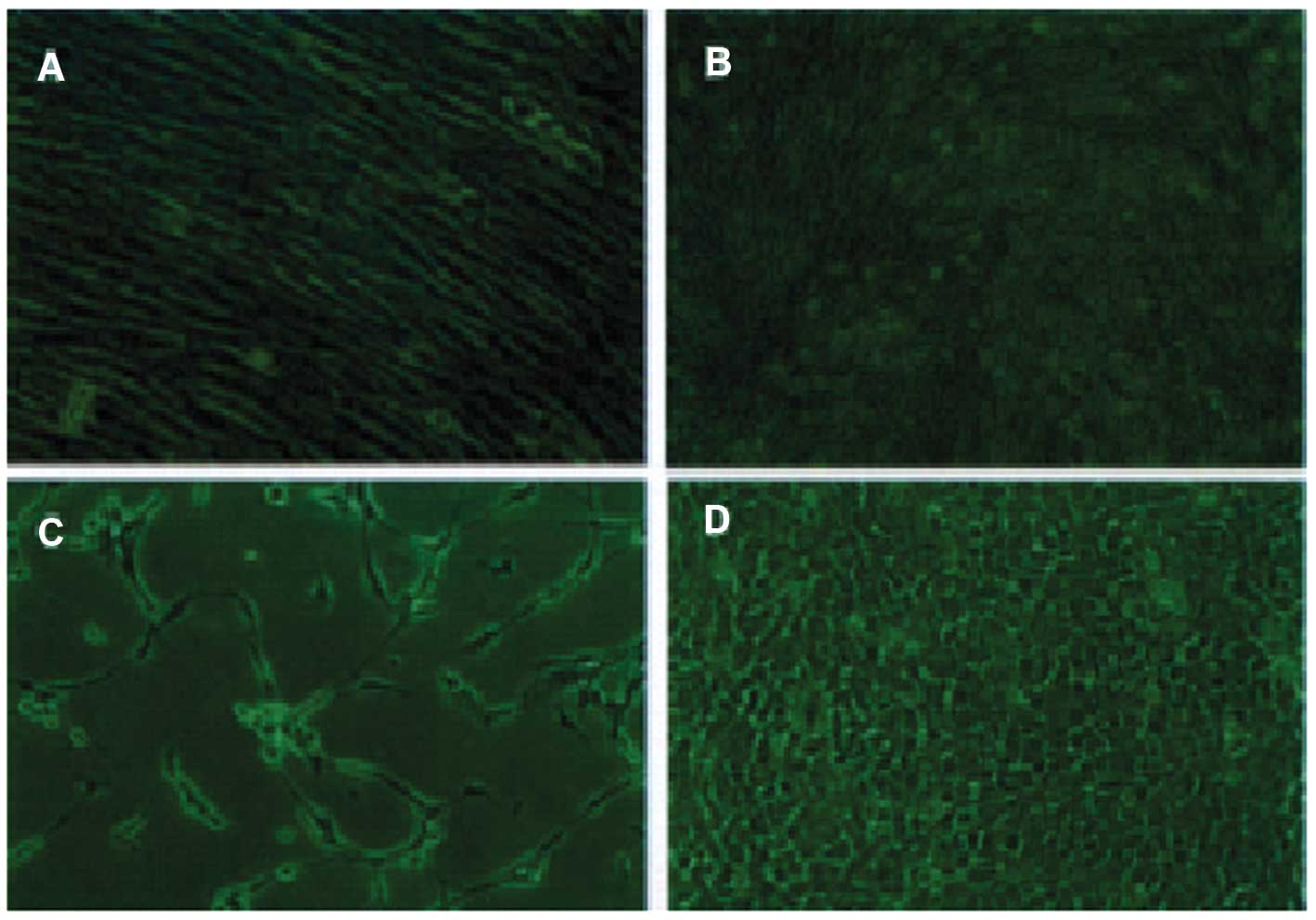
detected 72 h and 144 h post induction (Fig. 6C–D).
The gene microarray analysis of P7 and P57 hBMMSCs
demonstrated that 20 genes were upregulated >2-fold and
40 genes were downregulated >2-fold in the P57 hBMMSCs.
The 10 most upregulated and 10 most downregulated genes are
summarized in Table I.
 | Table ITen most upregulated and downregulated
genes in cultured late human bone marrow mesenchymal stem
cells. |
Table I
Ten most upregulated and downregulated
genes in cultured late human bone marrow mesenchymal stem
cells.
| A, 10 most
upregulated genes |
|---|
|
|---|
| Probe set ID | Gene symbol | Chromosomal
location | HBHSC-60 vs
HBmSc-Signal log ratio |
|---|
| 201242_s_at | ATP1B1 | chr1q24 | 2.4 |
| 219567_s_at | FLJ21144 | chr1p34.2 | 2.3 |
| 241940_at | --- | --- | 2.2 |
| 237566_at | --- | --- | 2.1 |
| 204298_s_at | LOX | chr5q23.2 | 1.8 |
| 209427_at | SMTN | chr22q12.2 | 1.7 |
| 219778_at | ZFPM2 | chr8q23 | 1.6 |
| 204881_s_at | UGCG | chr9q31 | 1.5 |
| 209283_at | CRYAB | chr11q22.3-q23.1 | 1.5 |
| 201110_s_at | THBS1 | chr15q15 | 1.5 |
|
| B, 10 most
downregulated genes |
|
| Probe set ID | Gene symbol | Chromosomal
location | HBHSC-60 vs
HBmSc-Signal log ratio |
|
| 220822_at | --- | --- | −3.5 |
| 226677_at | ZNF521 | chr18q11.2 | −3.5 |
| 202227_s_at | BRD8 | chr5q31 | −3.2 |
| 230030_at | HS6ST2 | chrxq26.2 | −2.9 |
| 202213_s_at | CUL4B | chrxq23 | −2.8 |
| 208612_at | GRP58 | chr15q15 | −2.5 |
| 201444_s_at | ATP6AP2 | chrxq21 | −1.9 |
| 217572_at | --- | --- | −1.9 |
| 201310_s_at | C5orf13 | chr5q22.1 | −1.8 |
| 205350_at | CRABP1 | chr15q24 | −1.8 |
Discussion
In the present study, hBMMSCs were isolated from a
surgical patient and stably expanded ex vivo until P65. Flow
cytometric analysis of P25 hBMMSCs demonstrated that the cells
expressed CD105 and CD106 but not CD14, CD34 and CD45, and
exhibited apparent contact inhibition. Furthermore, these cells
were able to form cartilage and osteoid tissues in nude mice. In
addition, bone marrow cells were observed in the osteoid tissue and
formation of erythrocytes was also noted, suggesting there was
ongoing erythropoiesis in the osteoid tissue. Myogenesis in nude
mice was also observed following inoculation of hBMMSCs. H&E
staining revealed typical striations in the newly formed skeletal
muscles. These findings indicated that hBMMSCs are able to
differentiate into blood and muscle cells. Early infusion of
hBMMSCs allows hematopoiesis in the early stage of bone marrow
failure, which provides time for recovery of the bone marrow from
the infusion of autologous hematopoietic stem cells. This is
consistent with the microenvironment induction theory, stating that
stem cells form tissue-specific cells depending on the
microenvironment (18).
The gene expression profiles of P7 and P57 hBMMSCs
were investigated and it was identified that 20 genes were
upregulated >2 fold and 40 genes were downregulated >2 fold.
These genes are involved in angiogenesis, cell cycle control and
the regulation of proliferation and apoptosis. Furthermore, the
late passages of hBMMSCs were associated with ploidy compared with
the early passages of hBMMSCs (data not presented). These findings
indicated that the passage of hBMMSCs may be associated with gene
mutations and gene expression changes. Therefore, early passages of
hBMMSCs should be used for stem cell therapy. These findings are
consistent with those by Maitra et al (17).
Mesenchymal stem cells possess pluripotent potential
and are induced to differentiate into cells of various types for
organ regeneration, including osteoblasts, chondrocytes,
cardiomyocytes, myocytes, adipocytes and hematopoietic cells. As a
result, they hold significant potential for broad clinical
applications in the treatment of numerous malignancies. The present
study found that it is feasible to isolate and long-term culture
hBMMSCs from patients, and that it is safe to use hBMMSCs within 30
passages. An hBMMSCs cell line was established which may be used
for further studying the biological properties of hBMMSCs for
tissue engineering.
Acknowledgements
This study was supported by the Chinese Ministry of
Science and Technology Sino-Swiss International Collaboration
project (grant no. 2012DFG31320) and the National Natural Science
Foundation of China project (grant no. 81371702).
References
|
1
|
Herzog EL, Chai L and Krause DS:
Plasticity of marrow-derived stem cells. Blood. 102:3483–3493.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sato Y, Araki H, Kato J, et al: Human
mesenchymal stem cells xenografted directly to rat liver are
differentiated into human hepatocytes without fusion. Blood.
106:756–763. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Dominici M, Le Blanc K, Mueller I, et al:
Minimal criteria for defining multipotent mesenchymal stromal
cells. The International Society for Cellular Therapy position
statement. Cytotherapy. 8:315–317. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Nakamura K, Ito Y, Kawano Y, et al:
Antitumor effect of genetically engineered mesenchymal stem cells
in a rat glioma model. Gene Ther. 11:1155–1164. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Tropel P, Noël D, Platet N, Legrand P,
Benabid AL and Berger F: Isolation and characterisation of
mesenchymal stem cells from adult mouse bone marrow. Exp Cell Res.
295:395–406. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Tocci A and Forte L: Mesenchymal stem
cell: use and perspectives. Hematol J. 4:92–96. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
da Meirelles LS and Nardi NB: Murine
marrow-derived mesenchymal stem cell: isolation, in vitro
expansion, and characterization. Br J Haematol. 123:702–711. 2003.
View Article : Google Scholar
|
|
8
|
Parr A, Tator C and Keating A: Bone
marrow-derived mesenchymal stromal cells for the repair of central
nervous system injury. Bone Marrow Transplant. 40:609–619. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bai L, Caplan A, Lennon D and Miller RH:
Human mesenchymal stem cells signals regulate neural stem cell
fate. Neurochem Res. 32:353–362. 2007. View Article : Google Scholar
|
|
10
|
Mangi AA, Noiseux N, Kong D, He H, Rezvani
M, Ingwall JS and Dzau VJ: Mesenchymal stem cells modified with Akt
prevent remodeling and restore performance of infarcted hearts. Nat
Med. 9:1195–1201. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Duan HF, Wu CT, Wu DL, et al: Treatment of
myocardial ischemia with bone marrow-derived mesenchymal stem cells
overexpressing hepatocyte growth factor. Mol Ther. 8:467–474. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Pittenger MF, Mackay AM, Beck SC, et al:
Multilineage potential of adult human mesenchymal stem cells.
Science. 284:143–147. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Mezey É and Chandross KJ: Bone marrow: a
possible alternative source of cells in the adult nervous system.
Eur J Pharmacol. 405:297–302. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Muraglia A, Cancedda R and Quarto R:
Clonal mesenchymal progenitors from human bone marrow differentiate
in vitro according to a hierarchical model. J Cell Sci.
113:1161–1166. 2000.PubMed/NCBI
|
|
15
|
Liechty KW, MacKenzie TC, Shaaban AF, et
al: Human mesenchymal stem cells engraft and demonstrate
site-specific differentiation after in utero transplantation in
sheep. Nat Med. 6:1282–1286. 2000. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Caterson EJ, Nesti LJ, Albert T, Danielson
K and Tuan R: Application of mesenchymal stem cells in the
regeneration of musculoskeletal tissues. MedGenMed. 5:E12001.
|
|
17
|
Maitra A, Arking DE, Shivapurkar N, et al:
Genomic alterations in cultured human embryonic stem cells. Nat
Genet. 37:1099–1103. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
Koç ON, Gerson SL, Cooper BW, Dyhouse SM,
Haynesworth SE, Caplan AI and Lazarus HM: Rapid hematopoietic
recovery after coinfusion of autologous-blood stem cells and
culture-expanded marrow mesenchymal stem cells in advanced breast
cancer patients receiving high-dose chemotherapy. J Clin Oncol.
18:307–316. 2000.PubMed/NCBI
|