Introduction
Colorectal cancer (CRC) is one of the most common
types of cancer worldwide, accounting for ~9% of all cancer
mortality in 2013 (1). Metastatic
disease develops in >50% of patients, exhibiting an ultimately
fatal prognosis, with 5-year survival rates of 10% (2). Although treatment with chemotherapy
and radiation therapy has improved and become more effective for
locally advanced mid to low rectal cancers and metastatic colon
lesions over the past three decades (3), to date, no major advance has been
observed in 5-year survival rates and the number of patients cured
from metastatic CRC remains low.
Metastasis is a complex process involving multiple
steps. Tumor cells are required to achieve the two rate-limiting
steps of invasion and distant colony formation. Metastatic spread
to the liver and lungs occurs most frequently in colorectal cancer
metastases in humans (4). Numerous
CRC patients experience metastasis to the liver or lung and fail to
respond to curative therapies, thus significant research efforts
have focused on identifying the molecular changes underlying CRC
metastasis (5). Several candidate
biomarkers have been reported, including ST6 galactose I and β1
integrin (6,7). However, the mechanism of distant
metastasis in CRC remains to be elucidated.
MicroRNAs (miRNAs) are a group of small, endogenous,
non-protein-coding RNA molecules that negatively regulate gene
expression (8,9). In previous decades, miRNAs were found
to be important in multiple biological processes and the metabolic
regulation of cancer (10). In
addition, multiple studies have indicated the importance of miRNAs
in managing the efficiency of chemotherapy in several types of
human cancer (11,12). In the present study, the CRC cell
line HCT116 was infected with a recombinant lentivirus carrying the
firefly luciferase gene and a stable cell line was obtained via
puromycin selection (HCT116/Luc). This cell line was selected as an
in vivo model of metastasis (13). The present study aimed to
investigate the differences of miRNA expression profiles between
the HCT116/Luc located in the colon and lung, respectively.
Materials and methods
Preparation of the lentivirus
All cell culture reagents were purchased from
Invitrogen Life Technologies (Carlsbad, CA, USA). The firefly
luciferase gene was inserted into the lentivirus vector
Ubi-MCS-IRES-Puromycin to obtain plasmid Ubi-Luc-IRES-Puromycin.
HEK293T cells, obtained from the Type Culture Collection of the
Chinese Academy of Sciences, (Shanghai, China), were plated at a
density of 1×106 cells per 100 mm dish 24 h prior to
transfection. The cells were transfected with
Ubi-Luc-IRES-Puromycin along with packaging vectors, pMD2G and
psPAX2 according to the manufacturer’s instructions for
Lipofectamine 2000. The transfection medium was removed the
following day and replaced with Dulbecco’s modified Eagle’s medium
supplemented with 10% fetal bovine serum (FBS). The supernatants
from these cells were collected 72 h after recovery from
transfection. The supernatants were filtered using a 0.45 μM
syringe filter, combined with 4 μg/ml of polybrene and added to the
cultured human CRC cell line HCT116.
Cell culture and transfection
The HCT116 cell line was obtained from the cell bank
of the Chinese Academy of Sciences (Beijing, China). Cells were
cultured in RPMI-1640 supplemented with 10% FBS (complete media).
They were infected with the recombinant lentivirus carrying the
firefly luciferase gene and a stable cell line (HCT116-Luc) was
obtained via puromycin selection. The expression of luciferase was
confirmed by a luciferase assay system kit purchased from Promega
Corporation (Madison, WI, USA) according to the manufacturer’s
instructions.
Cell line subselection using an in vivo
model of metastasis
HCT116-Luc cells were collected and resuspended in
phosphate-buffered saline at a concentration of 4×107
cells/ml. Balb/c null mice (Shanghai SLAC Laboratory Animal Co.,
Ltd., Shanghai, China) were inoculated with 0.1 ml cell suspension
through a tail vein. On day 28, tumor metastasis in each mouse was
monitored using the IVIS Lumina II in vivo imaging system
(Xenogen, Alameda, CA, USA). Tumor tissues were isolated from mice
that had significant lung and colon metastases. A single cell
suspension was prepared using a trypsin digestion method. Briefly,
tumor tissues were cut into small sections and digested with 0.25%
trypsin at 4°C overnight, followed by incubation at 37°C for 30
min. Following removal of any tissue residue by centrifugation at
300 × g for 5 min, cells were suspended with complete media
supplemented with 2 μg/ml of puromycin. Following three rounds of
subselection, HCT116/Luc-P (cells isolated from the lung) and
HCT116/Luc-I (cells isolated from the colon) were obtained. The
present study was approved by the ethics committee of Shanghai
University of Traditional Chinese Medicine (Shanghai, China)
RNA isolation
Cells were seeded at a density of 600,000 cells/well
in a flat-bottom 6-well plate in 2 ml of complete media. After 48
h, cells were collected and total RNA was prepared using TRIzol
(Invitrogen Life Technologies) according to the manufacturer’s
instructions with the exception that the precipitation was allowed
to remain for 12 h at −20°C, which efficiently recovers all RNA
species, including miRNAs. The RNA concentration was determined
using a Nanodrop™ spectrophotometer (NanoDrop Technologies, Inc.,
Rockland, DE, USA). RNA integrity was assessed using a
Tris-acetate-EDTA buffered agarose gel electrophoresis, analyzing
integrity and association between the 28S rRNA, 18S rRNA and 5S
rRNA bands.
Microarray miRNA profiling
The stacking-hybridized universal tag (SHUT) assay
presented by Duan et al (14) is an efficient technique for
high-throughput miRNA profiling using microarrays. Following the
techniques described by Duan et al, a full-spectrum
microarray was designed targeting each of the 2,017 mature human
miRNAs listed in the Sanger miRBase version 19 (Wellcome Trust
Sanger Institute, Hinxton, UK). Using the Agilent microarray
(Agilent Technologies, Inc., Santa Clara, CA, USA), miRNA profiles
of the HCT116/Luc-P and HCT116/Luc-I were obtained following the
procedures as described by Zhang and Li (15).
Image scanning and data analysis
Following hybridization and washing, slides were
scanned using a GenePix 4100A microarray scanner (Molecular
Devices, Inc., Sunnyvale, CA, USA) at constant power and
photomultiplier tube gain settings through a single-color channel
(wavelength of 532 nm). The raw pixel intensities were extracted
using the GenePix Pro 7.0 software (Molecular Devices, Inc.). Data
processing, including filtration, background-correction,
transformation and normalization as well as subsequent statistical
analysis was performed using the methods provided by Duan et
al (14). Finally, a heatmap
was plotted to demonstrate the differential expression level of the
mature miRNAs between HCT116/Luc-P and HCT116/Luc-I.
Results
Construction of the stable cell line
HCT116/Luc
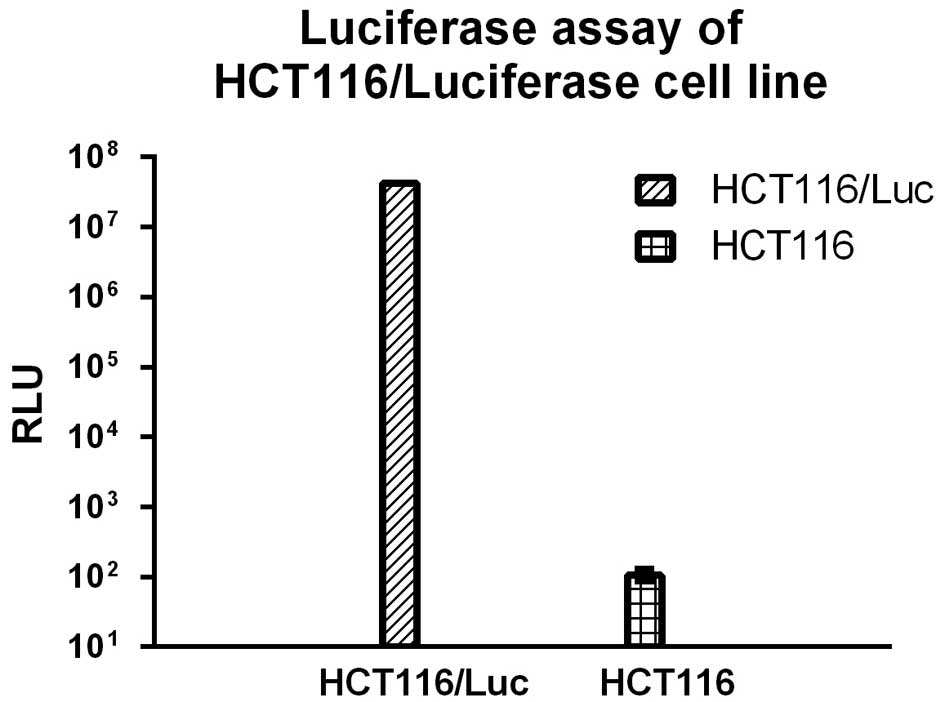
To obtain the stable cell line HCT116/Luc, the CRC
cell line HCT116 was transfected with lentivirus-mediated
luciferase, selected using puromycin and detected using a
luciferase assay kit (cat no. E4030, Promega, Shanghai, China). As
shown in Fig. 1, the luminescent
signal of the selected cells were significantly higher than for the
control HCT116 cells.
Selection of HCT116/Luc-P and
HCT116/Luc-I using an in vivo model of metastasis
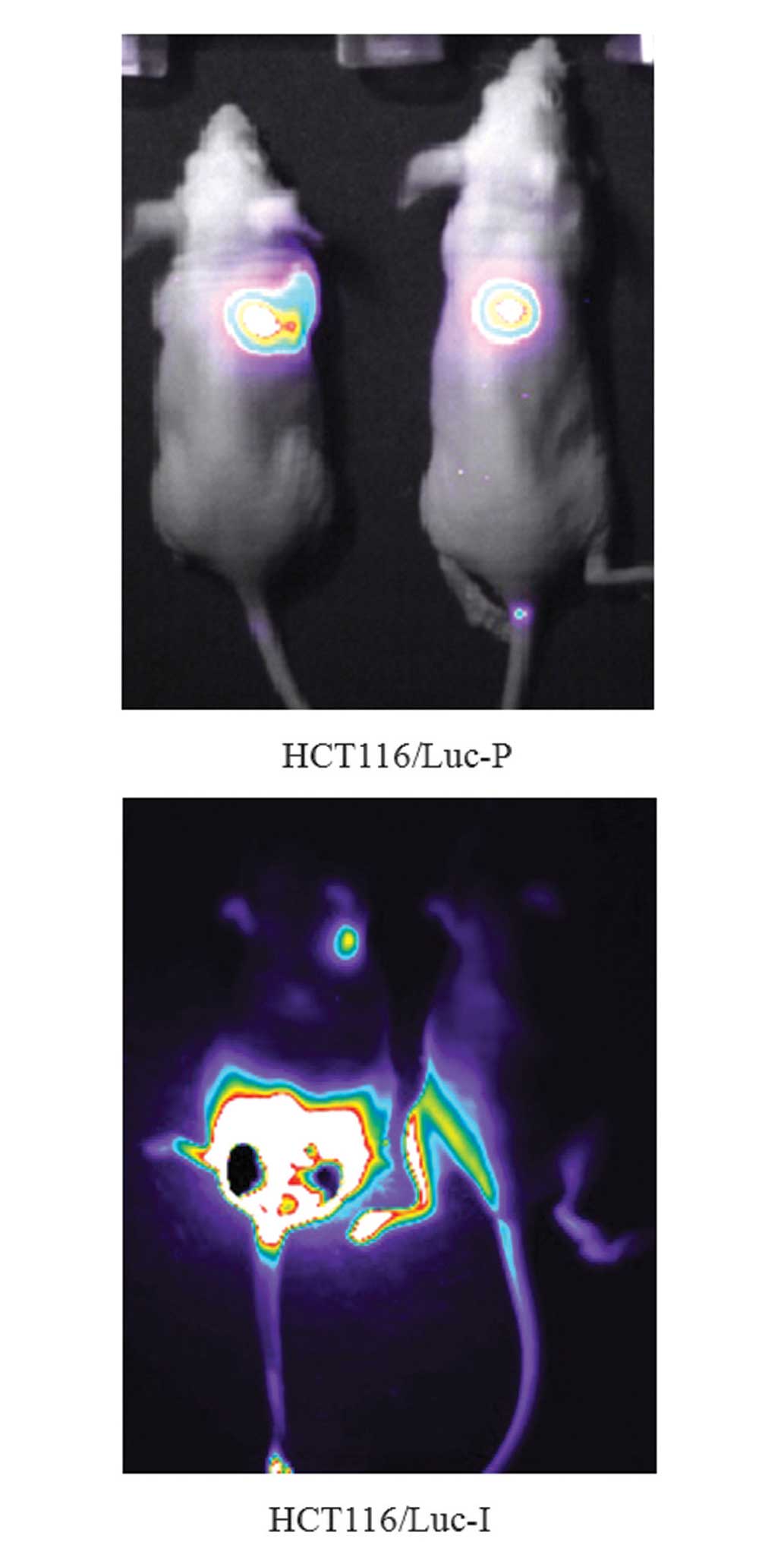
Four weeks after inoculation of the mice with
HCT116/Luc, tumor metastasis in each mouse was monitored using the
IVIS Lumina II in vivo imaging system (Caliper Life
Sciences, Hopkinton, MA, USA) as shown in Fig. 2. Mice that exhibited significant
lung and colon metastasis were selected and tumor tissues were
isolated and cultured. Following three rounds of subselection,
HCT116/Luc-P and HCT116/Luc-I were obtained and identified using a
luciferase assay system.
Differential miRNA expression profiles
between HCT116/Luc-P and HCT116/Luc-I
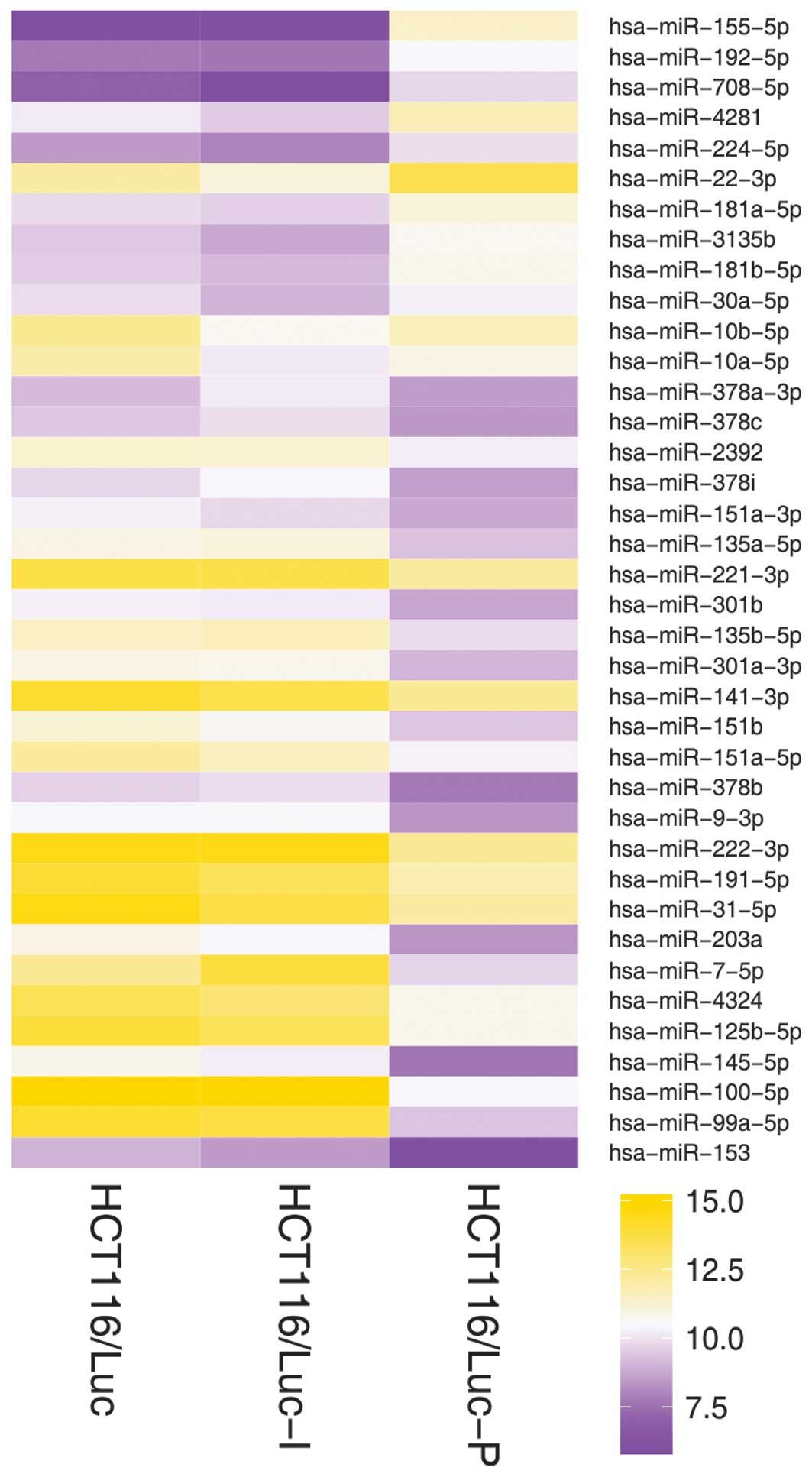
Based on the SHUT assay, the miRNA expression
profiles in HCT116/Luc, HCT116/Luc-P and HCT116/Luc-I were obtained
and the heat map of partial results is shown in Fig. 3. Of the 2,017 human miRNAs
analyzed, 38 miRNAs were detected to be expressed differentially
between cells of HCT116/Luc-I and HCT116/Luc-P (Table I; Fig.
3). Additionally, 26 of the 38 miRNAs were observed to be
expressed at a level >2-fold higher in HCT116/Luc-I than in
HCT116/Luc-P, while the remaining 12 miRNAs demonstrated
significantly lower expression.
 | Table IComparison of miRNA expression
profiles of HCT116 cells located in the lung and colon. |
Table I
Comparison of miRNA expression
profiles of HCT116 cells located in the lung and colon.
| miRNA category | Number | miRNAs |
|---|
| Upregulated in
HCT116/Luc-I | 26 | hsa-miR-153,
hsa-miR-99a-5p, hsa-miR-100-5p, hsa-miR-145-5p, hsa-miR-125b-5p,
hsa-miR-4324, hsa-miR-7-5p, hsa-miR-203a, hsa-miR-31-5p,
hsa-miR-191-5p, hsa-miR-222-3p, hsa-miR-9-3p, hsa-miR-378b,
hsa-miR-151a-5p, hsa-miR-151b, hsa-miR-141-3p, hsa-miR-301a-3p,
hsa-miR-135b-5p, hsa-miR-301b, hsa-miR-221-3p, hsa-miR-135a-5p,
hsa-miR-151a-3p, hsa-miR-378i, hsa-miR-2392, hsa-miR-378c,
hsa-miR-378a-3p |
| Downregulated in
HCT116/Luc-I | 12 | hsa-miR-10b-5p,
hsa-miR-10a-5p, hsa-miR-30a-5p, hsa-miR-181b-5p, hsa-miR-3135b,
hsa-miR-181a-5p, hsa-miR-22-3p, hsa-miR-224-5p, hsa-miR-4281,
hsa-miR-708-5p, hsa-miR-192-5p, hsa-miR-155-5p |
Discussion
miRNAs are important in several types of human
cancer. miRNAs have been demonstrated to be involved in the
regulation of expression of a number of cancer-associated genes
(8,16). Extensive research is currently
focused on identifying miRNAs that may be important in cancer
therapy and diagnosis. To date, more than 2,000 human miRNAs have
been identified. These small RNAs impact many cellular processes
and their deregulation is causative of many human cancers. In
addition, many miRNAs were reported as predictive biomarkers and
therapeutic targets in several cancers (17–20).
CRC is the third most common type of cancer in males
and females (21). One of the
hallmarks of CRC is the ability to invade adjacent structures and
to metastasize to remote organs (22,23).
CRC is the second leading cause of cancer-associated mortality in
the USA (24). The majority of
mortalities caused by colorectal cancer are due to metastatic
disease (4). As numerous CRC
patients experience metastasis to the liver or lung and fail to
respond to curative therapies, significant research efforts have
aimed to identify the molecular changes or regulatory mechanisms
underlying CRC metastasis (5). The
present study, focusing on the differences of miRNA expression in
HCT16 cell lines located in the lung and colon aimed to examine the
potential miRNA targets associated with the regulation of
colorectal lung metastases.
As shown in the results, the expression of 38 miRNAs
was significantly different (all P<0.05) in HCT116/Luc-P and
HCT116/Luc-I. Notably, when comparing the miRNA expression profile
of HCT116/Luc, 35 of the 38 miRNAs exhibited similar expression
levels in HCT116/Luc and HCT116/Luc-I while significantly different
levels were observed in HCT116/Luc-P. This phenomenon may suggest
that in the population of the CRC cell line HCT116, only a small
number demonstrate metastastic characteristics.
A total of 26 miRNAs (shown in Table I) were revealed to be expressed at
a level >2-fold greater in HCT116/Luc-I than in HCT116/Luc-P.
Among these miRNAs, several have been reported to be downregulated
in cancer cells or tissues in previous years (25–29).
Hsa-miR-99a-5p was demonstrated to be downregulated >2-fold in
squamous cell lung carcinoma tissues, compared with normal tissues
(25). In the present study, the
expression of Hsa-miR-99a-5p in HCT116/Luc-P was >20-fold lower
than that in HCT116/Luc-I, which suggests that colorectal lung
metastases cells may have partial lung carcinoma characteristics.
Consistent with the results reported in the present study, several
of these miRNAs have been previously reported to be tumor
suppressors or downregulated in cancer cells. Hsa-miR-145-5p was
reported to be downregulated in intracranial aneurysms (26). Knocking down hsa-miR-135b-5p in
cell lines led to the depletion of the pediatric cancer stem cell
fraction and impairment of sphere formation (27). In various breast cancer cell lines,
the expression of hsa-miR-9-3p was low and synthetic-enhancer tumor
suppressor characteristics were exhibited (28). Previously, Hsa-miR-7-5p expression
was reported to be reduced in metastatic melanoma-derived cell
lines compared with primary melanoma cells (29). This is consistent with the current
results and possibly indicates that hsa-miR-7-5p may be important
in the regulation of carcinoma cell line metastasis.
A total of 12 miRNAs (shown in Table I) were demonstrated to be
overexpressed in HCT116/Luc-P. Among these miRNAs, hsa-miR-30a-5p
has been previously reported to be overexpressed in glioma cell
lines and glioma samples and its expression level positively
correlated with the grade of malignancy of the tumor (30). Hsa-miR-181b-5p was also revealed to
be upreglulated in gastric cancer, which may correspond with lymph
node invasion, nerve invasion and vascular invasion (31).
Further to the reported miRNAs, the function of
other miRNAs in cancer require further investigation. The present
profiling may be the first step toward delineating the differential
expression of miRNAs in CRC cells located in colon and lung. This
may enable researchers to elucidate the regulation associated with
miRNAs in colorectal lung metastases. These miRNAs require further
validation and functional analysis to evaluate whether they are
important in the pathogenesis of colorectal lung metastases or are
adopted as markers for the prediction of colorectal metastasis.
Acknowledgements
The authors would like to thank Assistant
Investigator Kexiao Zheng and Professor Jiong Li (Suzhou Institute
of Nano-tech and Nano-Bionics, Chinese Academy of Sciences) for
providing the miRNA microarray assay support. The present study was
funded by Medicine and Healthcare in Zhejiang Province General
Studies Program (grant nos. 2012KYA027 and 2013KYA023).
References
|
1
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics. CA cancer J Clin. 63:11–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Cidón EU: The challenge of metastatic
colorectal cancer. Clin Med Insights Oncol. 4:55–60. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Janjan NA, Crane C, Feig BW, et al:
Improved overall survival among responders to preoperative
chemoradiation for locally advanced rectal cancer. Am J Clin Oncol.
24:107–112. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wang J, Rajput A, Kan JL, et al: Knockdown
of Ron kinase inhibits mutant phosphatidylinositol 3-kinase and
reduces metastasis in human colon carcinoma. J Biol Chem.
284:10912–10922. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Park JJ and Lee M: Increasing the α2, 6
sialylation of glycoproteins may contribute to metastatic spread
and therapeutic resistance in colorectal cancer. Gut Liver.
7:629–641. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lee HJ, Lee M, Kang CM, et al:
Identification of possible candidate biomarkers for local or whole
body radiation exposure in C57BL/6 mice. Int J Radiat Oncol Biol
Phys. 69:1272–1281. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Vassos N, Rau T, Merkel S, et al:
Prognostic value of β1 integrin expression in colorectal liver
metastases. Int J Clin Exp Pathol. 7:288–300. 2013.
|
|
8
|
Zhang BH, Pan XP, Cobb GP and Anderson TA:
microRNAs as oncogenes and tumor suppressors. Dev Biol. 302:1–12.
2007. View Article : Google Scholar
|
|
9
|
Zhang BH, Stellwag EJ and Pan XP:
Large-scale genome analysis reveals unique features of microRNAs.
Gene. 443:100–109. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Koturbash I, Zemp FJ, Pogribny I and
Kovalchuk O: Small molecules with big effects: the role of the
microRNAome in cancer and carcinogenesis. Mutat Res. 722:94–105.
2011. View Article : Google Scholar
|
|
11
|
Meng F, Henson R, Lang M, et al:
Involvement of human micro-RNA in growth and response to
chemotherapy in human cholangiocarcinoma cell lines.
Gastroenterology. 130:2113–2129. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Xia L, Zhang D, Du R, et al: miR-15b and
miR-16 modulate multidrug resistance by targeting BCL2 in human
gastric cancer cells. Int J Cancer. 123:372–379. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Khanna C and Hunter K: Modeling metastasis
in vivo. Carcinogenesis. 26:513–523. 2005. View Article : Google Scholar
|
|
14
|
Duan D, Zheng KX, Shen Y, et al:
Label-free high-throughput microRNA expression profiling from total
RNA. Nucleic Acids Res. 39:e1542011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang X and Li W: 5-Fluorouracil in
combination with cisplatin alters the microRNA expression profile
in the CNE nasopharyngeal carcinoma cell line. Mol Med Rep.
6:303–308. 2012.PubMed/NCBI
|
|
16
|
Zhang BH, Stellwag EJ and Pan X:
Large-scale genome analysis reveals unique features of microRNAs.
Gene. 443:100–109. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yau TO, Wu CW, Dong Y, et al: microRNA-221
and microRNA-18a identification in stool as potential biomarkers
for the non-invasive diagnosis of colorectal carcinoma. Br J
Cancer. 111:1765–1771. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Saumet A, Mathelier A and Lecellier CH:
The potential of microRNAs in personalized medicine against
cancers. Biomed Res Int. 2014:6429162014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Heneghan HM, Miller N and Kerin MJ: MiRNAs
as biomarkers and therapeutic targets in cancer. Curr Opin
Pharmacol. 10:543–550. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Olson EN: MicroRNAs as therapeutic targets
and biomarkers of cardiovascular disease. Sci Transl Med.
6:239ps32014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chang GJ, Kaiser AM, Mills S, et al:
Practice parameters for the management of colon cancer. Dis Colon
Rectum. 55:831–843. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Portera CA Jr, Berman RS and Ellis LM:
Molecular determinants of colon cancer metastasis. Surg Oncol.
7:183–195. 1998. View Article : Google Scholar
|
|
23
|
Yokota J: Tumor progression and
metastasis. Carcinogenesis. 21:497–503. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Jemal A, Siegel R, Ward E, et al: Cancer
statistics, 2007. CA Cancer J Clin. 57:43–66. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Gao W, Shen H, Liu L, et al: MiR-21
overexpression in human primary squamous cell lung carcinoma is
associated with poor patient prognosis. J Cancer Res Clin Oncol.
137:557–566. 2011. View Article : Google Scholar
|
|
26
|
Jiang Y, Zhang M, He H, et al:
MicroRNA/mRNA profiling and regulatory network of intracranial
aneurysm. BMC Med Genomics. 6:362013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Sanchez-Diaz PC, Hsiao TH, Chang JC, et
al: De-regulated microRNAs in pediatric cancer stem cells target
pathways involved in cell proliferation, cell cycle and
development. PLoS One. 8:e616222013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zawistowski JS, Nakamura K, Parker JS, et
al: MicroRNA 9–3p targets β1 integrin to sensitize claudin-low
breast cancer cells to MEK inhibition. Mol Cell Biol. 33:2260–2274.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Giles KM, Brown RA, Epis MR, et al:
miRNA-7–5p inhibits melanoma cell migration and invasion. Biochem
Biophys Res Commun. 430:706–710. 2013. View Article : Google Scholar
|
|
30
|
Wang K, Jia Z, Zou J, et al: Analysis of
hsa-miR-30a-5p expression in human gliomas. Pathol Oncol Res.
19:405–411. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Chen G, Shen ZL, Wang L, et al:
Hsa-miR-181a-5p expression and effects on cell proliferation in
gastric cancer. Asian Pac J Cancer Prev. 14:3871–3875. 2013.
View Article : Google Scholar : PubMed/NCBI
|