Introduction
Poly(ADP-ribose) polymerases (PARPs) comprise a
superfamily containing 17 members in humans (1). Numerous PARP-1, -2, -14 and
tankyrases have previously been shown to mediate inflammatory
reactions (2–6), and PARP-1 has been associated with
inflammatory or oxidative stress-related skin pathologies (7–9).
PARP-1 is an important pro-inflammatory mediator with important
roles in the maturation of immune cells (10–12),
activation of pattern recognition receptors (13–19),
activation of pro-inflammatory transcription factors (6), and reactive species (20).
The contact hypersensitivity (CHS) reaction is a
T-cell mediated, delayed type hypersensitivity reaction that is
elicited by low molecular weight molecules, such as oxazolone (OXA)
(21,22). Upon sensitization, infiltrating
cells and keratinocytes produce free radicals, pro-inflammatory
cytokines, adhesion molecules and matrix metalloproteinases in a
PARP-1 and nuclear factor-κB-dependent manner, resulting in the
induction of inflammation (6,7,20,23–25).
PARP-1 expression inhibition, or knock-out, has been shown to
reduce the severity of experimental CHS (7,26).
The present study aimed to characterize whether alterations of
eicosanoid and docosanoid metabolism and signaling may contribute
to the anti-inflammatory effects of PARP-1 expression ablation.
Materials and methods
Materials
OXA and the other chemicals used in the present
study were obtained from Sigma-Aldrich (St. Louis, MO, USA), unless
stated otherwise.
Animal experiments
All of the experiments performed on mice were
approved by the Local Ethical Committee (9/2008/DE MÁB) and were
performed according to EU and national guidelines.
PARP-1+/+ and PARP-1−/− mice, derived from
heterozygote-to-heterozygote breeding, were kept in the animal
facility of the Life Science Building, University of Debrecen
(Debrecen, Hungary) (27). A CHS
model was constructed as described by previous methods (7). Briefly, the mice were randomized into
four groups: PARP-1+/+ vehicle sensitized (control),
PARP-1−/− vehicle sensitized (control),
PARP-1+/+ OXA sensitized (CHS) and PARP-1−/−
OXA sensitized (CHS) groups (n=5/5/6/5). Sensitization was carried
out on the shaved abdominal wall skin of the mice, with an
administration of 100 μl 2% OXA solubilised in acetone: olive oil
(4:1; CHS groups), or 100 μl vehicle of acetone:olive oil (4:1;
control groups). Seven days after sensitization, all of the mice
were challenged with 100 μl 0.5% OXA, which was applied to the
epidermis of the shaved back. After 24 h, the mice were sacrificed
and the skin was harvested.
Histology and microscopy
Hematoxylin-eosin staining and immunohistochemistry
was performed on paraffin-fixed 7 μm tissue sections, as described
previously (7).
High performance liquid
chromatography-electrospray tandem mass spectrometry
(HPLC-ESI-MS-MS) analysis
The procedure for the HPLC-ESI-MS-MS analysis The
analysis of free fatty acids, eicosanoids and docosanoids in the
skin was performed, according to previously described methods
(28), with minor
modifications.
Sample preparation
The whole analytical sample preparation procedure
used was based on an established method used for retinoid
quantification (29). Briefly, 50
mg skin biopsy, was treated with 150 μl acetonitrile and the skin
samples were cut into small pieces with scissors, on ice. If less
than 50 mg of skin biopsy was present, water was added to yield 50
mg of sample weight. The mixtures were agitated for 3 min, and the
precipitated protein was centrifuged at 16060 g (13,000 rpm), 4°C
for 6 min. A total of 130 μl resulting supernatant was mixed with
10 μl isotope supernatant mix and evaporated in Eppendorf reaction
vials (Eppendorf, Hamburg, Germany) using an Eppendorf concentrator
at 30°C for ~60 min, until the sample volume was ~10 μl. The
Eppendorf concentrator was vented with argon, in order to prevent
degradation of eicosanoids and docosanoids. The dried extract was
resuspended in ~25 μl HPLC solvent A [64.3% water (Chromasolv Plus;
Sigma-Aldrich, Budapest, Hungary), 35.5% acetonitrile (Merck KGaA,
Darmstadt, Germany) and 0.2% formic acid (Sigma-Aldrich, Hungary)]
to yield 35 μl. The sample was then vortexed (15 sec), agitated (3
min) and transferred into micro injection inserts vials (Waters,
Budapest, Hungary). The glass vials containing 35 μl extract were
transferred into brown screw top vials with PTFE/silicone septa
(Waters), and placed into the pre-cooled (15°C) autosampler of the
Waters 2695XE separation module.
Chromatographic system
The HPLC system consisted of a Waters 2695XE
separation module (Waters) including a gradient pump, autosampler,
degasser and a heated column compartment. A MS-MS detector, with an
ESI ionizing option, (Micromass Quattro Ultima PT; Waters, Elstree,
UK; a gift from Biosystems Int., France) was used as the detector.
The system was controlled using MassLynx software (Waters).
HPLC conditions
The eluents were degassed in the Waters 2695XE
separation module prior to mixing, and passed through an in-line
filter (1–2 μm; Knauer, Berlin, Germany) before reaching the
analytical column (LiChroCART, 125×2 mm; Superspher 100, RP-18,
endcapped; Merck KgaA), which was embedded in the column
compartment. A multilinear gradient was formed from solvent A (as
mentioned previously) and solvent B (methanol; Merck KGaA). The
gradient consisted of the following steps: 0.0 min 20% B, 3.0 min
20% B, 5.0 min 60% B, 15.0 min 100% B, 15.9 min 100% B and 16.0 min
5% B. The flow rate was adjusted to 0.4 ml/min and the column was
heated to 40°C. From the same biological extract, 10 μl was used
for each HPLC analysis. This step was performed twice, using the
same HPLC conditions, and two different MS-MS analysis options were
conducted for better resolution and quantification of the various
analytes.
MS options
The Micromass Quattro Ultima PT was controlled using
MassLynx software. Argon was used with an inlet pressure of 0.8
bar. ESI (electro spray ionization source; Waters) was vented by
nitrogen, which was continuously produced by a nitrogen generator
(Peak Scientific NM30 Nitrogen generator; Peak Scientific,
Billerica, MA, USA) including a compressor (Waters), the inlet flow
was set at 3.6×10−3 mbar.
Multiple reaction monitoring
settings
ESI, with a negative ESI-setting, was performed with
the HPLC eluent, following the ion source temperature of 85°C. The
desolvation gas flow was 780 l/h, the desolvation temperature was
400°C, the cone gas flow was 10 l/h, the capillary current was 3
μA, and the cone voltage was 50 V. The aperture voltage was set at
0 V and the range-finder lens voltage was set at 35 V (for one) and
0.2 V (for two). The analyzer settings were LM1 resolution, 14.5;
HM1 resolution, 14.5; ion energy, 10.7; entrance, −1; collision, 0
(collision parameters were set for each substance at the MS-method
parameters); exit, 2; LM2 resolution, 14.5; HM2 resolution, 14.5;
ion energy, 25.0 and multiplier energy, 650 V.
Multiple reaction monitoring settings for
polyunsaturated fatty acids (PUFA), eicosanoids/docosanoids
semi-quantification
The following methods were used, as described in a
previous study (27): Method A
from 0.0–9.0 min for PGF2 349.0 -> 192.7, collision
energy 22 eV; PD1 and PD1 isomers, like PDX, 359.0 -> 153.3,
collision energy 17 eV; TXB2 369.0 -> 195.0,
collision energy 13 eV; PGE3 349.0 -> 233.0,
collision energy 17 eV; LXA4/LXB4 351.0 ->
115.0, collision energy 12 eV; 8iPGF3 351.0 -> 193.0,
collision energy 22 eV; 20-COOH-LTB4 365.0 -> 195.0,
collision energy 13 eV; RvD1, RvD2 375.0 -> 141.3, collision
energy 13 eV; RvE1 375.1 -> 141.3, collision energy 13 eV. From
9.0–12.5 min for 13-HODE 294.7 -> 170.7, collision energy 16 eV;
9-HODE 294.7 -> 194.7, collision energy 16 eV; 5-HEPE 317.0
-> 115.0, collision energy 17 eV; 12-HEPE 317.0 -> 179.0,
collision energy 17 eV; 15-HEPE 317.0 -> 219.0, collision energy
17 eV, LTC4 623.9 -> 272.0, collision energy 14 eV.
From 12.5–16.0 min for LA 279.3 -> 279.0, collision energy 10
eV; 8-HEPE 317.0 -> 255.0, collision energy 17 eV; 18-HEPE 317.0
-> 259.0, collision energy 17 eV; 5-oxoETE, 12-oxoETE and
15-oxoETE 317.0 -> 273.0, collision energy 17 eV; 20-HETE 319.0
-> 245.0, collision energy 10 eV; LTC4 623.9 ->
272.0, collision energy 14 eV; LTE4 438.0 -> 333.0,
collision energy 13 eV. From 12.5–16.0 min for LA 279.3 -> 59.2,
collision energy 25 eV; EPA 301.0 -> 203.2, collision energy 12
eV; AA 303.0 -> 259.3, collision energy 14 eV; DHA 327.1 ->
29.3, collision energy 14 eV.
Method B (27) from
0.0–9.8 min for PGE2, d15d12PGD2,
PGD2, d15d12PGJ2 and PGJ2 315.0
-> 271.3, collision energy 13 eV; LTB5 333.0 ->
195.0, collision energy 13 eV; LTB4 335.0 -> 195.0,
collision energy 13 eV; RvE1 349.1 -> 195.3,
collision energy 13 eV; HXA3, HXB3 and
20-COOH-AA 335.0 -> 273.3, collision energy 13 eV;
LXA5 349.0 -> 115.0, collision energy 12 eV;
20-OH-LTB4 351.0 -> 195.0, collision energy 13 eV;
MaR 359.0 -> 250.0, collision energy 13 eV. From 9.8–12.5 min
for 5-HETE 318.7 -> 115.0, collision energy 14 eV; 8-HETE 319.0
-> 155.0, collision energy 14 eV; 11-HETE 319.0 -> 167.0,
collision energy 14 eV; 12-HETE 319.0 -> 179.0, collision energy
14 eV; 15-HETE 319.0 -> 218.9, collision energy 11 eV; 4-HDHA
343.0 -> 101.0, collision energy 10 eV; 10-HDHA 343.0 ->
181.0, collision energy 10 eV; 14-HDHA 343.0 -> 205.0, collision
energy 10 eV; 17-HDHA 343.0 -> 245.0, collision energy 14 eV,
20-HDHA 343.0 -> 285.0, collision energy 10 eV; 13-oxoODE 293.0
-> 249.0, collision energy 17 eV. And from 12.5–16.0 min for LA
279.3 -> 59.2, collision energy 25 eV; EPA 301.0 -> 203.2,
collision energy 12 eV; AA 303.0 -> 259.3, collision energy 11
eV; and DHA 327.1 -> 229.3, collision energy 14 eV.
Standard solutions
Stock solutions of the PUFAs, eicosanoids and
docosanoids were prepared by dissolving the solutions in methanol
to yield a final concentration of 10 μg/ml. The solutions were
obtained from Cayman-Chemicals (Tallinn, Estonia), BioMol
International (Kastel-Med KFT, Budapest, Hungary), Sigma-Aldrich
(Hungary), Larodan Lipids (Malmö, Sweden) and Dr. Charles Serhan
(Harvard, MA, USA). All stock solutions were stored in the dark, at
−80°C until further use. The reference PUFAs, eicosanoids and
docosanoids (BioMol International/Enzo Life Sciences, Farmingdale,
NY, USA) were used for assay validation.
Quantification
Individual eicosanoids and docosanoids were
quantified based on the determination of the area under the curve
(AUC), which was compared with the AUC of the standard compounds.
To ensure optimal extraction, isotope-labelled standard compounds
were used. This analytical procedure was previously established for
liquids and tissue analysis (28).
mRNA preparation, reverse transcription
and quantitative polymerase chain reaction (qPCR)
Total RNA extraction, reverse transcription, and a
subsequent qPCR, were performed as described previously (7). The primers used are listed in
Table I.
 | Table IOligonucleotides used in quantitative
polymerase chain reactions. |
Table I
Oligonucleotides used in quantitative
polymerase chain reactions.
| Gene | Sequence |
|---|
| Mouse 36B4 | 5′-AGA TTC GGG ATA
TGC TGT TGG-3′
5′-AAA GCC TGG AAG AAG GAG GTC-3′ |
| Mouse GAPDH | 5′-CAA GGT CAT CCA
TGA CAA CTT TG-3′
5′-GGC CAT CCA CAG TCT TCT GG-3′ |
| Mouse
cyclophyllin | 5′-TGG AGA GCA CCA
AGA CAG ACA-3′
5′-TGC CGG AGT CGA CAA TGA T-3′ |
| Mouse FABP7 | 5′-GAG TAC ATG AAA
GCT CTG GGC-3′
5′-AAC CGA ACC ACA GAC TTA CAGT T-3′ |
| Mouse COX2 | 5′-GTG AAG GGA AAT
AAG GAG CTT CC-3′
5′-GTG ATT TAA GTC CAC TCC ATG GC-3′ |
| Human 36B4 | 5′-CCA TTG AAA TCC
TGA GTG ATG TG-3′
5′-GTC GAA CAC CTG CTG GAT GAC-3′ |
| Human actin | 5′-GAC CCA GAT CAT
GTT TGA GAC C-3′
5′-CAT CAC GAT GCC AGT GGT AC-3′ |
| Human
cyclophyllin | 5′-GTC TCC TTT GAG
CTG TTT GCA GAC-3′
5′-CTT GCC ACC AGT GCC ATT ATG-3′ |
| Human PARP-1 | 5′-CAC TGG TAC CAC
TTC TCC TGC TTC-3′
5′-CTT TGC CTG TCA CTC CTC CAG-3′ |
Establishment of a PARP-1 knockdown
HEK293T cell line and luciferase reporter assays
HEK293T human embryonic kidney cells (American Type
Culture Collection, Manassas, VA, USA) were maintained in
Dulbecco’s modified Eagle’s medium, supplemented with 4.5 g/l
glucose, 10% fetal calf serum, 2.5 μg/ml puromycin. These cells
stable expressing a small hairpin (sh)RNA specifically targeting
PARP-1 (pLKO.1-PARP1). pLKO.1 was used in the control cells
(Sigma-Aldrich, USA). PARP-1 expression was subsequently
suppressed, as confirmed by qPCR.
pLightSwitch_PromFABP7 (Switchgear Genomics,
Carlsbad, CA, USA) and pCMV-βgal were transfected into the above
cell line using jetPEI (PolyPlus Transfections SA, Illkirch,
France). Following a 24 h incubation, the luciferase and
β-galactosidase activities were determined. Luciferase activity was
normalized to β-galactosidase activity.
Statistical analysis
The results of the cell assays are expressed as the
means ± standard deviation, and statistical significance between
the groups was determined by Student’s t-test. The results of the
in vivo studies are expressed as the means, and statistical
significance was determined using the Kruskal-Wallis and
Mann-Whitney tests. A P<0.05 was considered to indicate a
statistically significant difference.
Results
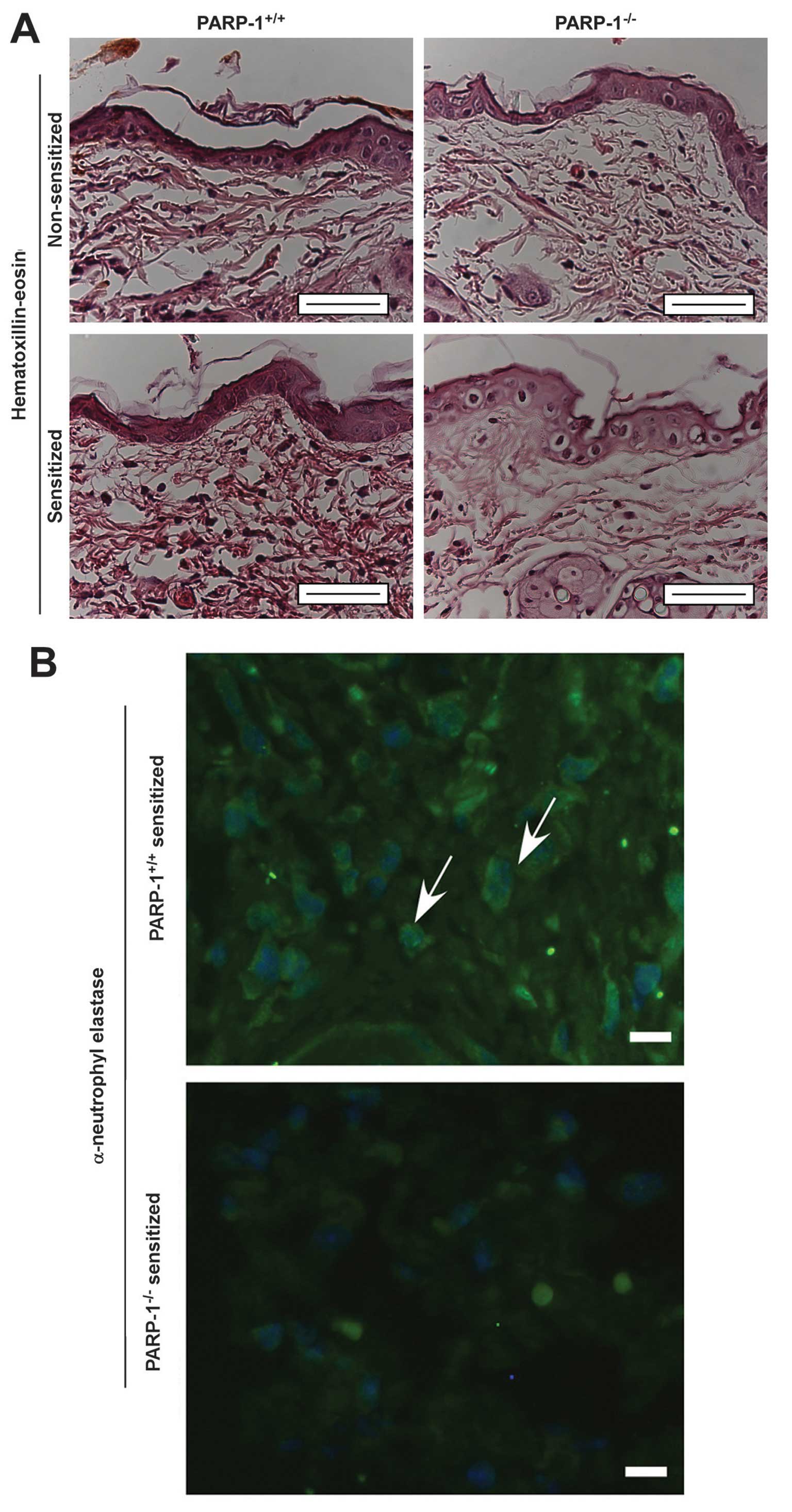
Neutrophil infiltration and edema at the site of the
CHS reaction was observed in the histological examinations;
however, the levels of these were reduced in the
PARP-1−/− mice (Fig. 1A and
B). This finding indicates that PARP-1 may contribute to
inflammation in CHS, therefore PARP-1−/− mice may be
protected against CHS. These results are concordant with our
previous observations (7,26,30).
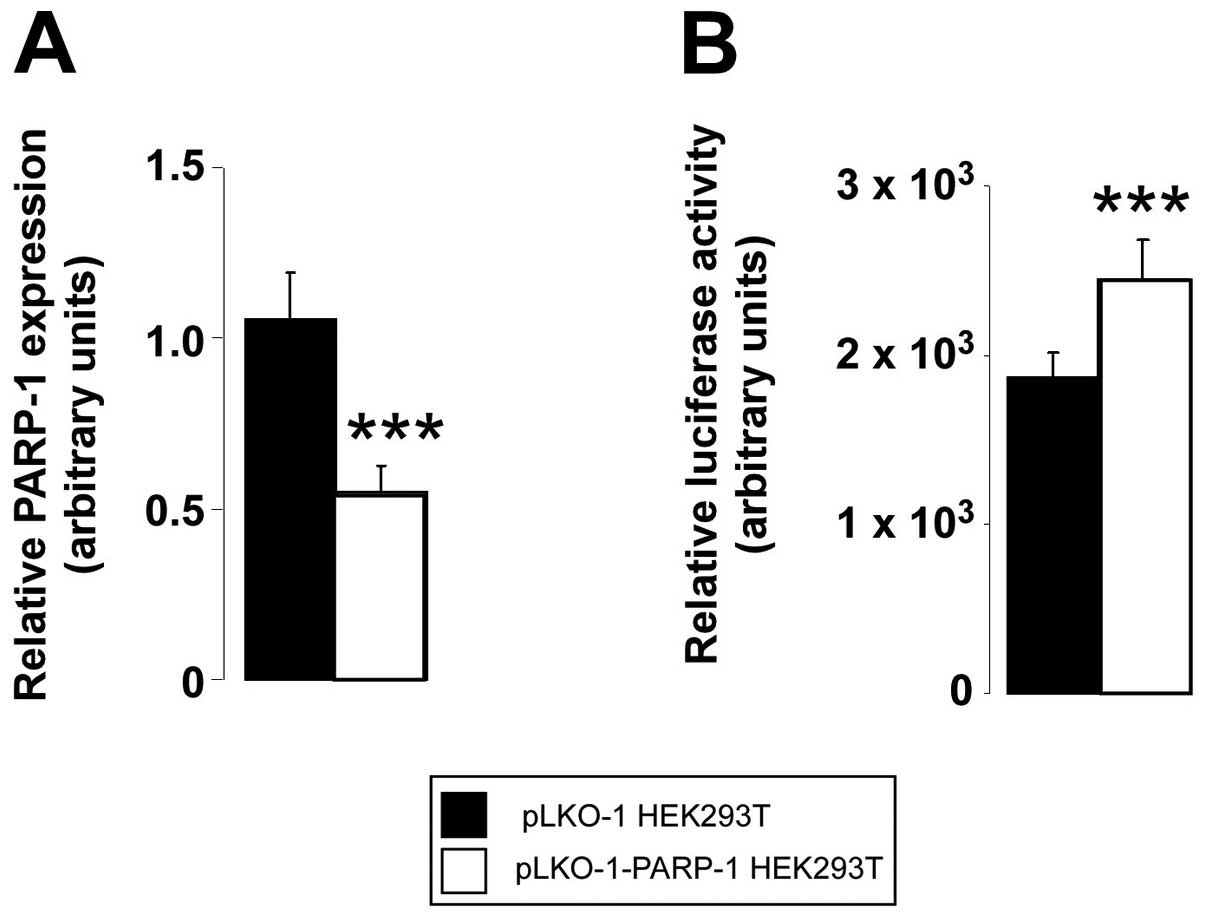
The qPCR detected higher mRNA expression levels of
fatty acid binding protein (FABP)7 in the PARP-1−/− mice
(Fig. 2). The depletion of PARP-1
expression (Fig. 3A) induced the
activity of the FABP7 promoter (Fig.
3B), suggesting that PARP-1 may have direct effects on the
FABP7 promoter.
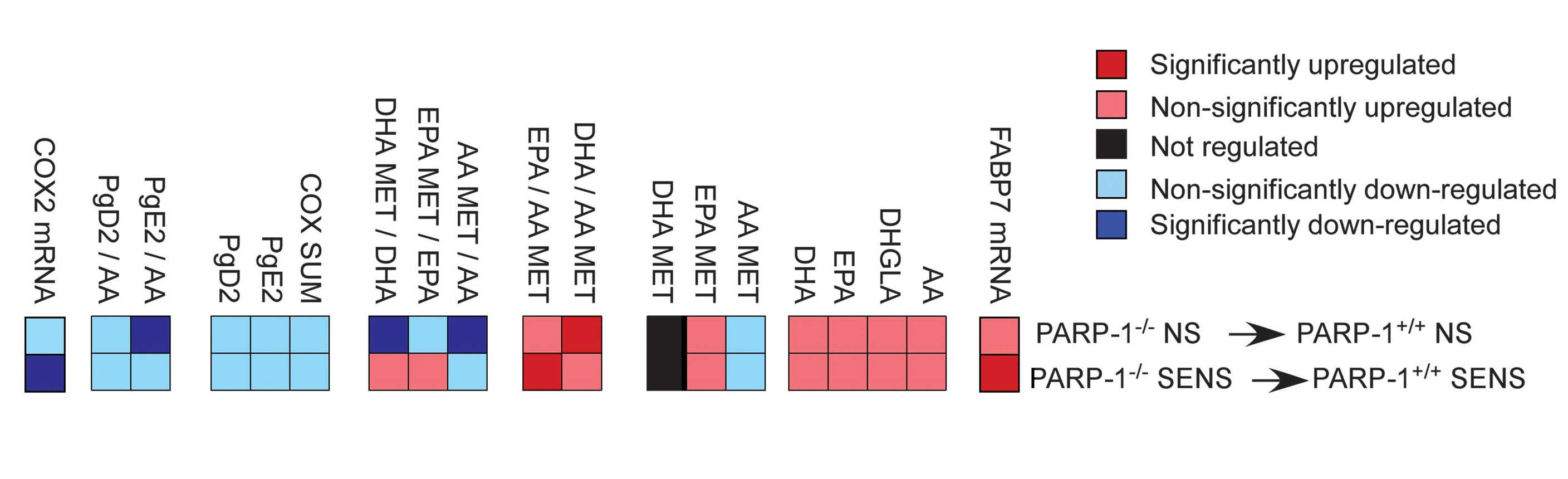 | Figure 2Eicosanoid and docosanoid signaling
alterations in the skin of poly(ADP-ribose) polymerase-1
(PARP-1)−/− mice, due to enhanced fatty acid binding
protein (FABP)7 expression. The FABP7 mRNA expression levels, and
eicosanoid and docosanoid concentrations, were determined in the
skin of control (PARP-1+/+ NS), OXA-sensitized
PARP-1+/+ (PARP-1+/+ SENS) and respective
PARP-1−/− mice (n=5/5/6/5). AA, arachidonic acid; DHGLA,
dihomo-gamma-linoleic acid; EPA, eicosapentaenoic acid; DHA,
docosahexaenoic acid; MET, metabolites; COX, cyclooxygenase; COX2,
cyclooxygenase 2; Pg, prostaglandin. |
Using a lipidomic approach, the composition of
eicosanoids and docosanoids in the skin was determined. There was
an increase in four of the major PUFAs, which are precursors of
eicosanoids and docosanoids: Arachidonic acid (AA; 20:4 n-6),
dihomo-gamma-linoleic acid (DHGLA; 20:3 n-6), docosahexaenoic acid
(DHA; 22:5 n-3) and eicosapentaenoic acid (EPA; 20:5 n-3). These
free fatty acids had increased levels in the skin of the control
and OXA-challenged PARP-1−/− mice, as compared with
their respective PARP-1+/+ cohorts (Fig. 2).
The levels of AA-metabolites (AA MET), as compared
with the mother compound AA (AA MET/AA), as well as EPA MET/EPA and
DHA MET/DHA, had lower ratios in the non-sensitized animals, and
the n3-PUFAs ratios were increased in the sensitized animals
(Fig. 2). The sum of the
cyclooxygenase (COX) metabolites (COX SUM), and the levels of COX
metabolites prostaglandin E2 (PgE2) and PgD2, as well as the
PgE2/AA and PgD2/AA ratios, were all decreased showing reduced
synthesis of pro-inflammatory lipid mediators. The levels of
anti-inflammatory or pro-resolving n3-PUFA metabolites were mainly
increased, resulting from EPA and DHA-metabolism (31) in the
PARP-1−/− animals (Fig. 2). These data are concordant with
the decreased expression levels of COX-2 in the
PARP-1−/− mice(Fig.
2).
Discussion
In the present study, it was determined that
ablation of PARP-1 expression had characteristic effects on skin
eicosanoid and docosanoid signaling, and metabolism. The two n-3
PUFAs (DHA and EPA), DHA- and EPA-metabolites, and the ratios of
DHA- and EPA-metabolites vs. AA-metabolites were increased,
indicating a pro-resolving, anti-inflammatory environment (31) in the PARP-1−/− mice,
especially following OXA-sensitization. The formation of the
pro-inflammatory COX-metabolites was lower in the skin of the
PARP-1−/− mice, which is concordant with a previously
determined anti-inflammatory environment (6). PARP-1 has previously been shown to
regulate COX-2 expression (32)
which may explain the observed decrease in COX-2 metabolites.
It may be suggested that increased FABP7-levels are
responsible for the observed alterations of lipid mediator
signaling. FABP7 has preference towards n-3, as compared with n-6,
PUFAs (33), this may be a
plausible explanation for the accumulation of EPA-derived
metabolites, and increased ratios of PUFA-metabolites (DHA/AA MET,
EPA/AA MET) in the PARP-1+/+ mice.
The results of the present study widen the spectrum
of altered lipid metabolism upon PARP-1 expression ablation
(34,35), by implicating skin eicosanoid and
docosanoid metabolism and signaling. A reduction in PARP-1
expression results in a reduced pro-inflammatory and increased
pro-resolving or anti-inflammatory environment in the skin. Higher
levels of the DHA and EPA metabolites, and reduced COX-pathway
metabolites were previously implicated in a reduction of
DNFB-induced CHS (36),
peritonitis (31), atopy (37) or allergy (38).
These data are concordant with our previous
findings, which showed that genetic or pharmacological inhibition
of PARP-1 is capable of protecting against CHS (7,26,39).
Notably, differences in eicosanoid and docosanoid metabolism and
signaling are also present in unchallenged mice and it may be
speculated that these minor changes may render PARP-1−/−
mice less susceptible to CHS. Furthermore, the changes to gene
expression levels and lipid mediator composition are associated
with inflammatory pathologies that are mediated not only by
T-helper (Th) 1 or Th2 cells, but also the newly defined Th17
cells, which have previously been associated with the induction of
FABPs in psoriasis (40). In
conclusion, alteration in skin eicosanoid and docosanoid metabolism
and signaling may modify the barrier function of the skin, wherein
the role of PARPs remains relatively unexplored.
Acknowledgements
The present study was supported by grants from the
National Innovation Office (Baross program Seahorse grant; no.
TÁMOP-4.2.2. A-11/1/KONV-2012-0025), OTKA (nos. PD83473, K105872,
K108308, CNK80709, K104720 and K108308), and the Medical and Health
Science Center (Mecenatura no. Mec-8/2011). PB and MS are
recipients of the Bolyai fellowship from the Hungarian Academy of
Sciences. The authors of the present study would like to
acknowledge the technical assistance of Mrs Erzsébet Herbály.
Abbreviations:
|
AA
|
arachidonic acid
|
|
CHS
|
contact hypersensitivity
|
|
COX
|
cyclooxygenase
|
|
DAPI
|
4,6-diamidino-2-phenylindole
|
|
DHA
|
docosahexaenoic acid
|
|
DHGLA
|
dihomo-γ-linoleic acid
|
|
EPA
|
eicosapentaenoic acid
|
|
FABP
|
fatty acid binding proteins
|
|
MET
|
metabolite
|
|
OXA
|
oxazolone
|
|
PARP
|
poly(ADP-ribose) polymerase
|
|
PUFAs
|
polyunsaturated fatty acids
|
|
Pg
|
prostaglandin
|
References
|
1
|
Amé JC, Spenlehauer C and de Murcia G: The
PARP superfamily. Bioessays. 26:882–893. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Szántó M, Brunyánszki A, Kiss B, et al:
Poly(ADP-ribose) polymerase-2: emerging transcriptional roles of a
DNA-repair protein. Cell Mol Life Sci. 69:4079–4092. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Mehrotra P, Hollenbeck A, Riley JP, et al:
Poly (ADP-ribose) polymerase 14 and its enzyme activity regulates
T(H)2 differentiation and allergic airway disease. J Allergy Clin
Immunol. 131:521–531. 2013. View Article : Google Scholar
|
|
4
|
Mehrotra P, Riley JP, Patel R, et al:
PARP-14 functions as a transcriptional switch for Stat6-dependent
gene activation. J Biol Chem. 286:1767–1776. 2011. View Article : Google Scholar :
|
|
5
|
Levaot N, Voytyuk O, Dimitriou I, et al:
Loss of Tankyrase-mediated destruction of 3BP2 is the underlying
pathogenic mechanism of cherubism. Cell. 147:1324–1339. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bai P and Virág L: Role of
poly(ADP-ribose) polymerases in the regulation of inflammatory
processes. FEBS Lett. 586:3771–3777. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Brunyánszki A, Hegedus C, Szántó M, et al:
Genetic ablation of PARP-1 protects against oxazolone-induced
contact hypersensitivity by modulating oxidative stress. J Invest
Dermatol. 130:2629–2637. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Virág L, Szabó E, Bakondi E, et al: Nitric
oxide-peroxynitrite-poly (ADP-ribose) polymerase pathway in the
skin. Exp Dermatol. 11:189–202. 2002. View Article : Google Scholar
|
|
9
|
Bakondi E, Gönczi M, Szabó E, et al: Role
of intracellular calcium mobilization and cell-density-dependent
signaling in oxidative-stress-induced cytotoxicity in HaCaT
keratinocytes. J Invest Dermatol. 121:88–95. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Selle A, Ullrich O, Harnacke K and Hass R:
Retrodifferentiation and rejuvenation of senescent monocytic cells
requires PARP-1. Exp Gerontol. 42:554–562. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Aldinucci A, Gerlini G, Fossati S, et al:
A key role for poly(ADP-ribose) polymerase-1 activity during human
dendritic cell maturation. J Immunol. 179:305–312. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mocchegiani E, Muzzioli M, Giacconi R, et
al: Metallothioneins/PARP-1/IL-6 interplay on natural killer cell
activity in elderly: parallelism with nonagenarians and old
infected humans. Effect of zinc supply. Mech Ageing Dev.
124:459–468. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zerfaoui M, Errami Y, Naura AS, et al:
Poly(ADP-ribose) polymerase-1 is a determining factor in
Crm1-mediated nuclear export and retention of p65 NF-kappa B upon
TLR4 stimulation. J Immunol. 185:1894–1902. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Rolli J, Rosenblatt-Velin N, Li J, et al:
Bacterial flagellin triggers cardiac innate immune responses and
acute contractile dysfunction. PLoS One. 5:e126872010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Eaves-Pyles T, Murthy K, Liaudet L, et al:
Flagellin, a novel mediator of Salmonella-induced epithelial
activation and systemic inflammation: I kappa B alpha degradation,
induction of nitric oxide synthase, induction of proinflammatory
mediators, and cardiovascular dysfunction. J Immunol.
166:1248–1260. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Liaudet L, Deb A, Pacher P, et al: The
Flagellin-TLR5 axis: Therapeutic opportunities. Drug News Perspect.
15:397–409. 2002. View Article : Google Scholar
|
|
17
|
Liaudet L, Murthy KG, Mabley JG, et al:
Comparison of inflammation, organ damage, and oxidant stress
induced by Salmonella enterica serovar Muenchen flagellin and
serovar Enteritidis lipopolysaccharide. Infect Immun. 70:192–198.
2002. View Article : Google Scholar
|
|
18
|
Liaudet L, Szabó C, Evgenov OV, et al:
Flagellin from gram-negative bacteria is a potent mediator of acute
pulmonary inflammation in sepsis. Shock. 19:131–137. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Murthy KG, Deb A, Goonesekera S, Szabó C
and Salzman AL: Identification of conserved domains in Salmonella
muenchen flagellin that are essential for its ability to activate
TLR5 and to induce an inflammatory response in vitro. J Biol Chem.
279:5667–5675. 2004. View Article : Google Scholar
|
|
20
|
Pacher P, Beckman JS and Liaudet L: Nitric
oxide and peroxynitrite in health and disease. Physiol Rev.
87:315–424. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Grabbe S and Schwarz T: Immunoregulatory
mechanisms involved in elicitation of allergic contact
hypersensitivity. Immunol Today. 19:37–44. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Grabbe S, Steinert M, Mahnke K, et al:
Dissection of antigenic and irritative effects of epicutaneously
applied haptens in mice. Evidence that not the antigenic component
but nonspecific proinflammatory effects of haptens determine the
concentration-dependent elicitation of allergic contact dermatitis.
J Clin Invest. 98:1158–1164. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Olmos A, Giner RM, Recio MC, et al:
Effects of plant alkylphenols on cytokine production, tyrosine
nitration and inflammatory damage in the efferent phase of contact
hypersensitivity. Br J Pharmacol. 152:366–373. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Haskó G, Mabley JG, Németh ZH, et al:
Poly(ADP-ribose) polymerase is a regulator of chemokine production:
relevance for the pathogenesis of shock and inflammation. Mol Med.
8:283–289. 2002.PubMed/NCBI
|
|
25
|
Soriano F, Virág L, Jagtap P, et al:
Diabetic endothelial dysfunction: the role of poly(ADP-ribose)
polymerase activation. Nat Med. 7:108–113. 2001. View Article : Google Scholar
|
|
26
|
Bai P, Hegedus C, Szabó E, et al:
Poly(ADP-ribose) polymerase mediates inflammation in a mouse model
of contact hypersensitivity. J Invest Dermatol. 129:234–238. 2009.
View Article : Google Scholar
|
|
27
|
de Murcia JM, Niedergang C, Trucco C,
Ricoul M, Dutrillaux B, Mark M, Oliver FJ, Masson M, Dierich A,
LeMeur M, Walztinger C, Chambon P and de Murcia G: Requirement of
poly(ADP-ribose) polymerase in recovery from DNA damage in mice and
in cells. Proc Natl Acad Sci USA. 94:7303–7307. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Szklenar M, Kalkowski J, Stangl V, Lorenz
M and Rühl R: Eicosanoids and docosanoids in plasma and aorta of
healthy and atherosclerotic rabbits. J Vasc Res. 50:372–382. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Rühl R: Method to determine 4-oxo-retinoic
acids, retinoic acids and retinol in serum and cell extracts by
liquid chromatography/diode-array detection atmospheric pressure
chemical ionisation tandem mass spectrometry. Rapid Commun Mass
Spectrom. 20:2497–2504. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Szabó E, Virág L, Bakondi E, et al:
Peroxynitrite production, DNA breakage, and poly(ADP-ribose)
polymerase activation in a mouse model of oxazolone-induced contact
hypersensitivity. J Invest Dermatol. 117:74–80. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Elabdeen HR, Mustafa M, Szklenar M, et al:
Ratio of pro-resolving and pro-inflammatory lipid mediator
precursors as potential markers for aggressive periodontitis. PLoS
One. 8:e708382013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lin Y, Tang X, Zhu Y, Shu T and Han X:
Identification of PARP-1 as one of the transcription factors
binding to the repressor element in the promoter region of COX-2.
Arch Biochem Biophys. 505:123–129. 2011. View Article : Google Scholar
|
|
33
|
Hanhoff T, Lücke C and Spener F: Insights
into binding of fatty acids by fatty acid binding proteins. Mol
Cell Biochem. 239:45–54. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Bai P and Cantó C: The role of PARP-1 and
PARP-2 enzymes in metabolic regulation and disease. Cell Metab.
16:290–295. 2012.PubMed/NCBI
|
|
35
|
Szántó M, Brunyánszki A, Márton J, et al:
Deletion of PARP-2 induces hepatic cholesterol accumulation and
decrease in HDL levels. Biochem Biophys Acta. 1842:594–602.
2014.
|
|
36
|
Tomobe YI, Morizawa K, Tsuchida M, et al:
Dietary docosahexaenoic acid suppresses inflammation and
immunoresponses in contact hypersensitivity reaction in mice.
Lipids. 35:61–69. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Abba C, Mussa PP, Vercelli A and Raviri G:
Essential fatty acids supplementation in different-stage atopic
dogs fed on a controlled diet. J Anim Physiol Anim Nutr (Berl).
89:203–207. 2005. View Article : Google Scholar
|
|
38
|
Rühl R, Koch C, Marosvölgyi T, et al:
Fatty acid composition of serum lipid classes in mice following
allergic sensitisation with or without dietary docosahexaenoic
acid-enriched fish oil substitution. Br J Nutr. 99:1239–1246. 2008.
View Article : Google Scholar
|
|
39
|
Virág L and Szabó C: The therapeutic
potential of poly(ADP-ribose) polymerase inhibitors. Pharmacol Rev.
54:375–429. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Madsen P, Rasmussen HH, Leffers H, Honoré
B and Celis JE: Molecular cloning and expression of a novel
keratinocyte protein (psoriasis-associated fatty acid-binding
protein [PA-FABP]) that is highly up-regulated in psoriatic skin
and that shares similarity to fatty acid-binding proteins. J Invest
Dermatol. 99:299–305. 1992. View Article : Google Scholar : PubMed/NCBI
|