Introduction
β-lactam is an effective antibiotic eliciting
marginally toxic effects. This antibiotic has been widely used for
a number of years. As such, certain bacteria produce hydrolases
that induce resistance. Numerous factors contributing to antibiotic
resistance have been determined; these factors include
Capnocytophaga species, one of the most frequently
identified organisms in the oral microbiome (1). A clinical survey has demonstrated
that the overall prevalence of extended-spectrum β-lactamase
(ESBL)-producing Klebsiella pneumoniae (K.
pneumoniae) is 11.8% (2).
AmpC β-lactamase belongs to group I and class C
according to the functional classification by Bush et al and
the molecular classification by Ambler et al, respectively
(3,4). Due to high AmpC enzyme expression
levels, bacteria are resistant to penicillin, cephalosporin
generations I, II and III, cephamycins, monobactams and enzyme
inhibitors (5); therefore,
numerous studies have focused on this enzyme.
AmpC β-lactamases may be divided according to
chromosome- and plasmid-mediated types. Although the mechanism of
AmpC enzyme synthesis remains unclear, this enzyme is possibly
regulated by an Amp composite operon, which is present in numerous
Gram-negative bacteria containing AmpC as a structural gene, as
well as AmpC, AmpR, AmpG, AmpD and AmpE as regulatory genes. The
AmpC gene codes for the AmpC enzyme, which is then regulated by
AmpR, AmpG, AmpD and AmpE (6–9).
Although the AmpC enzyme may be translated from the AmpC gene, this
enzyme has numerous mutations (10). AmpR and AmpC are arranged
contiguously and are transcribed reversely to produce an AmpR
protein; this protein is a negative regulatory factor of AmpC
(11). With the emergence of
β-lactamase antibiotics, AmpR activates AmpC to produce an AmpC
enzyme; otherwise, AmpR functions as an AmpC suppressor (6,11).
AmpG is a transmembrane protein, a permease, which is able to
transfer substances between the periplasmic space and the cytoplasm
to AmpR by other proteins; therefore, AmpR may also function as an
activator to promote AmpC (6).
AmpD is translated from AmpD as a new type of N-acetyl
glucosamine-L-alanine amidase. AmpD is a negative regulator of
β-lactamases and is located at the distal end next to the AmpE gene
(7,11,12).
AmpE is situated at the downstream region of AmpD, which is
expressed coordinately with AmpD; however, the function of this
gene in AmpC enzyme induction remains unclear (13).
Studies have demonstrated that strains expressing
the AmpC enzyme contain high levels of AmpD, AmpR and AmpG; AmpD is
a regulator of AmpC by AmpD protein, which negatively regulates the
AmpC enzyme (14,15). One cross-sectional study dealt with
the isolation of bacteria from critically ill patients who had
undergone tracheal intubation and mechanical ventilation; 32.9% of
the bacterial strains were Acinetobacter spp. and 25.1% were
K. pneumoniae, with Acinetobacter spp. the
predominant AmpC β-lactamase producer (16). AmpC enzymes that are produced in
plasmids also exhibit the same biochemical properties and
resistance as chromosome types, as well as the same active sites
identified by amino acid sequences analysis, including Ser-X-X-Lys
at position 64 (the essential site of serine), Lys-Ser/Thr-Gly at
position 315–317 (the essential site in active center tertiary
structure) and tyrosine residue at position 150 (one important site
of Thy-C-Asn in Class C enzyme that facilitates the hydrolysis of
β-lactamases). With the induction of AmpC enzyme in the plasmid,
certain cephalosporins cause resistance by inducing AmpC
expression. The resistance is associated with AmpC, AmpD, AmpE,
AmpR and AmpG, as chromosomal types; simultaneously, the resistance
became stronger with the multiplication of the resistant strains
that causes treatment failure clinically. The majority of the time,
the resistance genes are transformed between the chromosome and
plasmid by transposon, integron and insertion sequences (5); therefore, the resistance induced by
plasmid-mediated AmpC enzyme causes more harm than that of
chromosome-mediated AmpC.
Plasmid-mediated AmpC enzymes are commonly detected
in K. pneumoniae (17). In
a previous study, the levels of DHA-1 plasmid AmpC enzyme were
detected locally; the sequences of AmpC + AmpR (AmpCR), AmpD, AmpE
and AmpG genes in the DHA-1 plasmid of K. pneumoniae and the
effect of the AmpD repressor gene on pAmpC were also identified.
With the establishment of the pACYC184-X plasmid system, pAmpC
regulation was revealed; meanwhile, AmpD mutation in the
plasmid-mediated AmpC enzyme was detected by examining the sequence
of the structural and promoter genes, as well as the whole gene. To
understand the effect of AmpR on the AmpC enzyme, the
transcriptional activity, space structure and AmpIR sequence of Kp1
AmpR were detected, and the topological structure of AmpE and AmpG
were simultaneously predicted. In conclusion, the resistance of
strains that produce the AmpC enzyme was further understood, along
with the development of a theory regarding the plasmid Amp
operon.
Materials and methods
Strains and plasmid
Ten K. pneumoniae strains of DHA-1 plasmid
(i.e., Kp1, Kp4, Kp11, Kp13, Kp17, Kp18, Kp19, Kp20, Kp21 and Kp22)
were collected by the laboratory of the Second Affiliated Hospital
of Harbin Medical University (Harbin, China), but only Kp1 and Kp17
were selected for AmpD and AmpR regulation. The plasmids used were
the following: K. pneumoniae-sensitive strains (Kps) that
were collected from the laboratory of the Second Affiliated
Hospital of Harbin Medical University, standard strains of
Escherichia coli (E. coli) DH5α (Shanghai Shenyang
BoCai Company, Shanghai, China), depressed sustained
high-expressing AmpC Enterobacter cloacae (E. cloacae
029M; Zhejiang University, Hangzhou, Zhejiang, China), pGEM-T Easy
vector plasmid (Promega Corporation, Madison, WI, USA), pACYC184
plasmid expression vector (product of NEB company, collected by
Qingdao Marine Research Centre, Qingdao, Shangdong, China) and
pKK232-8 promoter probe plasmid (Megiddo Bio-Pharm Technology Co.,
Ltd., Shanghai, China).
Antibiotic susceptibility of strains
All strains were analyzed separately using the disk
diffusion method (Kirby-Bauer). Monoclonal colonies were mixed to
form a solution with 0.5 MCF concentration (1.5×108
CFU/ml). The solution was placed on an MH agar plate using a
sterile cotton swab, with cefoxitin (FOX) as an inducer in the
center; piperacilin (PRL), ceftazidime (CAZ), cefotaxime (CTX),
cefepime (CEP), cefoperazone/sulbactam (SCF), imipenem (IPM),
piperacillin/tazobactam (TZP) and aztreonam (ATM) were placed at a
distance of 20–22 mm from the center of the FOX disk. Following
this, the MH agar plates were incubated at 35°C for 18–20 h to
observe the bacterial inhibition zone diameters.
Phenotype examination of resistant
strains
Complex phenotype analysis was designed in reference
to Moland et al (18,19).
ESBL and AmpC induction may be observed on the same plate
simultaneously. FOX and AMC were induced at the center, whereas
PRL, CAZ, cefepim (FEP), CTX, ATM, TZP and ceftriaxone (CRO)
surrounded the periphery and were distributed evenly. The plate was
cultured at 35°C for 18–20 h. E. coli ATCC25922 was selected
as the negative control strain and E. cloacae 029 was
selected as the inducible AmpC enzyme positive control strain. The
results were categorized according to the following criteria:
ESBL-positive (the bacteriostasis circle close to the AMC side
expanded or widened), inducible AmpC enzymes (the bacteriostasis
circle around FOX or close to FOX became flat or depressed) and
sustainable yield AmpC enzyme (the edge of the bacteriostasis
circle became smooth).
Amplification of chromosomal DNA and
preparation of the plasmid template
The stains were inoculated on blood agar following
isolation and were cultured overnight at 37°C. A single colony was
selected to inoculate the 5 ml LB liquid medium of FOX (with a
final concentration of 16 μg/ml), then incubated in a shaking water
bath at 200 r/min and 37°C for 18–20 h. Subsequently, the
chromosomal DNA was extracted from 3 ml culture medium, and the
plasmid DNA was extracted from 1–2 ml culture medium. The procedure
was performed according to the manufacturer’s instructions of the
E.Z.N.A.® bacterial DNA kit and the E.Z.N.A. plasmid
mini kit I (Omega Bio-Tek Inc., Norcross, GA, USA),
respectively.
Sequence analysis of the AmpC enzyme
gene
For the AmpCR, AmpD, AmpE and AmpG genes, the
specific primers were designed in the DHA-1 plasmid AmpC enzyme
K. pneumoniae, according to the Morganella morganii
(M. morganii) strain on GenBank bacteria (AmpCR), K.
pneumoniae MTUH-K2044 (AmpD) and K. pneumoniae AP006725
(AmpE and AmpG) (Table I). The
plasmid AmpCR gene and chromosomal AmpD, AmpE and AmpG genes were
amplified and purified by polymerase chain reaction (PCR) using
primers P1–P2, P3–P4, KPGF-KPGR and KPEF-KPER, shown in Table I. AmpCR and AmpD PCR amplification
were performed for 30 cycles of 10 sec each denaturation at 98°C,
annealing at 56°C for 5 sec, extension at 72°C for 2 min and 30 sec
and a final extension at 72°C for 7 min. AmpG PCR amplification was
performed for initial denaturation at 94°C for 5 min, 35 cycles of
30 sec each denaturation at 94°C, annealing at 51°C for 30 sec,
extension at 72°C for 30 sec and a final extension at 72°C for 7
min. AmpE PCR amplification was performed for initial denaturation
at 95°C for 4 min, 30 cycles of 30 sec each denaturation at 95°C,
annealing at 58°C for 30 sec, extension at 72°C for 30 sec and a
final extension at 72°C for 7 min. The products of AmpCR (AmpC +
AmpR + AmpIR) and AmpD were conjugated to the pGEM-T Easy carrier
and transformed into E. coli DH5α-competent cells. The
recombinant colonies were screened by a blue-white plaque selection
to examine the sequences of the recombinants and compare these
colonies with those of the M. morganii strain. The sequences
of the AmpE and AmpG genes were compared with those of the AmpE and
AmpG genes in the antibiotic-sensitive Kp (Kps), and to detect
AmpCR, AmpD, AmpE and AmpG genes in the DHA-1 plasmid AmpC enzymes.
All sequence analysis was conducted by Beijing Sunbiotech Co., Ltd.
(Beijing, China), and the genes were all compared with the
sequences in GenBank by Blast (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
 | Table IPrimers, plasmid vectors and cloning
strains used in this study. |
Table I
Primers, plasmid vectors and cloning
strains used in this study.
| Gene | Primer | Length (bp) | GenBank no. | Cloning vector | Cloning
strains |
|---|
| AmpCR | P1 5′
CCCAAGCTTGGGAGGTGAGTGAGTTTTA CG 3′ | 2767 | AF055067 | pGEM-T
Easy/pACYC184 | E. coli
DH5α |
|
HindIII | | Morganella
morganii | | |
| P2 5′
GGGCTCTAGAGCGAAAAAATTATTCCAGTGCAC 3′ | | | | |
| XbaI | | | | |
| AmpIR | P5 5′
GTCTGACCATAATCCACCTGT 3′ | 159 | AF055067 | pGEM-T Easy | E. coli
DH5α |
| P6 5′
CAGAGCGGAAATCAGTGTTG 3′ | | Morganella
morganii | | |
| AmpD | P3 5′
GATAAGCTTCAGAAGTGGGAAACATGC 3′ | 622 | NC009648 | pGEM-T
Easy/pACYC184 | E. cloa
029M |
|
HindIII | | Klebsiella
pneumoniae | | |
| P4 5′
AGCGTCTAGAACAGCGTCATGAGGT 3′ | | | | |
| XbaI | | | | |
| AmpDP | P7 5′ CAGA
GGATCC GTTCCAGCAGCACGTCAC 3′ | 330 | AP006725 | pKK232-8 | E. coli
DH5α |
| BamHI | | Klebsiella
pneumoniae | | |
| P8 5′ GCTC
AAGCTT AATCCACGGACCACCAAA 3′ | | | | |
|
HindIII | | | | |
| AmpG | KP-G F
5′-CGGGATTGATAACGAAAGG-3′ | 1476 | AP006725 | | |
| KP-G R
5′-CCGATATGGCGCAGGACA-3′ | | Klebsiella
pneumoniae | | |
| AmpE | KP-E F
5′-GGAAAACCGATCCAGGACCG-3′ | 1059 | AP006725 | | |
| KP-E R
5′-TCAGGCAACAAAAGCCCCGC-3′ | | Klebsiella
pneumoniae | | |
The chromosomal AmpD structural gene was amplified
by PCR and conjugated to the pGEM-T Easy carrier to form pT-AmpD,
whose sequence was also detected. The site of the AmpD gene
promoter was predicted by Softberry Fgenesh (http://www.softberry.com) for sequence analysis. The
AmpDP PCR procedure was performed with initial denaturation at 94°C
for 5 min, 30 cycles of 30 sec each denaturation at 94°C, annealing
at 62°C for 30 sec, extension at 72°C for 45 sec and a final
extension at 72°C for 5 min. A recombinant carrier, pK-AmpDP, was
identified by the conjugation of the promoter sequence to pKK232-8
promoter probe plasmid, and transformed into E. coli DH5α,
which was selected from the double-resistant plates of ampicillin
and chloramphenicol.
Effect of exogenous AmpCR and AmpD on
AmpC enzyme induction of host bacteria
The AmpCR gene was cloned into pACYC184 carrier in
Kp1 and Kp17 strains to form the recombinant plasmid
pACYC184-AmpCR. The plasmid was then transformed into E.
coli DH5α (AmpCR−/AmpD+) to evaluate the
AmpC enzyme expression in recombinant E-AmpCR with the exogenous
AmpCR induced into the AmpCR−/AmpD+ strain.
The AmpD gene was cloned into the pACYC184 carrier to form the
recombinant plasmid pACYC184-AmpD. The plasmid was then transformed
into E. cloacae 029M (AmpCR+/AmpD−)
with the a high level of AmpC expression sustained, due to the AmpD
mutation to examine the AmpC enzyme expression in recombinant
Ea-AmpD with the exogenous AmpD+ induced into the
AmpCR+/AmpD− strain. The impact on AmpC
enzyme in the host strain by recombinant plasmid pACYC184-AmpCR and
pACYC184-AmpD was evaluated by examination of the induced
resistance phenotype and the FOX minimum inhibitory concentration
(MIC). All primers, cloning carriers and cloning strains are listed
in Table I.
Electromobility shift assay (EMSA) of
AmpIR and AmpR
The interaction between AmpIR and AmpR was detected
using EMSA with the mark of biotin probes and chemiluminescent
detection technology in Kp1 (DHA-1 plasmid AmpC enzyme K.
pneumoniae) and purified recombinant AmpR. The AmpC-R
(including AmpR, AmpIR and AmpC) was amplified using PCR using
primers P1 and P2, shown in Table
I, and conjugated with the pGEM-T Easy carrier, prior to
sequencing using an ABI Prism3100 automated DNA sequencer (Applied
Biosystems Life Technologies, Foster City, CA, USA). The nucleotide
and amino acid sequences were analyzed using the BLAST program of
the NCBI. The 3D structure of AmpR was established according to the
sequence of amino acids, with Pseudomonas aeruginosa (P.
aeruginosa) transcriptional regulator PAO477 as the template by
Swiss-Model server (http://www.swissmodel.expasy.org) and SPdviewer 4.0.1
(http://www.swissmodel.expasy.org).
Topology prediction analysis of AmpE and
AmpG
The transmembrane domains of all DHA-1 K.
pneumoniae and Kps were predicted by SOSUI engine ver. 1.1
(http://bp.nuap.nagoya-u.ac.jp/sosui/sosui_submit.html).
The AmpE and AmpG transmembrane domains in the experiment and
sensitive strains were also analyzed, as well as the functions of
these enzymes.
Ethical approval
The study was approved by the ethics committee of
the Second Affiliated Hospital of Harbin Medical University.
Results
Analysis of Amp protein expression and
gene mutation
All the electrophoretic bands were at their expected
positions following amplification by PCR with the specific
primers.
Mutations and amino acid changes were analyzed by
comparing them with the corresponding sequences of M.
morganii (Genbank No.AF055067) and K. pneumoniae strains
that were sensitive to all the antibiotics tested, using Align of
the BLAST program. All ten revealed the same AmpR mutations (Thr114
→Ala). AmpG mutations were identified in all K. pneumoniae
strains compared with the strains that were sensitive to all the
antibiotics. The changes in the amino acids were: i) Silent
mutations: Kp20, Kp21 and Kp22; ii) site 97 alanine mutated to
threonine (Ala97→Thr): Kp1, Kp4; iii) isoleucine transformed into
leucine (Ile19→Leu); iv) site 97 alanine mutated to threonine
(Ala97→Thr): Kp11, Kp13, Kp17, Kp18 and Kp19 (Table II).
 | Table IIMutations of AmpC, AmpR, AmpD, AmpG
and AmpE. |
Table II
Mutations of AmpC, AmpR, AmpD, AmpG
and AmpE.
| Strain | Phenotype | AmpR (compared with
Morganella morganii AF055067) | AmpC (compared with
Morganella morganii AF055067) | AmpD (compared with
Klebsiella pneumoniae NC009648) | AmpE (compared with
cefoxitin-susceptible Kps strains) | AmpG (compared with
cefoxitin-susceptible Kps strains) | Cefoxitin MIC
(μg/ml) |
|---|
| KP1 |
IB;ESBL− | Thr114→Ala | None | None | Glu20→Gly,
Ala31→Val, Val93→Ala, 465 deletion C | Ala97→Thr | 64 |
| KP4 |
IB;ESBL+ | Thr114→Ala | None | None | Glu20→Gly,
Ala31→Val, Val93→Ala, 465 deletion C | Ala97→Thr | 64 |
| KP11 |
IB;ESBL+ | Thr114→Ala | None | Leu143→Ile | Val93→Ala | Ile19→Leu,
Ala97→Thr | 64 |
| KP13 |
IB;ESBL+ | Thr114→Ala | None | Leu143→Ile | Ala31→Val,
Val93→Ala | Ile19→Leu,
Ala97→Thr | 1024 |
| KP17 |
IB;ESBL+ |
Thr114→Ala
Val72→Ala | Leu104→Pro | Leu143→Ile | Ala31→Val,
Val93→Ala | Ile19→Leu,
Ala97→Thr | 512 |
| KP18 |
IB;ESBL+ | Thr114→Ala | None | Leu143→Ile | Ala31→Val,
Val93→Ala | Ile19→Leu,
Ala97→Thr | 1024 |
| KP19 |
IB;ESBL+ | Thr114→Ala | None | Leu143→Ile | Ala31→Val,
Val93→Ala | Ile19→Leu,
Ala97→Thr | 1024 |
| KP20 |
IB;ESBL− | Thr114→Ala | None | None | Val93→Ala | None | 64 |
| KP21 |
IB;ESBL+ | Thr114→Ala | None | None | Ala31→Val,
Val93→Ala | None | 64 |
| KP22 |
IB;ESBL+ | Thr114→Ala | None | None | Val93→Ala | None | 64 |
Sequence alignment and location of AmpD
promoter
The DNA fragment, amplified by PCR of the AmpD
promoter with a length of ~330 bp, was linked to the pKK232-8 probe
plasmid with the CAT gene and was transfected into E. coli
DH5α-competent cells. The -10 box of the AmpD promoter in K.
pneumoniae was located upstream, at the -19 to -24 position,
and the sequence was TGAGTT (as predicted by Softberry Fgenesh gene
finder). Additionally, the -35 box was located upstream, at
approximately -53 to -58, and the sequence was TTCATC. No
nucleotide mutation was found in the internal sequence between the
structural and promoter genes of AmpD K. pneumoniae compared
with the FOX-susceptible strains, whereas the mutation at the
structural and promoter genes were the same.
FOX-resistant phenotype and MIC of
recombinant E-ampCR1 and E-ampCR17
Host strain E. coli DH5α was initially
sensitive to all of the antibiotics, and marked resistance was
induced by the transfection of exogenous AmpCR. E-AmpCR1 and
E-AmpCR17 (pAC YC184-AmpCR) recombinants exhibited inducible
resistance to all tested antibiotics, wherein the bacterial
inhibition zone was flattened with the FOX or IPM disk as an
inducer in the plate center. The MIC value of FOX increased by
eight-fold in E-AmpCR1 and E-AmpCR17 compared with the host strain
E. coli DH5α, from 16 to 128 μg/ml, and the MIC value
demonstrated no change when the free conversion of E-pACYC184
vector plasmid was the recombinant. Therefore, the susceptibility
of the host strain was not affected by the pACYC184 vector
(Table III).
 | Table IIICefoxitin MIC in recombinant and
strains. |
Table III
Cefoxitin MIC in recombinant and
strains.
| Strain | Plasmid
features | Cefoxitin MIC
(μg/ml) | Resistance
phenotype |
|---|
| E. coli
DH5α | AmpCR−,
AmpD+ | <16 | None |
| E. cloacae
029M | AmpCR+,
AmpD− | >1024 | Over
expression |
| Kp1 | AmpCR+,
AmpD+ | 256 | Induced |
| Kp17 | AmpCR+,
AmpD+ | 512 | Induced |
|
E-pACYC184 | pACYC184 | <16 | None |
| E-AmpCR1 |
pACYC184-AmpCR1 | 128 | Induced |
| E-AmpCR17 |
pACYC184-AmpCR17 | 128 | Induced |
| Ea-AmpD1 | pAC
YC184-AmpD1 | 256 | Induced |
| Ea-AmpD17 | pAC
YC184-AmpD17 | 256 | Induced |
Comparison of repressor gene sequences in
AmpD-induced strains by PCR and the enzyme digestion of cloning
vectors
The nucleotide and amino acid homology of the AmpD
gene with Kp1 and Kp sensitivity were both 100%; while the
nucleotide and amino acid homology with Kp17 were both 99%; the 143
leucine changed to isoleucine (Leu→Ile; data not shown).
Structure and resistant phenotype of the
pACYC184-AmpD plasmid vector
The AmpD of Kp1 and Kp17 was linked to XbaI
(at position 1425) and HindIII (at position 1524), and the
restriction sites of the pACYC184 plasmid vector, respectively;
therefore, the plasmid that carried AmpD became the repressor of
AmpC β-lactamase. The Ea-AmpD1 and Ea-AmpD17 (pAC YC184-AmpD)
recombinants revealed an inducible resistance to CAZ when FOX or
IPM were present as inducers. The MIC of FOX on Ea-AmpD1 and
Ea-AmpD17 decreased to 256 μg/ml, whereas the MIC of the E.
cloacae 029M host strain was >1,024 μg/ml. Therefore, the
AmpD gene, from K. pneumoniae, has a function in partial
compensation (Table III).
Alignment analysis of the AmpIR gene and
structural modeling of the AmpR protein
The DNA-protein interaction system, which was
established by an AmpIR probe (biotin-labeled and unlabeled
competing probe) and recombinant protein AmpR of K.
pneumoniae, demonstrated that the AmpR protein would bind to
the probe specifically, and a significant retardation phenomenon
would be observed. The suppression was more evident with the
increase in protein concentration (Fig. 1). The AmpCR sequence was 2186 bp,
amplified with primers P1 and P2, and the GenBank number was
HM568877.1. This sequence contains AmpR, AmpC and AmpIR genes,
which are the intercistronic sequences of the AmpC and AmpR genes.
Using Blast in NCBI, the homology of AmpIR and the M.
morganii strain (GenBank no. AF055067) was identified to be
97%, with three base mutations: 915T→A, 933T→C and 961C→T.
Similarity was observed between the DNA biting site of the 38-bp
AmpR protein in the Kp1 plasmid and the AmpIR, with a T-N11-A
sequence and a unique palindromic region. AmpR protein, which
comprises 291 amino acids (EU476911.1), demonstrated one amino acid
mutation (114T→A) when compared with the M. morganii strain
(AF055067). The structure of AmpR protein was modeled with P.
aeruginosa PAO477 regulatory protein (PDB: 2esnB) as the
homology template, and was modified by Swiss-Model server and
associated software. A helix-turn-helix domain was identified at
the AmpR amino terminus, and an auxiliary inducer binding domain
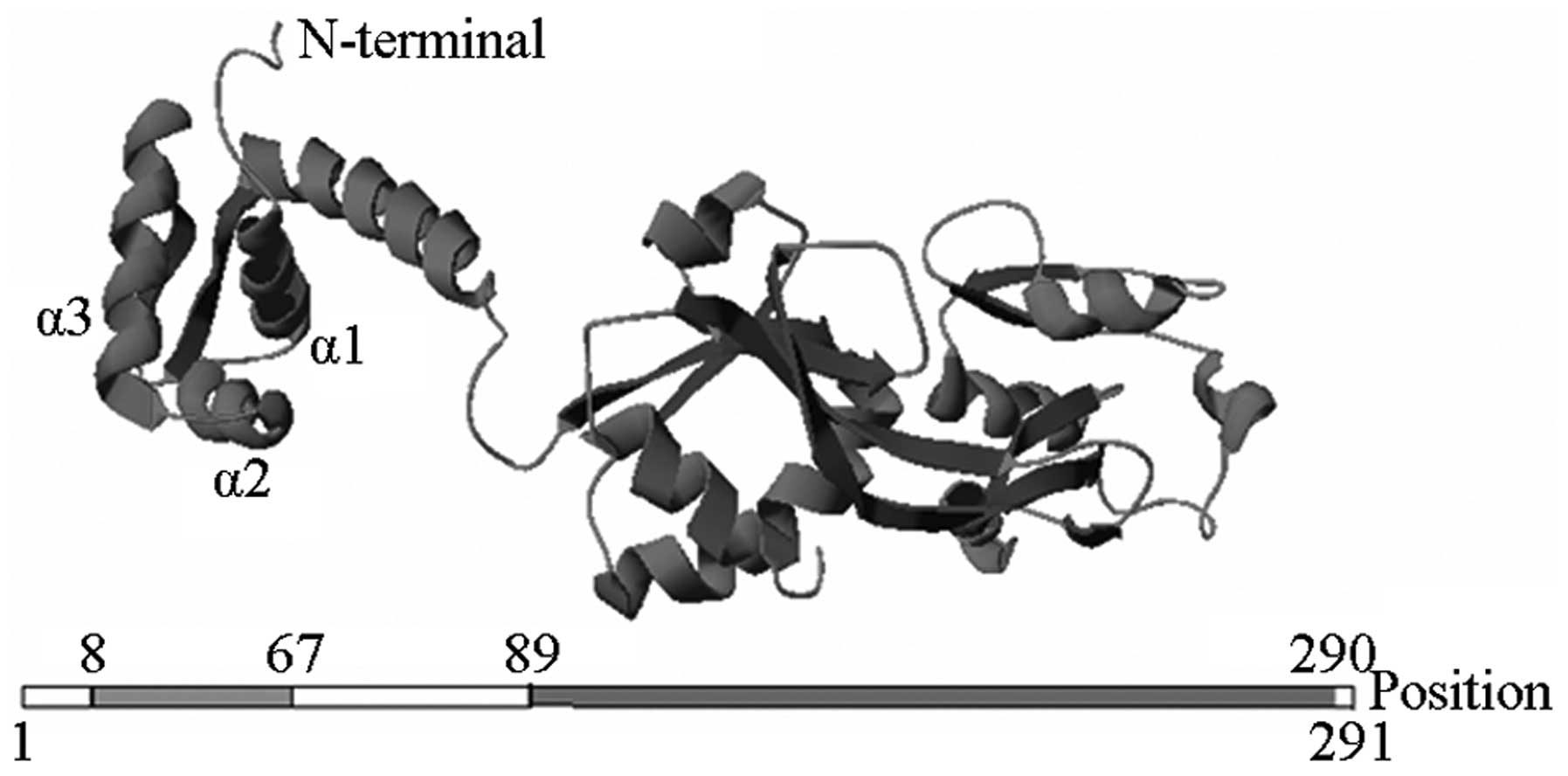
was found at the carboxyl terminus (Fig. 2).
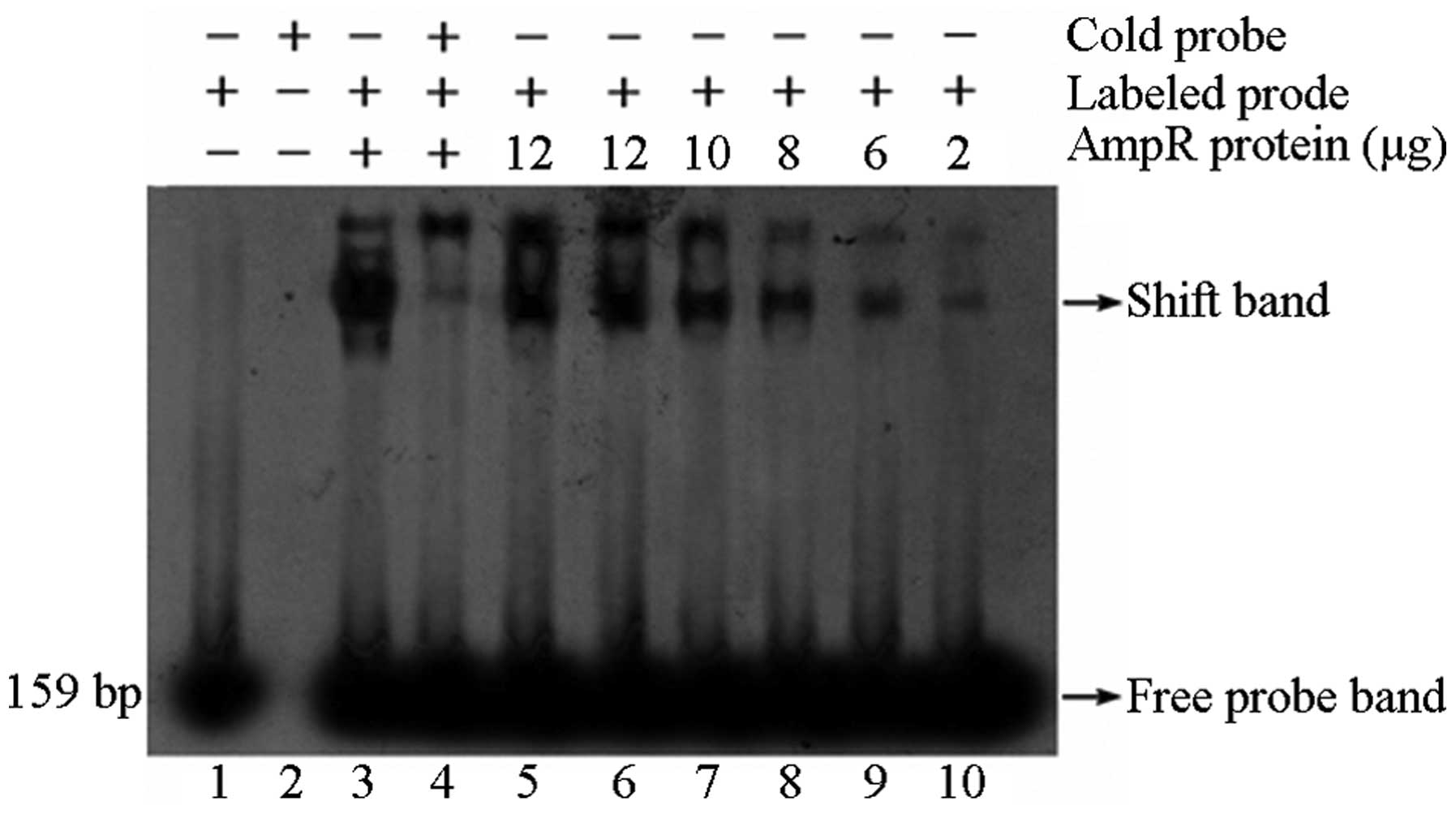 | Figure 1Retarded band of AmpIR probe and AmpR
recombinant protein. The 159-bp AmpIR probe joined with the AmpR
recombinant protein to form the retardation band, which was
inhibited by the cold probe (with a concentration 100-fold that of
the labeled probe). Lane 1, labeled probe; lane 2, unlabeled probe;
lane 3, labeled probe + AmpR protein; lane 4, unlabeled probe +
labeled probe + AmpR protein; lane 5–10, labeled probe + AmpR
protein (12, 12, 10, 8, 6 and 2 μg). |
Prediction of transmembrane domain of
AmpG in DHA-1 plasmid AmpC enzyme K. pneumoniae
Predictions were performed using Expasy online
software SOSUI engine Twelve transmembrane domains were identified
in the protein. The N-terminus and C-terminus were exposed outside
of the membrane. The transmembrane domains were located at
positions 11–33, 48–70, 82–104, 144–166, 174–196, 225–247, 278–300,
321–343, 352–374, 383–405, 426–448 and 461–483 (Table IV). The Ile19→Leu, Ala 97→Thr,
Val441→Ala and Ala444→Gly mutations were found in the transmembrane
domains (Table IV).
 | Table IVPrediction of the AmpG transmembrane
regions of Klebsiella pneumoniae encoding DHA-1-type
plasmid-mediated AmpC β-lactamase. |
Table IV
Prediction of the AmpG transmembrane
regions of Klebsiella pneumoniae encoding DHA-1-type
plasmid-mediated AmpC β-lactamase.
| Transmembrane
domain | N-terminal | Sequence of
transmembrane domain | C-terminal | Length (AA) |
|---|
| 1 | 11 |
QPKSAILLILGFASGLPLALTSG | 33 | 23 |
| 2 | 48 |
TIGFFSLVGQAYVFKFLWSPLMD | 70 | 23 |
| 3 | 82 |
GWLLTTQVLLLLAIAAMGFLEPV | 104 | 23 |
| 4 | 144 |
GAAISVLGYRLGMLVSGGLALWL | 166 | 23 |
| 5 | 174 |
QGMYWLMAALLVPCIIATLLAPE | 196 | 23 |
| 6 | 225 |
WLILLLIVLYKLGDAFAMSLTTT | 247 | 23 |
| 7 | 278 |
YGGVLMQRLTLFRALLIFGLLQG | 300 | 23 |
| 8 | 321 |
TAVFFENLCGGMGTAAFVALLMT | 343 | 23 |
| 9 | 352 |
TQFALLSALSAVGRVYVGPIAGW | 374 | 23 |
| 10 | 383 |
TFYLFSVVAAVPGIALLLLCRQT | 405 | 23 |
| 11 | 426 |
ALALGILTLGCLLLAVWLALLIL | 448 | 23 |
| 12 | 461 |
GLLEVAALTAVGGILFGGLLDYL | 483 | 23 |
Prediction of transmembrane domain of
AmpE in DHA-1 plasmid AmpC enzyme K. pneumoniae
The transmembrane domains of AmpE were predicted by
SOSUI and six transmembrane domains in the strains were found.
However, Kp1 and Kp4 had a frameshift mutation (155) caused by a
base deletion (465 ΔC) and, therefore, only three transmembrane
domains were identified (Table
V).
 | Table VPrediction of the AmpE transmembrane
regions of Klebsiella pneumoniae encoding DHA-1-type
plasmid-mediated AmpC β-lactamase. |
Table V
Prediction of the AmpE transmembrane
regions of Klebsiella pneumoniae encoding DHA-1-type
plasmid-mediated AmpC β-lactamase.
| Transmembrane
domain | N-terminal | Sequence of
transmembrane domain | C-terminal | Length (AA) |
|---|
| 1 | 1 |
MTLFTTLLVLIAERLFKLGEHWQ | 23 | 23 |
| 2 | 38 |
FSLPGTLLMTLVAVAVVYIIQRL | 60 | 23 |
| 3 | 65 |
LFNIPLLVFWILLGLLCIGAGK | 86 | 23 |
| 4 | 149 |
FWFVVGGAWGPLTLIAYSVLRAW | 171 | 23 |
| 5 | 189 |
IDGILHIVDWLPVRLVGVVYALV | 211 | 23 |
| 6 | 266 |
TSLVVVVIMALLTIYGTLV | 284 | 19 |
Discussion
K. pneumoniae is one of the most important
pathogens that causes clinical infections and nosocomial
infections. The wild type of K. pneumoniae is resistant to
ampicillin. With the abuse of antibiotics, such as cephalosporins
III, bacteria produce ESBLs, which may be suppressed by β-lactamase
inhibitors, such as clavulanic acid (20). In the present study, the gene and
amino acid mutations of AmpCR, AmpD, AmpE and AmpG of the DHA-1
plasmid AmpC enzyme were observed in ten K. pneumoniae
strains. A high homology was found in the base sequence of AmpCR
between the plasmid and chromosome types; and the conserved region
of AmpR and its cistron fragment was notably longer than that for
AmpC. The interval region of the cistron in the plasmid contained
AmpR and AmpC promoters, and a number of the base pairs belonged to
both of the promoters, which coincides with the regulation of the
AmpC enzyme chromosome (21,22)
and plasmids (23). High homology
was noted between the interval region of the cistron in the AmpC of
the Kp strain and chromosome of M. morganii bacteria, where
DNA binding sites of AmpR protein that contains T-N11-A sequence
and palindrome structure are found. AmpR belongs to LysR regulatory
protein family, which is one of the transcription factors of AmpC
enzymes (24) that may inhibit
AmpC synthesis when β-lactamase enzyme is absent, and promote AmpC
synthesis, with β-lactam antibiotics as the inducer. To gain
structural insights into AmpR regulation, a crystal structure of
the effector binding domain (EBD) was determined; the base of the
interdomain pocket of this crystal structure inhibited AmpR from
inducing AmpC (Thr103Val, Ser221Ala and Tyr264Phe) or resulted in
constitutive AmpC expression (Gly102Glu) (25). The resistance gene may be
transfected by transposons, integrons and insertion sequences, for
example, from a chromosome to the plasmid or a plasmid to a
plasmid. For example, in producing a plasmid-mediated CFE-1 type
AmpC enzyme in E. coli KU640, an insertion sequence 26
(IS26) was found in addition to AmpR and AmpC, which was able to
transfer the Amp gene (26). From
the FOX-resistant K. pneumoniae strains, the majority of
AmpC genes were Miriam or DHA type, which transfer the resistance
carried by the plasmid to the recipient strain by conjugation
(27).
The resistance to drugs that is spread between
bacterial species by plasmids was the most important mechanism, and
marginal differences were noted on the sites and functions of the
genes, either encoded by plasmid or chromosome; however, the
observations remained different among species (28). For example, the DHA-2-type AmpC
enzyme plasmid-mediated K. pneumoniae is the point-mutant
strain of the DHA-1 type AmpC enzyme gene, and has a 99% homology
with M. morganii (29). In
the present study, EMSA demonstrated that AmpR binding with AmpIR
is the transcription factor of the plasmid AmpC enzyme. The
structure of the AmpR protein is similar to the LysR family with a
helix-turn-helix domain at the N terminus, which was also
constructed in the three-dimensional model of AmpR.
No deletion mutations of the regulatory gene were
observed among the 10 strains in the present study, including AmpD,
AmpE and AmpG. No AmpD or AmpG mutation was observed in some
strains that were used in the present study; however, all of the
strains with AmpE mutation were compared with the FOX-susceptible
strains. Although mutation occurred in the strains, no strain was
identified with the phenotype and MIC of FOX.
AmpD is a cytoplasmic peptidoglycan amidase that is
involved in baterial cell-wall recycling and is a key enzyme in
β-lactamase induction (30). AmpD
represses the inducible expression of the AmpC enzyme. Mutations in
the promoter or the structural gene of AmpD are able to abrogate
the repression of AmpC enzyme transcription, and inactivation of
AmpD significantly increases AmpC expression. The role of AmpD in
entry into the cell wall, recycling events and in reversal of the
induction production of β-lactamase has been proposed as a possible
factor in the development of antibiotic resistance (31). At present, the chromosomal AmpD
gene was found in K. pneumonia strains. The effect of
AmpD on the inducible expression in the presence of DHA-1 type AmpC
enzyme with AmpR was examined with the pACYC184-X plasmid
evaluation system. Transformation resulted in a decrease in
cefoxitin MICs and an inducible phenotype of β-lactams. Thus, the
AmpD gene of the Kp1 strain may complement the AmpD mutant
background of E. cloacae 029M, in part. These findings
highlight the importance of AmpD in the regulation of the K.
pneumonia AmpC cephalosporinase production. The location of the
AmpD promoter region remains unknown. The present study identified
putative promoter -10 box and -35 box regions located −19 – −24 bp
and −53 – 58 bp upstream of AmpD using Softberry FGENESH online, by
constructing the recombinant carrier pK-AmpDP which contained the
upstream nucleotide sequence of AmpD.
Currently, the majority of studies regarding AmpE
focus on the polymorphism, topology prediction and polar
characteristics, but not on the function of the gene. AmpE is an
intracellular membrane protein composed of 284 amino acids, with a
molecular weight of 31.2 kDa. The transmembrane domain suggests
that AmpE is a receptor on the membrane, such as anchored protein
or ion channel protein (32). In
1989, Lindquist et al (33)
found that AmpE was adjacent to AmpD in the chromosome of E.
coli K12, forming the AmpDE composite operon, and AmpR,
AmpR-AmpC, AmpD, AmpE, and AmpD-AmpE cloning plasmids were
constructed with E. coli JRG582 Δ
(AmpD+/AmpE+) 2; the cloned plasmids were
then transformed into E. coli JRG582, an AmpDE deletion
strain, to find the expression and induction of β-lactamase based
on the Amp compound operon with the deletion of different
regulation genes. The results demonstrated that induced β-lactam
expression in AmpC enzyme was not affected by AmpE in E.
coli JRG582
(AmpR+/AmpC+/AmpD+/AmpE−),
i.e., AmpD was suppressed independently; in E. coli JRG582
(AmpR+/AmpC+/AmpD−/AmpE+),
the endogenic β-lactamase expression was effected by AmpE, which
means AmpE enhanced the repression effect of AmpD. The results
overturned the hypothesis of Honoré et al (34), who proposed that AmpE is the signal
transduction protein in the process of AmpC enzyme induction, and
the induction of AmpC is completely disrupted without AmpE although
AmpR is present. Only three transmembrane segments of AmpE are
present in Kp1 and Kp4, but no changes in induction and resistance
to FOX are observed. Therefore, AmpC enzyme would not be induced by
AmpE independently with the expression of AmpD, AmpG and AmpCR, and
no effect on AmpC enzyme was observed with the AmpE mutation. With
the cloned normal and mutation sequences of AmpE on the chromosome
of K. pneumoniae, AmpE was predicted as a transmembrane
protein, which may be used in further investigations on the
regulation and molecular biological characteristics of AmpC.
The AmpG gene encodes a transmembrane protein
(permease) that transports peptide murein debris from the periplasm
to the cytoplasm, which serve as signal molecules for the induction
of AmpC β-lactamase (35). In the
present study, the MIC values was extremely high in four strains,
with two point mutations (Ile19→Leu and Ala97→Thr) in five strains,
but had little association with the AmpG mutation.
The structure of AmpG in K. pneumoniae, as
predicted by the SOSUI system, was different from the AmpG in E.
coli predicted by Chahboune et al (36). A total of 12 transmembrane segments
and a large cytoplasmic loop B, with N-terminus and C-terminus
outside the cytomembrane were found; whereas, 10 transmembrane
segments and two large cytoplasmic loops of the AmpG were predicted
in E. coli. Schmidt et al (37) obtained three mutant strains of AmpG
by nitrosoguanidine E. coli SN0301 (with E. cloacae
AmpCR), which were Gly151→Asp151, Gly268→Asp268 and Gly373→Asp373,
and all the mutations led to the loss of inducibility and could be
recovered with AmpG recovery. AmpG stability was affected by
Gly268→Asp268, which was at the beginning of a transmembrane
segment; and the inducing signal transduction was affected by
Gly151→Asp151 and Gly373→Asp373, which were located at the two
large cytoplasmic loop. The amino acid mutations found in the
present study (Ile19→Leu, Ala 97→Thr) were in the inside surface of
the transmembrane domains, which had no effect on the function of
AmpG. Mutations of AmpG, affecting the inducible AmpC β-lactamase
production, have been demonstrated, through the use of chemical
mutagenesis methods described in previous studies (37,38).
However, in the present study, spontaneous mutations of the AmpG
gene in K. pneumoniae isolates did not have a significant
effect on the induction of AmpC. Further investigations are thus
required in order to clarify the role of the regulatory genes,
AmpR, AmpD, AmpE and AmpG, and their corresponding gene products,
in the induction of plasmid-mediated AmpC β-lactamase expression.
Additional areas of future research include the role of
penicillin-binding proteins or transmembrane proteins as sensors
for induction, as well as the interaction of AmpC, AmpR, AmpD, AmpE
and AmpG. With the association between all mutations of the AmpG
and AmpC enzymes induced, the resistance to β-lactam antibiotics
may be partially overcome.
Acknowledgements
This study was supported by the Natural Science
Foundation of Heilongjiang Province of China (grant no.
H2013100).
References
|
1
|
Ehrmann E, Handal T, Tamanai-Shacoori Z,
Bonnaure-Mallet M and Fosse T: High prevalence of β-lactam and
macrolide resistance genes in human oral Capnocytophaga species. J
Antimicrob Chemother. 69:381–384. 2014. View Article : Google Scholar
|
|
2
|
Raji MA, Jamal W, Ojemhen O and Rotimi VO:
Point-surveillance of antibiotic resistance in Enterobacteriaceae
isolates from patients in a Lagos Teaching Hospital, Nigeria. J
Infect Public Health. 6:431–437. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bush K, Jacoby GA and Medeiros AA: A
functional classification scheme for beta-lactamases and its
correlation with molecular structure. Antimicrob Agents Chemother.
39:1211–1233. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ambler RP, Coulson AF, Frère JM, et al: A
standard numbering scheme for the class A beta-lactamases. Biochem
J. 15:269–270. 1991.
|
|
5
|
Philippon A, Arlet G and Jacoby GA:
Plasmid-determined AmpC-type beta-lactamases. Antimicrob Agents
Chemother. 46:1–11. 2002. View Article : Google Scholar
|
|
6
|
Garcia DL and Dillard JP: Mutations in
ampG or ampD affect peptidoglycan fragment release from Neisseria
gonorrhoeae. J Bacteriol. 190:3799–3807. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Jacobs C: Pharmacia Biotech & Science
prize. 1997 grand prize winner Life in the balance: cell walls and
antibiotic resistance. Science. 278:1731–1732. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wiedemann B, Pfeifle D, Wiegand I and
Janas E: beta-Lactamase induction and cell wall recycling in
gram-negative bacteria. Drug Resist Updat. 1:223–226. 1998.
View Article : Google Scholar
|
|
9
|
Jacobs C, Joris B, Jamin M, et al: AmpD,
essential for both β-lactamase regulation and cell wall recycling,
is a novel cytosolic N-acetylmuramyl-L-alanine amidase. Mol
Microbiol. 15:553–559. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yu WL, Ko WC, Cheng KC, et al:
Institutional spread of clonally related Serratia marcescens
isolates with a novel AmpC cephalosporinase (S4): a 4-year
experience in Taiwan. Diagn Microbiol Infect Dis. 61:460–467. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Roh IK, Kim IJ, Chung JH and Byun SM:
Affinity purification and binding characteristics of Citrobacter
freundii AmpR, the transcriptional regulator of the ampC
beta-lactamase gene. Biotechnol Appl Biochem. 23:149–154.
1996.PubMed/NCBI
|
|
12
|
Jacobs C, Huang LJ, Bartowsky E, Normark S
and Park JT: Bacterial cell wall recycling provides cytosolic
muropeptides as effectors for beta-lactamase induction. EMBO J.
13:4684–4694. 1994.PubMed/NCBI
|
|
13
|
Li JB, Cheng J, Yin J, et al: Progress on
AmpC beta-lactamases. Curr Bioinform. 4:218–225. 2009. View Article : Google Scholar
|
|
14
|
Jacoby GA: AmpC beta-lactamases. Clin
Microbiol Rev. 22:161–182. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Schmidtke AJ and Hanson ND: Model system
to evaluate the effect of ampD mutations on AmpC-mediated
beta-lactam resistance. Antimicrob Agents Chemother. 50:2030–2037.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Khanal S, Joshi DR, Bhatta DR, Devkota U
and Pokhrel BM: β-lactamase-producing multidrug-resistant bacterial
pathogens from tracheal aspirates of intensive care unit patients
at National Institute of Neurological and Allied Sciences, Nepal.
ISRN Microbiol. 2013:8475692013. View Article : Google Scholar
|
|
17
|
Pai H, Kang CI, Byeon JH, et al:
Epidemiology and cahnical features of blood stream infections
caused by AmpC-type-beta-lactamase-producing Klebsiella pneumoniae.
Antimiciob Agents Chemother. 48:3720–3728. 2004. View Article : Google Scholar
|
|
18
|
Moland ES: Newer β-Lactamases: clinical
and laboratory implications, Part II. Clinical Microbiology
Newsletter. 30:79–85. 2008. View Article : Google Scholar
|
|
19
|
Moland ES: Newer β-Lactamases: clinical
and laboratory implications, Part I. Clinical Microbiology
Newsletter. 30:71–77. 2008. View Article : Google Scholar
|
|
20
|
Gniadkowski M: Evolution and epidemiology
of extended-spectrum beta-lactamases (ESBLs) and ESBL-producing
microorganisms. Clin Microbiol Infect. 7:597–608. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lodge J, Busby S and Piddock L:
Investigation of the Pseudomonas aeruginosa ampR gene and its role
at the chromosomal ampC beta-lactamase promoter. FEMS Microbiol
Lett. 111:315–320. 1993.PubMed/NCBI
|
|
22
|
Campbell JI, Ciofu O and Høiby N:
Pseudomonas aeruginosa isolates from patients with cystic fibrosis
have different beta-lactamase expression phenotypes but are
homogeneous in the ampC-ampR genetic region. Antimicrob Agents
Chemother. 41:1380–1384. 1997.PubMed/NCBI
|
|
23
|
Poirel L, Guibert M, Girlich D, Naas T and
Nordmann P: Cloning, sequence analyses, expression, and
distribution of ampC-ampR from Morganella morganii clinical
isolates. Antimicrob Agents Chemother. 43:769–776. 1999.PubMed/NCBI
|
|
24
|
Balasubramanian D, Schneper L, Merighi M,
et al: The regulatory repertoire of Pseudomonas aeruginosa AmpC
ß-lactamase regulator AmpR includes virulence genes. PLoS One.
7:e340672012. View Article : Google Scholar
|
|
25
|
Balcewich MD, Reeve TM, Orlikow EA, et al:
Crystal structure of the AmpR effector binding domain provides
insight into the molecular regulation of inducible ampc
beta-lactamase. J Mol Biol. 400:998–1010. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Nakano R, Okamoto R, Nakano Y, et al:
CFE-1, a novel plasmid-encoded AmpC beta-lactamase with an ampR
gene originating from Citrobacter freundii. Antimicrob Agents
Chemother. 48:1151–1158. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lee Y, Choi H, Yum JH, et al: Molecular
mechanisms of carbapenem resistance in Enterobacter cloacae
clinical isolates from Korea and clinical outcome. Ann Clin Lab
Sci. 42:281–286. 2012.PubMed/NCBI
|
|
28
|
Fortineau N, Poirel L and Nordmann P:
Plasmid-mediated and inducible cephalosporinase DHA-2 from
Klebsiella pneumoniae. J Antimicrob Chemother. 47:207–210. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Decré D, Verdet C, Raskine L, et al:
Characterization of CMY-type beta-lactamases in clinical strains of
Proteus mirabilis and Klebsiella pneumoniae isolated in four
hospitals in the Paris area. J Antimicrob Chemother. 50:681–688.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Carrasco-López C, Rojas-Altuve A, Zhang W,
et al: Crystal structures of bacterial peptidoglycan amidase AmpD
and an unprecedented activation mechanism. J Biol Chem.
286:31714–31722. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lee M, Zhang W, Hesek D, et al: Bacterial
AmpD at the crossroads of peptidoglycan recycling and manifestation
of antibiotic resistance. J Am Chem Soc. 131:8742–8743. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Bauvois B: Transmembrane proteases in
focus: diversity and redundancy? J Leukoc Biol. 70:11–17.
2001.PubMed/NCBI
|
|
33
|
Lindquist S, Galleni M, Lindberg F and
Normark S: Signalling proteins in enterobacterial AmpC β-lactamase
regulation. Mol Microbiol. 3:1091–1102. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Honoré N, Nicolas MH and Cole ST:
Regulation of enterobacterial cephalosporinase production: the role
of a membrane-bound sensory transducer. Mol Microbiol. 3:1121–1130.
1989. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhang Y, Bao Q, Gagnon LA, et al: ampG
gene of Pseudomonas aeruginosa and its role in β-Lactamase
expression. Antimicrob Agents Chemother. 54:4772–4779. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Chahboune A, Decaffmeyer M, Brasseur R and
Joris B: Membrane topology of the Escherichia coli AmpG permease
required for recycling of cell wall anhydromuropeptides and AmpC
beta-lactamase induction. Antimicrob Agents Chemother.
49:1145–1149. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Schmidt H, Korfmann G, Barth H and Martin
HH: The signal transducer encoded by ampG is essential for
induction of chromosomal AmpC β-lactamase in Escherichia coli by
beta-lactam antibiotics and ‘unspecific’ inducers. Microbiology.
141:1085–1092. 1995. View Article : Google Scholar
|
|
38
|
Korfmann G and Sanders CC: ampG is
essential for high level expression of AmpC β-lactamase in
Enterobacter cloacae. Antimicrob Agents Chemother. 33:1946–1951.
1989. View Article : Google Scholar : PubMed/NCBI
|