Introduction
Abnormal alterations in Wnt genes are
associated with neural tube defects (NTDs) (1,2).
NTDs occur when the neural tube fails to close at the early stage
of neural development, which can result in spina bifida,
myelomeningocele (also known as spina bifida aperta), exencephaly
or craniorachischisis (3). A
previous study demonstrated that a null mutation of Wnt3a, a
canonical Wnt, led to spina bifida aperta in mice (4). Wnt5a−/−, a
non-canonical Wnt, mice did not suffer from NTDs; however,
mice with double deletions of the Wnt5a−/− and
Ltap/Vangl2 genes exhibited craniorachischisis
(5). Furthermore, knockout
mutations of the canonical Wnt3 or non-canonical Wnt
genes, Wnt5a and Wnt7a, induce deformities of the
anteroposterior (A/P) axis of the neural tube (6). These findings suggest that, in
addition to roles of several known Wnt genes in the etiology
of NTDs, other Wnt members may be involved in the
pathogenesis of NTDs.
Wnt2b is a canonical Wnt, which is
expressed in the primitive streak at embryonic day (E) 7–7.75,
followed by the dorsal midline of the telencephalon and
mesencephalon at E8.5–9.5, and in the cortical hem at E11.5–17.5 in
murine embryos. This specific spatio-temporal expression of
Wnt2b suggests that it functions in gastrulation,
neurulation and cerebral cortex patterning (7,8). In
addition, Wnt7b, which is associated with canonical and
non-canonical Wnt signaling, is expressed at E5.5, and is
expressed in the forebrain, and the ventral and intermediate spinal
cord at the developmental stages during which patterning and neural
specification occur (9). Notably,
Wnt7b can assist in ensuring correct A/P guidance, and is
involved in dorsoventral patterning of the amphibian embryonic
neural tube (10). Although
Wnt2b and Wnt7b mutations have not induced NTDs in
mice (6), they are essential
during neural tube patterning; therefore, the present study
hypothesized that the expression levels of these two genes are
likely to be disrupted in NTDs.
Epigenetic modifications typically regulate gene
expression; histone 3 lysine 4 acetylation (H3K4ac) and
trimethylation of H3 lysine 4 (H3K4me3), are commonly located in
active promoters and are associated with gene activation, whereas
H3K27me3 suppresses transcription (11). DNA methylation is predominantly
catalyzed by DNA methyltransferase through the addition of a methyl
group to cytosine residues on dinucleotide CpG islands, and is
frequently distributed in the core promoter close to the
transcription start site (TSS); once the promoter CpG islands are
methylated, these genes are often transcriptionally inactivated
(12). Our previous studies
demonstrated that epigenetic modifications were significantly
altered in patients with NTDs (13,14);
however, the detailed mechanism and target genes remain to be
elucidated.
The present study aimed to investigate dysregulation
of epigenetic modification of Wnt genes in NTDs, using a
mouse model of spina bifida. It was hypothesized that in NTDs,
dysregulation of epigenetic modification is involved in the
regulation of Wnt2b and Wnt7b gene expression.
Materials and methods
Animals
All animals experiments were performed in accordance
with the standards of the Capital Institute of Pediatrics Ethics
Committee (Beijing, China; permit no. SYXK 2008-0011). A total of
40 adult C57BL/6J mice (20 male and 20 female) were housed in
light- and temperature-controlled rooms (12/12 h 1ight/dark cycle;
23±1°C) and maintained on pure mouse feed and tap water provided
ad libitum. Adult virgin females (above 9 weeks of age and
21 g in weight) were mated for 2 h with males (1 male per 2
females, 8:00–10:00 am) of the same stock. A vaginal plug observed
at 10:00 AM was considered as embryonic day 0 (E0). The mice were
administered with a 20 mg/kg body weight dose of retinoic acid (RA;
suspended in olive oil; Sigma-Aldrich, St. Louis, MO, USA) on
E8.0–0 h, E8.0–6 h and E8.0–12 h. Mice treated with isovolumetric
pure olive oil (Agric, Crete, Greece) at these same time points
served as control groups, as described in our previous study
(15). E18.0 fetuses were
delivered by cesarean section, and 21 cases of spina bifida were
selected (Fig. 1A and B). As the
spinal cord of individual mice provides insufficient material for
RNA extraction, the mice were randomly divided into three groups
and the samples in each group were pooled. A total of 10 normal
spinal cords were randomly pooled to create three control groups,
two of which contained 3 mice and one contained 4 mice.
Human subjects
Aborted NTD case subjects were obtained from Fenyang
Hospital and Linxian County Hospital (Shanxi, China). The surgical
details were, as previously described (14). The spinal cord tissues were used in
the following experiments. A total of four NTD-affected fetuses and
four age-matched controls were used in the present study. The
clinical phenotypes of the four cases of NTD were as follows: One
case of thoracic meningomyelocele (24 weeks gestation; female), one
case of anencephalic and occipital encephalomeningocele (17 weeks
gestation; female), and two cases of thoracic and lumbar spina
bifida (20 and 26 weeks gestation, respectively; female). The
present study was approved by the Committee of Medical Ethics at
the Capital Institute of Pediatrics (SHERLLM2014002). Written
informed consent was obtained from the parents on behalf of the
fetuses.
RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was isolated from the samples using
TRIzol® (cat. no. 15596-026; Invitrogen; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) and was reverse transcribed
using a First Strand cDNA Synthesis kit (cat. no. K1612; Beijing
TransGen Biotech Co., Ltd., Beijing, China). The cDNA samples were
analyzed using an Applied Biosystems 7500 Real-Time PCR system
(Thermo Fisher Scientific, Inc.) and a 2X PCR UltraSYBR Mixture kit
(cat. no. CW0956; CWBIO, Beijing, China) containing
carboxy-X-rhodamine, according to the manufacturer's protocol. The
expression levels of the target genes were normalized to
glyceraldehyde 3-phosphate dehydrogenase (Gapdh). The fold
change in expression was determined using the 2−ΔΔCq
method (16). The primer sequences
designed using Primer 5 and provided by Thermo Fisher Scientific,
Inc. are listed in Table I. The
PCR thermocycling steps were as follows: 95°C for 10 min, then 40
cycles of 95°C for 15 sec, 60°C for 1 min and 72°C for 5 min.
 | Table IPrimers for reverse
transcription-quantitative polymerase chain reaction analyses of
Wnt2b and Wnt7b mRNA. |
Table I
Primers for reverse
transcription-quantitative polymerase chain reaction analyses of
Wnt2b and Wnt7b mRNA.
| Gene | Primer (5′→3′) |
|---|
| mWnt2b | F:
CGAGGTGGCAAACATCCTAT |
| R:
CTTTGAAGGCTCCACTCCTG |
| mWnt7b | F:
GGCTCCTTCCTACTCGCTCT |
| R:
GAGTGTGACTGGTGGGTGTG |
| Gapdh | F:
TGTGTCCGTCGTGGATCTGA |
| R:
CCTGCTTCACCACCTTCTTGA |
Chromatin immunoprecipitation (ChIP)
assays
A ChIP assay kit was purchased from Invitrogen
(Thermo Fisher Scientific, Inc.; cat. no. 49–2024). Chromatin was
prepared from targeted tissues and sonicated to DNA lengths between
300 and 500 bp. The following antibodies were used: Polyclonal
anti-rabbit H3K4me (cat. no. ab106165; Abcam, Cambridge, UK),
polyclonal anti-rabbit H3K4ac (cat. no. ab113672; Abcam) and
anti-H3K27me3 (cat. no. 17–622; EMD Millipore, Billerica, MA, USA)
antibodies. Alkaline phosphate-conjugated rabbit
anti-immunoglobulin G (Beijing Zhongshan Golden Bridge
Biotechnology Co., Ltd., Beijing, China)-treated chromatin
immunoprecipitated samples served as a negative control. The
antibodies were coupled with Dynabeads® Protein A/G
(Thermo Fisher Scientific, Inc.; cat. no. 49–2024) at 4°C for 1 h,
then chromatin was bound to the antibody-dynabeads complex at 4°C
for 2 h. ChIP-qPCR analysis was performed using an ABI 7500 system
(Applied Biosystems). The primers (Thermo Fisher Scientific, Inc.)
used for ChIP-PCRs are listed in Table II. The relative enrichment of the
histone modification was determined using the
2(input-Cq)NTDs/2(input-Cq)control
method (14).
 | Table IIPrimer sequences for chromatin
immunoprecipitation. |
Table II
Primer sequences for chromatin
immunoprecipitation.
| Gene | Primer (5′→3′) | RefSeq
accession | Product size
(bp) | Target region |
|---|
| Wnt2b-1 | F:
CGAGAGTGGGTGGGAGAAG | NM_009520 | 125 |
chr3:104765115-104765239 |
| R:
GGACTCCTTTGGCCACACTG | | | |
| Wnt2b-2 | F:
GAAGTGTCCGCAACCCTCTC | NM_009520 | 103 |
chr3:104764911-104765013 |
| R:
TCCTAGCGCCCTTCACTCA | | | |
| Wnt7b | F:
CGGAGCCAATACGCAGCA | NM_001163633 | 100 |
chr15:85411511-85411610 |
| R:
CCAAACAGGTGAAAGTACGCG | | | |
| WNT2B-1 | F:
GGGCGGTGATAGAAGTTGCT | NM_004185 | 122 |
chr1:113008796-113008917 |
| R:
TCAGCTTTCCGAAGAAGGGC | | | |
| WNT2B-2 | F:
CCCAGTGGAGTCAGGAAAGG | NM_004185 | 141 |
chr1:113009674-113009814 |
| R:
TGGCAACTTGGTGCACTGTA | | | |
| WNT7B | F:
GAGACCACCCACTCCCATTG | NM_058238 | 145 |
chr22:46373432-46373576 |
| R:
CCTGGGGTTGTGGGAATCTC | | | |
DNA methylation analyses
Following ultrasonic sonication (P-141008; Diagenode
SA, Seraing, Belgium) of mouse and human spinal cord tissues,
genomic DNA was extracted from the homogenates using a Genomic DNA
Miniprep kit (Axygen Scientific, Inc., Union City, CA, USA). A
total of 500 ng genomic DNA from each sample was bisulfite-treated
using an EZ DNA Methylation-Gold™ kit (cat. no. D5005; Zymo
Research, Irvine, CA, USA). The Sequenom MassARRAY platform
(CapitalBio Corporation, Beijing, China) was used to perform
quantitative methylation analysis of Wnt2b and Wnt7b
(13). The primers used to detect
DNA methylation levels of the target regions are presented in
Table III.
 | Table IIIPrimer sequences for DNA
methylation. |
Table III
Primer sequences for DNA
methylation.
| Gene | Primer (5′→3′) | RefSeq
accession | Product size
(bp) | Target region |
|---|
| Wnt2b-1 | F:
aggaagagagGGGAAAGTAGTTTAGTTGTTTTTGA | NM_009520 | 349 |
chr3:104765115-104765463 |
| R:
cagtaatacgactcactatagggagaaggctAAACTCCTTTAACCACACTACCCTC | | | |
| Wnt2b-2 | F:
aggaagagagAGGGTAGTGTGGTTAAAGGAGTTTT | NM_009520 | 280 |
chr3:104764859-104765138 |
| R:
cagtaatacgactcactatagggagaaggctCTTCCCTACCTACAACCCCCTAT | | | |
| Wnt7b | F:
aggaagagagTGAAGTGGTTAGGTTGGTTGGTAT | NM_001163633 | 205 |
chr15:85411502-85411706 |
| R:
cagtaatacgactcactatagggagaaggctACCTAAAACCCAAACAAATAAAAAT | | | |
| WNT2B-1 | F:
aggaagagagGAAGGATAGGGTTAGTGGTTAGGAA | NM_004185 | 340 |
chr1:113008996-113009335 |
| R:
cagtaatacgactcactatagggagaaggctAACCCCTTTTTAACAATTATAAACATCT | | | |
| WNT2B-2 | F:
aggaagagagTTTTTTTTAGGGTTTTGTGAGAATTT | NM_004185 | 338 |
chr1:113008683-113009020 |
| R:
cagtaatacgactcactatagggagaaggctTTCCTAACCACTAACCCTATCCTTC | | | |
| WNT2B-3 | F:
aggaagagagGATGGGGTTTTTAAGATTTAGGAGA | NM_004185 | 340 |
chr1:113008435-113008774 |
| R:
cagtaatacgactcactatagggagaaggctACAAAAAAAACAACAAACCCATAAA | | | |
| WNT2B-4 | F:
aggaagagagTTGTTATATTTGTTTGTGGAGGAGG | NM_004185 | 268 |
chr1:113007526-113007793 |
| R:
cagtaatacgactcactatagggagaaggctAATCAAAATCTTCCTAAAAACACCC | NM_004185 | | |
| WNT2B-5 | F:
aggaagagagTTTTGTTTTTATGTTTTTGAATAGG | NM_004185 | 328 |
chr1:113007314-113007641 |
| R:
cagtaatacgactcactatagggagaaggctACCCCTTCCCAACAAAACTATTAT | | | |
| WNT7B-1 | F:
aggaagagagGTTTTGGGGAGTAGGGGTTATTTA | NM_058238 | 326 |
chr22:46374152-46374477 |
| R:
cagtaatacgactcactatagggagaaggctACTACCCTATCCCACCTAACCAAAC | | | |
| WNT7B-2 | F:
aggaagagagGGGAGTTATTTAGGGTTGAGAAAGT | NM_058238 | 318 |
chr22:46373742-46374059 |
| R:
cagtaatacgactcactatagggagaaggctACACTAAAAAAAACTCCTAACCCCC | NM_058238 | | |
| WNT7B-3 | F:
aggaagagagGGTTAATGGGTTTTGTAGGAGGTTA | NM_058238 | 265 |
chr22:46373430-46373694 |
| R:
cagtaatacgactcactatagggagaaggctAAAAAACCACCCACTCCCATTAC | NM_058238 | | |
Statistical analysis
Statistical analyses of the transcripts, histone
modification enrichment and DNA methylation levels of Wnt
genes were performed at least three times. Data were compared
between the test and control groups using Student's t-test with
SPSS software, version 20 (IBM SPSS, Armonk, NY, USA). P<0.05
was considered to indicate a statistically significant
difference.
Results
Altered transcription levels of Wnt2b and
Wnt7b in a mouse model of RA-induced spina bifida
To determine into the possible changes occurring in
the expression levels of Wnt2b and Wnt7b during the
development of RA-induced NTDs, the mRNA expression levels were
determined. The results indicated that, in the mice with spina
bifida, the mRNA expression levels of Wnt2b and Wnt7b
were 3.1- and 2.8-fold higher, respectively, compared with the
control mice (Fig. 1C). These
results were consistent with those of a previous study, which
detected the transcriptional upregulation of WNT2 (including
WNT2B) and WNT7B in NT2/NTera2 cells following RA
treatment (17). These results
indicated that the transcription of Wnt2b and Wnt7b
were abnormally activated in RA-induced spina bifida.
Abnormal histone modifications are
present in the promoter regions of Wnt2b and Wnt7b
Due to the ectopic alterations in mRNA expression
levels, epigenetic factors in the GC-rich promoter regions of
Wnt2b and Wnt7b were examined, according to the
current UCSC database (July 2007 NCBI37/mm9; http://genome.ucsc.edu/). As shown in Fig. 2A and B, H3K4me3 and H3K27me3
modifications target Wnt2b and Wnt7b in embryonic
stem cells and the brain in the central nervous system, whereas
H3K9me3 modifications are weak. Furthermore, in the cerebellum and
mouse embryonic fibroblasts, these histone modification enrichments
are markedly lower, indicating that the histone modifications,
which occur in Wnt2b and Wnt7b are stable and
spatio-temporal specific. Therefore, the present study aimed to
determine whether H3K27me3 or H3K4me bind to the target regions of
samples by performing ChIP assays. As H3K4ac is also a
transcriptional activation marker (18), the enrichment of this modification
was also assessed. In the target regions of Wnt2b and
Wnt7b in the RA-treated NTD mice, enrichments of H3K4ac were
upregulated, whereas H3K27me3 enrichments were significantly
downregulated in the chr3: 104765115–104765239 segment of
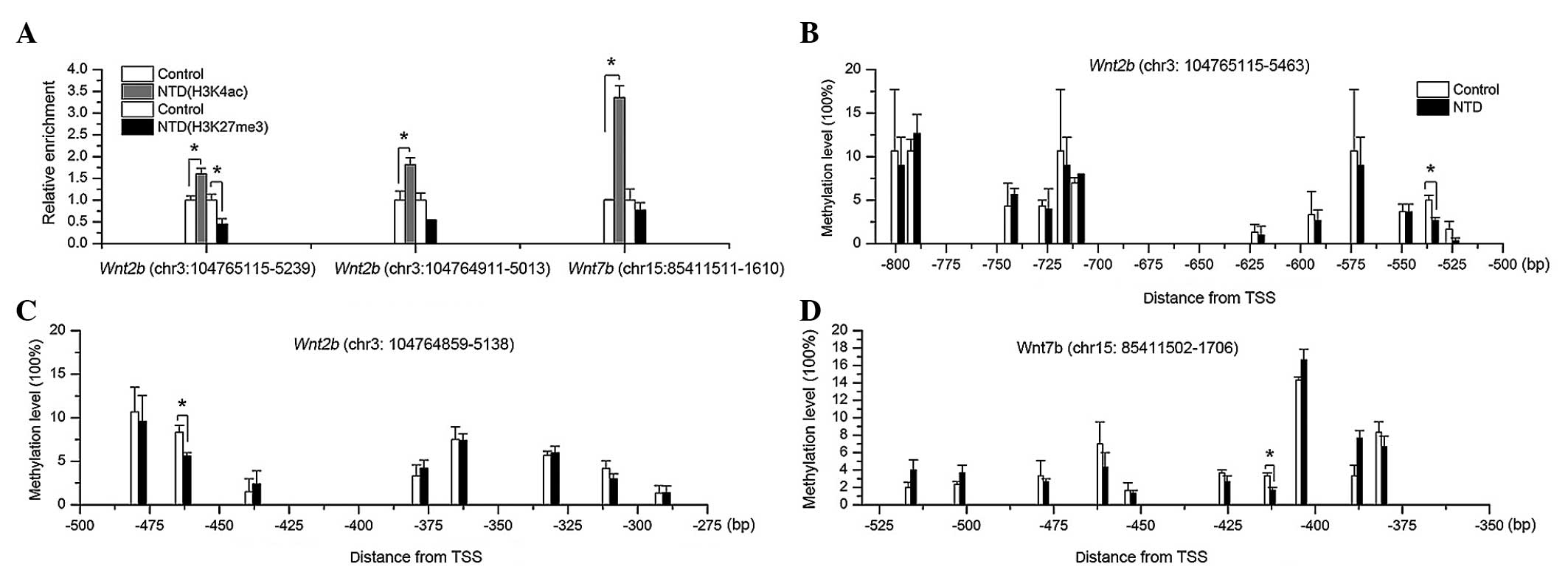
Wnt2b (Fig. 3A). The
enrichment of H3K4me was undetectable in the target regions of the
two genes (data not shown). The abnormally attenuated H3K27me3 and
enhanced H3K4ac enrichment of Wnt2b and Wnt7b
indicated their roles in the dysregulated increased transcription
observed in the mice with NTDs.
DNA methylation levels of Wnt2b and Wnt7b
are abnormal in histone modification-enriched regions
Alterations in the histone modification profiles of
the Wnt genes prompted the present study to examine DNA
methylation in the histone modification-enriched regions. No
difference was observed in the mean methylation levels of the
target Wnt2b and Wnt7b regions between the NTD
samples (Wnt2b, 5.36%; Wnt7b, 5.13%) and the control
samples (Wnt2b, 5.79%; Wnt7b,4.93%; P=0.70 and
P=0.92, respectively), as determined using the MassARRAY platform.
However, the methylation levels of Wnt2b at the CpG-units
chr3: 104765161-62 (−535 bp upstream from TSS) and chr3:
104765089-90 (−463 bp upstream from TSS) were significantly
hypomethylated in the mice with spina bifida, compared with the
controls (Fig. 3B and C). For
Wnt7b, the chr15: 85411571-72 CpG site (407 bp upstream of
the TSS) was also hypomethylated (Fig.
3D). These results suggested that, in NTDs, the increased mRNA
expression levels of Wnt2b and Wnt7b may also be
partially due to hypomethylation of specific CpG-units in the
promoter regions.
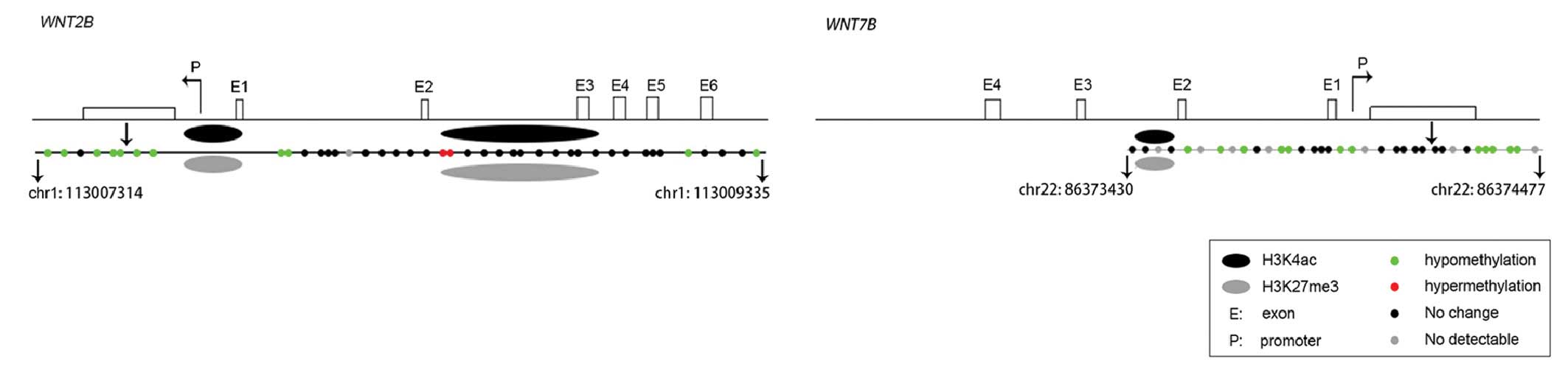
The combination of the ChIP assay and DNA
methylation assay data suggested that, in the mouse model of
RA-induced spina bifida, increased H3K4ac and decreased H3K27me3
enrichment, and hypomethylation of CpG islands in the promoter
regions potentially led to enhanced mRNA expression (Fig. 4).
Aberrant epigenetic modification of WNT2B
and WNT7B in human NTDs
As abnormal epigenetic modifications of Wnt2b
and Wnt7b were detected in the mice with NTDs, the present
study subsequently aimed to validate whether similar changes were
present in the human NTD samples. In H1 embryonic stem cells, at
the early stage prior to neural development, the GC-rich promoters
of the two WNT genes were enriched for H3K27me3 (Fig. 5A and B), and were bound to a series
of enzymes for its catalysis, including enhancer of zeste homolog 2
(EZH2) and suppressor of zeste homolog 12 (SUZ12). However, low
levels of HDAC2-6 were detected, which is known to regulate the
deacetylation of H3. Furthermore, astrocytes (NH-A), which are a
type of mature neuronal cell associated with the later stage of
neural development, exhibited reduced H3K27me3 and EZH2 enrichment
(19), indicating that H3K27me3
and H3K4ac may preferentially function in the early stage of neural
development.
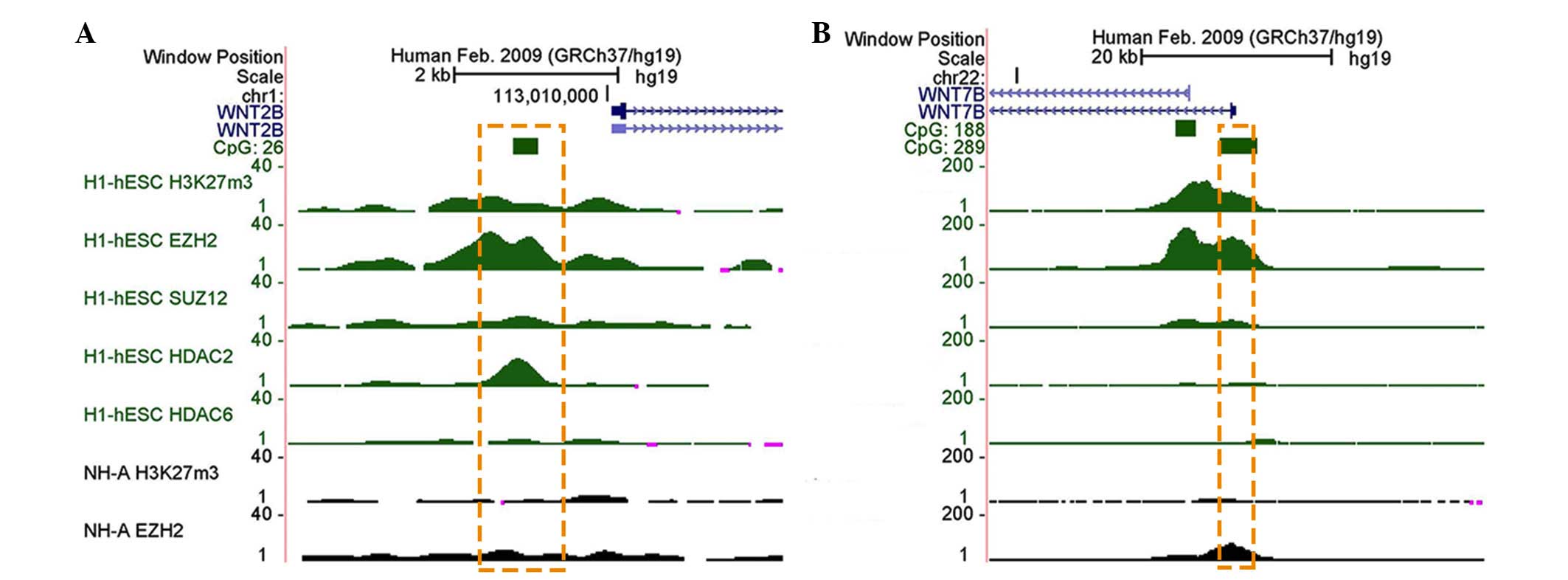 | Figure 5Histone modification enrichment
profiles of WNT2B and WNT7B in humans. Browser shots
from the UCSC genome browser showing H3K4me3, H3K9me3 and H3K27me3
ChIP-seq data in various tissues and cells. The positions of the
UCSC CpG islands (green) are shown. ChIP-seq data is presented as
the number of reads that overlap genomic windows. In humans, stable
H3K27me3 modifications were detected in (A) WNT2B and (B)
WNT7B in the ESCs, but not in NH-A. Orange dotted box
indicates the target regions of genes in the present study. H3K,
histone 3 lysine; me3, trimethylation; ChIP, chromatin
immunoprecipitation; hESC, human embryonic stem cell; NH-A,
astrocyte; EZH2, enhancer of zeste homolog 2; SUZ, suppressor of
zeste homolog 12; HDAC, histone deacetylase. |
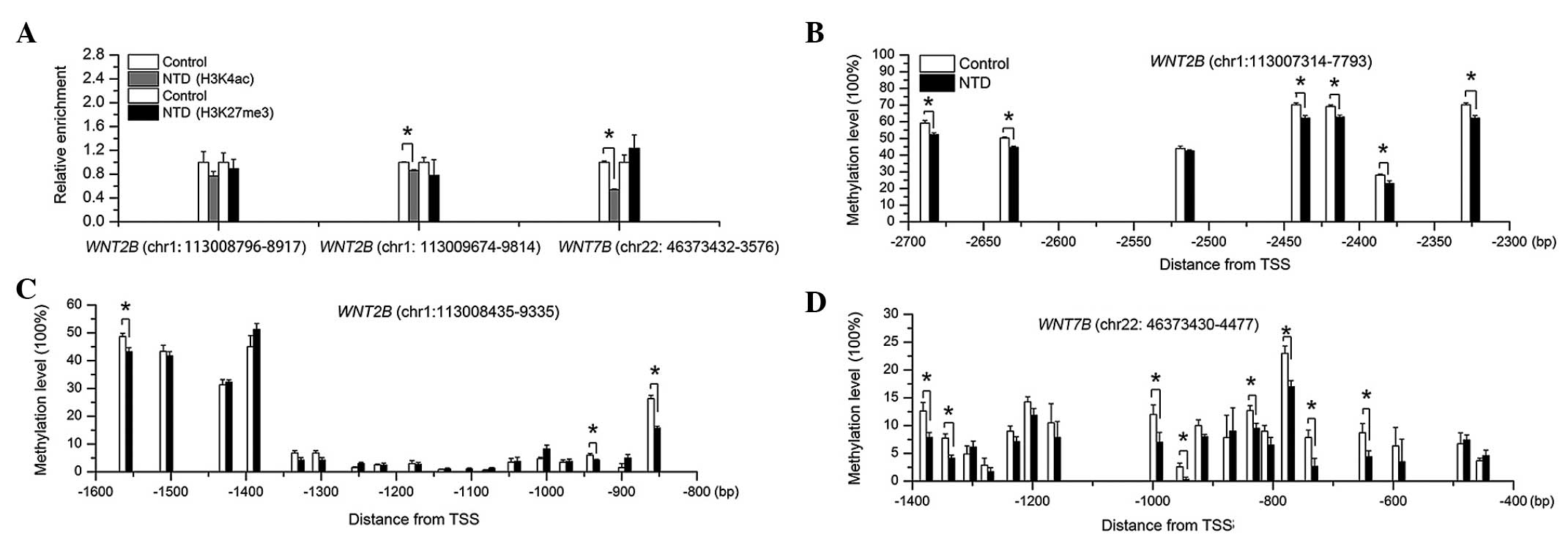
In the human spinal cord samples, the GC-rich
promoter regions of WNT2B and WNT7B were enriched for
H3K4ac and H3K27me3. However, in the NTD samples, H3K4ac enrichment
was downregulated, compared with the controls, and there were no
differences in H3K27me3 enrichment between the groups (Fig. 6A). Furthermore, several CpG-units
of WNT2B and WNT7B were significantly hypomethylated,
compared with the control group, particularly those close to the
TSS (Figs. 6B–D and 7). These results indicated that aberrant
epigenetic modification of WNT2B and WNT7B occurred
in the human NTDs.
Discussion
Previous studies have demonstrated that H3K27me3
binding assists in maintaining gene silencing in multi-cellular
organisms, and is associated with the timed suppression of
developmental regulatory genes (20,21).
By contrast, H3K4ac is considered to be involved in the activation
of gene transcription (18),
whereas methylated DNA makes it difficult for histones to form
nucleosomes, resulting in chromosomal instability and modified
accessibility of regulatory proteins, which alter the pattern of
gene expression (12,22). The present study hypothesized that
the aberrant DNA methylation and histone modification profiles
observed in NTDs may affect the binding ability of RNA polymerase
and key transcription factors to promoter regions on genes,
including EZH2, SUZ12, HDAC2, HDAC6, which in turn regulate histone
modification and affect transcription initiation (19). In addition, H3K4ac and H3K27me3
modifications of the target genes in the present study were
observed to function preferentially at the early stage of neural
development, however not in the later stages. Furthermore, taking
the data from our previous study into account, which reported DNA
hypomethylation of long interspersed nuclear element-1 and low
expression levels of H3K79me2 in human NTDs (13,14),
the present study reiterates the hypothesis that abnormal
epigenetic modifications have a destructive role in the etiology of
NTDs.
It remains uncertain how alterations in Wnt2b
and Wnt7b affect normal neural development. As ligands,
Wnt2b and Wnt7b can function through the canonical
β-catenin pathway, which maintains the proliferative state of
neural progenitor cells (23,24).
In addition, Wnt7b can signal through the non-canonical
pathway (planar cell polarity), affect convergent extension
movements prior to the closure of the neural tube, and control A/P
axis formation and neural patterning (25). Therefore, the upregulated
expression levels of Wnt2b and Wnt7b may lead to
excessive signaling and result in disordered neural development,
suggesting that aberrant Wnt gene expression may be involved
in the etiology of NTDs. However, the Wnt5a genetic knockout
mice demonstrated that individual deficiencies in Wnt genes
do not induce an NTD phenotype (5). This suggests that, when multiple
Wnt ligands are altered at one time, they may potentially
contribute to the etiology of disease. However, further studies are
required to confirm this hypothesis.
In conclusion, the present study provided novel
insight into the synergistic function of transcriptional
dysregulation and epigenetic modifications. Specifically, abnormal
epigenetic modifications in the Wnt genes were detected
following exposure to RA and may significantly contribute to the
development and etiology of NTDs.
Acknowledgments
The present study was supported by the National
Natural Science Foundation of China (Beijing, China; grant. nos.
81471163 and 81300489) and the National '973' project (grant. no.
2013CB945404). The authors would like to thank all participating
hospitals and medical staff for their assistance in sample
collection and recording of clinical information, and would like to
thank all subjects and their family members for their cooperation
in providing clinical information and samples for this study.
References
|
1
|
Mulligan KA and Cheyette BN: Wnt signaling
in vertebrate neural development and function. J Neuroimmune
Pharmacol. 7:774–787. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Carter M, Chen X, Slowinska B, Minnerath
S, Glickstein S, Shi L, Campagne F, Weinstein H and Ross ME:
Crooked tail (Cd) model of human folate-responsive neural tube
defects is mutated in Wnt coreceptor lipoprotein receptor-related
protein 6. Proc Natl Acad Sci USA. 102:12843–12848. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Copp AJ and Greene ND: Genetics and
development of neural tube defects. J Pathol. 220:217–230.
2010.
|
|
4
|
Greco TL, Takada S, Newhouse MM, McMahon
JA, McMahon AP and Camper SA: Analysis of the vestigial tail
mutation demonstrates that Wnt-3a gene dosage regulates mouse axial
development. Genes Dev. 10:313–324. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Qian D, Jones C, Rzadzinska A, Mark S,
Zhang X, Steel KP, Dai X and Chen P: Wnt5a functions in planar cell
polarity regulation in mice. Dev Biol. 306:121–133. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
van Amerongen R and Berns A: Knockout
mouse models to study Wnt signal transduction. Trends Genet.
22:678–689. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kemp C, Willems E, Abdo S, Lambiv L and
Leyns L: Expression of all Wnt genes and their secreted antagonists
during mouse blastocyst and postimplantation development. Dev Dyn.
233:1064–1075. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tsukiyama T and Yamaguchi TP: Mice lacking
Wnt2b are viable and display a postnatal olfactory bulb phenotype.
Neurosci Lett. 512:48–52. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Beretta CA, Brinkmann I and Carl M: All
four zebrafish Wnt7 genes are expressed during early brain
development. Gene Expr Patterns. 11:277–284. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Fenstermaker AG, Prasad AA, Bechara A,
Adolfs Y, Tissir F, Goffinet A, Zou Y and Pasterkamp RJ: Wnt/planar
cell polarity signaling controls the anterior-posterior
organization of monoaminergic axons in the brainstem. J Neurosci.
30:16053–16064. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ha M, Ng DW, Li WH and Chen ZJ:
Coordinated histone modifications are associated with gene
expression variation within and between species. Genome Res.
21:590–598. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Weber M, Hellmann I, Stadler MB, Ramos L,
Pääbo S, Rebhan M and Schübeler D: Distribution, silencing
potential and evolutionary impact of promoter DNA methylation in
the human genome. Nat Genet. 39:457–466. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang L, Wang F, Guan J, Le J, Wu L, Zou J,
Zhao H, Pei L, Zheng X and Zhang T: Relation between
hypomethylation of long interspersed nucleotide elements and risk
of neural tube defects. Am J Clin Nutr. 91:1359–1367. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang Q, Xue P, Li H, Bao Y, Wu L, Chang
S, Niu B, Yang F and Zhang T: Histone modification mapping in human
brain reveals aberrant expression of histone H3 lysine 79
dimethylation in neural tube defects. Neurobiol Dis. 54:404–413.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chen S, Bai B, Wang X, Li H and Zhang T:
Retinoic acid induced neural tube defects in C57 mice in a
dose-dependent and time-specific way. Chinese Journal of Birth
Health & Heredity. 122–124. 2014.
|
|
16
|
Zhang M, Shi J, Huang Y and Lai L:
Expression of canonical WNT/β-CATENIN signaling components in the
developing human lung. BMC Dev Biol. 12:212012. View Article : Google Scholar
|
|
17
|
Katoh M: Regulation of WNT signaling
molecules by retinoic acid during neuronal differentiation in NT2
cells: Threshold model of WNT action (review). Int J Mol Med.
10:683–687. 2002.PubMed/NCBI
|
|
18
|
Xhemalce B and Kouzarides T: A
chromodomain switch mediated by histone H3 Lys 4 acetylation
regulates heterochromatin assembly. Genes Dev. 24:647–652. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ram O, Goren A, Amit I, Shoresh N, Yosef
N, Ernst J, Kellis M, Gymrek M, Issner R, Coyne M, et al:
Combinatorial patterning of chromatin regulators uncovered by
genome-wide location analysis in human cells. Cell. 147:1628–1639.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Nie Y, Liu H and Sun X: The patterns of
histone modifications in the vicinity of transcription factor
binding sites in human lymphoblastoid cell lines. PLoS One.
8:e600022013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Voigt P, Tee WW and Reinberg D: A double
take on bivalent promoters. Genes Dev. 27:1318–1338. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Eden A, Gaudet F, Waghmare A and Jaenisch
R: Chromosomal instability and tumors promoted by DNA
hypomethylation. Science. 300:4552003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Cajánek L, Ribeiro D, Liste I, Parish CL,
Bryja V and Arenas E: Wnt/beta-catenin signaling blockade promotes
neuronal induction and dopaminergic differentiation in embryonic
stem cells. Stem Cells. 27:2917–2927. 2009.PubMed/NCBI
|
|
24
|
Wexler EM, Paucer A, Kornblum HI, Palmer
TD and Geschwind DH: Endogenous Wnt signaling maintains neural
progenitor cell potency. Stem Cells. 27:1130–1141. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Freese JL, Pino D and Pleasure SJ: Wnt
signaling in development and disease. Neurobiol Dis. 38:148–153.
2010. View Article : Google Scholar :
|