Introduction
Endometriosis is a gynecological disorder, which is
characterized by the combined and multifocal localization of
ectopic endometrial implants in the abdominal organs and abdominal
cavity (1), which are frequently
accompanied by clinical manifestations that include severe chronic
pelvic pain and infertility (1-3). It
is considered that endometrial lesions arise from additive
interactions between genetic and environmental factors (2).
The ability of endometrial implants to survive
outside of the uterus results from their increased estrogen
activity, overactive oncogenic pathways and increased production of
prostaglandins, cytokines and metalloproteinases (4,5).
Additionally, the survival of endometrial implants can be prolonged
due to abnormal immune system functioning, such that the ectopic
endometrium does not get flagged for removal from the body
(4,5).
There is increasing evidence to suggest that
endometriosis is an epigenetic disease (6–9).
Epigenetic regulators of gene transcription include DNA methylation
patterns and histone modifications, which do not require direct
changes to the DNA sequence (10).
Several genes have been identified to possess abnormal methylation
and expression patterns in women presenting with endometriosis
(11–18). DNA methylation is produced by DNA
methyltransferase enzymes (DNMTs) (10). These include the maintenance
methyltransferase, DNMT1, as well as the de novo DNMT3A and
DNMT3B enzymes (10). Aberrant
expression levels of DNMT1, DNMT3A and DNMT3B have been
demonstrated in patients with endo metriosis (17). There is an additional member of the
human DNMT3 family, DNMT3 like (DNMT3L), which does not possess
methyltransferase activity itself, but acts in a co-operative
manner with DNMT3A and DNMT3B (19).
The presence of subtelomeric hypermethylated regions
of DNA, as well as regions of hypomethylated DNA, which are
distributed along the chromosomes, has been demonstrated in all
types of endometriosis (20).
Additionally, it has been demonstrated that the DNMT3L
variant, rs113593938, which is situated within exon 10, makes an
important contribution to subtelomeric hypomethylation (21). Previous reports have also
identified an association between the DNMT3L rs8129776 gene
variant and the development of endometrioma (22). Therefore, the present study aimed
to determine the relative contributions of the DNMT3L
c.910-635A/G (rs8129776), c.832C/T (rs7354779), c.812C/T
(rs113593938) and c.344+62C/T (rs2276248) single nucleotide
polymorphisms (SNPs) on the development of stage I II
endometriosis. To address this question, women were selected from
the Polish population with defined stage I–II endometriosis,
according to the revised American Society for Reproductive Medicine
classification (23).
Materials and methods
Study subjects
Peripheral blood samples were extracted from
infertile women with endometriosis (n=154) and from healthy control
individuals (n=383). The patients were recruited through the
Gynecologic and Obstetrical University Hospital, Division of
Reproduction at Poznan University of Medical Sciences (Poznan,
Poland). Suspected cases of pelvic endometrioma were investigated
laparoscopically (Table I).
Visualization of endometriotic lesions and histopathological
criteria were used to diagnose minimal endometriosis in infertile
women. Each case of endometriosis was staged according to the
revised classification of the American Society for Reproductive
Medicine (23). The patients in
the experimental cohort in the present study exhibited regular
menses, an anatomically intact reproductive tract and infertility
spanning at least 1 year, despite the desire for conception. The
infertility was confirmed not to be a result of male factor
infertility. The control patient cohort in the present study
included fertile women of reproductive age, who were identified not
to have any malignant disease, endometriosis or adenomyosis
following surgical examinations performed during cesarean section.
Each of the women in the control group had regular menses and an
anatomically intact reproductive tract. Additional inclusion and
exclusion criteria for the patient cohorts have been previously
described in detail (24). The
study subjects were matched by age, and were all Caucasians of
Polish descent (Table I). Written
informed consent was obtained from all the individuals involved in
the present study, and all procedures were approved by the ethics
committee of Poznan University of Medical Sciences (Poznan,
Poland).
 | Table IClinical characteristics of the
patients with endome triosis and control subjects. |
Table I
Clinical characteristics of the
patients with endome triosis and control subjects.
| Characteristic | Endometriosis | Control |
|---|
| Number | 154 | 383 |
| Age (years) | 31 (20–42)a | 31 (21–39)a |
| Parity | NA | 1 (1–2)a |
| Duration of
infertility (years) | 3 (1–6)a | NA |
| rASRM (stage) | Stage I (n=85) | |
| Stage II
(n=69) | NA |
Evaluation of the presence of DNMT3L
SNPs
The genomic DNA was isolated from peripheral blood
leukocytes by salt extraction. The SNPs used in the present study
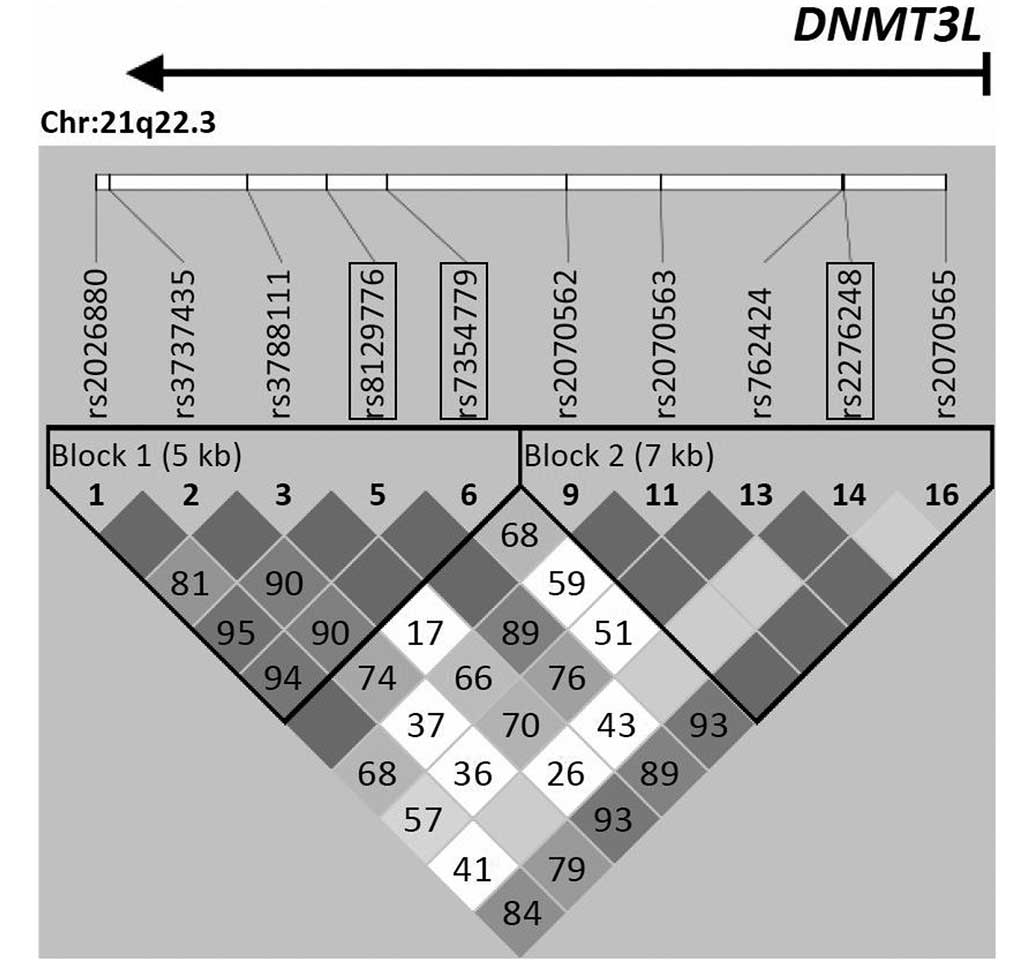
were selected on the basis of previous results (24), and are shown in Fig. 1. DNA samples were genotyped for
four DNMT3L SNPs, including c.910-635A/G (rs8129776), c.832C/T
(rs7354779), c.812C/T (rs113593938) and c.344+62C/T (rs2276248;
Table I and Fig. 1). Genotyping was performed via high
resolution melting (HRM) curve analysis using the LightCycler 480
system (Roche Diagnostics, Mannheim, Germany) with 5X HOT FIREPol
EvaGreen HRM mix (Solis BioDyne, Tartu, Estonia). The PCR program
consisted of an initial step at 95°C for 15 min to activate HOT
FIREPol DNA polymerase, followed by 50 amplification cycles of
denaturation at 95°C for 10 sec, primer-dependent annealing at
60.6°C for 10 sec, and elongation at 72°C for 15 sec. Amplified DNA
fragments were then subjected to HRM with 0.1°C increments in
temperature ranging from 76–97°C. Primer sequences and conditions
for HRM analysis are presented in Table II. The quality of genotyping was
evaluated by repeating measurements on a randomly selected 10% of
the samples.
 | Table IIHigh resolution melting curve
conditions for genotyping of DNMT3L polymorphic
variants. |
Table II
High resolution melting curve
conditions for genotyping of DNMT3L polymorphic
variants.
| rs no. | Chromosome
location | SNP function |
Alleles3 | MAFb | Primers for PCR
amplification (5′–3′) | PCR product length
(bp) | Annealing temp.
(°C) | Melt temp, range
(°C) |
|---|
| rs8129776 | Chr21:45669629 | Intron | A/G | 0.38 | F:
GAACAGAGGTCGTAAGTTCCA
R: GTTATGGAGGAGCGGTGA | 85 | 57.8 | 76–91 |
| rs7354779 | Chr21:45670770 | Missense
p.Arg278Gly | C/T | 0.25 | F:
CACCAGATTGTCCACGAAC
R: GGTACCTGTTCCAGTTCCAC | 95 | 57.8 | 80–95 |
| rs113593938 | Chr21:45670790 | Missense
p.Arg271Gln | C/T | 0.01 | F:
GGGGCTGCCTGGCTTGGGC
R: CCTCAGCCCTGCCCCCTCACC | 92 | 70.0 | 82–97 |
| rs2276248 | Chr21:45679258 | Intron | C/T | 0.02 | F:
AAATCCACCCACACTCCAGA
R: CTGCGGAAACCCTGATTG | 111 | 57.8 | 80–95 |
Statistical analysis
For each SNP, the Hardy Weinberg equilibrium (HWE)
was assessed using Pearson's goodness-of-fit χ2
statistic. Differences in allele and genotype frequencies between
the case and control subjects were evaluated using Fisher's test.
The associations between the SNPs selected for the present study
and the development of endometriosis were evaluated using the
Cochran-Armitage trend test. Statistical analyses were conducted
using Statistica version 10 (Stat Soft, Inc., Tulsa, USA). Odds
ratios (ORs) and associated 95% confidence intervals (95% CIs) were
also determined. The data were analyzed using recessive and
dominant inheritance models. Pair wise linkage disequilibrium (LD)
between the selected SNPs was computed as D′ and r2
values using HaploView 4.0 software (http://www.broadinstitute.org//scientific-community/software).
HaploView 4.0 software was also used for haplotype assessment.
Significant P-values were corrected using a 1,000 fold permutation
test.
Meta-analysis of the c.910- 635A/G
(rs8129776) SNP
A meta-analysis of two independent datasets, of
Borghese et al (22) and
the present study, was performed using either fixed- or
random-effect modeling. A fixed-effect model was used when no
evidence of significant heterogeneity was identified between the
two datasets, and a random-effect model was used when heterogeneity
was observed. The heterogeneity between the two studies was
assessed using χ2 based Cochrane Q and I2
statistics (25,26). P values generated using Cochrane's
Q test that were <0.10 were considered to indicate the presence
of heterogeneity between the two datsets. I2 values of
25, 50 and 75% were considered to indicate low, moderate and high
levels of heterogeneity, respectively. The effects of contrast
between alleles (G, vs. A) and between the dominant (AG+GG, vs. AA)
and recessive (GG, vs. AG+AA) models were also estimated. All
statistical calculations were conducted using Comprehensive
Meta-Analysis version 2.0 software (www.MetaAnalysis.com).
Results
Distribution of DNMT3L c.910-635A/G
(rs8129776), c.832C/T (rs7354779), c.812C/T (rs113593938) and
c.344+62C/T (rs2276248) polymorphisms in patients with
endometriosis-associated infertility
No divergence from the HWE was identified in the
frequency of any of the genotypes examined in any of the groups
(P>0.05). The number of genotypes, OR and 95% CI calculations
for each of the four DNMT3L SNPs are shown in Table III. The lowest P values
determined by the trend test were observed in the DNMT3L
c.832C/T (rs7354779) SNP (Ptrend=0.114). No association
was identified between any of the four DNMT3L SNPs and
endometriosis associated infertility (Table III). In the dominant and
recessive inheritance models for the c.910-635A/G polymorphism, the
ORs were 1.150 (95% CI=0.783–1.689; P=0.476) and 1.085 (95%
CI=0.633-1.860; P=0.767), respectively. In evaluating the same
inheritance models for the c.832C/T SNP, the ORs were 1.346 (95%
CI=0.925–1.961; P=0.120) and 1.320 (95% CI=0.700–2.491; P=0.390),
respectively. In the dominant model for the c.812C/T and the
c.344+62C/T SNPs, the ORs were measured as 2.215 (95%
CI=0.351–18.011; P=0.324) and 0.826 (95% CI=0.295–2.313; P=0.806),
respectively.
 | Table IIIAssociation between DNMT3L
polymorphic variants and the risk of endometriosis. |
Table III
Association between DNMT3L
polymorphic variants and the risk of endometriosis.
| rs no. | Position |
Alleles3 | Genotype
casesb | Genotype
controlsb | Ptrend
value |
ORdommnt(95%CI)c; P-value |
ORrecessive (95%
CI)d; P-value |
|---|
| rs8129776 |
c.910–635A>G | A/G | 22/74/58 | 51/175/157 | 0.509 | 1.150
(0.783–1.689); 0.476 | 1.085
(0.633–1.860); 0.767 |
| rs7354779 | c.832T>C | C/T | 16/70/68 | 31/155/198 | 0.114 | 1.346
(0.925–1.961); 0.120 | 1.320
(0.700–2.491); 0.390 |
| rs 113593938 | c.812C>T | C/T | 0/2/152 | 0/2/382 | 0.342 | 2.215
(0.351–18.011); 0.324f | NA |
| rs2276248 | c.344+62T>C | C/T | 0/5/149 | 0/15/369 | 0.715 | 0.826
(0.295–2.313); 0.806f | NA |
Association between DNMT3L haplotypes and
endometriosis-associated infertility
The haplotype analyses of the DNMT3L SNPs did
not suggest any polymorphism combination to be a risk factor for
the development of endometriosis associated infertility (Table IV). The lowest overall P values;
P=0.046, (Pcorr=0.094) and P=0.046,
(Pcorr=0.109), were observed for haplotypes ATC
(rs8129776/rs7354779/rs113593938) and AT (rs8129776/rs7354779). The
DNMT3L SNPs featured in the present study ranged between
complete and weak pairwise LD values. The D′ and r2
values, as calculated from the control samples, ranged between
0.001 and 1.000 (Table V).
 | Table IVHaplotype analysis of DNMT3L
polymorphic variants. |
Table IV
Haplotype analysis of DNMT3L
polymorphic variants.
| Polymorphism | Haplotype | Frequency | Case, control
ratio | χ2 | P value | Pcorr
valuea |
|---|
|
rs8129776_rs7354779 | GT | 0.364 | 0.374, 0.359 | 0.214 | 0.644 | 0.887 |
| AT | 0.340 | 0.295, 0.358 | 3.994 | 0.046 | 0.109 |
| AC | 0.291 | 0.322, 0.278 | 2.112 | 0.146 | 0.301 |
|
rs7354779_rs113593938 | TC | 0.703 | 0.666, 0.717 | 2.837 | 0.092 | 0.097 |
| CC | 0.294 | 0.328, 0.280 | 2.445 | 0.118 | 0.122 |
|
rs113593938_rs2276248 | CT | 0.978 | 0.977, 0.978 | 0.004 | 0.953 | 1.000 |
| CC | 0.019 | 0.016, 0.020 | 0.131 | 0.717 | 0.930 |
|
rs8129776_rs7354779_rs113593938 | GTC | 0.362 | 0.371, 0.359 | 0.139 | 0.709 | 0.941 |
| ATC | 0.340 | 0.295, 0.358 | 3.992 | 0.046 | 0.094 |
| ACC | 0.288 | 0.319, 0.276 | 2.027 | 0.155 | 0.331 |
|
rs7354779_rs113593938_rs2276248 | TCT | 0.684 | 0.649, 0.698 | 2.402 | 0.121 | 0.188 |
| CCT | 0.294 | 0.328, 0.280 | 2.442 | 0.118 | 0.185 |
| TCC | 0.019 | 0.016, 0.020 | 0.131 | 0.717 | 0.992 |
|
rs8129776_rs7354779_rs113593938_rs2276248 | GTCT | 0.348 | 0.356, 0.345 | 0.124 | 0.725 | 0.985 |
| ATCT | 0.339 | 0.294, 0.358 | 3.965 | 0.047 | 0.131 |
| ACCT | 0.285 | 0.318, 0.272 | 2.364 | 0.124 | 0.361 |
| GTCC | 0.014 | 0.015, 0.014 | 0.007 | 0.933 | 1.000 |
 | Table VLinkage disequilibrium between
polymorphic variants of the DNMT3L gene in the control
samples. |
Table V
Linkage disequilibrium between
polymorphic variants of the DNMT3L gene in the control
samples.
| Polymorphism | Polymorphism
|
|---|
| rs8129776 | rs7354779 | rs113593938 | rs2276248 |
|---|
| rs8129776 | – | 0.960a | 1.000a | 0.554a |
| rs7354779 | 0.209 | – | 1.000 | 0.603a |
| rs113593938 | 0.001b | 0.007b | – | 1.000a |
| rs2276248 | 0.011b | 0.003b | 0.000b | – |
Meta-analysis
The association between the DNMT3L rs8129776
polymorphism and the risk of endometriosis is shown in Table VI. The meta-analysis was performed
between two datasets, comprising a French population from a study
by Borghese et al (22) and
the Polish population in the present study. Under the assumption of
a dominant model, no heterogeneity was identified between the
studies (Q test P value=0.255; I2=22.7%). The OR
(fixed-effect model) for patients with endometriosis with AG+GG,
vs. AA was 1.007 (95% CI: 0.739–1.370; P=0.967). Under the
assumption of a recessive and allelic model, a high level of
heterogeneity was observed between the two datasets (Q test P
value=0.006 and 0.013, respectively; I2>83%). The OR
(random effect model) for patients with endometriosis with the GG,
vs. AG+AA was 0.596 (95% CI: 0.176–2.010; P=0.404). The OR (random
effect model) for the G allele in patients with endometriosis was
0.836 (95% CI: 0.480–1.456; P=0.527).
 | Table VIMeta-analysis of the association
between the DNMT3L rs8129776 polymorphism and endometriosis
risk. |
Table VI
Meta-analysis of the association
between the DNMT3L rs8129776 polymorphism and endometriosis
risk.
| Model | Model and
study | Odds ratio | Lower limit | Upper limit | Z value | Heterogeneity
|
|---|
| P value | Q valuea | P valuea | I2
value |
|---|
| Dominant model
(AG+GG, vs. AA) | | | | | | | | |
| Borghese et
al (22) | 0.791 | 0.472 | 1.327 | 0.888 | 0.375 | 1.293 | 0.255 | 22.675 |
| Present study | 1.150 | 0.783 | 1.689 | 0.712 | 0.476 | | | |
| Fixed effect | | 1.007 | 0.739 | 1.370 | 0.042 | 0.967 | | | |
| Recessive model
(GG, vs. AG+AA) | | | | | | | | |
| Borghese et
al (22) | 0.313 | 0.155 | 0.632 | 3.242 | 0.001 | 7.569 | 0.006 | 86.789 |
| Present study | 1.085 | 0.633 | 1.86 | 0.296 | 0.767 | | | |
| Random effect | | 0.596 | 0.176 | 2.010 | −0.835 | 0.404 | | | |
| Allelic model (G,
vs. A) | | | | | | | | |
| Borghese et
al (22) | 0.622 | 0.436 | 0.889 | 2.610 | 0.009 | 6.120 | 0.013 | 83.659 |
| Present study | 1.096 | 0.834 | 1.440 | 0.661 | 0.509 | | | |
| Random effect | | 0.836 | 0.480 | 1.456 | −0.633 | 0.527 | | | |
Discussion
Endometriosis is an estrogen dependent inflammatory
disease, which is considered to arise in response to epigenetic
changes (13,16,27).
Such changes lead to enhancements in the estrogen activity and
invasiveness of endometriotic cells, and may be associated with
hypomethylation of the steroidogenic factor 1 (SF-1),
aromatase, estrogen receptor 2 and E-cadherin (13,16,27,28)
genes. By contrast, infertility in women presenting with
endometriosis may be associated with the hypermethylation of the
homeobox A10 (HOXA10), HOXA11 and progesterone
receptor (PR-B) genes (14,15,28,29).
DNMT3L is one of the essential molecules involved in
de novo DNA methylation during the epigenetic reprogramming
of cells (30). DNMT3L interacts
with either DNMT3A or DNMT3B to effect de novo DNA
methylation. DNMT3L may also bind to selective chromatin regions
that have specific histone modifications capable of modulating
DNMT3A action (31–33). The ability of DNMT3L to bind to
histone deacetylase 1 further suggests the active role that DNMT3L
has in regulating transcriptional repression at the chromatin level
(34). It has also been suggested
that hyperacetylation of histones may account for the
overexpression of G protein coupled estrogen receptor 1,
SF-1 and hypoxia inducible factor 1α in endometrial lesions
(35–38). Furthermore, decreased expression
and hypoacetylation of histones have been observed in endometriotic
cells for the CCAAT/enhancer binding protein, cyclin dependent
kinase (CDK) inhibitor 2A, CDK inhibitor 1A, CDK inhibitor
1B, checkpoint kinase 2, death receptor 6, E-cadherin and
HOXA10 genes (8,35,38,39).
The DNMT3L c.812C/T transition (rs113593938),
associated with subtelomeric hypomethylation, leads to the
replacement of an arginine residue with glutamine at position 271
of the DNMT3L protein, which can effect the stimulation of DNMT3A
(21). No significant associations
between the DNMT3L rs8129776, rs7354779, rs113593938 or
rs2276248 polymorphisms and stage I II endometriosis associated
infertility were identified in the present study, either on an
individual SNP basis or in combinations of SNPs. By contrast,
Borghese et al (22)
demonstrated an association between rs8129776 and the development
of endometrioma (22), and it was
further demonstrated that the ACCC+T haplotypes for rs8129776,
rs7354779, rs113593938 and rs2276248 served as risk factors for
endometrioma (22). Such
discrepancies between studies may be due to the use of patient
cohorts presenting with different classes of endometriosis. Neither
the meta-analyses performed by Borghese et al (22) nor the meta-analysis performed in
the present study revealed any association between the rs8129776
SNP and the development of endometriosis.
In addition, two genome wide association studies
(GWAS), which were performed in Caucasian and Japanese populations
failed to demonstrate an association between DNMT3L SNPs and
endometriosis (40,41). However, the Caucasian patients
included in these studies presented with an assortment of disparate
categories of endometriosis (40),
although no histological analyses were performed to confirm the
presence of systematic endometriosis in the Japanese patient cohort
(41). The GWAS also eliminated
several of the genetic variants, which were demonstrated to be
associated with disease, due to low significance.
In conclusion, the present study demonstrated that
the DNMT3L SNPs; rs8129776, rs7354779, rs113593938 and
rs2276248, are not risk factors for endometriosis. The present
study was performed on patients presenting with infertility and
stage I II endometriosis, and further investigations are required
in the future to include additional categories of endometriosis,
using a larger groups of patients from disparate cohorts.
Acknowledgments
This study was supported by Poznan University of
Medical Sciences (grant. no. 502-01-01124182-07474).
References
|
1
|
Chapron C, Fauconnier A, Vieira M, Barakat
H, Dousset B, Pansini V, Vacher Lavenu MC and Dubuisson JB:
Anatomical distribution of deeply infiltrating endometriosis:
Surgical implications and proposition for a classification. Hum
Reprod. 18:157–161. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Dun EC, Taylor RN and Wieser F: Advances
in the genetics of endometriosis. Genome Med. 2:752010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
de Ziegler D, Borghese B and Chapron C:
Endometriosis and infertility: Pathophysiology and management.
Lancet. 376:730–738. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bulun SE: Endometriosis. N Engl J Med.
360:268–279. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Berbic M and Fraser IS: Regulatory T cells
and other leukocytes in the pathogenesis of endometriosis. J Reprod
Immunol. 88:149–155. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zheng Y, Tabbaa ZM, Khan Z, Schoolmeester
JK, El Nashar S, Famuyide A, Keeney GL and Daftary GS: Epigenetic
regulation of uterine biology by transcription factor KLF11 via
posttranslational histone deacetylation of cytochrome p450
metabolic enzymes. Endocrinology. 155:4507–4520. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Izawa M, Taniguchi F, Terakawa N and
Harada T: Epigenetic aberration of gene expression in
endometriosis. Front Biosci (Elite Ed). 5:900–910. 2013. View Article : Google Scholar
|
|
8
|
Kai K, Nasu K, Kawano Y, Aoyagi Y,
Tsukamoto Y, Hijiya N, Abe W, Okamoto M, Moriyama M and Narahara H:
Death receptor 6 is epigenetically silenced by histone
deacetylation in endometriosis and promotes the pathogenesis of
endometriosis. Am J Reprod Immunol. 70:485–496. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Fung JN, Rogers PA and Montgomery GW:
Identifying the biological basis of GWAS hits for endometriosis.
Biol Reprod. 92:872015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Turek Plewa J and Jagodziński PP: The role
of mammalian DNA methyltransferases in the regulation of gene
expression. Cell Mol Biol Lett. 10:631–647. 2005.PubMed/NCBI
|
|
11
|
Lee B, Du H and Taylor HS: Experimental
murine endometriosis induces DNA methylation and altered gene
expression in eutopic endometrium. Biol Reprod. 80:79–85. 2009.
View Article : Google Scholar
|
|
12
|
Wu Y, Strawn E, Basir Z, Halverson G and
Guo SW: Promoter hypermethylation of progesterone receptor isoform
B (PR B) in endometriosis. Epigenetics. 1:106–111. 2006. View Article : Google Scholar
|
|
13
|
Xue Q, Lin Z, Cheng YH, Huang CC, Marsh E,
Yin P, Milad MP, Confino E, Reierstad S, Innes J and Bulun SE:
Promoter meth ylation regulates estrogen receptor 2 in human
endometrium and endometriosis. Biol Reprod. 77:681–687. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wu Y, Starzinski Powitz A and Guo SW:
Prolonged stimulation with tumor necrosis factor alpha induced
partial methylation at PR B promoter in immortalized epithelial
like endometriotic cells. Fertil Steril. 90:234–237. 2008.
View Article : Google Scholar
|
|
15
|
Wu Y, Halverson G, Basir Z, Strawn E, Yan
P and Guo SW: Aberrant methylation at HOXA10 may be responsible for
itsaberrant expression in the endometrium of patients with
endometriosis. Am J Obstet Gynecol. 193:371–380. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Xue Q, Lin Z, Yin P, Milad MP, Cheng YH,
Confino E, Reierstad S and Bulun SE: Transcriptional activation of
steroidogenic factor 1 by hypomethylation of the 5′ CpG island in
endometriosis. J Clin Endocrinol Metab. 92:3261–3267. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wu Y, Strawn E, Basir Z, Halverson G and
Guo SW: Aberrant expression of deoxyribonucleic acid
methyltransferases DNMT1, DNMT3A and DNMT3B in women with
endometriosis. Fertil Steril. 87:24–32. 2007. View Article : Google Scholar
|
|
18
|
Borghese B, Mondon F, Noël JC, Fayt I,
Mignot TM, Vaiman D and Chapron C: Gene expression profile for
ectopic versus eutopic endometrium provides new insights into
endometriosis oncogenic potential. Mol Endocrinol. 22:2557–2562.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kareta MS, Botello ZM, Ennis JJ, Chou C
and Chédin F: Reconstitution and mechanism of the stimulation of de
novo methylation by human DNMT3L. J Biol Chem. 281:25893–25902.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Borghese B, Barbaux S, Mondon F, Santulli
P, Pierre G, Vinci G, Chapron C and Vaiman D: Research resource:
Genome wide profiling of methylated promoters in endometriosis
reveals a subtelomeric location of hypermethylation. Mol
Endocrinol. 24:1872–1885. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
El Maarri O, Kareta MS, Mikeska T, Becker
T, Diaz-Lacava A, Junen J, Nüsgen N, Behne F, Wienker T, Waha A, et
al: A systematic search for DNA methyltransferase polymorphisms
reveals a rare DNMT3L variant associated with subtelomeric
hypomethylation. Hum Mol Genet. 18:1755–1768. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Borghese B, Santulli P, Héquet D, Pierre
G, de Ziegler D, Vaiman D and Chapron C: Genetic polymorphisms of
DNMT3L involved in hypermethylation of chromosomal ends are
associated with greater risk of developing ovarian endometriosis.
Am J Pathol. 180:1781–1786. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Revised american society for reproductive
medicine classification of endometriosis: 1996.Fertil Steril.
67:817–821. 1997.
|
|
24
|
Szczepańska M, Wirstlein P, Skrzypczak J
and Jagodziński PP: Polymorphic variants of CYP17 and CYP19A and
risk of infertility in endometriosis. Acta Obstet Gynecol Scand.
92:1188–1193. 2013.
|
|
25
|
Ioannidis JP, Patsopoulos NA and Evangelou
E: Uncertainty in heterogeneity estimates in meta-analyses. BMJ.
335:914–916. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Higgins JP, Thompson SG, Deeks JJ and
Altman DG: Measuring inconsistency in meta-analyses. BMJ.
327:557–560. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Izawa M, Taniguchi F, Uegaki T, Takai E,
Iwabe T, Terakawa N and Harada T: Demethylation of a nonpromoter
cytosine phosphate guanine island in the aromatase gene may cause
the aberrant up regulation in endometriotic tissues. Fertil Steril.
95:33–39. 2011. View Article : Google Scholar
|
|
28
|
Szczepańska M, Wirstlein P, Skrzypczak J
and Jagodziński PP: Expression of HOXA11 in the mid luteal
endometrium from women with endometriosis associated infertility.
Reprod Biol Endocrinol. 10:12012. View Article : Google Scholar
|
|
29
|
Wu Y, Starzinski Powitz A and Guo SW:
Trichostatin A, a histone deacetylase inhibitor, attenuates
invasiveness and reactivates E-cadherin expression in immortalized
endometriotic cells. Reprod Sci. 14:374–382. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Liao HF, Tai KY, Chen WS, Cheng LC, Ho HN
and Lin SP: Functions of DNA methyltransferase 3-like in germ cells
and beyond. Biol Cell. 104:571–587. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Hu JL, Zhou BO, Zhang RR, Zhang KL, Zhou
JQ and Xu GL: The N-terminus of histone H3 is required for de novo
DNA methylation in chromatin. Proc Natl Acad Sci USA.
106:22187–22192. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ooi SK, Qiu C, Bernstein E, Li K, Jia D,
Yang Z, Erdjument-Bromage H, Tempst P, Lin SP, Allis CD, et al:
DNMT3L connects unmethylated lysine 4 of histone H3 to de novo
methylation of DNA. Nature. 448:714–717. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhang Y, Jurkowska R, Soeroes S, Rajavelu
A, Dhayalan A, Bock I, Rathert P, Brandt O, Reinhardt R, Fischle W
and Jeltsch A: Chromatin methylation activity of Dnmt3a and
Dnmt3a/3 L is guided by interaction of the ADD domain with the
histone H3 tail. Nucleic Acids Res. 38:4246–4253. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Deplus R, Brenner C, Burgers WA, Putmans
P, Kouzarides T, de Launoit Y and Fuks F: DNMT3L is a
transcriptional repressor that recruits histone deacetylase.
Nucleic Acids Res. 30:3831–3838. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Monteiro JB, Colón Díaz M, García M,
Gutierrez S, Colón M, Seto E, Laboy J and Flores I: Endometriosis
is characterized by a distinct pattern of histone 3 and histone 4
lysine modifications. Reprod Sci. 21:305–318. 2014. View Article : Google Scholar :
|
|
36
|
Samartzis N, Samartzis EP, Noske A, Fedier
A, Dedes KJ, Caduff R, Fink D and Imesch P: Expression of the G
protein coupled estrogen receptor (GPER) in endometriosis: A tissue
microarray study. Reprod Biol Endocrinol. 2012:10–30. 2012.
|
|
37
|
Imesch P, Samartzis EP, Dedes KJ, Fink D
and Fedier A: Histone deacetylase inhibitors down regulate G
protein coupled estrogen receptor and the GPER antagonist G 15
inhibits proliferation in endometriotic cells. Fertil Steril.
100:770–776. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Nasu K, Kawano Y, Kai K, Aoyagi Y, Abe W,
Okamoto M and Narahara H: Aberrant histone modification in
endometriosis. Front Biosci (Landmark Ed). 19:1202–1214. 2014.
View Article : Google Scholar
|
|
39
|
Kawano Y, Nasu K, Hijiya N, Tsukamoto Y,
Amada K, Abe W, Kai K, Moriyama M and Narahara H:
CCAAT/enhancer-binding protein α is epigenetically silenced by
histone deacetylation in endometriosis and promotes the
pathogenesis of endometriosis. J Clin Endocrinol Metab.
98:E1474–E1482. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Painter JN, Anderson CA, Nyholt DR,
Macgregor S, Lin J, Lee SH, Lambert A, Zhao ZZ, Roseman F, Guo Q,
et al: Genome wide association study identifies a locus at 7p15.2
associated with endometriosis. Nat Genet. 43:51–54. 2011.
View Article : Google Scholar :
|
|
41
|
Uno S, Zembutsu H, Hirasawa A, Takahashi
A, Kubo M, Akahane T, Aoki D, Kamatani N, Hirata K and Nakamura Y:
A genome-wide association study identifies genetic variants in the
CDKN2BAS locus associated with endometriosis in Japanese. Nat
Genet. 42:707–710. 2010. View
Article : Google Scholar : PubMed/NCBI
|