Introduction
Sudden sensorineural hearing loss (SSHL) is defined
as a sensorineural loss of hearing function, generally in one ear,
occurring over a short period of time due to uncertain causes
(1). SSHL is characterized by a
loss of >30 dB in at least three audiometric frequencies over a
period of 12–72 h or more (2).
SSHL predominantly occurs in age groups ranging between 50 and 60
years and the morbidity, due to varied underlying causes, reaches
5–20 per 100,000 individuals each year with no difference in gender
or region (3). The risk factors of
SSHL include hypertension, hypotension, diabetes mellitus, stroke
and acquired and inherited cardiopathy (3,4).
Unhealthy lifestyle habits, including smoking, alcohol consumption,
a sedentary lifestyle and sleep deprivation can also lead to SSHL
(5,6), however, the exact etiology of SSHL
remains to be elucidated. Previous studies have identified a few
underlying events, including vascular compromise, cochlear membrane
rupture and viral infection, in SSHL (7–9). In
previous years, genetic predisposition to SSHL susceptibility has
been actively investigated. In this context, nitric oxide synthase
3, caveolin 1 and grainyhead-like 2 (GRHL2) are suggested to
be involved in the etiology of SSHL (10–12).
GRHL2, also called brother of mammalian grainyhead
or transcription factor cellular promoter 2-like 3, is a
transcription factor that belongs to the grainyhead-like family
(13). GRHL2 was initially
identified in Drosophila and is important in the
organization of septate junctions, and thus is critical for
maintaining apical barrier functions in the epithelium (14). In humans, GRHL2 is also
predominantly expressed in the epithelial tissues and is important
in embryonic development, terminal differentiation of epithelial
cells, establishment and maintenance of human mucociliary airway
epithelium and neural tube closure (15,16).
Multiple diseases are associated with GRHL2, including gastric
diseases, breast cancer and sensorineural hearing loss (SHL)
(15–17). The GRHL2 gene in humans is
located on chromosome 8q22.3, and includes 16 exons and 15 introns
(15,18). Genetic polymorphisms in
GRHL2 are associated with the development of SHL, including
noise-induced hearing loss (NIHL) and age-related hearing
impairment (ARHI) (15,19,20).
However, the connection between the GRHL2 gene and
susceptibility to SSHL, which is a very important category of SHL,
has not been thoroughly investigated.
In the present study, SSHL patients and healthy
individuals were compared for genetic variations in GRHL2.
Polymerase chain reaction-restriction fragment length polymorphism
(PCR-RFLP) was used to detect GRHL2 genotypes. The
polymorphisms in GRHL2 and their association with
susceptibility to SSHL in a Chinese population was thus
investigated in order to determine the etiology of SSHL and to
provide valuable clinical diagnostic tools for SSHL.
Subjects and methods
Study subjects
Between January 2009 and April 2014, 190 patients
with SSHL, referred to the Departments of Otorhinolaryngology Head
and Neck Surgery at Kaihua People's Hospital (Quzhou, China) and
Hangzhou First People's Hospital (Hangzhou, China), were selected,
and included 108 males and 82 females with an average age of
38.5±4.8 years. A total of 210 age- and gender-matched healthy
subjects were selected, including 115 males and 95 females with an
average age of 38.9±5.3 years. Pure-tone audiometry was performed
and tobacco smoking and alcohol consumption status were recorded in
all participants. The selection criteria were based on the Sudden
Sensorineural Hearing Loss Diagnosis and Treatment Guidelines
published by the Chinese Medical Association Archives of
Otolaryngology Head and Neck Surgery Branch (21). The diagnostic standards were as
follows: SSHL occurring in a few minutes, hours or within 3 days;
non fluctuating sensorineural hearing loss (categorized into mild,
moderate severe or even complete deafness), sensorineural hearing
loss of 20 dB or more over three contiguous audiometric
frequencies; unknown cause due to systemic or local factors;
accompanying with tinnitus, ear blockage sensation, and
non-recurrent dizziness, nausea and vomiting; no other cranial
nerve injury with the exception of eighth cranial nerve injury
(http://d.wanfangdata.com.cn/Periodical_zhebyhk200608003.aspx).
The study protocol was approved by the Institutional Review Board
of the Kaihua People's Hospital (Quzhou, China). Written informed
consent was signed by each subject.
DNA extraction from blood clots
A commercially available blood DNA purification kit
(Beijing CWBio Co., Ltd., Beijing, China) was used to extract
genomic DNA from blood clots. The eluted DNA solution was collected
and stored at −20°C.
DNA content and purity detection
A UV 260 spectrophotometer (Shimadzu Corp., Kyoto,
Japan) was used to measure the absorbance at wavelengths of 260 and
280 nm. The Lambert-Beer law was used to calculate sample
concentration based on the c = A260/(ε × b) equation, where ε is
the molar absorption coefficient, b is the optical path length and
c is the molar concentration. The A260/A280 ratio was used to
determine the sample purity. DNA concentration was adjusted at
>15 ng/µl and the purity of DNA was between 1.6 and
1.9.
Selection of GRHL2 SNPs
SNPs of the human GRHL2 gene on chromosome 8
(8q22.3) were retrieved and the data packet was downloaded from
NCBI-dbSNP (http://www.ncbi.nlm.nih.gov/SNP/) and HapMap
(http://SNP.cshl.org/cgi-Perl/gbrowse/haPmaP27_B36/)databases.
Haploview 4.2 (http://www.broad.mit.edu/mpg/haploview/) statistical
software was used to select the tag SNPs of GRHL2 and the
parameters were set for Han Chinese in Beijing with a minor allele
frequency (MAF) >10%, r2>0.8, D'=1. The base sequences of the
selected site were suitable for primers designed for PCR
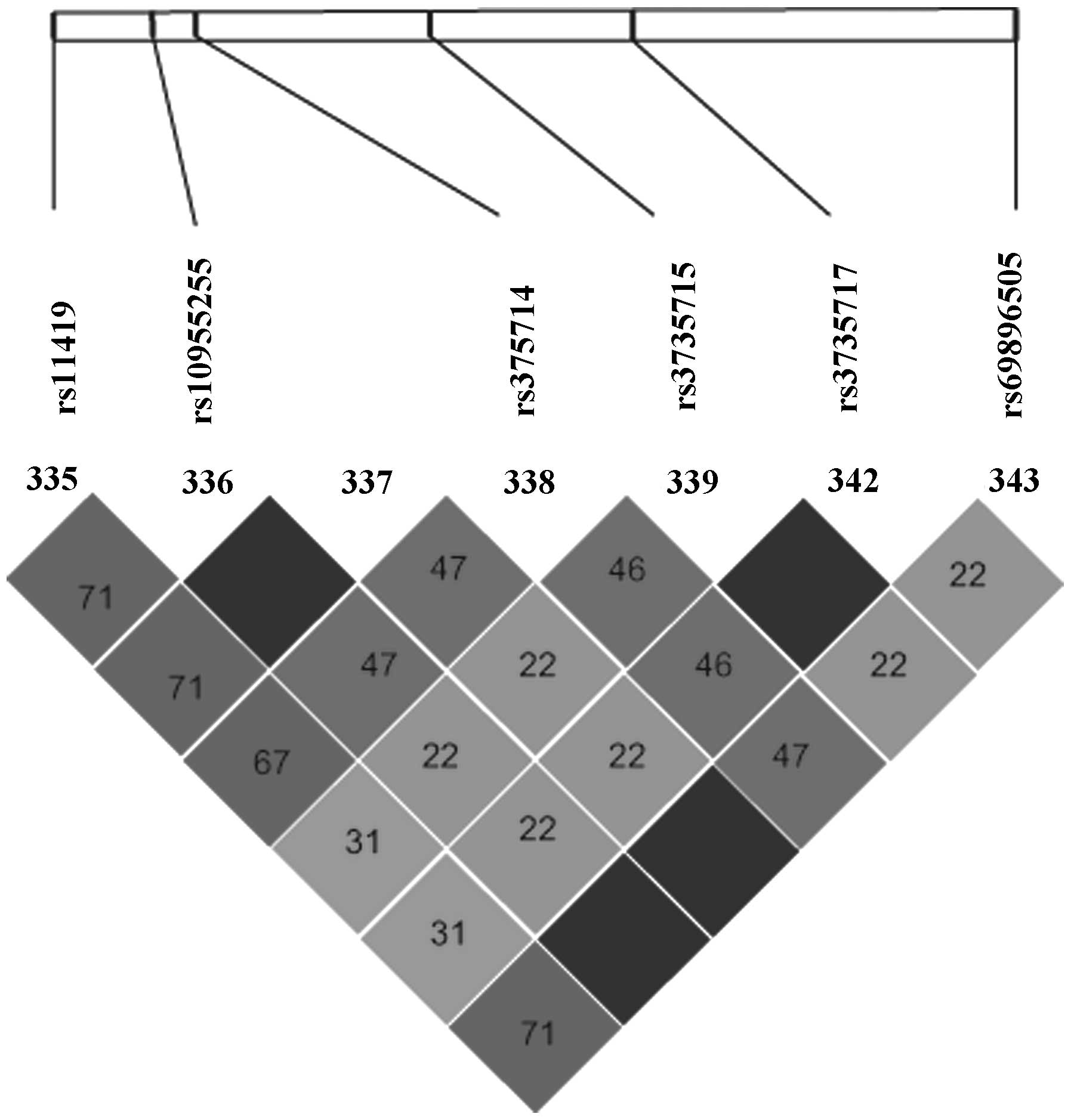
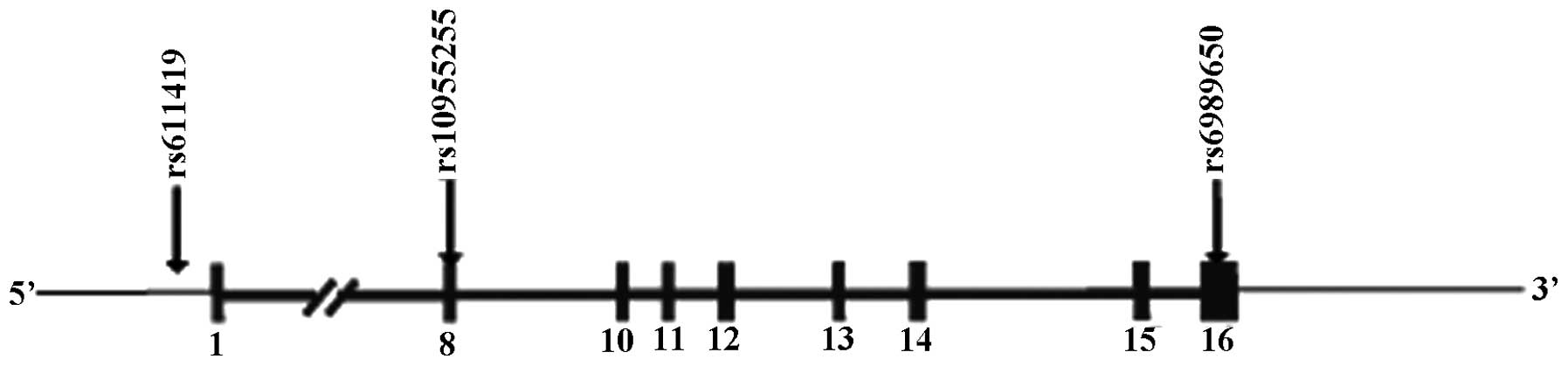
amplification. Three tag SNPs termed rs611419, rs10955255 and
rs6898650, were selected to stand for 100% SNP sites of the
GRHL2 gene (Fig. 1). The
loci of the three SNPs is present in Fig. 2.
Primer design
The PCR amplification primers for the three SNPs
were designed using Assay Designer 3.1 software (Sequenom, San
Diego, CA, USA) and their feasibility was based on the following
criteria: i) Primers and the templates should be complementary; ii)
stable dimers or hairpins between primers should be avoided; iii)
DNA mismatch with the template at non-target sites should be
avoided. Upstream and downstream primers for PCR reaction should
strictly comply with the design principles of primers. Three
primers are shown in Table I.
 | Table IPrimers for PCR amplification of
GRHL2 gene polymorphisms at rs611419, rs10955255 and
rs6898650. |
Table I
Primers for PCR amplification of
GRHL2 gene polymorphisms at rs611419, rs10955255 and
rs6898650.
| SNP | Primers for PCR
amplification (5′-3′) | Length (nt) | Molecular
weight (g/M) |
|---|
| rs611419 | F:
5′-GGAAATACTGGCACTCTCG-3′ | 19 | 5,809 |
| R: 5′
ACCTTCTCGTTCATCATCC 3′ | 19 | 5,650 |
| rs10955255 | F: 5′
CCGTGAATTGCTTGAGCACA 3′ | 20 | 6,113 |
| R: 5′
GGTTTGCAAAGTGAACATCAG 3′ | 21 | 6,489 |
| rs6898650 | F: 5′
GGATTTCACTGGTTTAGGG 3′ | 19 | 5,884 |
| R: 5′
AGCGTAGACTTCAAGTGAGC 3′ | 20 | 6,160 |
Genotyping
PCR-RFLP was used to determine the SNP genotype. PCR
reaction conditions were as follows: 5 min initial denaturation at
94°C, 36 cycles of 30 sec denaturation at 94°C, 40 sec annealing at
60°C and 45 sec extension at 72°C, followed by 7 min extension at
72°C. The amplification products were isolated and identified by
agarose gel electrophoresis at a voltage of 120 V for 30 min. PCR
products (15 µl) were digested with shrimp alkaline
phosphatase and the restriction enzyme ExoI at 60°C for 37
min and at 75°C for 15 min, respectively. The resulting restriction
fragments were electrophoresed in 2% agarose gels and visualized
under UV light. An ABI Prism 3130xl Genetic Analyzer (Applied
Biosystems, Foster City, CA, USA) was used to compare the resulting
restriction fragments and PCR products to identify the
genotype.
Statistical analysis
SPSS 17.0 statistical software package (SPSS, Inc.,
Chicago, IL, USA) was applied to analyze the data. Hardy-Weinberg
equilibrium was used to confirm the sample representation in the
population. Data are expressed as the mean ± standard deviation and
χ2 test was used to compare group differences. Logistic
regression analysis was used to calculate the odds ratios and 95%
confidence interval of each genotype was used to represent the
relative risk. P<0.05 was considered to indicate a statistically
significant difference.
Results
Clinical data in the SSHL group and the
control group
Table II shows the
profiles of the SSHL group and the control group. The auditory
threshold in the SSHL group (36.6±10.5 dB) was significantly higher
than the control group (13.2±3.0 dB) (P<0.01). No significant
differences in age, gender, smoking status and drinking status were
found between the SSHL group and the control group.
 | Table IICharacteristics of SSHL patients and
controls. |
Table II
Characteristics of SSHL patients and
controls.
| Variable | SSHL group | Control group | P-valuea |
|---|
| Age (years) | 38.5±4.8 | 38.9±5.3 | 0.431 |
| <40 (n, %) | 96 (50.5) | 98 (46.7) | 0.441 |
| ≥40 (n, %) | 94 (49.5) | 112 (53.3) | |
| Gender | | | |
| Male (n, %) | 108 (56.8) | 115 (54.8) | 0.676 |
| Female (n, %) | 82 (43.2) | 95 (45.2) | |
| Threshold (dB) | 36.6±10.5 | 13.2±3.0 | <0.0001 |
| Smoking status | | | |
| Non-smoker (n,
%) | 80 (42.1) | 90 (42.9) | 0.879 |
| Smoker (n, %) | 110 (57.9) | 120 (57.1) | |
| Drinking
status | | | |
| Non-drinker (n,
%) | 105 (55.3) | 116 (55.2) | 0.996 |
| Drinker (n,
%) | 85 (44.7) | 94 (44.8) | |
Basic information of the three SNPS in
the GRHL2 gene
Table III
describes the basic information of rs611419, rs10955255 and
rs6898650. rs611419 is located in the 5′ region with the A allele
and T allele. rs10955255 is located in intron 8 with the G allele
and A allele. rs6898650 is located in the 3′UTR with the C allele
and T allele. χ2 goodness-of-fit test was used to detect
the genotype frequency distribution of rs611419, rs10955255 and
rs6898650 in the SSHL group and the control group, and the results
demonstrated that the three sites in the two groups conformed to
the Hardy-Weinberg equilibrium (all P>0.05).
 | Table IIIBasic information and HWE results of
the three SNPs in the GRHL2 gene. |
Table III
Basic information and HWE results of
the three SNPs in the GRHL2 gene.
| SNP (rs no.) | Base change | Location | MAF
|
HWE(χ2/P)
|
|---|
| HapMapa | SSHL group | Control group | SSHL group | Control group |
|---|
| rs611419 | A>T | 5′ gene site | 0.356 | 0.425 | 0.482 | 0.048/0.826 | 0.075/0.785 |
| rs10955255 | G>A | Intron 8 | 0.116 | 0.090 | 0.103 | 1.305/0.253 | 0.081/0.776 |
| rs6989650 | C>T | 3′UTR | 0.233 | 0.208 | 0.222 | 0.224/0.636 | 1.343/0.247 |
Genotype and allele frequency
distributions in the GRHL2 gene
Table IV shows the
results from logistic regression regarding the risk of SSHL.
rs611419 of the GRHL2 gene may be a protective factor for
SSHL (AT+TT vs. AA: OR=0.63, 95% CI=0.41–0.98, P=0.038). Similarly,
rs10955255 site polymorphisms may reduce the risk of SSHL (AA vs.
GG: OR=0.54, 95% CI=0.31–0.95, P=0.32; GA+AA vs. GG: OR=0.58, 95%
CI=0.38–0.89, P=0.012). However, genotype and allele frequencies of
rs6989650 demonstrated no statistical differences in the SSHL group
and the control group (P>0.05).
 | Table IVAssociation of genotypes and allele
frequencies of the GRHL2 genetic polymorphisms and risk of
SSHL. |
Table IV
Association of genotypes and allele
frequencies of the GRHL2 genetic polymorphisms and risk of
SSHL.
| Genotype | SSHL group (n=190)
| Control group
(n=210)
| P-valuea | OR (95% CI)b |
|---|
| n | % | n | % |
|---|
| rs611419 | | | | | | |
| AA | 65 | 34.1 | 52 | 24.8 | 0.046 | 1.00 (Ref.) |
| AT | 91 | 48.1 | 103 | 49.0 | 0.140 | 0.71
(0.45–1.12) |
| TT | 34 | 17.8 | 55 | 26.2 | 0.014 | 0.49
(0.28–0.87) |
| AT+TT | 125 | 65.9 | 158 | 75.2 | 0.038 | 0.63
(0.41–0.98) |
| rs10955255 | | | | | | |
| GG | 71 | 37.5 | 54 | 25.7 | 0.040 | 1.00 (Ref.) |
| AG | 84 | 44.1 | 107 | 51.0 | 0.026 | 0.60
(0.38–0.94) |
| AA | 35 | 18.4 | 49 | 23.3 | 0.032 | 0.54
(0.31–0.95) |
| AG+AA | 119 | 62.5 | 156 | 74.3 | 0.012 | 0.58
(0.38–0.89) |
| rs6989650 | | | | | | |
| CC | 123 | 64.7 | 134 | 63.8 | 0.462 | 1.00 (Ref.) |
| CT | 61 | 32.2 | 64 | 30.6 | 0.863 | 1.04
(0.68–1.59) |
| TT | 6 | 3.1 | 12 | 5.6 | 0.233 | 0.54
(0.20–1.50) |
| CT+TT | 67 | 35.3 | 76 | 36.2 | 0.847 | 0.96
(0.64–1.45) |
Combined analysis between the three SNPs
and SSHL risk
In order to examine the interaction between these
polymorphisms, the three polymorphisms were combined for the
analysis. There was a marked association between the combined
genotypes and SSHL risk (P=0.035). Subjects carrying 3–8 variant
alleles demonstrated a lower SSHL risk compared with subjects
carrying 0–2 variant alleles (OR=0.59, 95% CI=0.36–0.96, P=0.034;
Table V).
 | Table VAssociation of frequency distribution
of the combined genotypes of GRHL2 polymorphisms and the
risk of SSHL. |
Table V
Association of frequency distribution
of the combined genotypes of GRHL2 polymorphisms and the
risk of SSHL.
| Number of
variantsa | SSHL group (n=190)
| Control group
(n=210)
| P-valueb | OR (95% CI)c |
|---|
| n | % | n | % |
|---|
| 0 | 4 | 2.1 | 1 | 0.5 | 0.035 | |
| 1 | 18 | 9.5 | 8 | 3.8 | | |
| 2 | 25 | 13.2 | 25 | 11.9 | | |
| 3 | 34 | 17.9 | 60 | 28.6 | | |
| 4 | 39 | 20.5 | 45 | 21.4 | | |
| 5 | 43 | 22.6 | 37 | 17.6 | | |
| 6 | 27 | 14.2 | 34 | 16.2 | | |
| Combined
genotype | | | | | | |
| 0–2 | 47 | 24.7 | 34 | 16.2 | 0.034 | 1.00 (Ref.) |
| 3–8 | 143 | 75.3 | 176 | 83.8 | | 0.59
(0.36–0.96) |
Stratification analysis of the combined
genotypes of the GRHL2 polymorphisms and SSHL risk
Stratification analysis based on age, gender,
smoking status and drinking status was conducted to analyze the
effect of the combined genotypes of the three SNPs on SSHL risk. In
individuals who consumed alcohol, subjects carrying 3–8 variant
alleles were more resistant to SSHL compared with subjects carrying
0–2 variant alleles (OR=0.40, 95% CI=0.21–0.76, P=0.004). However,
no significant differences were found between the frequency of
combined genotypes and age, gender and smoking status (all
P>0.05; Table VI).
 | Table VIStratification analyses between the
combined genotypes of the GRHL2 polymorphisms and risk of
SSHL. |
Table VI
Stratification analyses between the
combined genotypes of the GRHL2 polymorphisms and risk of
SSHL.
| Variable | SSHL/control
group | Combined genotypes
| P-valuea | OR (95% CI)b |
|---|
| 0–2 | 3–8 |
|---|
| Age (years) | | | | | |
| <40 | 96/98 | 21/13 | 75/85 | 0.115 | 1.83
(0.86–3.91) |
| ≥40 | 94/112 | 26/21 | 68/91 | 0.129 | 1.66
(0.86–3.19) |
| Gender | | | | | |
| Male | 108/115 | 36/28 | 133/159 | 0.121 | 1.54
(0.89–2.65) |
| Female | 82/95 | 11/6 | 10/17 | 0.074 | 3.12
(0.88–11.04) |
| Smoking status | | | | | |
| Non-smoker | 80/90 | 20/14 | 60/76 | 0.124 | 1.81
(0.84–3.88) |
| Smoker | 110/120 | 27/20 | 83/100 | 0.139 | 1.63
(0.85–3.11) |
| Drinking
status | | | | | |
| Non-drinker | 105/116 | 21/13 | 75/38 | 0.621 | 0.82
(0.37–1.81) |
| Drinker | 85/94 | 26/21 | 68/138 | 0.004 | 0.40
(0.21–0.76) |
Discussion
The etiology of SSHL remains to be elucidated.
Vascular occlusion is a potential cause of impaired cochlear
perfusion, and risk factors, including factor V Leiden and
prothrombin G20210A that lead to vascular perfusion defects, are
also suspected to be important in SSHL (22–24).
By contrast, inner ear damage was also associated with SSHL and
Yamamoto et al reported that insulin-like growth factor-1, a
growth factor involved in the development of the human inner ear,
enhanced the regeneration of hair cells damaged in SSHL (25). Genetic factors associated with SSHL
have generated significant interest. The present study investigated
the association between GRHL2 genetic polymorphisms and
susceptibility to SSHL. The exact protective function of
GRHL2 in reducing susceptibility to SSHL remains to be
elucidated. As a transcription factor, GRHL2 is important in
embryonic development and otic epithelial tissue differentiation by
promoting apical junction maturation (14,26,27).
GRHL2 could also be involved in apical barrier formation and
activate adult antimicrobial defense, which may be associated with
SSHL observed in viral infections (28,29).
GRHL2 could also regulate the expression of Rho GEF 19,
which is involved in wound healing, and thus the protective role of
GRHL2 may involve tissue repair processes (30).
In the present study, SSHL patients with the
rs611419 AT/TT genotype demonstrated a lower susceptibility to SSHL
than patients with the AA genotype, suggesting that the T allele in
rs611419 polymorphism may be a protective factor to SSHL in the
Chinese population. In addition, the significant difference in SSHL
risk between genotype AA and GG in rs10955255 demonstrated that the
A allele in rs10955255 reduced the risk of SSHL. Nevertheless, no
association between the rs6989650 polymorphism and SSHL was
identified, suggesting that rs6989650 may not be a risk factor for
SSHL. Van Laer et al analyzed 703 SNPs and found that
rs10955255 and rs2127034 in GRHL2 are ranked the top two in
association with ARHI (13). In
addition, Li et al also observed that rs611419 was
significantly associated with NIHL (15), in agreement with the results of the
present study.
In the present study, a marked association was found
between the combined genotypes and SSHL risk. The results
demonstrated that the combined genotypes correlated with lower SSHL
incidence and subjects carrying 3–8 variant alleles demonstrated a
significantly lower SSHL risk than subjects carrying 0–2 variant
alleles. Consistent with our results, Li et al also found
the combined genotypes with 3–8 variant alleles were associated
with a decreased risk of NIHL compared with those with 0–2 variant
alleles (15). In regards to
alcohol consumption, when the combined genotype consisted of 3–8
variant alleles, the risk of SSHL was lower compared with in
subjects carrying 0–2 variant alleles. No association was found
between the combined genotype frequency and age, gender and smoking
status. Bibulosity is one of the potential risk factors of SSHL
(31). Additionally, alcohol
consumption can psychologically and physiologically interact with
hearing loss (32).
The limitations of the present study are worth
mentioning. The accumulation of retrospective data was not under
control of the researchers analyzing the data, leading to
inevitable bias. In addition, other types of gene should be taken
into consideration when analyzing the genetic causes of SSHL.
Taken together, the GRHL2 genetic
polymorphisms, rs611419 and rs10955255, may confer protection
against SSHL and reduce the risk of SSHL. The combination of the
genotypes rs611419, rs10955255 and rs6989650 in the GRHL2
gene is associated with a reduced risk of SSHL with more variant
alleles. The present study provides fundamental genetic data for
the GRHL2 gene and demonstrates its association with SSHL,
and thus the polymorphisms could be potential genetic biomarkers
for investigating the mechanisms underlying SSHL.
Acknowledgments
This study was supported by grants from the Science
Technology Department of Zhejiang Province (grant no. 2014C33144)
and Science Technology Bureau of Quzhou (grant no. 2013128). The
authors would like to thank the researchers for their hard work and
reviewers for their valuable advice.
References
|
1
|
Lee HS, Lee YJ, Kang BS, Lee BD and Lee
JS: A clinical analysis of sudden sensorineural hearing loss cases.
Korean J Audiol. 18:69–75. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Na SY, Kim MG, Hong SM, Chung JH, Kang HM
and Yeo SG: Comparison of sudden deafness in adults and children.
Clin Exp Otorhinolaryngol. 7:165–169. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kuhn M, Heman-Ackah SE, Shaikh JA and
Roehm PC: Sudden sensorineural hearing loss: A review of diagnosis,
treatment and prognosis. Trends Amplif. 15:91–105. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chau JK, Lin JR, Atashband S, Irvine RA
and Westerberg BD: Systematic review of the evidence for the
etiology of adult sudden sensorineural hearing loss. Laryngoscope.
120:1011–1021. 2010.PubMed/NCBI
|
|
5
|
Talaat HS, Metwaly MA, Khafagy AH and
Abdelraouf HR: Dose passive smoking induce sensorineural hearing
loss in children? Int J Pediatr Otorhinolaryngol. 78:46–49. 2014.
View Article : Google Scholar
|
|
6
|
Nomura K, Nakao M and Yano E: Hearing loss
associated with smoking and occupational noise exposure in a
Japanese metal working company. Int Arch Occup Environ Health.
78:178–184. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Seo JH, Jeon EJ, Park YS, Kim J, Chang KH
and Yeo SW: Meteorological conditions related to the onset of
idiopathic sudden sensorineural hearing loss. Yonsei Med J.
55:1678–1682. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ryu OH, Choi MG, Park CH, Kim DK, Lee JS
and Lee JH: Hyperglycemia as a potential prognostic factor of
idiopathic sudden sensorineural hearing loss. Otolaryngol Head Neck
Surg. 150:853–858. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jun HJ, Chang J, Im GJ, Kwon SY, Jung H
and Choi J: Analysis of frequency loss as a prognostic factor in
idiopathic sensorineural hearing loss. Acta Otolaryngol.
132:590–596. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Teranishi M, Uchida Y, Nishio N, Kato K,
Otake H, Yoshida T, Suzuki H, Sone M, Sugiura S, Ando F, et al:
Polymorphisms in genes involved in the free-radical process in
patients with sudden sensorineural hearing loss and Meniere's
disease. Free Radic Res. 47:498–506. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bovo R, Ciorba A and Martini A:
Environmental and genetic factors in age-related hearing
impairment. Aging Clin Exp Res. 23:3–10. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Stew BT, Fishpool SJ and Williams H:
Sudden sensorineural hearing loss. Br J Hosp Med (Lond). 73:86–89.
2012. View Article : Google Scholar
|
|
13
|
Van Laer L, Van Eyken E, Fransen E, Huyghe
JR, Topsakal V, Hendrickx JJ, Hannula S, Mäki-Torkko E, Jensen M,
Demeester K, et al: The grainyhead like 2 gene (GRHL2), alias
TFCP2L3, is associated with age-related hearing impairment. Hum Mol
Genet. 17:159–169. 2008. View Article : Google Scholar
|
|
14
|
Werth M, Walentin K, Aue A, Schönheit J,
Wuebken A, Pode-Shakked N, Vilianovitch L, Erdmann B, Dekel B,
Bader M, et al: The transcription factor grainyhead-like 2
regulates the molecular composition of the epithelial apical
junctional complex. Development. 137:3835–3845. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li X, Huo X, Liu K, Li X, Wang M, Chu H,
Hu F, Sheng H, Zhang Z and Zhu B: Association between genetic
variations in GRHL2 and noise-induced hearing loss in Chinese high
intensity noise exposed workers: A case-control analysis. Ind
Health. 51:612–621. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Xiang J, Fu X, Ran W, Chen X, Hang Z, Mao
H and Wang Z: Expression and role of grainyhead-like 2 in gastric
cancer. Med Oncol. 30:7142013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Werner S, Frey S, Riethdorf S, Schulze C,
Alawi M, Kling L, Vafaizadeh V, Sauter G, Terracciano L, Schumacher
U, et al: Dual roles of the transcription factor grainyhead-like 2
(GRHL2) in breast cancer. J Biol Chem. 288:22993–33008. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lin YH, Wu CC, Hsu CJ, Hwang JH and Liu
TC: The grainyhead-like 2 gene (GRHL2) single nucleotide
polymorphism is not associated with age-related hearing impairment
in Han Chinese. Laryngoscope. 121:1303–1307. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Fransen E, Topsakal V, Hendrickx JJ, Van
Laer L, Huyghe JR, Van Eyken E, Lemkens N, Hannula S, Mäki-Torkko
E, Jensen M, et al: Occupational noise, smoking and a high body
mass index are risk factors for age-related hearing impairment and
moderate alcohol consumption is protective: A European
population-based multicenter study. J Assoc Res Otolaryngol.
9:264–276; discussion 1–3. 2008. View Article : Google Scholar :
|
|
20
|
Konings A, Van Laer L, Wiktorek-Smagur A,
Rajkowska E, Pawelczyk M, Carlsson PI, Bondeson ML, Dudarewicz A,
Vandevelde A, Fransen E, et al: Candidate gene association study
for noise-induced hearing loss in two independent noise-exposed
populations. Ann Hum Genet. 73:215–224. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chinese Medical Association Archives of
Otolaryngology Head and Neck Surgery Magazine Editorial Committee,
Chinese Medical Association Archives of Otolaryngology Head and
Neck Surgery Branch: Sudden sensorineural hearing loss diagnosis
and treatment guidelines. Chinese Journal of Otolaryngology Head
and Neck Surgery. 6:443–447. 2015.In Chinese.
|
|
22
|
Görür K, Tuncer U, Eskandari G, Ozcan C,
Unal M and Ozsahinoglu C: The role of factor V Leiden and
prothrombin G20210A mutations in sudden sensorineural hearing loss.
Otol Neurotol. 26:599–601. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lovato A, Tormene D, Staffieri C, Breda S,
Staffieri A and Marioni G: Sudden hearing loss followed by deep
vein thrombosis and pulmonary embolism in a patient with factor V
Leiden mutation. Int J Audiol. 53:625–628. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lan MY, Shiao JY, Hsu YB, Lin FY and Lin
JC: A preliminary study on the role of inherited prothrombotic risk
factors in Taiwanese patients with sudden sensorineural hearing
loss. Eur Arch Otorhinolaryngol. 268:817–822. 2011. View Article : Google Scholar
|
|
25
|
Yamamoto N, Nakagawa T and Ito J:
Application of insulin-like growth factor-1 in the treatment of
inner ear disorders. Front Pharmacol. 5:2082014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Vona B, Nanda I, Neuner C, Müller T and
Haaf T: Confirmation of GRHL2 as the gene for the DFNA28 locus. Am
J Med Genet A. 161A:2060–2065. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang S and Samakovlis C: Grainy head and
its target genes in epithelial morphogenesis and wound healing.
Curr Top Dev Biol. 98:35–63. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Paré A, Kim M, Juarez MT, Brody S and
McGinnis W: The functions of grainy head-like proteins in animals
and fungi and the evolution of apical extracellular barriers. PLoS
One. 7:e362542012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kikidis D, Nikolopoulos TP, Kampessis G,
Stamatiou G and Chrysovergis A: Sudden sensorineural hearing loss:
Subclinical viral and toxoplasmosis infections as aetiology and how
they alter the clinical course. ORL J Otorhinolaryngol Relat Spec.
73:110–115. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Boglev Y, Wilanowski T, Caddy J, Parekh V,
Auden A, Darido C, Hislop NR, Cangkrama M, Ting SB and Jane SM: The
unique and cooperative roles of the Grainy head-like transcription
factors in epidermal development reflect unexpected target gene
specificity. Dev Biol. 349:512–522. 2011. View Article : Google Scholar
|
|
31
|
Lin RJ, Krall R, Westerberg BD, Chadha NK
and Chau JK: Systematic review and meta-analysis of the risk
factors for sudden sensorineural hearing loss in adults.
Laryngoscope. 122:624–635. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Pinquart M and Pfeiffer JP: Alcohol use
among students with and without hearing loss. J Deaf Stud Deaf
Educ. 20:82–90. 2015. View Article : Google Scholar
|