Introduction
Sterol regulatory element binding protein-1c
(SREBP-1c) is one of SREBP family member and regulate the
expression of lipogenic genes (1,2).
Recent in vivo studies suggest that SREBP-1c is central to
most hepatic lipogenic genes. SREBP-1c is involved in encoding
enzymes that catalyze various steps in the fatty acid (FA) and
triglyceride (TG) synthesis pathway, including fatty acid synthase
(FAS) (3) and acetyl-CoA
carboxylase α (ACCα) (4). SREBP-1c
facilitates FAS synthesis and incorporates FA into TG (5). Abnormally high SREBP-1c levels may
cause hepatic TG accumulation and potentially induce other lipid
disorders (6).
SREBP-1c is also a target of insulin, which
activates transcription of the gene encoding SREBP1 by partially
increasing the activity of liver X receptor α (LXRα) (7). SREBP-1c promoter activity is
partially upregulated through LXRα/RXR heterodimer binding to the
promoter area of LXR elements (LXREs) (8). However, a study by Kamei's revealed
that a LXRα/RXR heterodimer binding to LXREs promotes was
antagonized by forkhead Box O1 (FoxO1) (9). FoxO1, which belongs to the a FoxO
transcription factor family, is typically regarded as a tumor
suppressor and lies downstream of the phosphoinositide 3-kinase
(PI3K)/AKT signaling pathway. A study by Deng et al
(10) indicated that FoxO1 was
associated with the SREBP-1c promoter and negatively regulated
srebp1 gene expression via multiple mechanisms including
modification of the promoter binding sites of Sp1 and SREBP-1c
itself. In addition, the present study supported the hypothesis
that increased FoxO1 levels decreased the level of SREBP-1c.
Long noncoding RNA (lncRNA) are transcribed RNA
molecules that lack an open reading frame and are longer than 200
nucleotides (11). LncRNA regulate
gene expression by diverse mechanisms (12). Recent evidence also suggests that
lncRNA are abnormally expressed in liver cancer (13); an example is highly upregulated in
liver cancer (HULC) (14).
Long non-coding RNA lncHR1 (HCV regulated 1, termed
lncHR1), was first reported as upregulated in Huh7 cell infected by
hepatitis C virus (HCV). As a new long non-coding RNA, lncHR1
exhibited obviously regulatory functions via SREBP-1c, the
accumulation of TG and lipid droplets in cells, and in transgenic
mouse model (15).
Metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) has
been reported to regulate SREBP-1c expression at a
post-transcriptional level had been verified (16). However, the molecular mechanisms
behind the regulating of lncHR1 by SREBP-1c have yet to be
determined.
In this study, we initially studied the molecular
mechanisms behind the regulation of SREBP-1c by lncHR1. The results
showed that lncHR1 may affect the phosphorylation of the
PDK1/AKT/FoxO1 signaling pathway, subsequently regulating SREBP-1c
protein levels in hepatocellular carcinoma lines. Thus, our study
offered a new information regarding lncRNA regulation of SREBP-1c
through the AKT/FoxO1 signaling pathway and provided a practical
and efficient platform for studying the function of lncRNA in lipid
metabolism.
Materials and methods
Cells and reagents
The Huh7 human hepatoma cell line was purchased from
Apath, Inc. (Brooklyn, NY, USA) (17). The cells were cultured in DMEM
medium from Thermo Fisher Scientific, Inc. (Waltham, MA, USA),
supplemented with 10% FBS, 1% NEAA, 1% penicillin, and 1%
streptomycin at 37°C and 5% CO2. Antibodies for
phospho-AKT (no. 9275), FoxO1 (no. 2280) and p-FoxO1 (no. 84192)
were obtained from Cell Signaling Technology, Inc. (Danvers, MA,
USA), antibodies for SREBP-1c (sc-13551) were from Santa Cruz
Biotechnology, Inc., (Dallas, TX, USA), β-actin (A3854) was from
Sigma-Aldrich (Merck KGaA, Darmstadt, Germany), and the Histone 3
(AF0009), IGF-1 (P5502) and LY294002 (S17375502) were from Beyotime
Institute of Biotechnology (Shanghai, China). Horseradish
peroxidase (HRP)-conjugated secondary antibodies (323-001-021) were
from Jackson Immuno-Research (West Grove, PA, USA).
Cell culture, plasmids and transient
transfection
LncHR1 was cloned into the BglII and
XhoI sites of a pMIR-Empty-DFT overexpression vector. shRNA
targeting lncHR1 were cloned into the BglII and XhoI
sites of the pQSUP-R plasmid vector. The efficiency of
overexpression or knockdown plasmids has been verified as
previously described (15).
Cultured cells were inoculated in 24 well plates
with 10% FBS medium and incubated overnight at 37°C and 5%
CO2 until the cells grew to 80% confluence. Before
transfection, cell medium was replaced with fresh medium not
containing with FBS for 1 h. DNA (1 µg) plasmids were diluted with
serum-free DMEM medium and 2 µl of the transfection reagent D293,
which was diluted with serum-free DMEM medium, and then mixed well.
The diluted transfection reagents were added to the diluted plasmid
by drop by drop, gently mixed and then kept at room temperature for
15 min. The suspension of the plasmid and the transfection reagent
was added into the transfected cells evenly with gently agitation
to ensure even distribution. The medium was replaced with fresh
medium containing 10% FBS after 5–8 h of transfection. After 48 h
of continuous culture, cells were collected and then used for
experiments.
Oil red O staining
Oil red O staining was done using the
following steps
Huh7 cells were inoculated in the 24 well plates
with the sterilized round glass slices and then carried out routine
cell processing. Pretreated cells were fixed with 3%
paraformaldehyde for 10 min at room temperature and then were
permeabilized with 0.4% Triton X-100 for 10 min at room
temperature. Pretreated cells were stained for 1 h with freshly
diluted 0.5 mM oil red O dissolved in 60% isopropanol. Stained
cells were washed with 50% ethyl alcohol twice for 5 min and then
was washed with PBS twice for 5 min. When washing the second times,
PBS with 4′,6-diamidino-2-phenylindole (DAPI) was added to stain
the nuclei. Last, at least 5 random fields were observed under a
Leica TCS-SL confocal microscope. The intensity was quantified with
Image J software (NIH).
Western blotting and
immunofluorescence analysis
Treated cells were collected and washed twice with
PBS. For cell lysate preparation, the monolayer of plates cells was
lysed with lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.5%
Nonidet P-40, 50 mM NaF, 1 mM Na3VO4, 5 mM
β-glycerophosphate, 1 mM dithiothreitol, and 1 mM
phenylmethylsulfonyl fluoride). The lysate was clarified by
centrifuging at 14,000 × g for 20 min. Samples were mixed with 2X
SDS loading buffer, boiled, and loaded onto a 10–12% polyacrylamide
gel. After electrophoresis, the proteins were transferred onto a
polyvinylidene difluoride membrane (Pall Corporation, East Hills,
NY, USA). The resulting blots were blocked with 5% bovine serum
albumin (BSA) for phosphoprotein antibodies, or 10% skim milk for
non-phosphoprotein antibodies for 1 h and incubated with the
selected primary antibody overnight at 4°C. The secondary antibody
used in the immunoblot was a 1:2,000 or 1:5,000 dilution of
HRP-linked anti-IgG. ECL reagent (Amersham Biosciences, England,
UK) was used as the substrate for detection, and the membrane was
exposed to an x-ray film for visualization. The density of the
bands was quantified using the NIH image software and were
normalized to the density of the control band.
The nuclear-cytosol fractionation experiments were
performed using a Nuclear-Cytosol Extraction kit (Applygen
Technologies, Inc., Beijing, China). Western blotting was performed
to quantify protein according to standard protocols published in
the literature. Cells were mounted on glass slides and the
immunofluorescence assay was performed as previously described
(2). Images were captured with a
Leica TCS-SL confocal microscope.
TG quantification
Intracellular TG were measured with a TG Assay kit
from Applygen Technologies, Inc., according to the manufacturer's
instructions. TG values are expressed as µM (at the cellular
level).
Statistical analysis
Bar graphs depict means ± standard deviation of at
least three independent experiments. A Student's t-test was used to
analyze differences between means for two independent samples, and
P<0.05 was considered to indicate a statistically significant
difference.
Results
It has been reported that lncHR1 regulated SREBP-1c
levels in Huh7 cells and transgenic mice fed a high fat diet
(15). However, the detailed
mechanism behind the regulation of SREBP-1c by lncHR1 has not been
further studied. Therefore, we utilized a cells model of the TG
accumulation induced by oleic acid (OA) and then studied the
effects of lncHR1 on SREBP-1c.
LncHR1 inhibited SREBP-1c through
regulation of AKT phosphorylation levels in the TG accumulation
model
The cell model of OA-induced TG accumulation was
used. The results showed that, in Huh7 cells, OA treatment for 24 h
obviously increased SREBP-1c protein (Fig. 1A) and mRNA level (Fig. 1B) compared to control cells. In
addition, there was no obvious change in lncHR1mRNA levels (data
not shown). In hepatic cells, higher SREBP-1c is conductive to the
synthesis and accumulation of TG. Therefore, we analyzed the TG
levels in the hepatic cell model. The results revealed an increase
of TG in OA treated cells (Fig.
1C). In the liver, SREBP-1c is an activator of FAS, which
enhances FA synthesis and accelerates TG accumulation (18). TG are normally stored in the form
of neutral lipid droplets in hepatocytes or secreted as TG-enriched
lipoproteins into the bloodstream. Through the oil red O staining
in morphology, we observed that OA treatment significantly
increased lipid droplets compared with untreated controls (Fig. 1D). Consistently, the volume of
intracellular lipid droplets was also increased (Fig. 1E), as determined via
immunofluorescence assay. These data verified that the model of
OA-induced hepatic cell TG accumulation. Therefore, this model can
be used for subsequent studied into the regulation of SREBP-1c
protein by lncHR1. The efficiency of overexpression or knockdown
lncHR1 vectors was previously verified (15) and the two vectors were used in this
experiment.
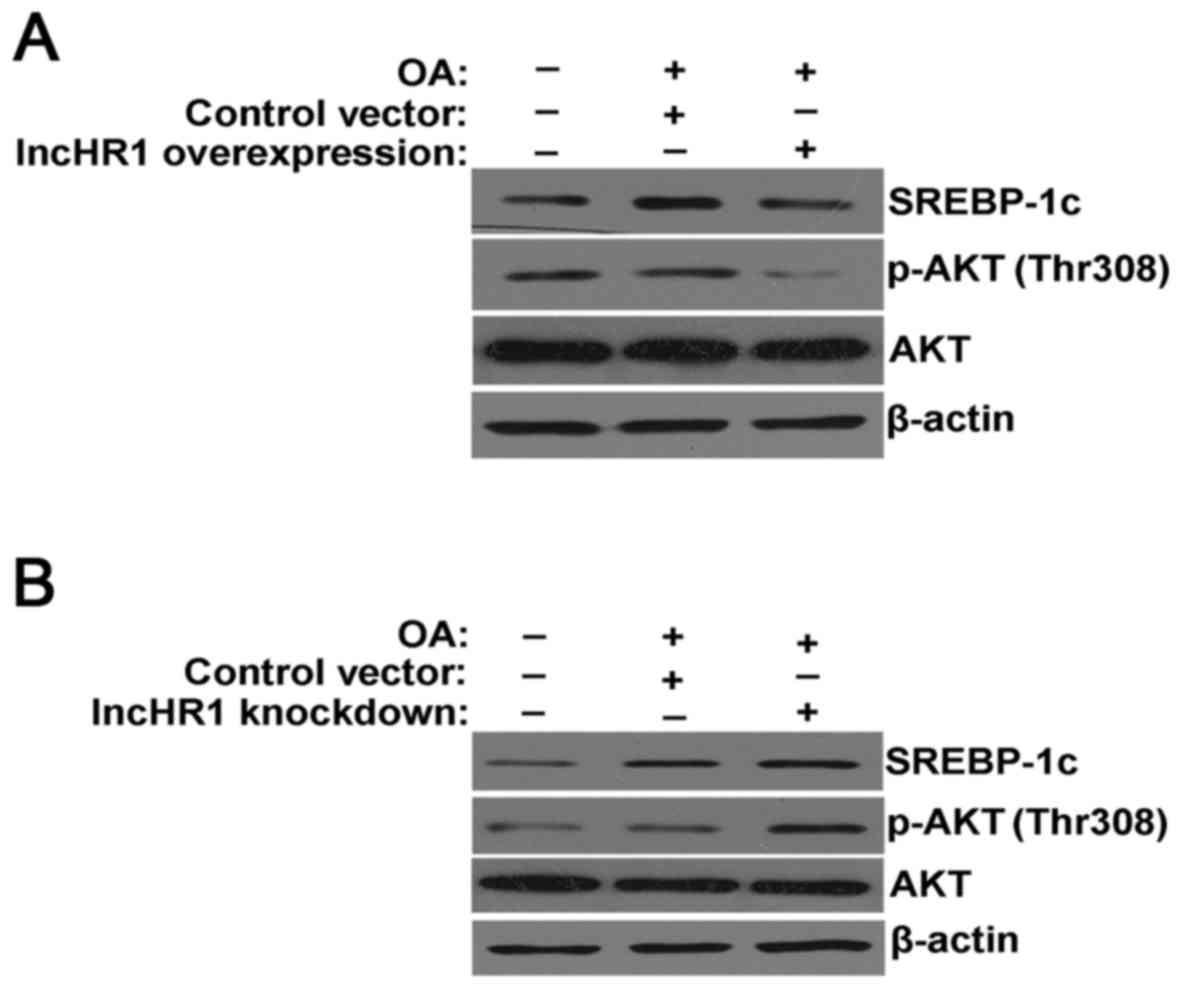
In the model of the TG accumulation, overexpression
of lncHR1 inhibited SREBP-1c protein expression, suppressed the
phosphorylation of AKT (Thr308) and total AKT protein levels.
β-actin was used as a loading control (Fig. 2A). Alternatively, knockdown of
lncHR1 elevated SREBP-1c protein levels and activated the
phosphorylation of AKT (Thr308) (Fig.
2B). Therefore, our results suggested that lncHR1 may inhibit
SREBP-1c through regulation of AKT phosphorylation in the model of
the TG accumulation. As previously reported, FoxO1 is a member of
the family of a forkhead box class-O transcription factor family
and is a downstream target of serine/threonine protein kinase B
(AKT). Research has confirmed that, in the promoter region of the
srebp-1c gene, FoxO1 weakens the combinatorial capacity of LXRα/RXR
binding to LXREs, subsequently inhibiting srebp1 gene expression
(10).
LncHR1 affected the phosphorylation of
PDK1/AKT/FoxO1 in Huh7 cells
To study how lncHR1 regulates the phosphorylation of
AKT, an activator (IGF-1) and inhibitor (LY294002) of the PI3K/AKT
pathway were used in this study. As shown in Fig. 3A, IGF-1 simultaneously increased
the phosphorylation level of AKT (Thr308) and FoxO1 (Ser256), but
this elevated phosphorylation was significantly reversed by
overexpression of lncHR1. Meanwhile, induced expression of SREBP-1c
by IGF-1 was also reversed after lncHR1 overexpression (Fig. 3A). As expected, the inhibitor
LY294002 suppressed AKT (Thr308) and FoxO1 (Ser256) phosphorylation
comparing to total AKT and β-actin. Nevertheless, lncHR1 knockdown
rescued LY2940002 inhibition of AKT and FoxO1 phosphorylation, as
well as SREBP-1c protein (Fig.
3B). Thus, AKT (Thr308) and FoxO1 (Ser256) may be the key site
of phosphorylation in regulation of the AKT/FoxO1/SREBP-1c axis by
lncHR1. Therefore, the molecule upstream of AKT may be the targets
of lncHR1. AKT is phosphorylated and activated by PI3K, which is
composed of a heterodimer between a p110 catalytic subunit and a
p85 regulatory subunit. The results showed that there was no change
in the protein level of p110β or p85, regardless of whether lncHR1
was overexpressed or knocked down (data not shown). Therefore, the
key site for lncHR1 regulation of the AKT/FoxO1/SREBP-1c axis is
likely independent of PI3K.
The major biological function of phosphatase and
tensin homolog (PTEN) relies on its phosphatase activity and PTEN
exerts tumor suppressive functions by suppressing the PI3K pathway
(19). In this study, the results
showed that lncHR1 expression had no effect on PTEN levels, similar
to the PI3K subunits (data not shown).
3-phosphoinositide-dependent protein kinase 1 (PDK1)
is downstream of PI3K and activated PDK1 can stimulates the
phosphorylation of Threonine 308 in the central catalytic domain
(20). Western blotting data
confirmed that PDK1 phosphorylation at Ser241 was suppressed by
lncHR1 overexpression (Fig. 3C)
and was activated by lncHR1 knockdown (Fig. 3D). However, the detailed mechanisms
of how lncHR1 regulate the phosphorylation of PDK1 at site Ser241
requires further study.
LncHR1 affected the distribution of
FoxO1 in Huh7 cells
A study by Kamei (9) also indicated that SREBP-1c gene
expression was up-regulated by the combination of LXRα/RXR bound to
LXRE, and that FoxO1 antagonized this combination to inhibit
srebp-1c gene expression.
Phosphorylation of FoxO1 (Ser256) (p-FoxO1) by AKT
occurs in the cytoplasm. Non-phosphorylated FoxO1 located in the
nucleus inhibits SREBP-1c expression. Therefore, Western blotting
and immunofluorescence assays were used to measure the
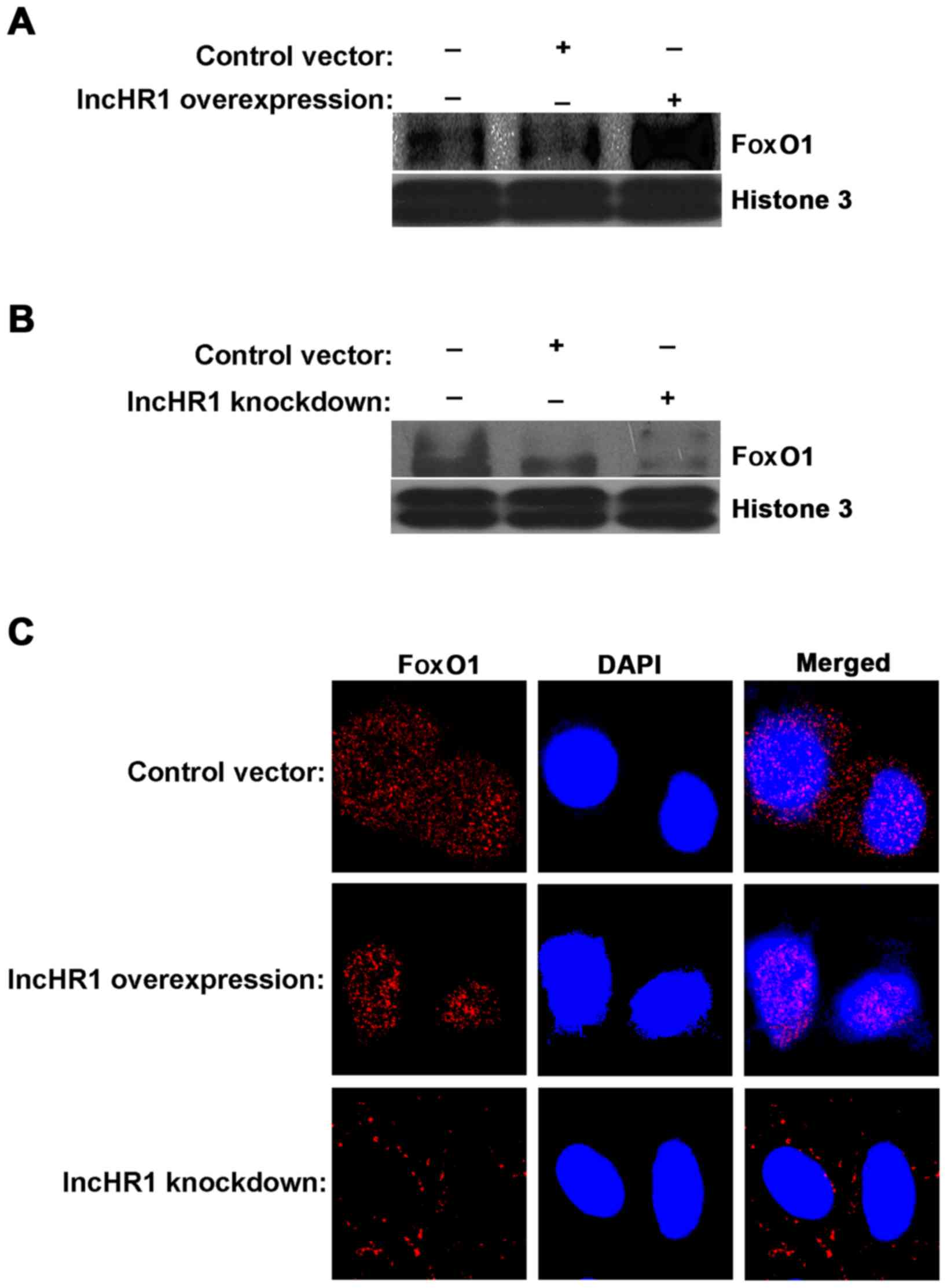
lncHR1-regulated translocation of FoxO1. First, we separated the
nuclear protein fraction for each treatment and then quantified
FoxO1 accumulation. The results indicated that a greater amount of
FoxO1 accumulated in the nucleus when lncHR1 was overexpressed
(Fig. 4A) and the opposite results
were observed after lncHR1 knockdown (Fig. 4B). This effect on the subcellular
translocation of FoxO1 was confirmed by immunofluorescent staining
(Fig. 4C). Thus, lncHR1 suppressed
phosphorylation of the AKT/FoxO1 level, increased the quantity of
FoxO1 in the nucleus, and then downregulated SREBP-1c expression.
LncHR1 may be a negative regulatory factor of SREBP-1c
transcription.
Above all, these results consolidated our findings
that lncHR1 suppressed SREBP-1c levels through decreased the
phosphorylation level of PDK1/AKT/FoxO1 and increased expression of
intranuclear FoxO1. According to these results, lncHR1 may inhibit
the expression of SREBP-1c through modulation of the PDK1/AKT/FoxO1
pathway in Huh7 cells. Further studies will investigate how lncHR1
regulate the phosphorylation of PDK1.
Discussion
In this study, we found that lncHR1 inhibited
SREBP-1c and TG accumulation when the phosphorylation level of the
PDK1/AKT/FoxO1 pathway was reduced through increased lncHR1. Based
on the results of previous experiments (15), we initially concluded that lncHR1
participated in the balance metabolism of lipid through regulating
the phosphorylation level of the PDK1/AKT/FoxO1 signaling pathway,
thus affecting SREBP-1c and TG accumulation.
SREBPs are key transcriptional factors that control
lipogenesis and lipid uptake (21). SREBP-1c activates the transcription
of multiple genes encoding for enzymes involved in the synthesis of
cholesterol, TG, and phospholipids, which are related to FA
synthesis (22). Some small
molecular that regulate SREBP levels may affect lipid metabolism.
For example, miR-33a is a highly conserved intronic miRNA, the
level of which is increased in normal tissues by increased
transcription of SREBF2, resulting in coordinate regulation of
cholesterol and other lipid levels by SREBF2 and miR-33a (23). MALAT1 induced hepatic lipid
accumulation and insulin resistance by increasing SREBP-1c and
target genes expression (16). Our
previous study (15) had confirmed
that lncHR1 indeed simultaneously regulated the SREBP-1c levels and
TG and lipid droplets in vivo.
The long non-coding RNA (lncRNA) urothelial
carcinoma-associated 1 (UCA1) showed significantly higher
expression in advanced gastric cancer tissues, and the study
results indicated that UCA1 regulates PI3K-AKT-mTOR signaling
proteins in vitro and in vivo (24).
In particular, SREBP-1c is the chief factor
regulating the transcription of genes involved in fat synthesis,
and can be up-regulated by LXRα (21). LXRα heterodimerized with the
nuclear receptors retinoid X receptors (RXRs) and binds to the LXR
element (LXRE) in the upstream promoter of srebp1 genes, thus
controlling the transcription of SREBP-1c and further regulating
the expression of the downstream genes involved in de novo fatty
acid metabolism, TG synthesis and cholesterol homeostasis (25). As reported previously, FoxO1
antagonized the binding of LXRα/RXR to LXRE and lowered srebp-1c
gene expression to some extent. FoxO1, a member of the FoxO family
of transcription factors, is the direct downstream substrates for
AKT. When phosphorylated by AKT, FoxO1 translocate from the nucleus
to the cytoplasm. In the current study, our results indicated that
either overexpression or knockdown lncHR1 may affect the
distribution of FoxO1 inside and outside of the nucleus (Fig. 4).
Activation of the PI3K/AKT signaling pathway results
in AKT-dependent phosphorylation of FoxO1, reducing FoxO1 nuclear
translocation and inhibiting its transcriptional function. PI3K/AKT
signaling upregulates glucose uptake and glycolysis and controls
the metabolic flux from glucose and glutamine to de novo lipid
synthesis (26).
SREBP-1c may be activated by the PI3K/AKT oncogenic
signaling pathway either PI3K/AKT/GSK3-β/SREBP-1c (27) or PI3K/AKT/mTORC1 signaling
(28). However, our previous
studies did not confirm that lncHR1 was involved in the classical
pathway. Activated PDK1 stimulates AKT activity by direct binding
and phosphorylation of Threonine 308 in the central catalytic
domain (20). In our study, the
results (Fig. 3) indicated that
lncRH1 participated in the phosphorylation of PDK1, subsequently
regulating the AKT/FoxO1/SREBP-1c pathway. Multiple studies have
revealed that PI3K/AKT signaling promotes fatty acid synthesis
through upregulation of SREBP-1c (29). A study by Wang indicated that
lncRNA AB073614 may be useful as a novel prognostic or treatment
agent for colorectal cancer due to its effects on the PI3K/AKT
signaling pathway (30).
It is well established that the major biological
function of PTEN relies on its phosphatase activity. PTEN
dephosphorylates PIP3 to PIP2, thereby inhibiting the PI3K
signaling pathway (19). However,
our results did not support the theory that lncHR1 influences PTEN
or involvement in this phosphorylation process (data not
shown).
There are several limitations in this study. i) The
detailed molecular mechanism for the effect of lncHR1 on the
phosphorylation of PDK1 has not been elucidated. ii) The
phosphorylation of AKT is regulated by mTOR, but whether lncHR1 is
involved in the mTOR pathway was not verified. Therefore, these
areas will be the next focus of our research.
Taken together, these findings indicated that lncHR1
may induce a decrease in the phosphorylation of the PDK1/AKT/FoxO1
pathway and suppressed SREBP-1c protein levels. However, the
precise role of lncHR1 in the phosphorylation of PDK1 remains to be
elucidated, and further investigations is needed.
Acknowledgements
The authors would like to thank Dr Wei Yang of the
MOH Key Laboratory of Systems Biology of Pathogens, Institute of
Pathogen Biology, Chinese Academy of Medical Sciences and Peking
Union Medical College for providing technical and material
support.
Funding
This work was supported by the Collective grants
from the Programs for Science and Technology Development of Henan
(grant nos. 172102310499 and 182102310240), Open Program of Henan
Key Laboratory of Biological Psychiatry (grant no. ZDSYS2016007)
and Dr. scientific research start-up fund (grant no.
XYBSKYZZ201605).
Availability of data and materials
The analyzed data sets analyzed during the study are
available from the corresponding author on reasonable request.
Authors' contributions
DL and ZY are responsible for the study concept and
design. LG and BD performed part of the cell biology experiment.
ML, TY and FY performed the molecular biology experiment. DL and ZY
were involved in the data analysis and manuscript drafting.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
lncRNA
|
long non-coding RNA
|
|
SREBP-1c
|
sterol regulatory element binding
protein-1c
|
|
OA
|
oleic acids
|
|
LXRα
|
liver X receptor α
|
|
SCAP
|
SREBP cleavage-activating protein
|
|
DAPI
|
4′,6-diamidino-2-phenylindole
|
|
FA
|
fatty acid
|
|
FAS
|
fatty acid synthase
|
|
TG
|
triglyceride
|
|
ACCα
|
acetyl-CoA carboxylase α
|
|
RT-qPCR
|
reverse transcription-quantitative
polymerase chain reaction
|
References
|
1
|
Brown MS and Goldstein JL: The SREBP
pathway: Regulation of cholesterol metabolism by proteolysis of a
membrane-bound transcription factor. Cell. 89:331–340. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Si Y, Liu S, Liu X, Jacobs JL, Cheng M,
Niu Y, Jin Q, Wang T and Yang W: A human claudin-1-derived peptide
inhibits hepatitis C virus entry. Hepatology. 56:507–515. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Horton JD, Bashmakov Y, Shimomura I and
Shimano H: Regulation of sterol regulatory element binding proteins
in livers of fasted and refed mice. Proc Natl Acad Sci USA.
95:5987–5992. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Repa JJ, Liang G, Ou J, Bashmakov Y,
Lobaccaro JM, Shimomura I, Shan B, Brown MS, Goldstein JL and
Mangelsdorf DJ: Regulation of mouse sterol regulatory
element-binding protein-1c gene (SREBP-1c) by oxysterol receptors,
LXRalpha and LXRbeta. Genes Dev. 14:2819–2830. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Horton JD, Goldstein JL and Brown MS:
SREBPs: Activators of the complete program of cholesterol and fatty
acid synthesis in the liver. J Clin Invest. 109:1125–1131. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ferré P and Foufelle F: SREBP-1c
transcription factor and lipid homeostasis: Clinical perspective.
Horm Res. 68:72–82. 2007.PubMed/NCBI
|
|
7
|
Chen G, Liang G, Ou J, Goldstein JL and
Brown MS: Central role for liver X receptor in insulin-mediated
activation of Srebp-1c transcription and stimulation of fatty acid
synthesis in liver. Proc Natl Acad Sci USA. 101:11245–11250. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Deng X, Yellaturu C, Cagen L, Wilcox HG,
Park EA, Raghow R and Elam MB: Expression of the rat sterol
regulatory element-binding protein-1c gene in response to insulin
is mediated by increased transactivating capacity of specificity
protein 1 (Sp1). J Biol Chem. 282:17517–17529. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kamei Y, Miura S, Suganami T, Akaike F,
Kanai S, Sugita S, Katsumata A, Aburatani H, Unterman TG, Ezaki O
and Ogawa Y: Regulation of SREBP1c gene expression in skeletal
muscle: Role of retinoid X receptor/liver X receptor and
forkhead-O1 transcription factor. Endocrinology. 149:2293–2305.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Deng X, Zhang W, O-Sullivan I, Williams
JB, Dong Q, Park EA, Raghow R, Unterman TG and Elam MB: FoxO1
inhibits sterol regulatory element-binding protein-1c (SREBP-1c)
gene expression via transcription factors Sp1 and SREBP-1c. J Biol
Chem. 287:20132–20143. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Johnsson P, Lipovich L, Grandér D and
Morris KV: Evolutionary conservation of long non-coding RNAs;
sequence, structure, function. Biochim Biophys Acta.
1840:1063–1071. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gibb EA, Brown CJ and Lam WL: The
functional role of long non-coding RNA in human carcinomas. Mol
Cancer. 10:382011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Maass PG, Luft FC and Bahring S: Long
non-coding RNA in health and disease. J Mol Med (Berl). 92:337–346.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Panzitt K, Tschernatsch MM, Guelly C,
Moustafa T, Stradner M, Strohmaier HM, Buck CR, Denk H, Schroeder
R, Trauner M and Zatloukal K: Characterization of HULC, a novel
gene with striking up-regulation in hepatocellular carcinoma, as
noncoding RNA. Gastroenterology. 132:330–342. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li D, Cheng M, Niu Y, Chi X, Liu X, Fan J,
Fan H, Chang Y and Yang W: Identification of a novel human long
non-coding RNA that regulates hepatic lipid metabolism by
inhibiting SREBP-1c. Int J Biol Sci. 13:349–357. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yan C, Chen J and Chen N: Long noncoding
RNA MALAT1 promotes hepatic steatosis and insulin resistance by
increasing nuclear SREBP-1c protein stability. Sci Rep.
6:226402016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Cheng M, Si Y, Yang Y, Liu X, Gong Q, Zhao
J, Niu Y, Li X, Jin Q and Yang W: Recombinant human interleukin
28B: Anti-HCV potency, receptor usage and restricted cell-type
responsiveness. J Antimicrob Chemother. 67:1080–1087. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Latasa MJ, Moon YS, Kim KH and Sul HS:
Nutritional regulation of the fatty acid synthase promoter in vivo:
Sterol regulatory element binding protein functions through an
upstream region containing a sterol regulatory element. Proc Natl
Acad Sci USA. 97:10619–10624. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Worby CA and Dixon JE: PTEN. Annu Rev
Biochem. 83:641–669. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Franke TF, Kaplan DR, Cantley LC and Toker
A: Direct regulation of the Akt proto-oncogene product by
phosphatidylinositol-3,4-bisphosphate. Science. 275:665–668. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Jianhua L, Xueqin M and Jifen H:
Expression and clinical significance of LXRα and SREBP-1c in
placentas of preeclampsia. Open Med (Wars). 11:292–296.
2016.PubMed/NCBI
|
|
22
|
Horton JD, Shimomura I, Ikemoto S,
Bashmakov Y and Hammer RE: Overexpression of sterol regulatory
element-binding protein-1a in mouse adipose tissue produces
adipocyte hypertrophy, increased fatty acid secretion, and fatty
liver. J Biol Chem. 278:36652–36660. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Najafi-Shoushtari SH, Kristo F, Li Y,
Shioda T, Cohen DE, Gerszten RE and Näär AM: MicroRNA-33 and the
SREBP host genes cooperate to control cholesterol homeostasis.
Science. 328:1566–1569. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Li C, Liang G, Yang S, Sui J, Yao W, Shen
X, Zhang Y, Peng H, Hong W, Xu S, et al: Dysregulated lncRNA-UCA1
contributes to the progression of gastric cancer through regulation
of the PI3K-Akt-mTOR signaling pathway. Oncotarget. 8:93476–93491.
2017.PubMed/NCBI
|
|
25
|
Plösch T, Gellhaus A, van Straten EM, Wolf
N, Huijkman NC, Schmidt M, Dunk CE, Kuipers F and Winterhager E:
The liver X receptor (LXR) and its target gene ABCA1 are regulated
upon low oxygen in human trophoblast cells: A reason for
alterations in preeclampsia? Placenta. 31:910–918. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Metallo CM, Gameiro PA, Bell EL, Mattaini
KR, Yang J, Hiller K, Jewell CM, Johnson ZR, Irvine DJ, Guarente L,
et al: Reductive glutamine metabolism by IDH1 mediates lipogenesis
under hypoxia. Nature. 481:380–384. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yecies JL, Zhang HH, Menon S, Liu S,
Yecies D, Lipovsky AI, Gorgun C, Kwiatkowski DJ, Hotamisligil GS,
Lee CH and Manning BD: Akt stimulates hepatic SREBP1c and
lipogenesis through parallel mTORC1-dependent and independent
pathways. Cell Metab. 14:21–32. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Peterson TR, Sengupta SS, Harris TE,
Carmack AE, Kang SA, Balderas E, Guertin DA, Madden KL, Carpenter
AE, Finck BN and Sabatini DM: mTOR complex 1 regulates lipin 1
localization to control the SREBP pathway. Cell. 146:408–420. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Porstmann T, Santos CR, Griffiths B, Cully
M, Wu M, Leevers S, Griffiths JR, Chung YL and Schulze A: SREBP
activity is regulated by mTORC1 and contributes to Akt-dependent
cell growth. Cell Metab. 8:224–236. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang Y, Kuang H, Xue J, Liao L, Yin F and
Zhou X: LncRNA AB073614 regulates proliferation and metastasis of
colorectal cancer cells via the PI3K/AKT signaling pathway. Biomed
Pharmacother. 93:1230–1237. 2017. View Article : Google Scholar : PubMed/NCBI
|