Introduction
Breast cancer is one of the most frequently
diagnosed cancers and the leading contributing factor in cancer
death among females (1). In spite
of significantly improved diagnosis and clinical treatment
strategies, low survival and high recurrence rate persist in many
breast cancer patients. Invasion and metastasis are the primary
reasons for mortality caused by breast cancer. However, the precise
molecular mechanism of breast cancer invasion and metastasis
remains ambiguous.
The receptor for advanced glycation end products
(RAGE), which is a member of the immune globulin family, was
isolated from the bovine lung endothelium (2). RAGE is a multiple ligand and pattern
recognition receptor that has been implicated in the progression of
various human cancers because of its ability to drive tumor growth
and progression (3). Meanwhile,
many clinical studies have indicated that higher RAGE is related to
various malignant tumors, such as prostate cancer and glioma
(4,5). RAGE has been implicated as a
potential mechanism driving the development, progression, and
metastasis of breast cancer (6–8).
Epithelial mesenchymal transition (EMT) is
indispensable to increasing invasion and migration during the
development of a malignant tumor. EMT can be affected by various
transcription factors and is accompanied by changes in the EMT
marker protein. In EMT, cancer cells exhibit the property of
mesenchymal cells; that is, the cells diffuse from the primary
tumor and invade adjacent tissues and blood vessels (9,10).
Recent studies have demonstrated that microRNA (miRNA)s are key
factors in EMT-associated cancer invasion and metastasis.
miRNAs belong to a family of small non-coding RNAs
with approximately 22–25 nucleotides in length. Functional studies
have demonstrated that miRNA serves as a modulating gene that can
regulate target mRNA to repress translation. Several miRNAs, such
as miR-200c (11), miR-34
(12), and miR-9 (13), enhance or repress metastasis and
invasion by regulating EMT. miR-185-5p has been reported to be
downregulated in NSCLC (14),
gastric cancer (GC) (15), breast
cancer (16), and glioma (17). Plasma miR-185 is decreased in
patients with esophageal squamous cell carcinoma and might suppress
tumor migration and invasion by targeting RAGE (18). However, the functional mechanism of
miR-185-5p targeting the RAGE mRNA in breast cancer is unknown.
Thus, understanding the potential mechanism of the invasion and EMT
of breast cancer may be an effective therapeutic strategy.
In this study, we first identified a negative
correlation between miR-185-5p and RAGE expressed in breast cancer
tissues and cells. Upregulation or downregulation of miR-185-5p
in vitro and in vivo resulted in the significant
inhibition or promotion of invasion and EMT. Furthermore,
upregulation of miR-185-5p impacted F-actin polymerization caused
by S100A8/A9 of breast cancer cells. Finally, the results indicated
that miR-185-5p, as an anti-oncogene, played a pivotal role by
targeting RAGE in breast cancer. Hence, miR-185-5p could act as a
rationale for the diagnosis and therapy of breast cancer
patients.
Materials and methods
Clinical samples
Fresh frozen breast tissue specimens and adjacent
normal breast tissue samples were acquired from the Affiliated
Hospital of the Weifang Medical University (Shandong, China).
Patients did not undergo chemotherapy or radiotherapy before
surgery. Written informed consent was acquired from all patients,
and the study was approved by the Institute Research Ethics
Committee at the Cancer Center, Weifang Medical University.
Cell culture
All human breast cancer cell lines, namely,
MDA-MB-231 (MDA231), T47D, MDA-MB-453 (MDA453), SK-BR-3 and MCF-7,
and human normal breast epithelial cell MCF-10A were acquired from
ATCC (USA). The MCF-10A cells were cultured in DMEM/F12, which
contained 10% FBS, 20 ng/ml EGF, 0.1 mg/ml CT, 10 mg/ml insulin,
and 500 ng/ml hydrocortisone. MCF-7 cells were cultured in MEM with
10% FBS and 1% sodium pyruvate. T47D and SK-BR-3 cells were
cultured in DMEM with 10% FBS. MDA231 cells were cultured in
RPMI-1640. MDA453 cells were cultured in L-15 with 10% FBS. All
cell cultures were incubated at 37°C under 5% CO2
atmosphere.
Plasmid construction and cell
transfection
MDA231 cells or MCF-7 cells (2×105) were
planted in six-well plates overnight to ensure that cell confluence
could reach 60–80% at the time of transfection. Lipofectamine 2000
(Invitrogen; Thermo Fisher Scientific Inc., Waltham, MA, USA) was
employed in cell transfection according to the manufacturer's
protocol. miR-185-5p mimics, negative control mimics (NCs),
miR-185-5p inhibitor, and anti-NC were synthesized by Genechem,
Inc. (Daejeon, Korea). The siRNA sequence of RAGE and a control
vector were synthesized by Genechem Inc. (8). The transfected cells were selected
for 14 days using 600 µg/ml G418. We isolated single cell culture
and maintained the stable transfected cells using 300 µg/ml G418.
The stable transfected cells were used for subsequent studies.
Western blot analysis
The cells or tissues were lysed using RIPA buffer
with protein phosphatase inhibitors for western blot. Whole cell or
tissue protein lysates were electrophoresed on 10% SDS-PAGE and
transferred onto PVDF membranes (Millipore, Billerica, MA, USA).
The membranes were subsequently incubated with primary antibodies
overnight, washed thrice, incubated with secondary antibody, and
exposed to ECL. We analyzed the samples using ImageJ software (NIH,
Bethesda, Maryland). The relative protein levels were quantified
using three different western blots. The following antibodies were
employed: RAGE (1:1,000; cat. no. 6996), E-cadherin (1:1,000; cat.
no. 3195), vimentin (1:1,000; cat. no. 5741; all Cell Signaling
Technology, Inc. Cell Signaling Technology, Inc., Boston, MA, USA),
Snail (1:500; cat. no. ab53519), Slug (1:500; cat. no. ab27568),
twist-related protein (Twist; 1:500; cat. no. ab50581), zinc finger
E-box binding homeobox (Zeb)1 (1:500; cat. no. ab124512; all Abcam,
Cambridge, UK), Zeb2 (1:500; cat. no. sc48789; Santa Cruz
Biotechnology, Inc., Dallas, TX, USA), HRP-conjugated anti-rabbit
IgG, and anti-mouse IgG antibody (1:4,000; cat. nos. 7074 and 7076;
Cell Signaling Technology, Inc.). The western blot results were
from at least three repeated experiments.
Cell invasion
The Transwell chamber invasion assay was spread in
the Matrigel. Breast cancer cells were mixed and incubated at 37°C
for 24 h. Then, the cells were washed thrice and fixed with 4%
paraformaldehyde for 15 min and stained with Giemsa for 35 min.
Five view pictures were randomly selected, and the cells were
counted under a light microscope (magnification, ×200).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNAs were isolated using TRIzol reagent.
RT-qPCR was conducted using SYBR-Green on an Applied Biosystems
7500 (Applied Biosystems; Thermo Fisher Scientific, Inc.).
Expression of U6 was used as the internal control. The relative
mRNA expression was normalized using the 2−ΔΔCq method
(19). The thermocycling
conditions for RT-qPCR were as follows: 95°C for 20 sec, followed
by 35 cycles of 95°C for 5 sec, 63°C for 30 sec and 72°C for 5 sec.
The sequence of the RT primer was
5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTCAGGA-3′. Specific
primers for miR-185-5p were as follows: Forward,
5′-GCGCGATTGGAGAGAAAGGCAGT-3′ and reverse,
5′-ATCCAGTGCAGGGTCCGAGG-3′.
Luciferase reporter assay
TargetScan 6.0 and miRNA.org were
used to predict sequences of miR-185-5p and pair 3′-untranslated
region (UTR) sequences of RAGE. MDA231 cells or MCF-7 cells
(3×104) were planted in triplicates in 24-well plates
for 24 h before transfection. Then, 100 ng of pGL3-RAGE-3′UTR
wild-type (WT) or pGL3-RAGE-3′UTR-mutant (MUT), plus 1 ng of pRL-TK
(Promega, Madison, WI, USA) and the matched miR-185-5p, NC,
anti-NC, or anti-miR-185-5p plasmid, were co-transfected using
Lipofectamine 2000 in accordance with the manufacturer's protocol.
Firefly luciferase activity was measured after 24 h of
transfection.
Chemotaxis assay
S100A8/A9 was added into the bottom chamber, and
2×105 cells were added into the upper chamber. The
chambers were incubated at 37°C under 5% CO2.
Subsequently, the polycarbonate filter membranes were fixed and
stained using Giemsa. The numbers of migrating cells were
calculated in five random fields using light microscopy, and the
average number of cells was determined. This experiment was
repeated thrice.
Cellular F-actin measurement
Breast cancer cells were immobilized with 4%
paraformaldehyde and PBS. Then, the cells were washed thrice for 5
min at each instance. The cells were washed with PBS and blocked
with buffer, which included goat serum, for 45 min. The cells were
stained with Rhodamine phalloidin for 1 h, washed thrice, and
covered with fluorescence decay resistant sealing tablets. F-actin
content was measured with a fluorescence reader. The relative
F-actin content was calculated as follows: F-actin ∆t/F-actin
0=fluorescence ∆t/fluorescence 0.
Immunofluorescence
The cells were fixed for 20 min using 4%
paraformaldehyde. Then, the cells were permeabilized with 0.1%
Triton X-100 and blocked with 1% BSA for 45 min at 37°C. The
primary antibodies E-cadherin and vimentin were incubated overnight
at 4°C. Cy3 and FITC secondary antibodies and DAPI were used.
Animal studies
The Severe Combined Immune-deficiency (SCID) mice
were approved by the Animal Care and Use Committee of Wei Fang
Medical University. Female SCID mice (4–5 weeks old) were used.
MDA231/NC, Scr/MDA231, MDA231/miR-185-5p, and SiRAGE/MDA231 cells
were injected into the oxter of the SCID mice (n=10). When the
xenografts were evident, an intratumor injection of S100A8/A9 at
100 ng/kg was performed for 4 weeks. After 8 weeks, metastasis in
the lung tissues was examined by H&E staining.
Statistical analysis
Data were analyzed using SPSS v16.0 software (SPSS,
Inc., Chicago, IL, USA). The results were presented as the mean ±
standard deviation. Χ2-test was used to analyze the
associations between miR-185-5p and the clinicopathologic features.
Student's t-test or analysis of variance with Dunnett's post hoc
test were conducted to determine the statistical significance for
comparisons between the groups. P<0.05 was considered to
indicate a statistically significant difference.
Results
Downregulation of miR-185-5p in
metastatic patient tissues and cells was correlated with
clinicopathological features of breast cancer
miR-185-5p is known to play a fatal role in various
cancers, such as breast cancer. However, the biological role of
miR-185-5p has not been fully elucidated. In the present study, we
first analyzed the miR-185-5p expression in breast cancer cells by
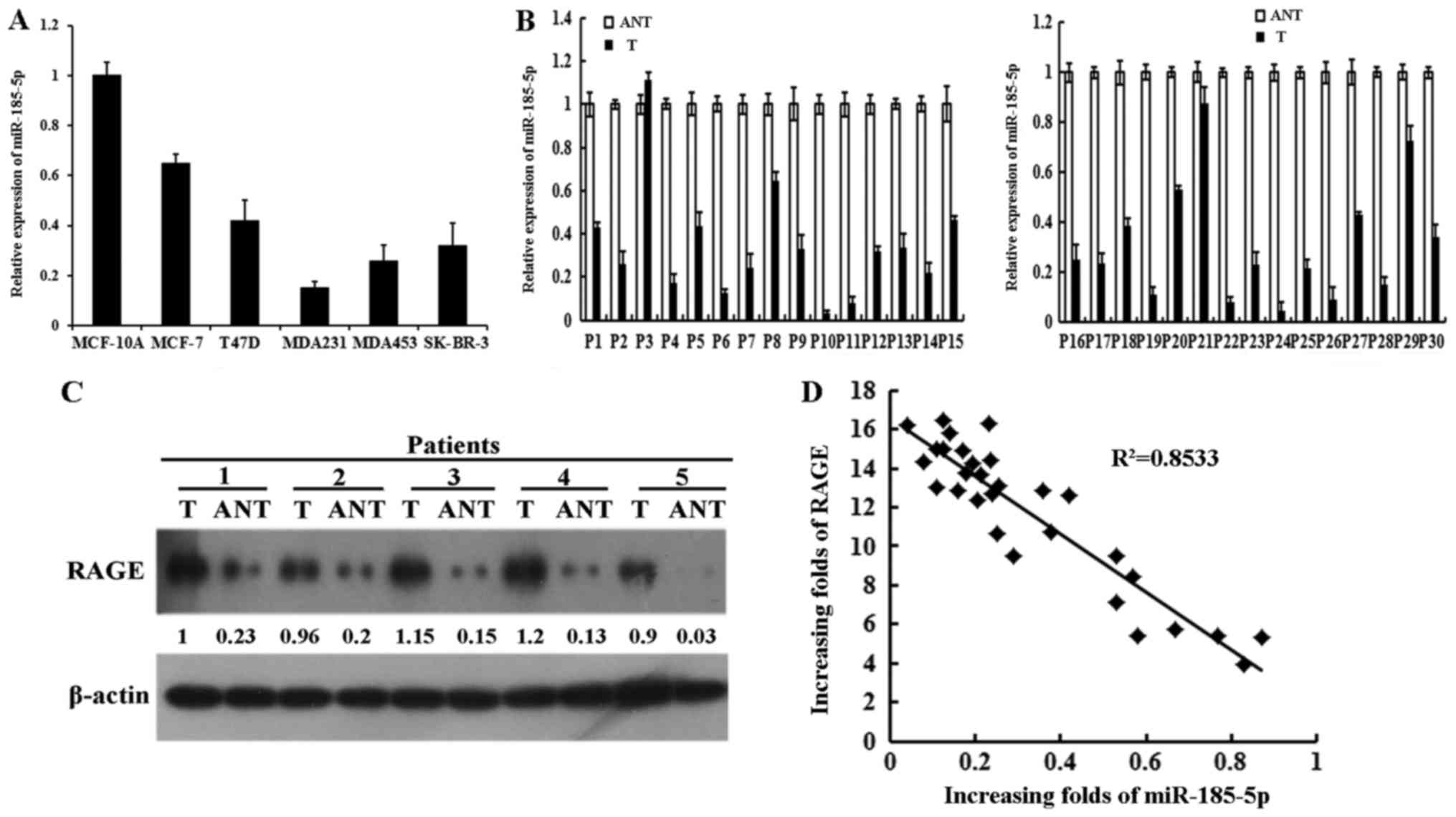
RT-qPCR. The results indicated that the expression of miR-185-5p
was markedly downregulated in breast cancer cells compared with
that in MCF-10A. The expression of miR-185-5p was higher in MCF-7
than in MDA231 (Fig. 1A). The
expression of miR-185-5p in 30 selected fresh breast cancer tissues
(T) and paired adjacent non-tumor tissues (ANTs) was tested using
RT-qPCR to further investigate the clinical relevance of
miR-185-5p. The results demonstrated that miR-185-5p was
significantly reduced in all 30 tumor tissues compared with the
paired ANTs (Fig. 1B).
Consequently, RAGE was markedly increased in most of the fresh
tumor tissues relative to the paired ANTs (Fig. 1C). Thus, the expression of
miR-185-5p was negatively associated with RAGE in the fresh tumor
tissues (Fig. 1D). RAGE was
previously associated with the clinical pathology of breast cancer
(8). Analysis of correlation
between miR-185-5p and clinical pathological features demonstrated
that downregulation of miR-185-5p in breast cancer was related to
clinical stage, tumor size (P=0.08) and grade (P=0.010), lymphatic
metastasis (P=0.014), and distant metastasis (P=0.022). However,
this downregulation was not associated with age (P=0.158), ER
(P=0.080), and PR (P=0.212) (Table
I). These data implied that miR-185-5p expression was
significantly related with the clinicopathological features of
breast cancer.
 | Table I.Associations between clinical
features and miRNA-185-5p expression in breast cancer patients. |
Table I.
Associations between clinical
features and miRNA-185-5p expression in breast cancer patients.
|
| miR-185-5p |
|
|---|
|
|
|
|
|---|
| Variables | High expression
(n) | Low expression
(n) | P-value |
|---|
| Age (years) |
|
|
|
|
≤50 | 29 | 23 | 0.158 |
|
≥51 | 21 | 27 |
|
| Tumor size
(cm) |
|
|
|
| ≤5 | 32 | 19 | 0.008 |
|
>5 | 18 | 31 |
|
| Tumor grade |
|
|
|
| I | 22 | 9 | 0.010 |
| II | 17 | 19 |
|
|
III | 11 | 22 |
|
| Lymph node
metastasis |
|
|
|
|
Yes | 20 | 32 | 0.014 |
| No | 30 | 18 |
|
| Distant
metastasis |
|
|
|
|
Yes | 17 | 28 | 0.022 |
| No | 33 | 22 |
|
| ER expression |
|
|
|
|
Positive | 27 | 19 | 0.080 |
|
Negative | 23 | 31 |
|
| PR expression |
|
|
|
|
Positive | 29 | 24 | 0.212 |
|
Negative | 21 | 26 |
|
| c-erbB-2
expression |
|
|
|
|
Positive | 22 | 26 | 0.274 |
|
Negative | 28 | 24 |
|
| E-cadherin
expression |
|
|
|
|
Positive | 31 | 20 | 0.022 |
|
Negative | 19 | 30 |
|
| Vimentin
expression |
|
|
|
|
Positive | 16 | 33 | 0.013 |
|
Negative | 34 | 17 |
|
miR-185-5p directly targeted RAGE
3′-UTR
miRNAs modulate gene expression via targeting the
3′-UTR. Bioinformatics analyses of 3′-UTR revealed one putative
binding site for miR-185-5p (Fig.
2A). To study the potential interaction of miR-185-5p and RAGE
in breast cancer cells, we predicted that miR-185-5p repressed RAGE
expression via targeting the 3′-UTR. The results suggested that the
expression of RAGE protein and mRNA was considerably reduced in
MDA231/miR-185-5p cells. By contrast, the expression of RAGE
protein and mRNA was markedly upregulated in the
MCF-7/anti-miR-185-5p cells than in MCF-7/anti-NC (Fig. 2B, C and D). We cloned the WT or MUT
RAGE 3′-UTR of a luciferase reporter gene to investigate whether
the predicted targeting site of miR-185-5p on 3′-UTR of RAGE
functioned in this regulation. WT or MUT RAGE vector and miR-185-5p
or NC were co-transfected into MDA231 cells. The results showed
that co-transfection of WT RAGE 3′-UTR and miR-185-5p vector into
MDA231 cells notably reduced luciferase activity compared with the
co-transfection of control vectors and miR-185-5p (Fig. 2E left). Contrasting results were
found in MCF-7 cells co-transfected with anti-miR-185-5p and WT
RAGE 3′-UTR (Fig. 2E right). All
results showed that miR-185-5p directly targeted 3′-UTR of RAGE and
repressed RAGE expression. Subsequently, the regulating
relationship between miR-185-5p and RAGE in S100A8/A9-induced EMT
was further confirmed. The expression of EMT-related marker was
detected after co-transfecting or only transfecting one plasmid.
The result evidently showed that the expression of E-cadherin and
vimentin was rescued (Fig. 2F).
The same results were observed in the MCF-7 cells (Fig. 2G). These findings indicated that
miR-185-5p inhibited EMT by modulating RAGE and suppressed
invasion.
 | Figure 2.miR-185-5p directly targets RAGE
3′-UTR. (A) Predicted targeting site of miR-185-5p to the 3′-UTRs
of RAGE. (B) RAGE protein expression was tested in the indicated
cells by western blotting. (C) and (D) Expression levels of RAGE
mRNA were detected by reverse transcription-quantitative polymerase
chain reaction in the indicated cells. (E) Luciferase activity of
pGL3-RAGE 3′-UTR WT or pGL3-RAGE 3′-UTR-MUT report in MDA231 and
MCF-7 cells. (F) and (G) Expression of vimentin and E-cadherin was
examined in (F) MDA231 and (G) MCF-7. For (B), (F) and (G), the
results of western blotting were from a representative of at least
three repeated experiments. *P<0.05, as indicated. miR,
microRNA; RAGE, advanced glycosylation end-product specific
receptor; WT, wild-type; MUT, mutant; NC, negative control; UTR,
untranslated region; si-, small interfering RNA; Scr, scramble. |
miR-185-5p inhibited invasion and EMT
of breast cancer cells
The expression of miR-185-5p was tested, and the
results are shown in Fig. 1A.
miR-185-5p was found to be a tumor suppresser gene. The function of
miR-185-5p on cell invasion and EMT was further detected. MCF-7 was
transfected with anti-NC and anti-miR-185-5p plasmid. Meanwhile,
MDA231 was transfected with miR-185-5p and NC plasmid. The invasion
ability of MDA231 and MCF-7 was detected by Transwell with or
without S100A8/A9 stimulation. The invasion capacity of
MDA231/miR-185-5p was notably lower than that of MDA231/NC
(Fig. 3A). By contrast, the
invasion ability of MCF-7/anti-miR-185-5p was significantly higher
than that of MCF-7/anti-NC (Fig.
3B). The results showed that the upregulation of miR-185-5p
notably inhibited the invasion. Moreover, the EMT biomarker changed
in the breast cancer cell lines after transfection with or without
stimulation of S100A8/A9. western blot results showed that vimentin
expression was significantly decreased with stimulation of
S100A8/A9 in the MDA231/miR-185-5p cells than in MDA231/NC.
However, E-cadherin expression in the MDA231/miR-185-5p cells was
significantly upregulated (Fig. 3C
left). Meanwhile, E-cadherin expression in the
MCF-7/anti-miR-185-5p cells was significantly downregulated. The
expression of vimentin was significantly upregulated in the
MCF-7/anti-miR-185-5p cells than in MCF-7/anti-NC with S100A8/A9
stimulation (Fig. 3C right). The
same results were observed by immunofluorescence staining (Fig. 3D). Hence, miR-185-5p inhibited
invasion and EMT.
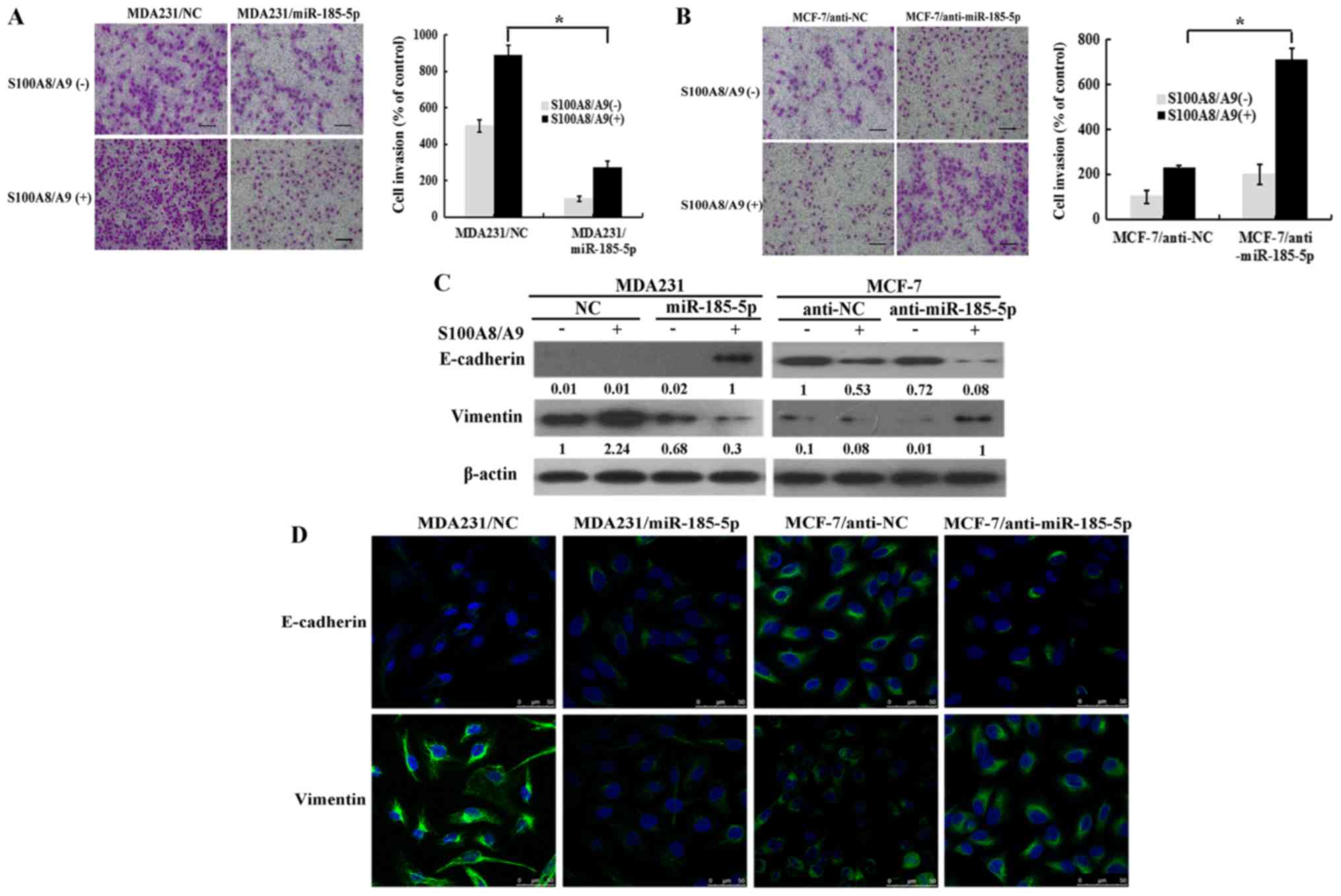 | Figure 3.miR-185-5p inhibits the invasion and
epithelial mesenchymal transition of breast cancer cells. (A)
Invasion capacity of MDA231/miR-185-5p and MDA231/NC cells was
analyzed. Left panel, invading cells (magnification, ×200); right
panel, quantification of penetrating cells. (B) Invasion capacity
of MCF-7/anti-miR-185-5p and MCF-7/anti-NC was analyzed. Left
panel, invading cells (magnification, ×200); right panel,
quantification of penetrating cells. (C) The expression of
E-cadherin and vimentin was examined in the indicated cells by
western blotting. The results of western blotting were from a
representative of at least three repeated experiments. (D)
Fluorescence microscopy of stained E-cadherin and vimentin in
indicated cells (scale bars 50 µm). *P<0.05, as indicated. miR,
microRNA; RAGE, advanced glycosylation end-product specific
receptor; NC, negative control. |
miR-185-5p inhibited S100A8/A9-induced
EMT of breast cancer cells via the NF-κB/Snail signaling
pathway
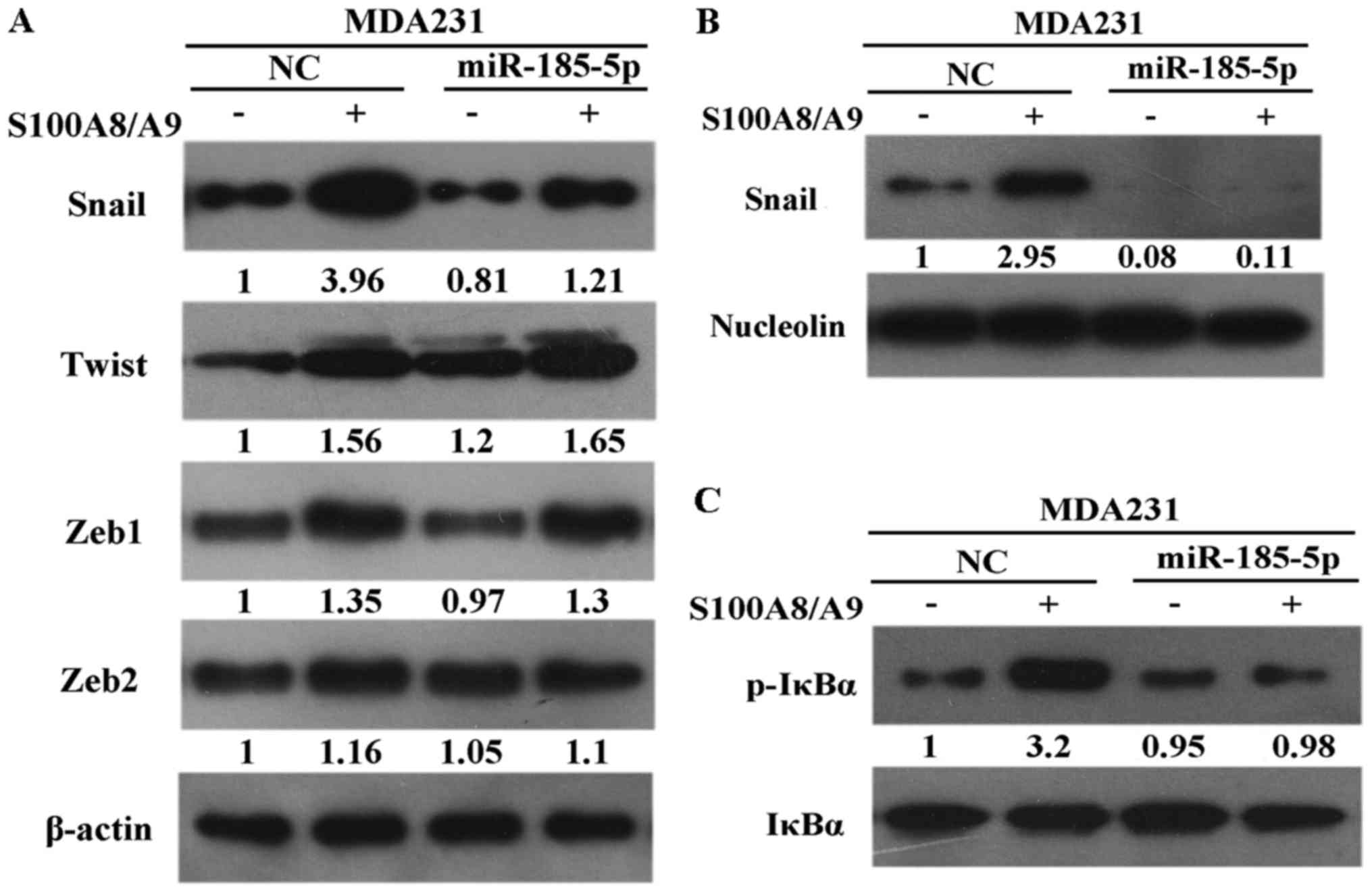
NF-κB is a transcription factor cytoplasmic
heterodimer protein of the IκB family and plays an important role
in tumor formation. The functional significance of miR-185-5p on
cell invasion was elucidated under S100A8/A9 stimulation. The
expression of Snail was remarkably downregulated in the
MDA231/miR-185-5p cells than in the MDA231/NC cells. However, the
expression levels of Twist, Zeb1, and Zeb2 were unchanged in the
MDA231/miR-185-5p cells (Fig. 4A).
In this study, Snail was downregulated in the nuclei of
MDA231/miR-185-5p cells than in the nuclei of MDA231/NC cells under
S100A8/A9 stimulation (Fig. 4B).
Afterward, IκBα phosphorylation was examined to determine whether
miR-185-5p mediated the stabilization of Snail through NF-κB
activity. The results indicated that IκBα phosphorylation was
considerably downregulated in MDA231/miR-185-5p cells than in
MDA231/NC cells (Fig. 4C). These
results proved that miR-185-5p modulated Snail stabilization
depending on the activation of NF-κB.
miR-185-5p inhibited S100A8/A9-induced
F-actin polymerization of breast cancer cells
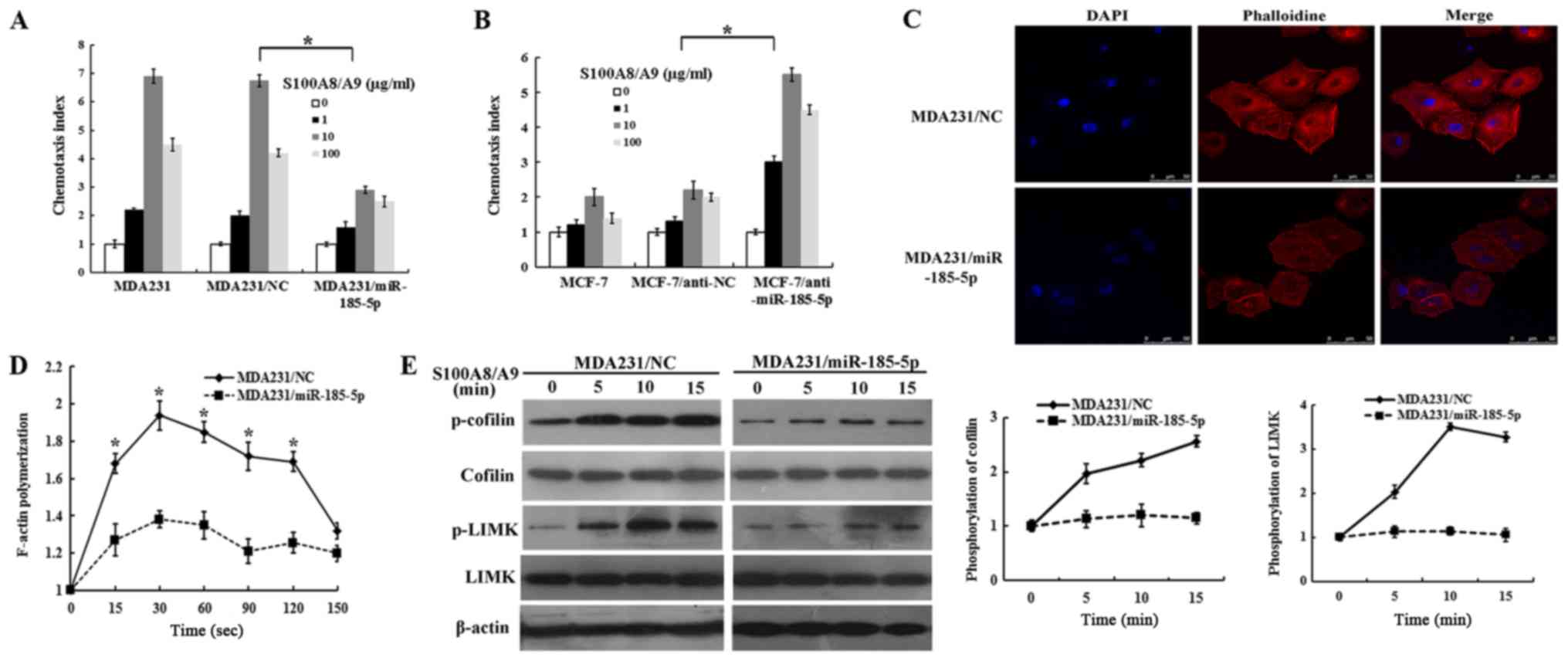
The cell chemotaxis assay was designed to detect
whether miR-185-5p could inhibit S100A8/A9 and induce chemotaxis of
breast cancer cells. No difference in chemotaxis ability was found
between MDA231 and MDA231/NC cells. However, this ability was
reduced in the MDA231/miR-185-5p cells than in the MDA231/NC cells
(Fig. 5A). By contrast,
MCF-7/anti-miR-185-5p cells showed an increase in chemotaxis
ability (Fig. 5B). The results
showed that miR-185-5p acted a vital part in inhibiting chemotaxis
of breast cancer cells. Varlet et al (20) found that fine tuning the actin
dynamics facilitated cytokinesis. Whether miR-185-5p could mediate
the reorganization of F-actin needed further investigation. Thus,
we detected F-actin polymerization under miR-185-5p upregulation in
breast cancer cells. The quantitative F-actin polymerization
experiment indicated that S100A8/A9 induced actin polymerization in
MDA231/NC cells at 20 and 60 sec, while F-actin polymerization was
notably decreased with S100A8/A9 stimulation in the
MDA231/miR-185-5p cells (Fig. 5C and
D). These results indicated that miR-185-5p strongly limited
the cytoskeleton rearrangement and could abolish the formation of
stress fibers. Cofilin is an important regulator to balance between
F-actin and G-actin. Dephosphorylated cofilin can promote G-actin
in forming F-actin, which contributes to cell migration through
lamellipodium (21). LIM kinase
(LIMK) regulates the structure of the F-actin cytoskeleton by
phosphorylation and inactivation of F-actin depolymerization
factors of the ADF/cofilin family (22). LIMK is a vital protein in tumor
formation and progression (23,24).
LIMK or cofilin phosphorylation was proved to be remarkably
increased in malignant melanoma (25) and prostate cancer (26). Further analysis indicated that
miR-185-5p markedly inhibited the activation of cofilin and LIMK in
MDA231/miR-185-5p cells. However, the levels of total LIMK and
cofilin remained unchanged (Fig.
5E). These results revealed that miR-185-5p played a vital role
in inhibiting F-actin polymerization of breast cancer cells.
Overexpression of miR-185-5p and
reduction of RAGE inhibited lung metastasis in SCID mice
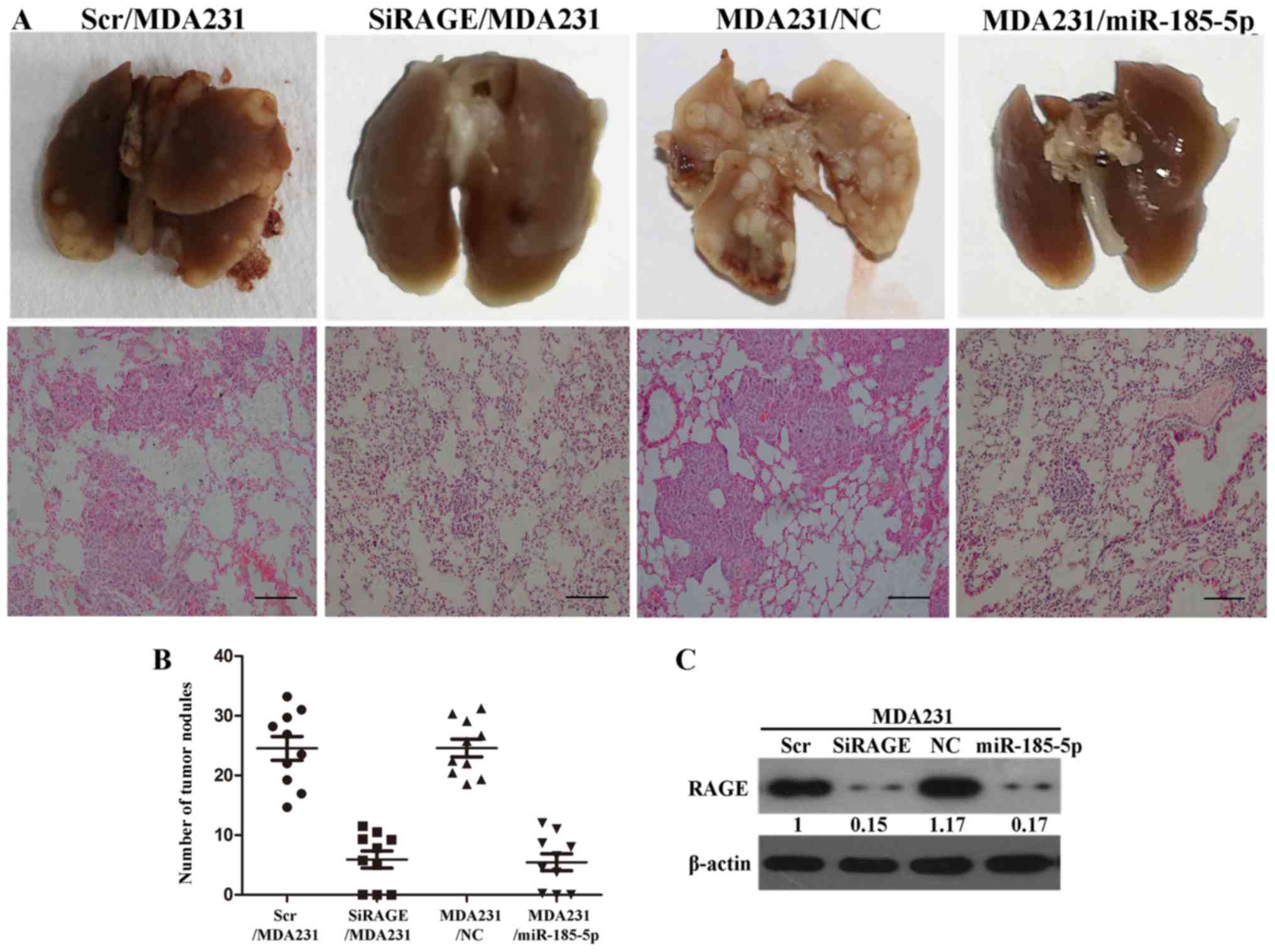
To verify the effect of miR-185-5p and RAGE on
breast cell invasion in vivo, we injected the axilla of SCID
mice with MDA231/NC and MDA231/miR-185-5p cells. Meanwhile, we
injected the axilla of SCID mice with SiRAGE/MDA231 and control
cells. The number of satellite tumors was counted in sections of
SCID mice sacrificed at 10 weeks. The tumor nodules were
considerably reduced in MDA231/miR-185-5p cells than in the
MDA231/NC group. Upregulation of miR-185-5p resulted in significant
inhibition of invasion in vivo. SiRAGE/MDA231 group showed
significantly reduced tumor nodules than the control group
(Fig. 6A and B). RAGE in sectioned
seeded tumors in SCID mice injected with MDA231/NC, Scr/MDA231,
SiRAGE/MDA231, and MDA231/miR-185-5p cells were analyzed by western
blot. The expression of RAGE was significantly reduced in
MDA231/miR-185-5p cells than in MDA231/NC (Fig. 6C). The results consistently
indicated that miR-185-5p could inhibit invasion of breast cancer
cell in vivo.
Discussion
An increasing number of studies have recently
manifested that the development and progression of malignant tumor
is a very complex process and affected by various factors. miRNAs
play a vital role in mediating gene by targeting the 3′-UTRs
(27), which are vital mediators
of cellular functions, such as proliferation (28) and G1/S transition (29,30).
Overexpression of miR-185-5p repressed TRIM29 in GC and
significantly inhibited malignant behavior (31). Meanwhile, miR-185 repressed
hepatocellular carcinoma tumor genesis by targeting the
DNMT1/PTEN/Akt signal pathway (32). miR-185-5p has been demonstrated to
directly target RAGE and inhibit the invasion and metastasis in
ESCC. However, the important molecular mechanism by which
miR-185-5p targets RAGE to repress breast cancer remains unknown.
In this study, the data revealed that miR-185-5p directly targeted
and modulated RAGE 3′-UTR and inhibited invasion and EMT. In
addition, this miRNA sufficiently reversed the mesenchymal
potentials and reduced the invasiveness of mesenchymal cells.
EMT is an important molecular mechanism that
determines the metastasis and invasion of carcinoma cells (33). Consistent with previous finding, we
previously showed that the combination of RAGE-S100A8/A9 promoted
the invasion and metastasis of breast cancer cells (8). Meanwhile, EMT regulated multiple
signaling pathways, such as TGF-β (34), PI3K, MAPK, and Wnt/β-catenin.
Numerous genes and transcription factors, such as Snail, Twist, and
ZEB1/2 (35,36), have been found as key regulators in
EMT. Zhai et al (37)
indicated that miR-143 inhibited migration and invasion by
downregulating ERK5 in breast cancer cells. In this study,
miR-185-5p inhibited the S100A8/A9-induced EMT via NF-κB/Snail
signaling pathway.
Among the multifaceted effects exerted by this
program, rearrangement of actin cytoskeleton is the major driving
mechanism for cell migration and invasion in the development of EMT
and cancer cells with invasive behavior (38). Wang et al (39) found that MIIP could inhibit
endometrial carcinoma migration via cytoskeleton reorganization
with markedly reduced formation of lamellipodia. Previous studies
indicated that HSPB8 regulated BAG3 levels through a mechanism that
involves effects on branched actin nucleation in cancer cells
(20). Phosphorylated cofilin
could process the polymerization of F-actin and formation of
lamellipodium, leading to the damage of lamellipodia-based
metastasis. Hence, these results indicated that miR-185-5p
inhibited F-actin polymerization. miR-185-5p has a significant part
in metastasis and invasion of breast cancer cells.
S100 family members have diverse functions in cancer
development. S100A8/A9, which is a member of the S100 protein
family, contributes to the progression of human cancers. Previous
studies showed that modulating the MMP-2 expression S100A8/A9
related to carcinoma cell phenotype and activity can regulate
carcinoma cell metastasis (40,41).
S100A8/A9 is released at the region of inflammation by phagocytes,
monocytes, epithelial cells, and endothelial cells (42). This protein regulates various
processes during chronic inflammation. The results show that the
S100A8/A9 protein dimers interact with RAGE at cytomembrane of
melanoma tumor cells (43,44). We previously reported the impact of
the RAGE-S100A8/A9 interaction on cell invasion and metastasis
formation in breast cancer (8,45).
Here, we demonstrated the miR-185-5p targeting of RAGE in
metastasis and invasive capabilities via combining to extracellular
S100A8/A9. Hence, miR-185-5p inhibited the invasion and migration
of breast cancer cells by targeting RAGE.
Thus, a relation between miR-185-5p and RAGE
expression was established in this study. The importance of
miR-185-5p regulation of F-actin polymerization and reversal of EMT
in breast cancer was determined. The NF-κB/Snail signaling pathway
was involved in the suppression of EMT. Furthermore, all findings
demonstrated that miR-185-5p played a role in inhibiting invasion
and migration in vivo. These results provided a new
rationale for using miR-185-5p to target RAGE, which is a novel
strategy for breast cancer therapy.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National
Natural Science Foundation of China (grant nos. 81702932, 81641111
and 81402389) and the Natural Science Foundation of Shandong
Province (grant no. ZR2015HL065).
Availability of data and materials
The datasets used and analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
CY and GZ performed the experiments and wrote the
manuscript. RS, XP and XW analyzed data. HL and YS assisted with
the design of the experiments. All authors read and approved the
final manuscript.
Ethics approval and consent to
participate
Written informed consent was acquired from all
patients, and the study was approved by the Institute Research
Ethics Committee at the Cancer Center, Weifang Medical
University.
Patient consent for publication
Written informed consent was acquired from all
patients.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
DeSantis C, Ma J, Bryan L and Jemal A:
Breast cancer statistics, 2013. CA Cancer J Clin. 64:52–62. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Malik P, Chaudhry N, Mittal R and
Mukherjee TK: Role of receptor for advanced glycation end products
in the complication and progression of various types of cancers.
Biochim Biophys Acta. 1850:1898–1904. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Jules J, Maiguel D and Hudson BI:
Alternative splicing of the RAGE cytoplasmic domain regulates cell
signaling and function. PLoS One. 8:e782672013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chen X, Zhang L, Zhang IY, Liang J, Wang
H, Ouyang M, Wu S, da Fonseca ACC, Weng L, Yamamoto Y, et al: RAGE
expression in tumor-associated macrophages promotes angiogenesis in
glioma. Cancer Res. 74:7285–7297. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Elangovan I, Thirugnanam S, Chen A, Zheng
G, Bosland MC, Kajdacsy-Balla A and Gnanasekar M: Targeting
receptor for advanced glycation end products (RAGE) expression
induces apoptosis and inhibits prostate tumor growth. Biochem
Biophys Res Commun. 417:1133–1138. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kalea AZ, See F, Harja E, Arriero M,
Schmidt AM and Hudson BI: Alternatively spliced RAGEv1 inhibits
tumorigenesis through suppression of JNK signaling. Cancer Res.
70:5628–5638. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kang R, Tang D, Schapiro NE, Livesey KM,
Farkas A, Loughran P, Bierhaus A, Lotze MT and Zeh HJ: The receptor
for advanced glycation end products (RAGE) sustains autophagy and
limits apoptosis, promoting pancreatic tumor cell survival. Cell
Death Differ. 17:666–676. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yin C, Li H, Zhang B, Liu Y, Lu G, Lu S,
Sun L, Qi Y, Li X and Chen W: RAGE-binding S100A8/A9 promotes the
migration and invasion of human breast cancer cells through actin
polymerization and epithelial-mesenchymal transition. Breast Cancer
Res Treat. 142:297–309. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gonzalez DM and Medici D: Signaling
mechanisms of the epithelial-mesenchymal transition. Sci Signal.
7:re82014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lamouille S, Xu J and Derynck R: Molecular
mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell
Biol. 15:178–196. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tseng JH, Bisogna M, Hoang LN, Olvera N,
Rodriguez-Aguayo C, Lopez-Berestein G, Sood AK, Levine DA and
Jelinic P: miR-200c-driven Mesenchymal-To-Epithelial Transition is
a Therapeutic Target in Uterine Carcinosarcomas. Sci Rep.
7:36142017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Siemens H, Jackstadt R, Hünten S, Kaller
M, Menssen A, Götz U and Hermeking H: miR-34 and SNAIL form a
double-negative feedback loop to regulate epithelial-mesenchymal
transitions. Cell Cycle. 10:4256–4271. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gwak JM, Kim HJ, Kim EJ, Chung YR, Yun S,
Seo AN, Lee HJ and Park SY: MicroRNA-9 is associated with
epithelial-mesenchymal transition, breast cancer stem cell
phenotype, and tumor progression in breast cancer. Breast Cancer
Res Treat. 147:39–49. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li S, Ma Y, Hou X, Liu Y, Li K, Xu S and
Wang J: MiR-185 acts as a tumor suppressor by targeting AKT1 in
non-small cell lung cancer cells. Int J Clin Exp Pathol.
8:11854–11862. 2015.PubMed/NCBI
|
|
15
|
Yoon JH, Choi YJ, Choi WS, Ashktorab H,
Smoot DT, Nam SW, Lee JY and Park WS: GKN1-miR-185-DNMT1 axis
suppresses gastric carcinogenesis through regulation of epigenetic
alteration and cell cycle. Clin Cancer Res. 19:4599–4610. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang R, Tian S, Wang HB, Chu DP, Cao JL,
Xia HF and Ma X: MiR-185 is involved in human breast carcinogenesis
by targeting Vegfa. FEBS Lett. 588:4438–4447. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tang H, Wang Z, Liu X, Liu Q, Xu G, Li G
and Wu M: LRRC4 inhibits glioma cell growth and invasion through a
miR-185-dependent pathway. Curr Cancer Drug Targets. 12:1032–1042.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Jing R, Chen W, Wang H, Ju S, Cong H, Sun
B, Jin Q, Chu S, Xu L and Cui M: Plasma miR-185 is decreased in
patients with esophageal squamous cell carcinoma and might suppress
tumor migration and invasion by targeting RAGE. Am J Physiol
Gastrointest Liver Physiol. 309:G719–G729. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Varlet AA, Fuchs M, Luthold C, Lambert H,
Landry J and Lavoie JN: Fine-tuning of actin dynamics by the
HSPB8-BAG3 chaperone complex facilitates cytokinesis and
contributes to its impact on cell division. Cell Stress Chaperones.
22:553–567. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Shi C, Cai Y, Li Y, Li Y, Hu N, Ma S, Hu
S, Zhu P, Wang W and Zhou H: Yap promotes hepatocellular carcinoma
metastasis and mobilization via governing
cofilin/F-actin/lamellipodium axis by regulation of
JNK/Bnip3/SERCA/CaMKII pathways. Redox Biol. 14:59–71. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Fife CM, McCarroll JA and Kavallaris M:
Movers and shakers: Cell cytoskeleton in cancer metastasis. Br J
Pharmacol. 171:5507–5523. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Manetti F: LIM kinases are attractive
targets with many macromolecular partners and only a few small
molecule regulators. Med Res Rev. 32:968–998. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Mali RS and Kapur R: Targeting Rho
associated kinases in leukemia and myeloproliferative neoplasms.
Oncotarget. 3:909–910. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Okamoto I, Pirker C, Bilban M, Berger W,
Losert D, Marosi C, Haas OA, Wolff K and Pehamberger H: Seven novel
and stable translocations associated with oncogenic gene expression
in malignant melanoma. Neoplasia. 7:303–311. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Mardilovich K, Gabrielsen M, McGarry L,
Orange C, Patel R, Shanks E, Edwards J and Olson MF: Elevated LIM
kinase 1 in nonmetastatic prostate cancer reflects its role in
facilitating androgen receptor nuclear translocation. Mol Cancer
Ther. 14:246–258. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhi Q, Zhu J, Guo X, He S, Xue X, Zhou J,
Hu B, Li H, Chen S, Zhao H and Kuang Y: Metastasis-related miR-185
is a potential prognostic biomarker for hepatocellular carcinoma in
early stage. Biomed Pharmacother. 67:393–398. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Osbourne A, Calway T, Broman M, McSharry
S, Earley J and Kim GH: Downregulation of connexin43 by
microRNA-130a in cardiomyocytes results in cardiac arrhythmias. J
Mol Cell Cardiol. 74:53–63. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Liao X, Xue H, Wang YC, Nazor KL, Guo S,
Trivedi N, Peterson SE, Liu Y, Loring JF and Laurent LC: Matched
miRNA and mRNA signatures from an hESC-based in vitro model of
pancreatic differentiation reveal novel regulatory interactions. J
Cell Sci. 126:3848–3861. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
James EN, Delany AM and Nair LS:
Post-transcriptional regulation in osteoblasts using localized
delivery of miR-29a inhibitor from nanofibers to enhance
extracellular matrix deposition. Acta Biomater. 10:3571–3580. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Qiu F, Xiong JP, Deng J and Xiang XJ:
TRIM29 functions as an oncogene in gastric cancer and is regulated
by miR-185. Int J Clin Exp Pathol. 8:5053–5061. 2015.PubMed/NCBI
|
|
32
|
Qadir XV, Han C, Lu D, Zhang J and Wu T:
miR-185 inhibits hepatocellular carcinoma growth by targeting the
DNMT1/PTEN/Akt pathway. Am J Pathol. 184:2355–2364. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ponnusamy MP, Seshacharyulu P, Lakshmanan
I, Vaz AP, Chugh S and Batra SK: Emerging role of mucins in
epithelial to mesenchymal transition. Curr Cancer Drug Targets.
13:945–956. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wu Y, Tran T, Dwabe S, Sarkissyan M, Kim
J, Nava M, Clayton S, Pietras R, Farias-Eisner R and Vadgama JV:
A83-01 inhibits TGF-β-induced upregulation of Wnt3 and epithelial
to mesenchymal transition in HER2-overexpressing breast cancer
cells. Breast Cancer Res Treat. 163:449–460. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Batlle E, Sancho E, Francí C, Domínguez D,
Monfar M, Baulida J and De Herreros García A: The transcription
factor snail is a repressor of E-cadherin gene expression in
epithelial tumour cells. Nat Cell Biol. 2:84–89. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yang J, Mani SA, Donaher JL, Ramaswamy S,
Itzykson RA, Come C, Savagner P, Gitelman I, Richardson A and
Weinberg RA: Twist, a master regulator of morphogenesis, plays an
essential role in tumor metastasis. Cell. 117:927–939. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhai L, Ma C, Li W, Yang S and Liu Z:
miR-143 suppresses epithelial-mesenchymal transition and inhibits
tumor growth of breast cancer through down-regulation of ERK5. Mol
Carcinog. 55:1990–2000. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Yilmaz M and Christofori G: EMT, the
cytoskeleton, and cancer cell invasion. Cancer Metastasis Rev.
28:15–33. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wang Y, Hu L, Ji P, Teng F, Tian W, Liu Y,
Cogdell D, Liu J, Sood AK, Broaddus R, et al: Erratum to: MIIP
remodels Rac1-mediated cytoskeleton structure in suppression of
endometrial cancer metastasis. J Hematol Oncol. 10:1632017.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Silva EJ, Argyris PP, Zou X, Ross KF and
Herzberg MC: S100A8/A9 regulates MMP-2 expression and invasion and
migration by carcinoma cells. Int J Biochem Cell Biol. 55:279–287.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Khammanivong A, Wang C, Sorenson BS, Ross
KF and Herzberg MC: S100A8/A9 (calprotectin) negatively regulates
G2/M cell cycle progression and growth of squamous cell carcinoma.
PLoS One. 8:e693952013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Gebhardt C, Németh J, Angel P and Hess J:
S100A8 and S100A9 in inflammation and cancer. Biochem Pharmacol.
72:1622–1631. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Ichikawa M, Williams R, Wang L, Vogl T and
Srikrishna G: S100A8/A9 activate key genes and pathways in colon
tumor progression. Mol Cancer Res. 9:133–148. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Saha A, Lee YC, Zhang Z, Chandra G, Su SB
and Mukherjee AB: Lack of an endogenous anti-inflammatory protein
in mice enhances colonization of B16F10 melanoma cells in the
lungs. J Biol Chem. 285:10822–10831. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Dahlmann M, Okhrimenko A, Marcinkowski P,
Osterland M, Herrmann P, Smith J, Heizmann CW, Schlag PM and Stein
U: RAGE mediates S100A4-induced cell motility via MAPK/ERK and
hypoxia signaling and is a prognostic biomarker for human
colorectal cancer metastasis. Oncotarget. 5:3220–3233. 2014.
View Article : Google Scholar : PubMed/NCBI
|