Introduction
Type 2 diabetes mellitus is a highly prevalent
disease worldwide and is associated with increased rates of
morbidity and mortality. It is estimated that 382 million adults
suffered from diabetes in 2013 and that this number will reach 592
million around the world by 2035 (1). It is a metabolic disorder
characterized by chronic hyperglycemia. Long-term untreatment or
improper treatment may lead to chronic complications of diabetes
mellitus, including diabetic nephropathy, diabetic microangiopathy
and diabetic neuropathy (2–4).
These chronic complications are also important factors, which may
cause the patient to succumb to the disease or result in
disability.
Long noncoding RNAs (lncRNAs) belong to a
heterogeneous class of regulatory ncRNAs with transcript lengths
>200 nucleotides and serve important roles in gene silencing and
posttranscriptional gene regulation (5,6).
During recent decades, lncRNAs have been identified as important
epigenetic factors in regulating various human diseases, including
cancer, neurodegenerative diseases, diabetes and cardiovascular
diseases (7–11).
For instance, inhibition of lncRNA insulin-like
growth factor 2 (IGF2) antisense RNA promoted angiogenesis in type
2 diabetes through upregulating IGF2 and vascular endothelial
growth factor (12). Zhou et
al (13) demonstrated that
lncRNA metastasis associated lung adenocarcinoma transcript 1
presented an extremely high expression level in pancreatic cancer
tissues and cells. It promoted the proliferation of AsPC-1 cells by
regulating Hippo-YAP signaling lncRNA HOX transcript antisense RNA
and colon cancer associated transcript 2 promoted the proliferation
of cervical cancer cells (14,15).
A study demonstrated that nine lncRNAs were then combined to form a
single prognostic signature for predicting metastatic risk in
breast cancer patients (16).
Another previous study reported that lncRNA AL049437 may contribute
to the risk of Parkinson's disease, while lncRNA AK021630 may
reduce the chance of Parkinson's disease (17). In recent years, there have been
numerous reports of diabetic central nervous system disease
(18–20). Roberts et al (21) considered the putative role(s) of
lncRNAs in neurodevelopment and brain function with an emphasis on
the epigenetic regulation of gene expression. Certain studies
demonstrated that the lncRNA TCL1 upstream neural
differentiation-associated RNA was demonstrated to be highly
conserved among vertebrates and was expressed within the developing
nervous system (22) and lncRNA
BC200 levels in Brodmann's area 9 [the area of brain affected in
Alzheimer's disease (AD)] were demonstrated to be higher in
age-matched AD brains compared with normal brains and the relative
levels of BC200 RNA in affected areas increased with the severity
of AD (23). In addition, the
six-lncRNA (AC005013.5, UBE2R2-AS1, ENTPD1-AS1, RP11-89C21.2,
AC073115.6 and XLOC_004803) signature may be involved in the
immune-associated biological processes and pathways, which are very
well known in the context of glioblastoma tumorigenesis (24). Increasing evidence also suggests
that long noncoding RNAs serve important regulatory roles in type 2
diabetes.
However, there remains controversy regarding the
results of pathophysiology and treatment in cognitive dysfunction
of type 2 diabetes. The aim of the present study was to analyze the
expression pattern of lncRNAs-mRNA in the hippocampus of type 2
diabetic mice through gene chip detection technology and to
investigate novel ideas for identifying abnormal gene expression of
cognitive dysfunction and pathophysiology in type 2 diabetes. This
study is a preliminary study, using the method of gene chip
analysis to investigate possible differences between genes in the
diabetic and non-diabetic group. Due to lots of differential gene,
further analysis and verification are needed.
Materials and methods
Mouse model
Twelve C57BL6J female mice (aged 8 weeks old) were
bred in a specific pathogen-free laboratory (temperature, 20–24°C;
humidity, 40–70%) at Shandong University (Jinan, China). Mice were
acclimated to the feed for 1 week prior to the initiation of
experimental intervention. All mice were housed in standard cages
and had free access to food and water. Following this, six mice
were randomly selected and labeled as control group. The other 6
mice were fasted for 12 h, then injected with 1% streptozotocin
(Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) solution which was
dissolved in 0.05 mol/l sodium citrate (pH 4.5; Beyotime Institute
of Biotechnology, Haimen, China), at a dose of 60 mg/kg and
injected for 5 days continuously. The control group had
intraperitoneal injections of buffer solution at the same dose.
After 10 days, the tail vein blood glucose level was detected
through a blood glucose meter (Bayer, Newbury, UK). All six mice
injected with 1% streptozotocin solution exhibited high blood
glucose levels (≥16.7 mmol/l) and were considered to be diabetic
model mice (25). The control
group and the diabetes group were administered daily 10 ml/kg
intragastric normal saline treatments for a total of 12 weeks. Mice
were weighed and were subjected to tail vein blood glucose testing
once each week.
Following the final saline treatment, the mice were
anaesthetized by intraperitoneal injection of 0.8% pentobarbital
(Sigma-Aldrich; Merck KGaA). The mouse limbs were fixed and the
abdomen was exposed, then the abdominal skin, peritoneum and
diaphragm were cut to expose the heart. Left ventricular blood was
withdrawn through a 1 ml syringe then this injected into a
coagulation + separation tube for extraction by 10 min
centrifugation at 900 × g at 20°C. Extracted serum was kept frozen
at −80°C within a cryopreservation tube. The brain tissue was then
obtained and then the hippocampus tissue was isolated and
subsequently stored at −80°C. The protocol for the present study
was approved by the Laboratory Animal Ethics committee of the Qilu
Hospital of Shandong University.
Reagents
TRIzol® Reagent (Invitrogen; Thermo
Fisher Scientific, Inc., Waltham, MA, USA; cat. no. 15596-026);
NucleoSpin® RNA clean-up (cat. no. 740.948.250);
Nucleospin® Extract II (cat. no. 740.609.250; both
Machery-Nagel, GmbH, Düren, Germany); GeeDom bio-chip universal
label reagent (Capital Biotechnology, Co., Ltd., Beijing, China;
cat. no. 360069); Cy3-dCTP (GE Healthcare, Chicago, IL, USA; cat.
no. PA53031); Cy5-dCTP (GE Healthcare; cat. no. PA55031); 10% SDS;
20 × sodium citrate buffer (both Capital Biotechnology, Co., Ltd.);
Agilent one-color RNA Spike-in kit (cat. no. 5188-5282); Agilent
two-color RNA Spike-in kit (cat. no. 5188-5279; both Agilent
Technologies Inc., Santa Clara, CA, USA); RNAiso Plus kit (Takara
Biotechnology Co., Ltd., Dalian, China); TransScript One-step gDNA
Removal and cDNA Synthesis SuperMix kits; TransStart Tip Green qPCR
Super MixSYBR; Reverse Transcriptase kit; and TaqMan MGB probes
(all Beijing Transgen Biotech Co., Ltd., Beijing, China) were
purchased for use in the present study.
Equipment
A 5810R centrifuge was purchased from Eppendorf
(Hamburg, Germany); the spectrophotometer ND-1000 was from
NanoDrop; Thermo Fisher Scientific, Inc. (Wilmington, DE, USA); the
concentrator (instrument model 5301) was from Eppendorf. The
Peltier Thermal Cycler PTC-225 (MJ Research, Inc., Waltham, MA,
USA). The hybridization Oven (G2545A) was from Agilent
Technologies, Inc., as was the Agilent G2565CA Microarray
Scanner.
Synthesis and labeling of cDNA
Total RNA samples were extracted from three healthy
mice (group A) and three type 2 diabetic mice (group B). Total RNA
was isolated using TRIzol® reagent (Invitrogen; Thermo
Fisher Scientific, Inc.) according to the manufacturer's protocol.
The first and second strand cDNA were synthesized with a
PrimeScript™ Double Strand cDNA Synthesis kit according to the
manufacturer's protocol (Takara Biotechnology Co., Ltd.).
Complementary RNA (cRNA) was transcribed via the T7 Enzyme Mix kit
using the second strand cDNA as a template and subsequently
purified using NucleoSpin® RNA clean-up (740.948.250;
Machery-Nagel, GmbH) to eliminate reagents like salt and enzymes
according to the manufacturer's protocol. In addition, the
aforementioned dNTPs and fluorophores were added to the mixture.
Following this, the purified RNA was reverse transcribed using a
Reverse Transcriptase kit (Beijing Transgen Biotech Co., Ltd.).
lncRNA and mRNA microarray expression
profiling
cDNA containing fluorescent labels Cy3-dCTP (GE
Healthcare; cat. nos. PA53031) or Cy5-dCTP (GE Healthcare; cat. no.
PA55031) or Cy3-dCTP and Cy5-dCTP together were hybridized on the
GeeDom human lncRNA+mRNA V4.0 array with GeeDom bio-chip universal
label reagent. The microarrays were washed and then scanned using
an Agilent G2565CA Microarray Scanner (Agilent Technologies, Inc.)
The resulting expression hybridization chip picture was analyzed
using Feature Extraction software 10.7 (Agilent Technologies,
Inc.). Data normalization and quality control analysis was
performed for each sample using GeneSpring GX 11.0 (Agilent
Technologies, Inc.). Gene expression differences and P-values were
assessed by GeneSpring GX 11.0. Cluster analysis was performed
using Cluster 3.0 software (Cluster Software, Inc.). Gene Ontology
Enrichment Analysis (GO analysis; http://geneontology.org/) of the target genes, Protein
ANalysis THrough Evolutionary Relationships (PANTHER; http://www.pantherdb.org/) and Kyoto Encyclopedia of
Genes and Genomes (KEGG; http://www.genome.jp/kegg/pathway.html) pathway
analyses, as well as BioCarta (https://cgap.nci.nih.gov/Pathways/BioCarta_Pathways),
Pathway Interaction Database (PID; http://pid.nci.nih.gov/), BioCyc (https://biocyc.org/) and Reactome (https://reactome.org/cgi-bin/frontpage?DB=gk_current)
analyses, were performed on differential mRNAs. lncRNA-mRNA
co-expression analysis was addressed along with target gene
prediction and transcription factor prediction for each biological
repeat.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was isolated using TRIzol®
reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to
the manufacturer's protocol. RNA was reverse transcribed to cDNA
with TransScript One-step gDNA Removal and cDNA Synthesis SuperMix
kits according to the manufacturer's protocol (Transgen
Biotechnology Co., Ltd.). RT-qPCR was performed using TransStart
Tip Green qPCR Super MixSYBR according to the manufacturer's
protocol (Transgen Biotechnology Co., Ltd.). The thermocycling
conditions for qPCR were as follows: Initial denaturation at 95°C
for 30 sec, followed by 40 cycles at 95°C for 5 sec, 60°C annealing
for 30 sec. Sequences of primers used for qPCR are presented in
Table I. Relative gene expression
data was analyzed using the 2−∆∆Cq method (26).
 | Table I.Sequences of primers used for reverse
transcription-quantitative polymerase chain reaction. |
Table I.
Sequences of primers used for reverse
transcription-quantitative polymerase chain reaction.
| Primer | Sequence
(5′->3′) |
|---|
| Prr32 forward |
CTTTCCACCAAGGGGTTCAC |
| Prr32 reverse |
GCAGTGGAGAAGCAAAAGCA |
| Lmx1a forward |
ACGGCCTGAAGATGGAGGA |
| Lmx1a reverse |
CAGAAACCTGTCCGAGATGAC |
| Slc5a7 forward |
ATGTCTTTCCACGTAGAAGGACT |
| Slc5a7 reverse |
TTGCCGCTGTTTTTGGTTTTC |
Statistical analysis
Statistical analysis was performed using SPSS v20
software (IBM Corp., Armonk, NY, USA). All data are expressed as
the mean ± standard deviation. A T-test indicated consistency
between microarray data and RT-qPCR results. P<0.05 was
considered to indicate a statistically significant difference. All
experiments were repeated in triplicate.
Results
LncRNA expression
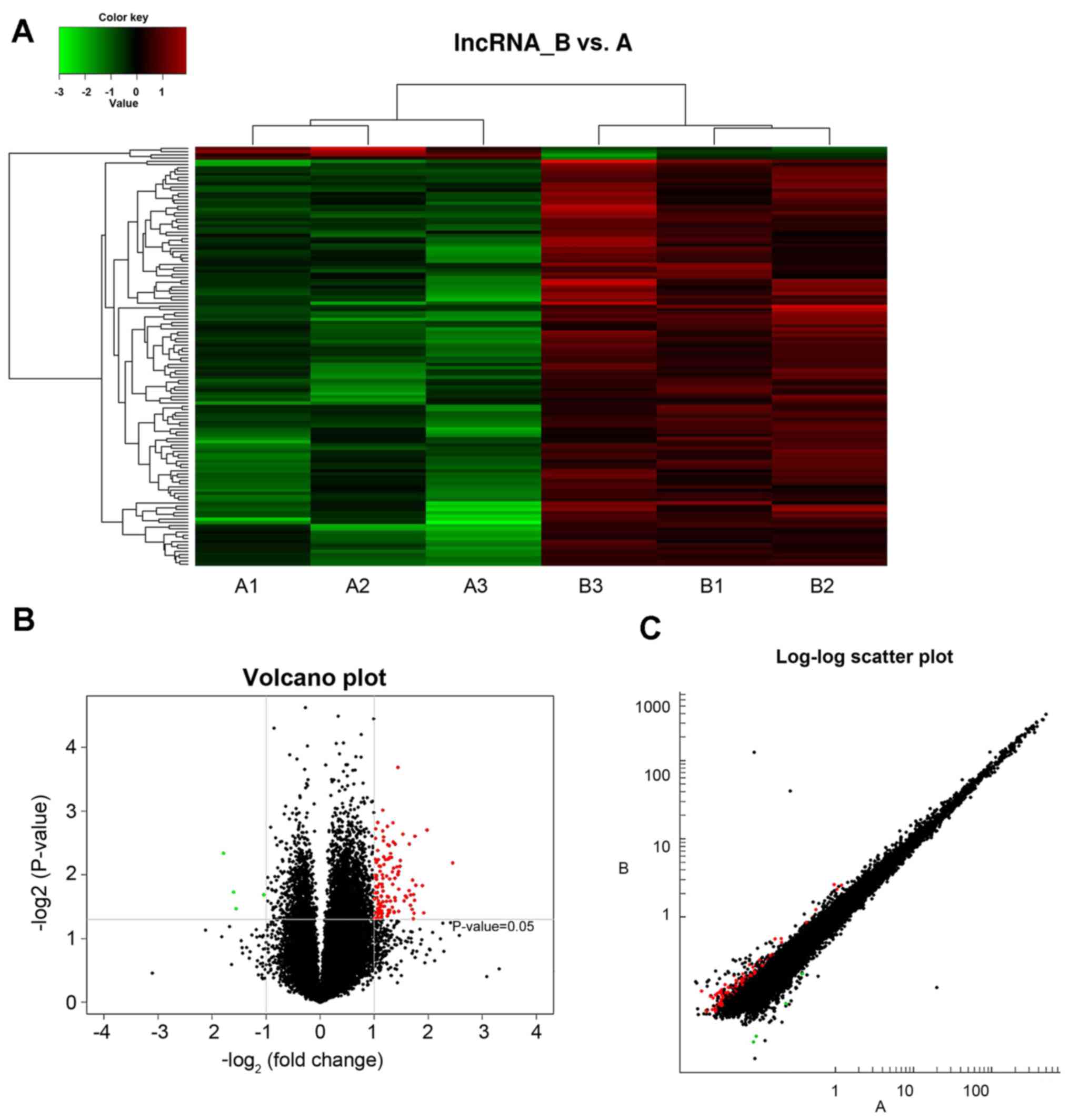
A total of 36,793 lncRNAs were detected within the
hippocampus of type 2 diabetic mice. LncRNA and mRNA data revealed
differential expression of lncRNA between healthy and diabetic mice
captured through data analysis, with differential expression
criteria set as fold-change ≥2.0 and P<0.05. Compared with the
control mice (group A), diabetic mice (group B) exhibited
differential expression of 130 lncRNAs, where 126 lncRNAs were
demonstrated to be upregulated and 4 lncRNAs were downregulated.
The expression levels of lncRNAs were visualized using clustering
analysis (Fig. 1A). The
differential expression ratios and P-values of lncRNAs in the two
groups were also compared using volcano (Fig. 1B) and scatter plots (Fig. 1C). The results revealed a marked
difference in the expression levels of lncRNAs exhibited by groups
A and B (red points), which suggested that the regulation of
lncRNAs is associated with the occurrence and development of type 2
diabetes.
mRNA expression
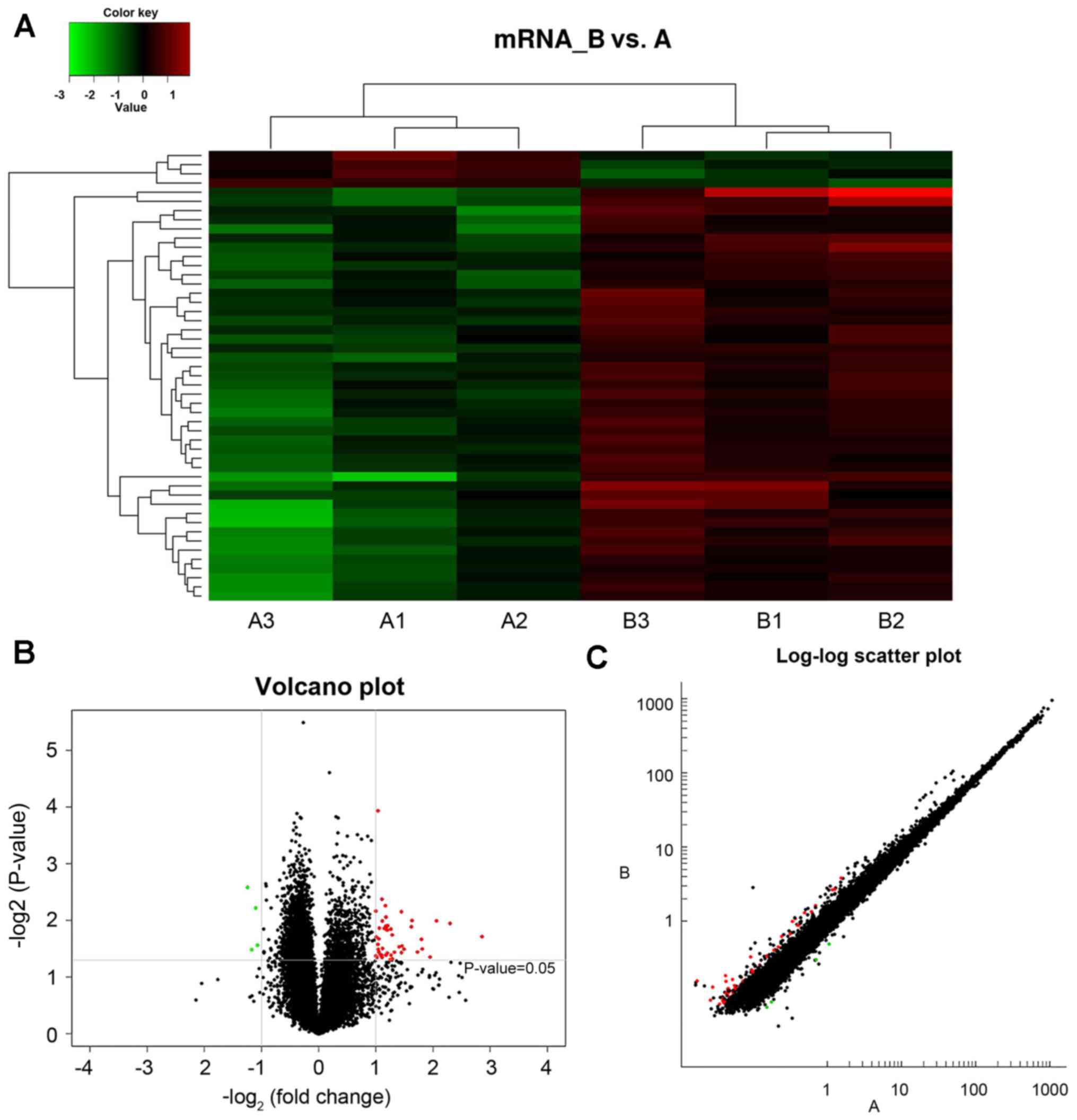
A total of 30,551 mRNAs were detected within the
hippocampus of type 2 diabetic mice. mRNA data was collected from
image and data analysis, where the differential expression criteria
was set as fold-change ≥2.0 and P<0.05. Compared with group A,
group B presented 49 mRNAs with altered expression, where 45
exhibited upregulation and 4 displayed downregulation. The
expression levels of mRNAs in groups A and B were visualized using
clustering analysis (Fig. 2A). The
differential expression ratios and P-values of mRNAs in the two
groups were also compared using volcano (Fig. 2B) and scatter plots (Fig. 2C). Fig. 2 demonstrates mRNA differential
expression association between different groups.
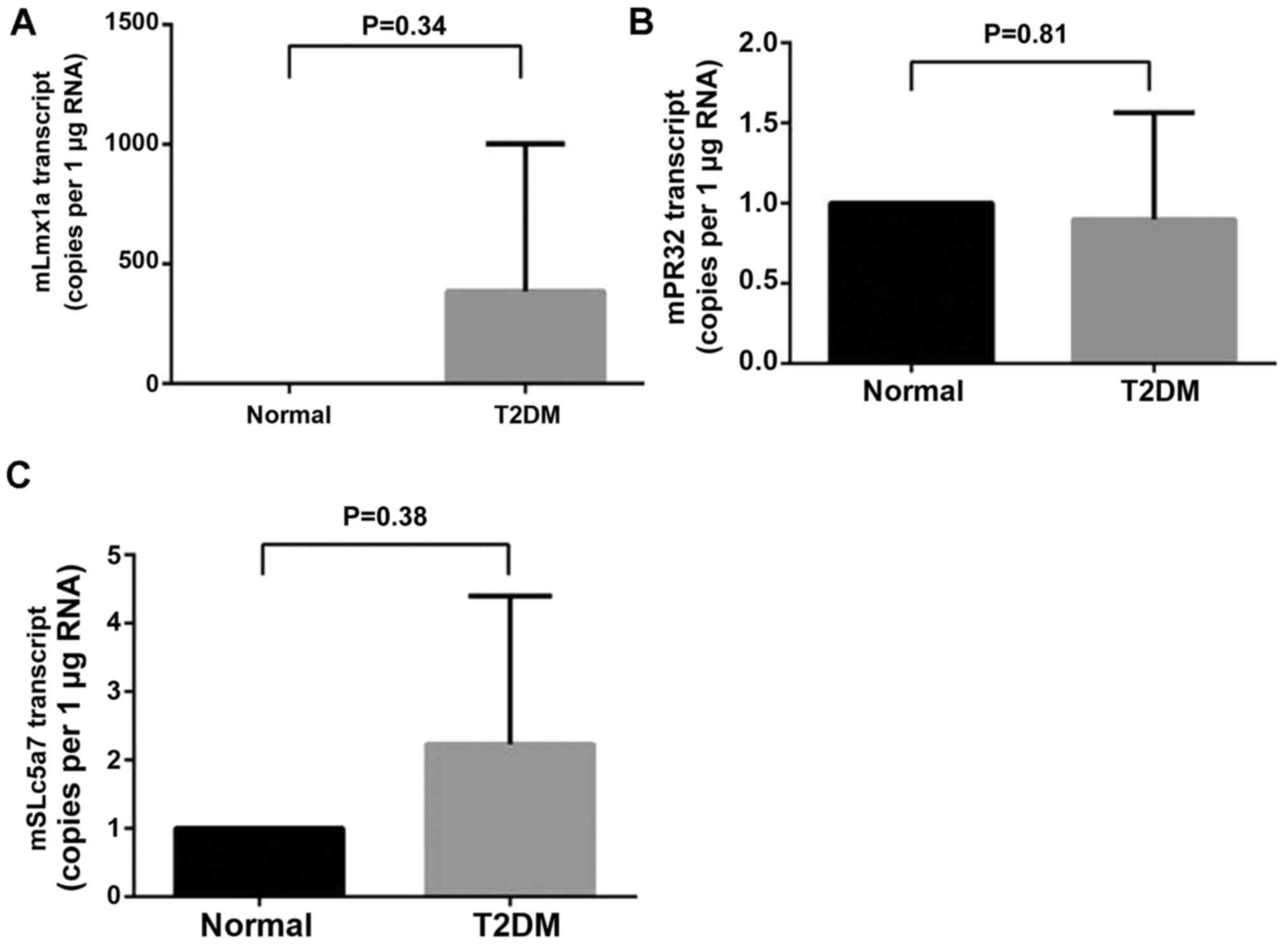
RT-qPCR validation array
RT-qPCR was performed in order to verify the
reliability of microarray data. A total of three differentially
expressed mRNAs (lmx1a, slc5a7 and prr32) with high expression in
the central nervous system were selected from the 49 differentially
expressed mRNAs as revealed in Fig.
2A. Alterations in expression levels of lmx1a (Fig. 3A), prr32 (Fig. 3B) and slc5a7 (Fig. 3C) revealed by RT-PCR were similar
to those observed in microarray data. A T-test indicated
consistency between microarray data and RT-qPCR results, and
demonstrated that our microarray data is reliable and can be used
for bioinformatics analysis in subsequent steps.
Functional analysis of differentially
expressed mRNA
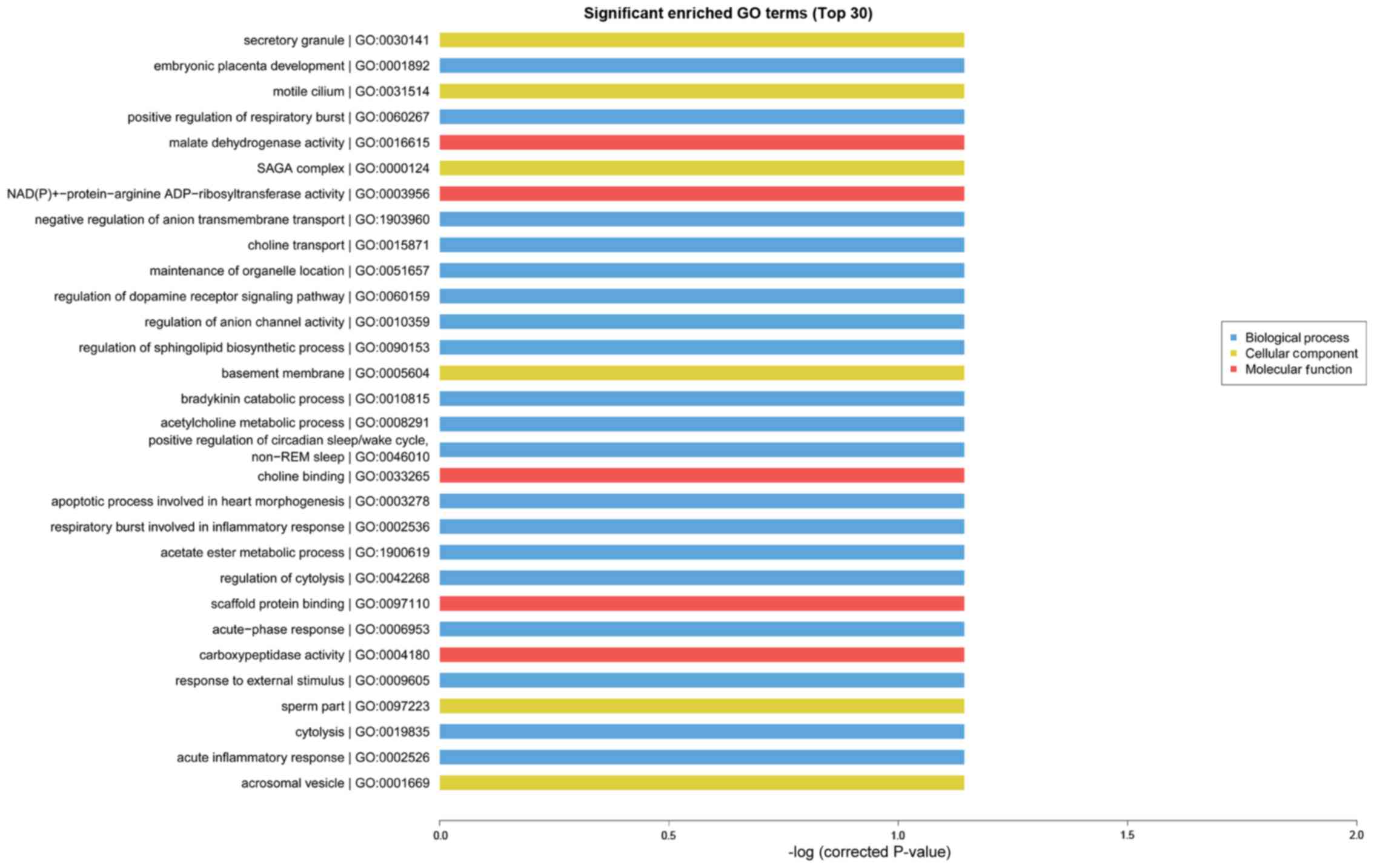
GO analysis is known as gene enrichment analysis
which can be used to confirm gene and gene production associated
bio-processes, cellular composition and molecular functions. GO and
KEGG analyses can be used to further evaluate the function of
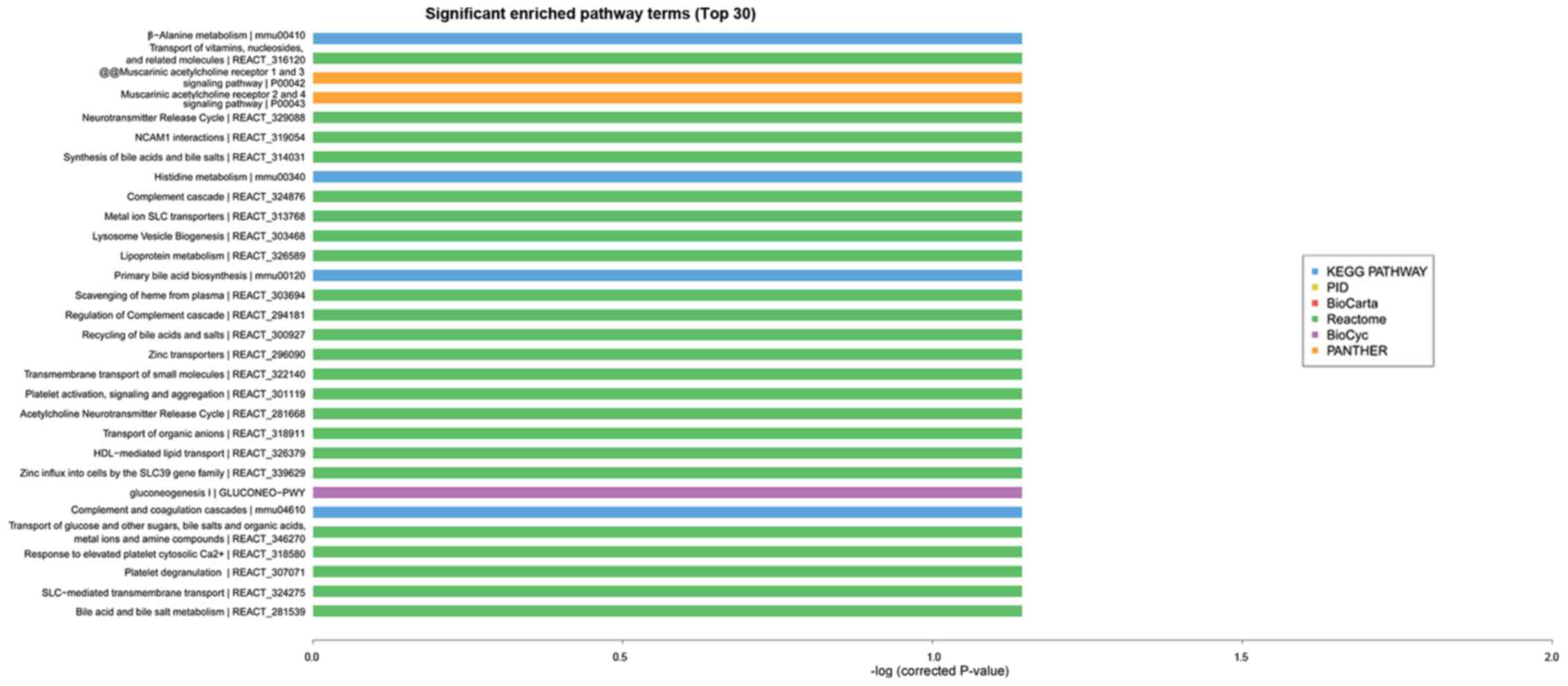
significantly expressing trend genes. As presented in Fig. 4, GO enrichment analysis
(identification of the biological processes involved in the
enrichment of transcripts) indicated that cognitive dysfunction in
type 2 diabetes was correlated with transport, cell adhesion, ion
transport, metabolic processes and other important biological
processes. In addition, KEGG and Reactome enrichment analysis,
which were performed to validate important module functions,
demonstrated that the pathology of cognitive dysfunction in type 2
diabetes is associated with the regulation of cholinergic synapses,
the NF-kB pathway, the Toll like receptor 4 (TLR4) cascade, and the
zinc transporter (Fig. 5).
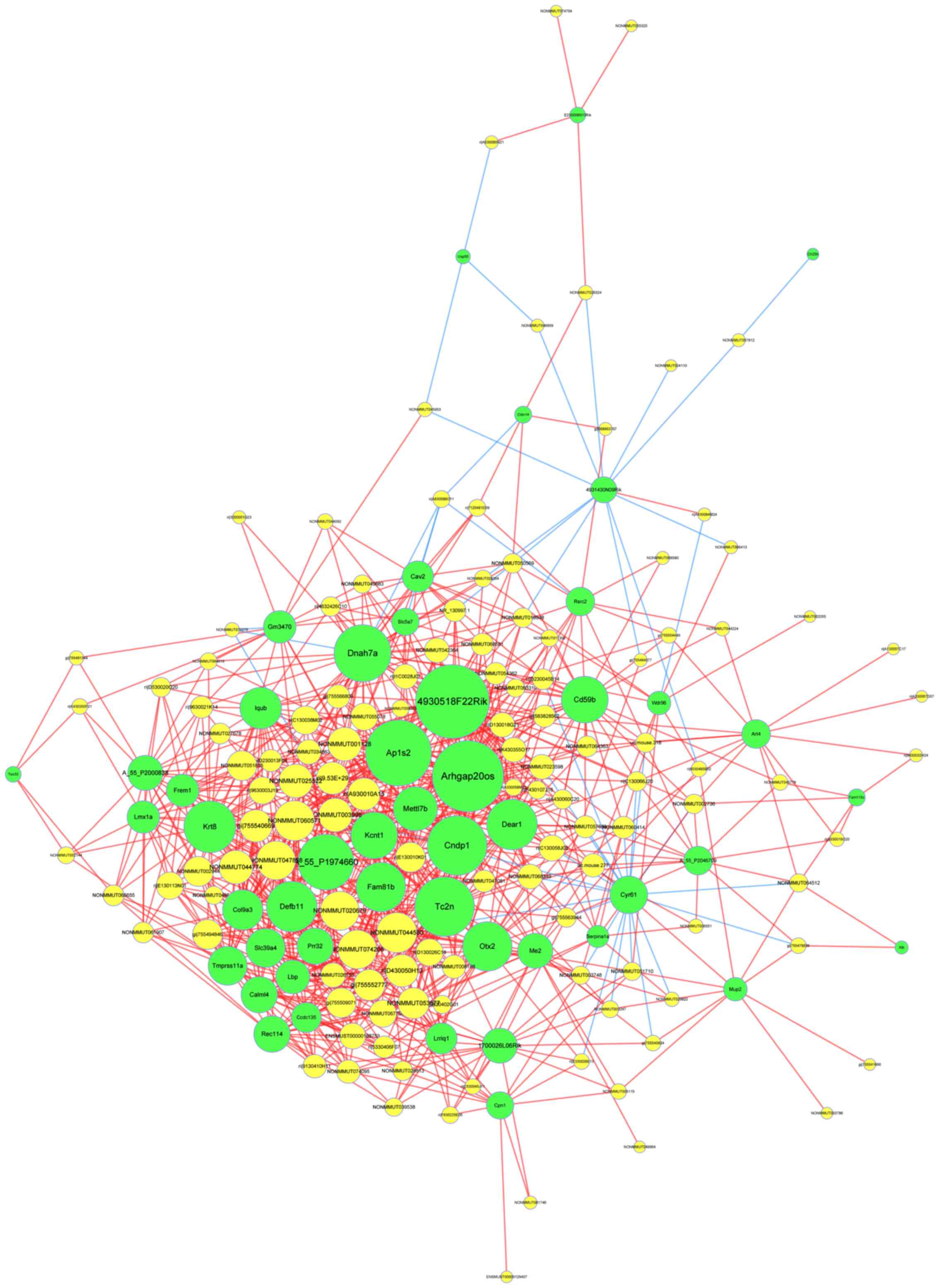
LncRNA-mRNA network
In order to identify important molecular mechanisms
of lncRNA associated with cognitive dysfunction in type 2 diabetes,
a dynamic lncRNA-mRNA network (Fig.
6) was constructed, which contained 123 lncRNA and 48 mRNA. To
address interactions between genes, a threshold of ≥0.997 was
implemented to evaluate the coexpression of lncRNA and mRNA via
quantization of the degree and the size of the circle. From the
figure, crucial lncRNAs were demonstrated, which may serve an
important role in the pathology of type 2 diabetes, including
lncRNA ENSMUST00000188753, lncRNA gi/755552777/ref/XR_875577.1/ and
lncRNA gi/755494846/ref/XR_387234.2/. In this figure, the size of a
circle indicates the ability of a gene to interact according to its
degree of quantification.
A co-expression network based on differential
lncRNAs and mRNA of healthy and type 2 diabetic mice is presented.
Green nodes represent mRNAs and yellow nodes represent lncRNAs. Red
lines express positive correlation and blue lines express negative
correlation.
Discussion
The present study is the first to the best of our
knowledge to report on lncRNA expression profiles in the
hippocampus of type 2 diabetic mice. Cognitive dysfunction in type
2 diabetes is a complex biological process involving a number of
molecular and cellular events. Although lncRNA was once regarded as
genome ‘noise’, lncRNAs have been demonstrated to be enriched in
the central nervous system and have important implications for
growth and development (27).
Therefore, understanding the lncRNA expression profile is critical
for investigating the potential function of lncRNAs in cognitive
dysfunction in type 2 diabetes mellitus.
Animal models serve an important role in the study
of type 2 diabetes. A reliable animal model was selected for the
extraction of whole cell RNA from the hippocampus of 3 animals in
each treatment condition. Experiments were conducted on triplicate
samples to ensure reliable and accurate microarray results were
associated with RT-qPCR results. After the microarray results were
determined to be reliable, RNA with a significant change in the
level of expression (multiples change ≥2) was selected for
bioinformatics analysis.
According to significantly altered mRNA expression,
potential LncRNA functions were studied through GO enrichment and
pathway analysis. GO analysis indicated altered expression in genes
involved in transport, cell adhesion, ion transport and metabolic
processes. KEGG and Reactome enrichment analysis indicated that
functionality of cholinergic synapses, the NF-kB pathway, TLR4
cascade and zinc transporter are associated with the pathological
alterations of cognitive dysfunction with type 2 diabetes.
Constructing lncRNA-mRNA network improved understanding of the
possible interactions and associations between differentially
expressed lncRNAs and mRNAs.
The cholinergic synaptic pathway is a classical
signaling pathway, which is involved in the synthesis and release
of acetylcholine. It is an important pathway for the transmission
of information in the nervous system. A previous study demonstrated
that cholinergic pathway damage is one of the mechanisms leading to
cognitive dysfunction in type 2 diabetes (28). Animal experiments demonstrated
decreased cholinergic system activity in the brain tissue of
diabetic rats and significantly decreased cholinergic fiber density
in the hippocampal CA1 region and molecular layer of the dentate
gyrus (29). Cognitive function
can be affected by damage of cholinergic nerves due to inflammation
and oxidative stress (29). A
previous study indicated that hypomnesia of zebra fish caused by
hyperglycemia exhibits a close association with
acetylcholinesterase activity elevation and galanthamine can help
with hypomnesia (30). The above
results suggested that the cholinergic synaptic pathway serves an
important role in the cognitive function of type 2 diabetic mice.
Iwamoto et al (31)
demonstrated the gene solute carrier family 5 member 7 (SLC5A7) has
a high affinity for the transport of acetylcholine in cholinergic
nerve terminals. In the present study the expression level of
SLC5A7 mRNA was upregulated and was significantly associated with
one lncRNA (gi|755552777|ref|XR_875577.1|) in the network, the two
were highly expressed. Therefore it was concluded that
gi|755552777|ref|XR_875577.1| may regulate the transport of
acetylcholine through upregulation of SLC5A7 expression and then
affect the expression of cognitive function, which provides us with
ideas for further research.
Additionally, there are a number of other signaling
pathways that can affect cell proliferation, cell differentiation,
gene expression regulation and cell survival. One of them is the
NF-κB signaling pathway. As an essential nuclear factor, NF-κB
participates in pathophysiological processes including inflammation
and apoptosis. In quiescent cells, NF-κB can combine with
inhibitor-κB (IκB) to form a tripolymer in the cytoplasm, with no
genetic transcription activity. When cells are stimulated by
proinflammatory cytokines, hyperglycemia, oxidative stress and
ultraviolet radiation IκB-α is phosphorylated and degraded, and
then NF-κB is subsequently released into the nucleus to combine
with the specific site of a nuclear target gene, thereby promoting
the expression of inflammation associated genes including
cytokines, adhesion molecules, chemotactic factor, which serve an
important role in diabetes and associated chronic complications
(32). A previous study (33) hypothesized that inflammatory
cytokines could induce apoptosis pathways, regulate the apoptosis
of mouse pancreatic β-cell factor, destroy the viability and
integrity of the islets and lead to lack of insulin secretion by
activating NF-κB. NF-κB sustained activation and inflammation
associated expression of cytokines, adhesion molecules, chemotactic
factors exists the hippocampus of diabetic mice treated with
streptozotocin, which can lead to cerebrovascular trauma and nerve
cell apoptosis (34). A study
reported that lncRNA-UCA1 dynamically regulates NF-κB in the
hippocampus of epilepsy rats (35). In addition, the NF-κB signaling
pathway mediated by TLR4 serves a key role in the activation and
regulation of inflammatory responses in brain tissues damaged by
intracerebral hemorrhage (36).
Previous studies have demonstrated that Lap is associated with
NF-κB, and NF-κB is associated with changes in hippocampal function
during diabetes mellitus. Therefore, our future studies will
investigate whether Lap affects cognitive impairment in diabetic
hippocampal tissues. In the present experiment the expression level
of this mRNA was upregulated and was significantly associated with
lncRNA gi/755552777/ref/XR_875577.1/ and lncRNA
gi/755494846/ref/XR_387234.2/ in the network, which were both
highly expressed. Therefore, in future studies we aim to
investigate the hypothesis that lncRNA
gi/755552777/ref/XR_875577.1/ and lncRNA
gi/755494846/ref/XR_387234.2/ can activate the NF-κB pathway
through upregulation of Lap expression, promote the inflammatory
process in the hippocampus, and then affect the expression of
cognitive function.
However, the present study also has certain
limitations. For example, data reliability is limited due to the
small sample size for the microarrays. In addition, with respect to
the research methods used, only differentially expressed lncRNA
functions can be predicted, but it is not possible to determine how
these lncRNA regulate target gene expression. The lncRNA function
can be validated by overexpression and RNA interference. Further
experiments are needed. There remain certain shortcomings in the
data analysis, such as the use of a small sample size. Furthermore,
genes with significantly different expression will be verified by
PCR and western blotting. The aim of future studies will be to
further study and pay attention to the molecular mechanism of
lncRNA. Further ingenuity pathway analyses and network analyses are
needed to provide more information about biological relevance and
the significance of the identified genes/pathways.
In conclusion, the present study demonstrated
differential expression profiles of lncRNA in the hippocampal
tissue of type 2 diabetes, which will help to study the
pathophysiology of cognitive impairment in type 2 diabetes.
Acknowledgements
The authors would like to thank Dr. Aubryanna
Hettingtonhouse of New York University for checking the
manuscript.
Funding
The present study was partly supported by the
National Natural Science Foundation of China (grant no. 81173250)
and the grants from the Project of Natural Foundation of Shandong
Province, China (grant no. ZR2017MH039).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
L-YC, YZL and CLL established the diabetic mice
model. L-YC, J-QY, XZ and CMM performed the experiments. L-YC, D-SL
and J-QY conceived the experimental design. L-YC, D-SL and RTH
analyzed the data and wrote the manuscript. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
The protocol for the present study was approved by
the Laboratory Animal Ethics committee of the Qilu Hospital of
Shandong University (Jinan, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Guariguata L, Whiting DR, Hambleton I,
Beagley J, Linnenkamp U and Shaw JE: Global estimates of diabetes
prevalence for 2013 and projections for 2035. Diabetes Res Clin
Pract. 103:137–149. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Agarwal SK, Saikia UK, Sarma D and Devi R:
Assessment of glomerular and tubular function in the evaluation of
diabetic nephropathy: A cross-sectional study. Indian J Endocrinol
Metab. 22:451–456. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Fi Z, Kovács G and Szentes V: Role of
trimetazidine in the treatment of diabetic microangiopathy in
ischaemic heart disease. Ore Hetil. 156:765–768. 2015.(In
Hungarian). View Article : Google Scholar
|
|
4
|
De Gregorio C, Contador D, Campero M,
Ezquer M and Ezquer F: Characterization of diabetic neuropathy
progression in a mouse model of type 2 diabetes mellitus. Biol
Open. 7:bio0368302018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
ENCODE Project Consortium, . An integrated
encyclopedia of DNA elements in the human genome. Nature.
489:57–74. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Mattick JS: RNA regulation: A new
genetics? Nat Rev Genetics. 5:316–323. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kataoka M and Wang DZ: Non-coding RNAs
including miRNAs and lncRNAs in cardiovascular biology and disease.
Cells. 3:883–898. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Liu JY, Yao J, Li XM, Song YC, Wang XQ, Li
YJ, Yan B and Jiang Q: Pathogenic role of lncRNA-MALAT1 in
endothelial cell dysfunction in diabetes mellitus. Cell Death Dis.
5:e15062014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ng SY, Lin L, Soh BS and Stanton LW: Long
noncoding RNAs in development and disease of the central nervous
system. Trends Genet. 29:461–468. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lalevee S and Feil R: Long noncoding RNAs
in human disease: Emerging mechanisms and therapeutic strategies.
Epigenomics. 7:877–879. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Prensner JR and Chinnaiyan AM: The
emergence of lncRNAs in cancer biology. Cancer Discov. 1:391–407.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhao Z, Liu B, Li B, Song C, Diao H, Guo
Z, Li Z and Zhang J: Inhibition of long noncoding RNA IGF2AS
promotes angiogenesis in type 2 diabetes. Biomed Pharmacother.
92:445–450. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhou Y, Shan T, Ding W, Hua Z, Shen Y, Lu
Z, Chen B and Dai T: Study on mechanism about long noncoding RNA
MALAT1 affecting pancreatic cancer by regulating Hippo-YAP
signaling. J Cell Physiol. 233:5805–5814. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wu L, Jin L, Zhang W and Zhang L: Roles of
long non-coding RNA CCAT2 in cervical cancer cell growth and
apoptosis. Med Sci Monit. 22:875–879. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sun J, Chu H, Ji J, Huo G, Song Q and
Zhang X: Long non-coding RNA HOTAIR modulates HLA-G expression by
absorbing miR-148a in human cervical cancer. Int J Oncol.
49:943–952. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sun J, Chen X, Wang Z, Guo M, Shi H, Wang
X, Cheng L and Zhou M: A potential prognostic long non-coding RNA
signature to predict metastasis-free survival of breast cancer
patients. Sci Rep. 5:165532015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ni Y, Huang H, Chen Y, Cao M, Zhou H and
Zhang Y: Investigation of long non-coding RNA expression profiles
in the substantia nigra of parkinson's disease. Cell Mol Neurobiol.
37:329–338. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chung JY, Kim HS and Song J: Iron
metabolism in diabetes-induced Alzheimer's disease: A focus on
insulin resistance in the brain. Biometals. 31:705–714. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Giatti S, Diviccaro S and Melcangi RC:
Neuroactive steroids and sex-dimorphic nervous damage induced by
diabetes mellitus. Cell Mol Neurobiol. 2018.(Epub ahead of print).
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhai Y, Meng X, Ye T, Xie W, Sun G and Sun
X: Inhibiting the NLRP3 inflammasome activation with MCC950
ameliorates diabetic encephalopathy in db/db mice. Molecules.
23:5222018. View Article : Google Scholar
|
|
21
|
Roberts TC, Morris KV and Wood MJ: The
role of long non-coding RNAs in neurodevelopment, brain function
and neurological disease. Philos Trans R Soc Lond B Biol Sci.
369:201305072014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lin N, Chang KY, Li Z, Gates K, Rana ZA,
Dang J, Zhang D, Han T, Yang CS, Cunningham TJ, et al: An
evolutionarily conserved long noncoding RNA TUNA controls
pluripotency and neural lineage commitment. Mol Cell. 53:1005–1019.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Mus E, Hof PR and Tiedge H: Dendritic
BC200 RNA in aging and in Alzheimer's disease. Proc Natl Acad Sci
USA. 104:10679–10684. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhou M, Zhang Z, Zhao H, Bao S, Cheng L
and Sun J: An immune-related six-lncRNA signature to improve
prognosis prediction of glioblastoma multiforme. Mol Neurobiol.
55:3684–3697. 2018.PubMed/NCBI
|
|
25
|
Heineke EW, Johnson MB, Dillberger JE and
Robinson KM: Antioxidant MDL 29,311 prevents diabetes in nonobese
diabetic and multiple low-dose STZ-injected mice. Diabetes.
42:1721–1730. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Livaka KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Guennewig B and Cooper AA: The central
role of noncoding RNA in the brain. Int Rev Neurobiol. 116:153–194.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Erkinjuntti T, Roman G, Gauthier S,
Feldman H and Rockwood K: Emerging therapies for vascular dementia
and vascular cognitive impairment. Stroke. 35:1010–1017. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kuhad A and Chopra K: Effect of sesamol on
diabetes-associated cognitive decline in rats. Exp Brain Res.
185:411–420. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Capiotti KM, De Moraes DA, Menezes FP,
Kist LW, Bogo MR and Da Silva RS: Hyperglycemia induces memory
impairment linked to increased acetylcholinesterase activity in
zebrafish (Danio rerio). Behav Brain Res. 274:319–325. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Iwamoto H, Calcutt MW and Blakely RD:
Differential impact of genetically modulated choline transporter
expression on the release of endogenous versus newly synthesized
acetylcholine. Neurochem Int. 98:138–145. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hao X, Ma B, Song G, Ye W, Li W and Tang
Y: The role of inflammatory factor NF-kappa B in the pathogenesis
and treatment of type 2 diabetes mellitus. Hebei Med J. 32:97–99.
2010.(In Chinese).
|
|
33
|
Park E, Lee SM, Lee JE and Kim JH:
Anti-inflammatory activity of mulberry leaf extract through
inhibition of NF-κB. J Funct Foods. 5:178–186. 2013. View Article : Google Scholar
|
|
34
|
Yao X and Jin X: NF-κB in expression of
hippocampal neurons with chronic diabetic rats. Chin J Med Pract.
4:5122005.(In Chinese).
|
|
35
|
Wang HK, Yan H, Wang K and Wang J: Dynamic
regulation effect of long non-coding RNA-UCA1 on NF-κB in
hippocampus of epilepsy rats. Eur Rev Med Pharmacol Sci.
21:3113–3119. 2017.PubMed/NCBI
|
|
36
|
Peng Q, Liu H, Shi S and Li M: Lycium
ruthenicum polysaccharide attenuates inflammation through
inhibiting TLR4/NF-κB signaling pathway. Int J Biol Macromol.
67:330–335. 2014. View Article : Google Scholar : PubMed/NCBI
|