Introduction
Diabetic nephropathy (DN), as a major microvascular
complication of diabetes, has become the leading cause of renal
decompensation and failure worldwide (1,2). The
global prevalence of type 2 diabetes contributes to an increasing
number of patients developing DN and eventually end-stage renal
disease, which requires renal replacement therapy. There are a
number of physiological and molecular alterations in the renal
microenvironment during DN, including oxidative stress, activation
of certain signaling pathways, inflammation and increasing matrix
accumulation (3–5). However, the underlying mechanism
leading to these changes has not been fully identified. Further
understanding of the molecular mechanisms involved in DN is of
clinical importance in order to improve the therapeutic strategies
for DN.
Long noncoding RNAs (lncRNAs) are a class of
transcripts longer than 200 nucleotides without protein coding
function. The mammalian genomes are replete with lncRNA genes that
are engaged in numerous biological processes (6). LncRNAs differ from protein-coding
genes in a number of ways. In particular, they are frequently not
as conserved compared with mRNAs at the level of the primary
sequence and the function of most lncRNAs remains unknown, which is
why they are dismissed to be the ‘dark matter’ of the
transcriptome. Recent studies have identified the regulatory role
of lncRNA in a number of gene expression processes, including
nuclear importation, alternative splicing, DNA methylation, mRNA
decay, as well as other transcriptional, post-transcriptional and
epigenetic regulatory roles (7,8).
Although a number of lncRNA molecules have been reported to serve
crucial roles in diverse diseases, only limited examples of lncRNAs
have been described in DN.
In the present study, the differentially expressed
genes (DEGs) were initially screened for lncRNAs and mRNAs, between
db/db mice and db/m mice by microarray technique. Bioinformatics
analysis identified several lncRNAs that putatively served an
important role in the regulation of mRNAs involved in DN onset of
db/db mice. By searching associated databases and analyzing the
results SOX2OT, which has never been reported in DN was selected
and focused on. SOX2OT was downregulated and highly conserved
between mice and humans. Gene-lncRNA co-expression networks were
constructed to investigate the putative function and interaction
patterns of SOX2OT. To study the role of SOX2OT in DN, human
podocyte and mesangial cells were also cultured to determine SOX2OT
expression pattern under high glucose conditions, aiming at
providing novel view on DN pathogenesis and identifying novel
directions for further research.
Materials and methods
Animal studies
C57BLKS/J background
Leprdb/Leprdb (db/db; n=3) and the
non-diabetic control Leprdb/m (db/m; n=3) male mice
(aged 8 weeks old; weight, 38–40 g) were bought from the Model
Animal Research Center of Nanjing University (Nanjing, China). Mice
were housed at 20–25°C, a humidity of 30–60% and a 12/12 h
light/dark cycle. db/db is a spontaneous diabetic model with
uncontrolled high blood glucose. All rats had unrestricted access
to food/water and were maintained for 20 weeks. Body weight,
fasting blood glucose, urinary protein/creatinine ratio, serum
creatinine and blood urea nitrogen were repeatedly measured to
monitor the onset and progression of DN. At the termination of the
study, mice were sacrificed under deep anesthesia prior to
collecting blood (0.5–1 ml) and tissue samples. Fasting blood
glucose was measured by quick sticks using tail blood. Urinary
protein, serum creatinine and blood urea nitrogen were measured by
the clinical laboratory of the First Affiliated Hospital of
Zhengzhou University. All protocols were approved by the
Institutional Animal Care and Use Committee of the First Affiliated
Hospital of Zhengzhou University and conducted in accordance with
the National Institutes of Health (NIH) Guide for the Care and Use
of Laboratory Animals.
Cell culture and treatment
Human podocytes cells (HPCs) and human mesangial
cells (HMCs) were a gift from Professor Fan Yi, Shandong University
(Jinan, China). HPCs and HMCs were cultured as previously described
(9,10). All cells were cultured in the
Dulbecco's modified Eagle's medium supplied with 10% fetal bovine
serum (both Gibco; Thermo Fisher Scientific, Inc., MA, USA) at 37°C
with 5% CO2. High glucose (HG, final concentration 30 mM
in culture medium) was used in the present study and the same
concentration of mannitol was used as an osmolality control.
Hematoxylin-eosin and periodic
Acid-Schiff staining
Tissue samples of renal cortex were fixed in
buffered formaldehyde solution for 24 h at room temperature,
dehydrated and embedded in paraffin wax. Sections were cut (4 µM)
and stained with hematoxylin-eosin (HE; OriGene Technologies, Inc.,
Beijing, China) and Periodic Acid-Shiffs (PAS, Solarbio Science and
Technology Co., Ltd., Beijing, China) reagent. In HE staining,
samples were treated with hematoxylin for 8 min and 1% ethanol
eosin for another 3 min. In PAS staining, slices were oxidized by
0.5% periodic acid for 7 min and soak in 0.5% sodium metabisulfite
for 1 min. Sections were mounted by neutral balsam (11,12).
Images were captured by Leica Microsystems GmbH, (Wetzlar,
Germany).
Transmission electron microscopy
observation
Ultra-microstructure of the basement membrane region
was observed by transmission electron microscopy (TEM) (13). Tissues for TEM observation were
fixed in 2% glutaraldehyde/2% paraformaldehyde in 0.1 mol/l
phosphate buffer for >24 h at room temperature, post-fixed in
buffered osmic acid, dehydrated in graded alcohols and embedded in
an Epon 812 mixture overnight at room temperature. Semi-thin
sections (2 µm) were rinsed overnight in 0.1 M phosphate buffer,
post-fixed for 2 h in 1% osmium tetroxide at room temperature,
dehydrated and then embedded in Araldite mixture. Ultrathin
sections were stained with uranyl acetate at room temperature for
15 min and lead citrate at room temperature for 25 min, and then
examined with a CM10 transmission electron microscope (Philips
Medical Systems B.V., Eindhoven, The Netherlands).
Microarray analysis
Agilent SurePrint G3 Mouse Gene Expression v2 8×60K
kit, which could scan the signals of 27122 Entrez Gene RNAs and
4578 lncRNAs, was used in the present study. Total RNA of the renal
cortex of the mice was extracted and purified using
mirVana™ miRNA Isolation kit (cat. no. AM1561; Ambion;
Thermo Fisher Scientific, Inc.) and checked for a RNA integrity
Number (RIN) to inspect RNA integration by an Agilent Bioanalyzer
2100 (Agilent Technologies, Inc., Santa Clara, CA, USA). Total RNA
was amplified and labeled by Low Input Quick Amp Labeling kit,
One-Color (cat. no. 5190-2305; Agilent Technologies, Inc.). Labeled
cRNA were purified using an RNeasy mini kit (cat. no. 74106;
Qiagen, GmBH, Germany). Each slide was hybridized with 600 ng
Cy3-labeled cRNA using Gene Expression Hybridization kit (cat. no.
5188-5242; Agilent Technologies, Inc.). Following 17 h
hybridization, slides were washed in staining dishes (cat. no. 121;
Thermo Fisher Scientific, Inc.) with Gene Expression Wash Buffer
kit (cat. no. 5188-5327, Agilent Technologies, Inc.). Slides were
scanned by Agilent Microarray Scanner (cat. no. G2565CA; Agilent
Technologies, Inc.) with default settings. Data were extracted with
Feature Extraction software 10.7 (Agilent Technologies, Inc.). All
experiments were performed according to the manufacturer's
protocols (14).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) analysis
Total RNA of the renal cortices of the mice was
extracted using TRIzol solution (Takara Biotechnology Co., Ltd.,
Dalian, China). Then total RNA was quantified using a Nanodrop 2000
spectrophotometer (Thermo Fisher Scientific, Inc.). A total of 1 µg
of RNA was used for cDNA synthesis using PrimeScript RT Master Mix
(Perfect Real Time) kit (Takara Biotechnology Co., Ltd.). The
temperature protocol for RT was 37°C for 15 min followed by 85°C
for 5 sec. SYBR Premix Ex Taq kit (Takara Biotechnology Co., Ltd.)
was used to evaluate the lncRNA and mRNA expression (15). RT-qPCR was performed in a
StepOnePlus Real-Time PCR System (Applied Biosystems; Thermo Fisher
Scientific, Inc.). The specific primers of target genes used were
as follows: Mus Klk1b7-ps forward, 5′-GTGTGAACCTCAAGCTCCTG-3′ and
reverse, 5′-CTGAGTCTCCCTTGCAAGTG-3′; Mus Suv39h2 forward,
5′-TGTGTGCCTTGCCTAGTTTC-3′ and reverse, 5′-CCACACCCTTTGCTACCTTG-3′;
Mus Ubtf forward, 5′-TGCGGTTCCTTGAGAGCTTG-3′ and reverse,
5′-CACCCTTGCCACCTTCCTG-3′; Mus Pkib forward,
5′-TGGCCGTGAAGGAAGATGC-3′ and reverse, 5′-ACTACCAGATCAAACACCCC-3′;
Mus 1700112J16Rik forward, 5′-TGACCCAGATATCAGTGCTC-3′ and reverse,
5′-TGTCTCTTCCTGTTCTGGTC-3′; Mus Gm15217 forward,
5′-TGGATCACAAGGACCCCAGT-3′ and reverse, 5′-GAACCAAATCCAACGGAGAC-3′;
Mus Pvt1 forward, 5′-AGAGCTGGTAGGAGACAGAC-3′ and reverse,
5′-TCTGCCAGCTCCTTCTTCAC-3′; Mus Hbegf forward,
5′-CTCCCTCTTGCAAATGCCTC-3′ and reverse, 5′-AGTACTACAGCCACCACAGC-3′;
Mus Hsd11b1 forward, 5′-GAGTTCAGACCAGAAATGCTC-3′ and reverse,
5′-AGACACTACCTTCTGGAGAC-3′; Mus Sox2ot forward,
5′-CCGAGAAGCAAACCTGACAG-3′ and reverse, 5′-AGTCTCTCCCATCAGCGTC-3′;
Homo LncR-SOX2-OT forward, 5′-AGACAATGAGCTGGCACCAC-3′ and reverse,
5′-AGGCAAGGTCAGAGACATAG-3′. Housekeeping gene GAPDH was used as an
endogenous control for normalization of RNA quantity differences.
The cycling conditions were as follows: 95°C for 30 sec, 40 cycles
at 95°C for 5 sec and then 60°C for 30 sec. All reactions were
carried out in triplicate. Relative expression of gene levels were
quantified using the 2−ΔΔCq method (16).
Gene ontology (GO) and pathway
enrichment analysis
GO enrichment analysis was performed to categorize
the differentially expressed mRNAs based on biological processes,
molecular functions and cellular components. The Kyoto Encyclopedia
of Genes and Genomes (KEGG) database was used to investigate the
enriched pathways of aberrantly expressed mRNAs (17,18).
P<0.05 was considered to indicate a statistically significant
difference.
Co-expression of lncRNAs and
mRNAs
Currently most lncRNAs have not been functionally
annotated. Therefore, their functions were predicted on the basis
of the functional annotations of their co-expressed mRNAs. Pearson
coefficients (PC) were used to construct a network of lncRNAs and
mRNAs. Pair-wise PC of genes (e.g., lncRNA and lncRNA, lncRNA and
mRNA, mRNA and mRNA) were used to construct networks using
Cytoscape 3.5.1 (www.cytoscape.org) using the following steps: i)
Choose interactions of lncRNA and mRNA to construct initial
networks (P<0.001), genes presented in network were considered
as ‘existed’. ii) Simultaneously import interactions of existed
‘lncRNA and lncRNA’, existed ‘mRNA and mRNA’ into initial networks
(P<0.001).
Fluorescence in situ hybridization
(FISH)
To detect lncRNA SOX2OT expression pattern of HPC
and HMC under normal or HG-stimulation conditions, FISH was
performed as previously described (19). The probe and kit for FISH was
purchased from Guangzhou RiboBio Co., Ltd., (Guangzhou, China).
Cultured HPCs and HMCs were fixed in paraformaldehyde for 10 min at
room temperature and incubated in Triton X-100 for 5 min.
Non-specific binding sites were blocked using pre-hybridization
buffer for 30 min at 37°C. A biotin-labeled SOX2OT probe was used
at 5 nM. Images were observed and captured using a fluorescence
microscope (Leica Microsystems GmbH).
Statistical analysis
Data are reported as the mean ± standard deviation.
Experiments were performed in triplicate. One-way analysis of
variance followed by Duncan's multiple range test were used to
check the significance of the differences in mean values.
Statistical analyses were performed with GraphPad Prism 5.0 (La
Jolla, CA, USA). P<0.05 was considered to indicate a
statistically significant difference in microarray analysis and if
fold-changes were >2 or <0.5, P values were corrected by the
Benjamini-Hochberg false discovery rate method using R software
(version 3.5.1; http://www.r-project.org/).
Results
Metabolic and pathological
characteristics of db/db mice
Table I described
detailed laboratory alterations of the mice at the end of the
study. The db/db mice demonstrated higher body weight, fasting
blood glucose and urinary protein level compared with the db/m
group (P<0.05). Serum creatinine level and blood urea nitrogen
also increased compared with the db/m mice. However, the serum
creatinine level was not statistically significant. HE staining,
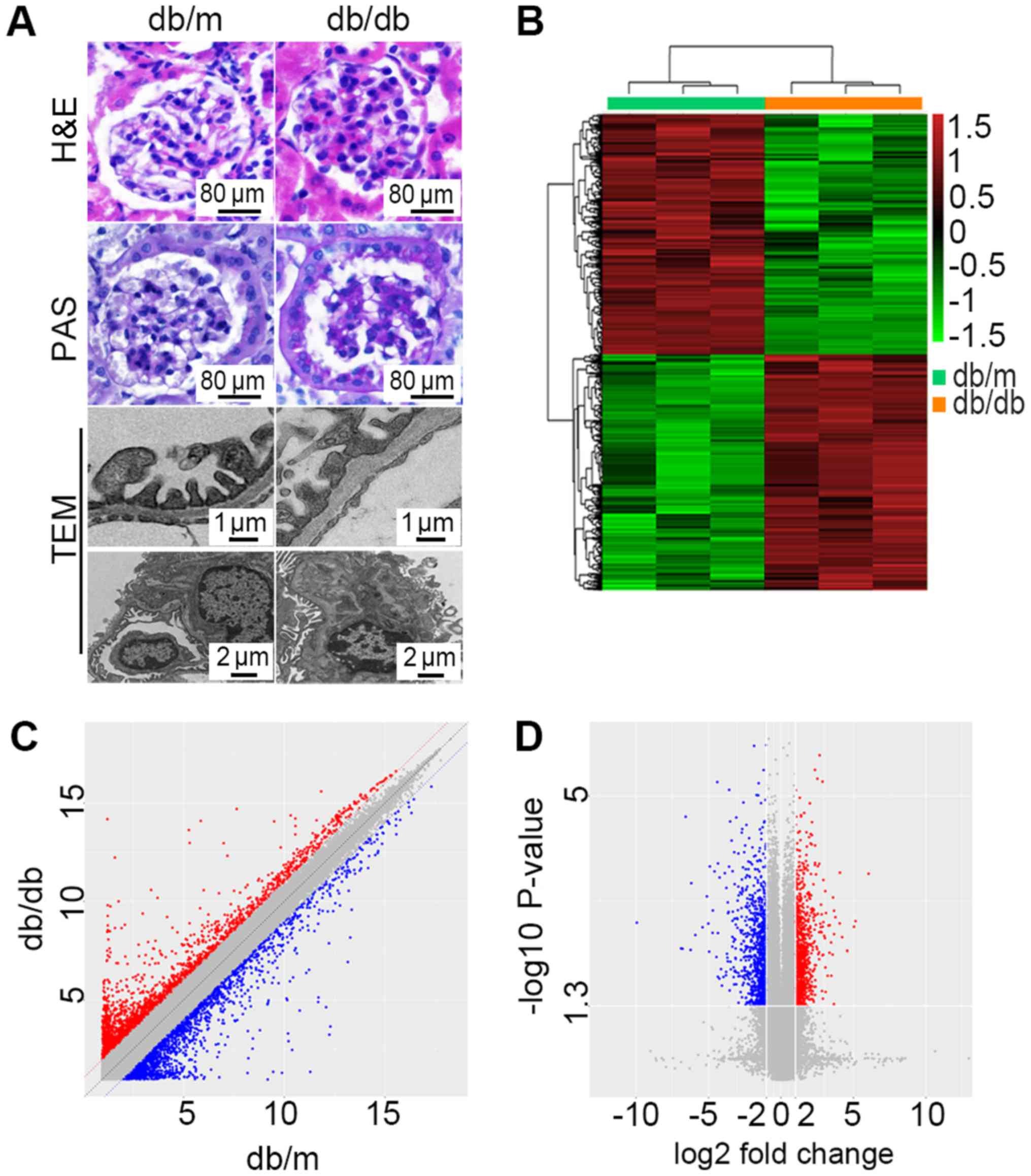
PAS staining and TEM (Fig. 1A)
were used for morphological diagnosis of DN. Db/db mice exhibited
foot process enfacement of podocytes, thickening of the basement
membrane and expansion of the mesangial matrix. All these results
indicated that a successful DN model of db/db mice had been
achieved.
 | Table I.Data of mice at the end point of the
experiment. |
Table I.
Data of mice at the end point of the
experiment.
| Parameter | Control (n=3) | DN (n=3) | P-value |
|---|
| Body weight
(g) | 23.27±0.30 | 39.20±0.35 | <0.0001 |
| Fasting blood
glucose (mmol/l) | 7.367±0.26 | 30.33±1.44 | <0.0001 |
| Urinary
protein/creatinine ratio (g/g) | 1.477±0.12 | 13.32±1.80 | 0.0028 |
| Serum creatinine
(µmol/l) | 33.30±3.30 | 41.47±1.38 | 0.0842 |
| Blood urea nitrogen
(mmol/l) | 8.627±0.47 | 13.38±0.25 | 0.0009 |
Microarray analysis reveals the DEGs
of db/db mice compared with db/m mice
The microarray gene chip data demonstrated 11,673
DEGs in db/db mice compared with db/m mice. A total of 5,233 genes
were upregulated while 6,440 were downregulated. Further study
demonstrated 1,568 DEGs with fold-change >2.0. Among those, 777
were consistently upregulated and 791 were consistently
downregulated. A hierarchically clustered heat map is presented in
Fig. 1B. Sec14l3 (NM_001029937)
and Gm6300 (NR_033591) were the most downregulated mRNA and lncRNA,
with an absolute fold-change of 118.84 and 59.97, respectively.
Hmgcs2 (NM_008256) and Rdh18-ps (NR_037604) were the most
upregulated mRNA and lncRNA with fold-changes of 23.81and 5.13.
Detailed DEGs of mRNAs and lncRNAs with the greatest change are
listed in Table II. The scatter
plot (Fig. 1C) presents variations
in DEGs expression between the two groups. Significant expression
differences can be seen. The volcano plot (Fig. 1D) was presented to visualize the
DEGs with statistical significance (fold-change ≥2.0,
P<0.05).
 | Table II.Top 10 down and upregulated mRNAs and
lncRNAs (db/db compared with db/m. Only genes with Accession number
started with ‘NM’ or ‘NR’ are listed here). |
Table II.
Top 10 down and upregulated mRNAs and
lncRNAs (db/db compared with db/m. Only genes with Accession number
started with ‘NM’ or ‘NR’ are listed here).
|
| mRNAs | lncRNAs |
|---|
|
|
|
|
|---|
| Gene symbol | Fold-change | Accession | Gene symbol | Fold-change | Accession |
|---|
| Sec14l3 | 0.008 | NM_001029937 | Gm6300 | 0.017 | NR_033591 |
| Slco1a1 | 0.010 | NM_013797 | Sox2ot | 0.031 | NR_015580 |
| Slc22a7 | 0.013 | NM_144856 | Gm1653 | 0.042 | NR_040591 |
| Masp2 | 0.014 | NM_010767 | 0610031O16Rik | 0.080 | NR_045760 |
| Cyp7b1 | 0.029 | NM_007825 | Carlr | 0.084 | NR_131254 |
| Col19a1 | 0.033 | NM_007733 | Gm15350 | 0.157 | NR_045775 |
| Mfsd2a | 0.038 | NM_029662 | B230323A14Rik | 0.172 | NR_040765 |
| Gm853 | 0.048 | NM_001034872 | Airn | 0.205 | NR_027773 |
| Alox15 | 0.049 | NM_009660 | Gm10433 | 0.255 | NR_045282 |
| Akr1c18 | 0.053 | NM_134066 | 5830418P13Rik | 0.260 | NR_040466 |
| Bhmt | 9.729 | NM_016669 | Gm16796 | 2.884 | NR_040367 |
| Qrfpr | 10.291 | NM_198192 | 1700110I01Rik | 2.920 | NR_038059 |
| Chrdl2 | 12.088 | NM_133709 | 2310069G16Rik | 2.954 | NR_040309 |
| Grem1 | 12.562 | NM_011824 | Zfp133-ps | 2.966 | NR_033459 |
| Kynu | 12.672 | NM_027552 | A530006G24Rik | 3.089 | NR_046014 |
| Gm10639 | 13.540 | NM_001122660 | A930001A20Rik | 3.101 | NR_040549 |
| Gsta2 | 16.617 | NM_008182 | I730030J21Rik | 3.270 | NR_045781 |
| Cry1 | 16.772 | NM_007771 | BB123696 | 3.466 | NR_027893 |
| Cyp4a14 | 18.634 | NM_007822 | Klk1b7-ps | 4.188 | NR_033120 |
| Hmgcs2 | 23.780 | NM_008256 | Rdh18-ps | 5.131 | NR_037604 |
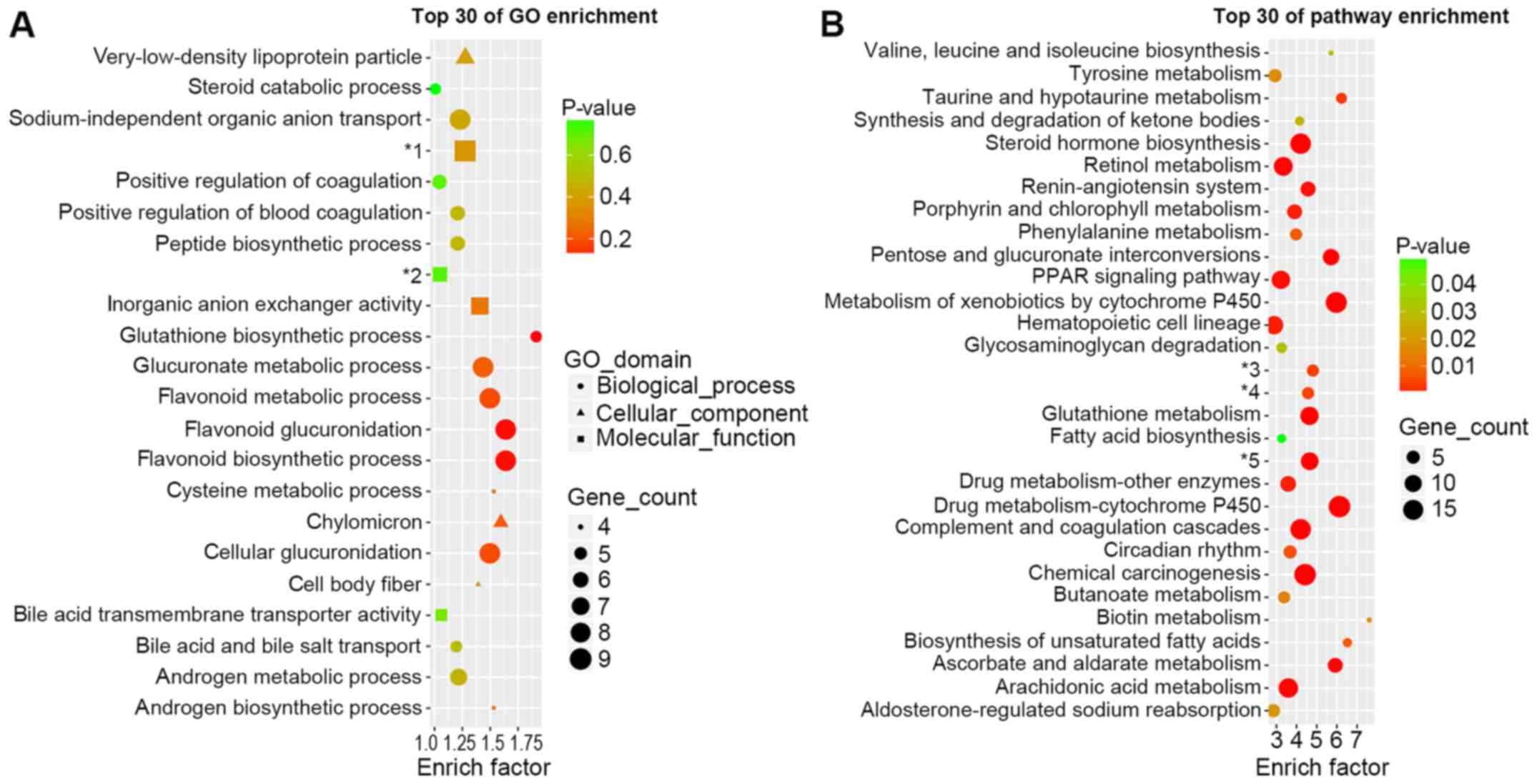
GO and pathway enrichment analysis of
differently expressed mRNAs
GO analysis (Fig.
2A) was performed in order to investigate the important
biological processes, cellular components and molecular functions
involved in the onset of DN. The most enriched biological process
was the glutathione biosynthetic process. Glucuronic and flavonoid
associated processes were also enriched, including glucoronate
metabolic processes, cellular glucuronidation, flavonoid metabolic
processes, flavonoid glucuronidation and flavonoid biosynthetic
processes. Chylomicron was the most enriched cellular component.
The most enriched molecular functions were inorganic anion
exchanger activity and sodium-independent organic anion
transmembrane transporter activity.
The top 30 enriched pathways are presented in
Fig. 2B. The most enriched pathway
was biotin metabolism. However, it only contained one DEG (OXSM).
Cytochrome P450-associated pathways were enriched, including
metabolism of xenobiotics by cytochrome P450 and drug
metabolism-cytochrome P450. Certain well-known pathways that are
associated with the onset and progression of DN could also been
observed, including the renin-angiotensin system, PPAR signaling
pathway, glutathione metabolism biosynthesis of unsaturated fatty
acids and arachidonic acid metabolism.
It is notable that in GO and pathway enrichment
analysis demonstrated alterations in steroid associated enrichment,
suggesting that steroid hormone disorder may also participate in
the onset of DN.
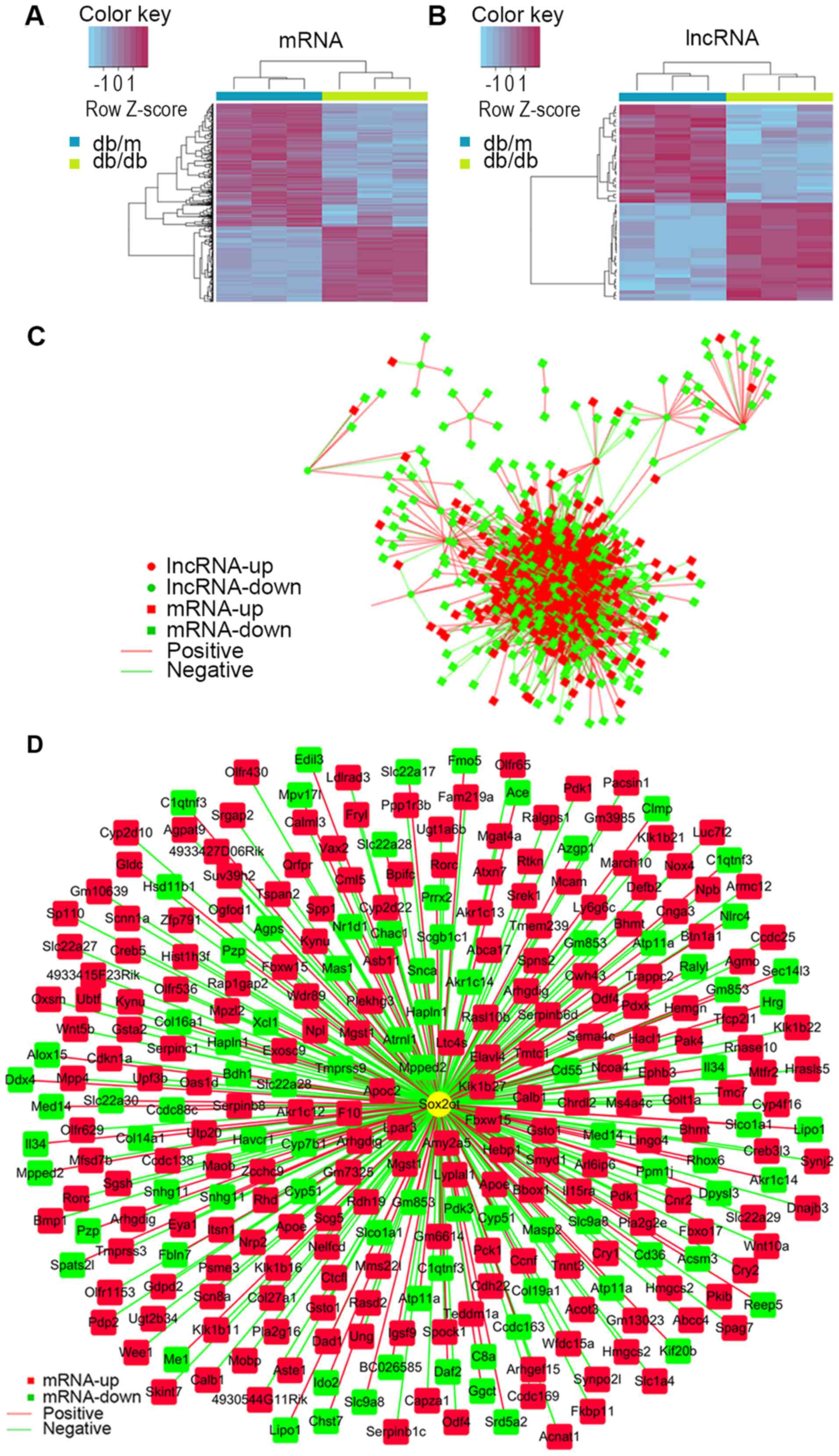
Differentially expressed lncRNA and
mRNA have close associations
Of all these DEGs, 880 mRNAs (335 were upregulated
and 545 were downregulated) and 60 lncRNAs (30 were upregulated and
30 were downregulated) were annotated. Hierarchically clustered
heat maps are presented in Fig. 3A and
B. To predict the function of differential lncRNAs, PCs of each
pair of differentially expressed lncRNAs and mRNAs were calculated
for the first step. The association of lncRNAs and mRNA were
constructed and intensive correlations (P≤0.001) between them are
demonstrated as network in Fig.
3C, indicating lncRNAs may perform their function via
interacting with associated mRNAs. To get a better view of the gene
network of SOX2OT the lncRNA of interest, a SOX2OT-centered network
was constructed (Fig. 3D).
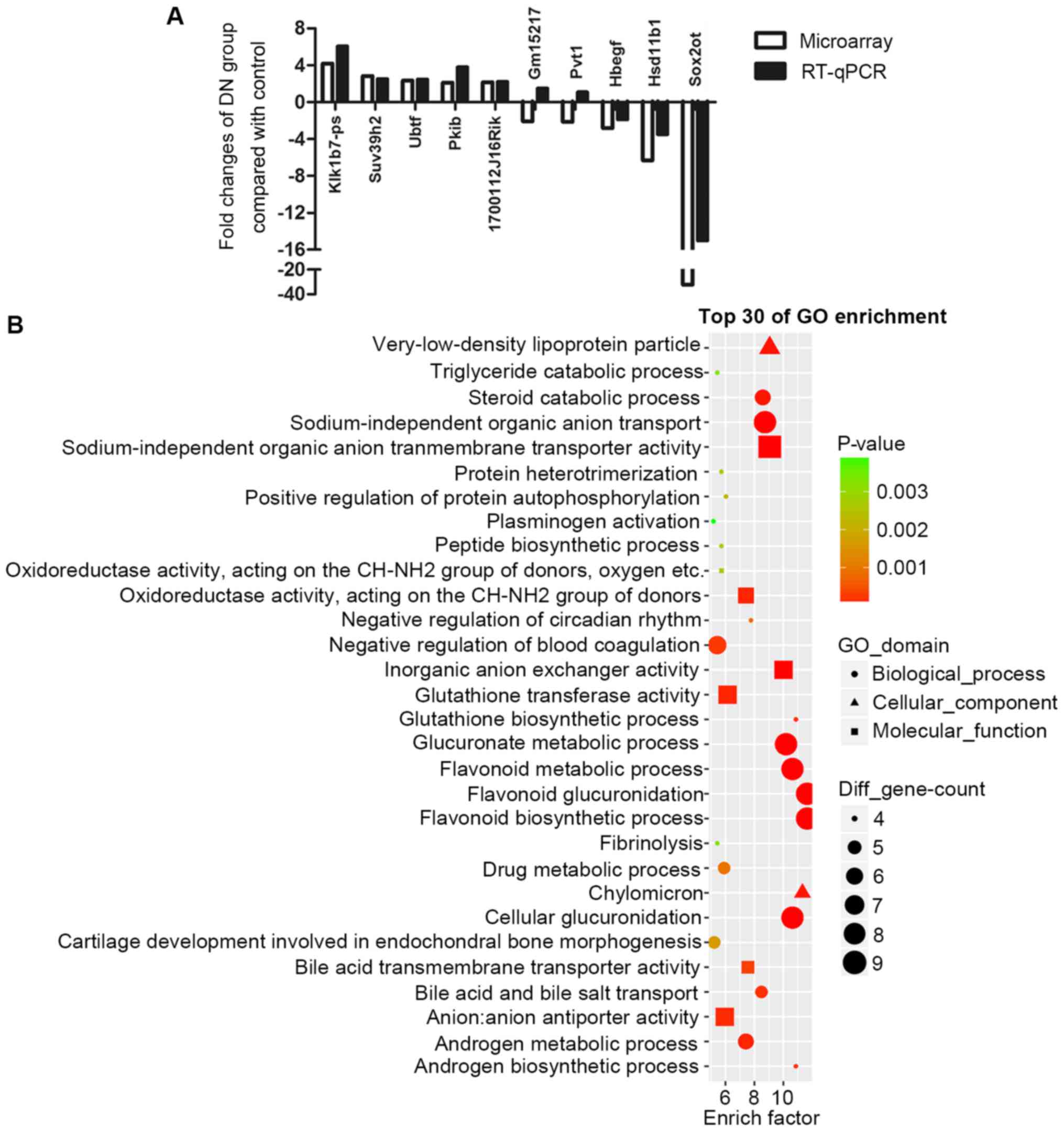
RT-qPCR validation of DEGs
First, the UCSC database was searched to check the
homology of all differentially expressed lncRNAs
(fold-change>2.0). Highly homologous lncRNAs were chosen for the
RT-qPCR validation, including Pvt1, SOX2OT, Gm15217, Klk1b7-ps and
1700112J16Rik. Certain other randomly picked differentially
expressed mRNAs, including: Ubtf, Pkib, Suv39h2, Hsd11b1 and Hbegf,
were also included in RT-qPCR validation. Fig. 4A demonstrates that most of the
results of RT-qPCR were consistent with microarray results. Out of
these 5 lncRNAs, Pvt1 and SOX2OT have already been reported to
exist in humans (20,21). Notably, SOX2OT is the most markedly
downregulated gene, with a fold-change of −32.
SOX2OT may have an important role in
DN
From Fig. 4A,
SOX2OT was identified to be the most dysregulated lncRNA which may
have an important function in DN. However, to the best of our
knowledge, its role in DN has never been reported previously. One
way of investigating the role of an lncRNA in a certain disease is
to study their putative co-expressed mRNAs. All the
SOX2OT-associated mRNAs were analyzed in GO enrichment (Fig. 4B). Notably, results demonstrated a
very similar expression pattern to that presented in Fig. 2A. Those enriched biological
processes in DN, including glutathione biosynthetic processes,
glucuronic and flavonoid associated processes, chylomicron, and
androgen metabolic processes were also enriched in
SOX2OT-associated mRNAs. These results indicated that, by serving a
central part in dysregulated biological processes, SOX2OT may have
a critical role in DN.
SOX2OT is highly conserved between
mice and humans
Since SOX2OT is evolutionarily conserved (22) and was the most altered lncRNA in
the present study, SOX2OT was investigated in the present study.
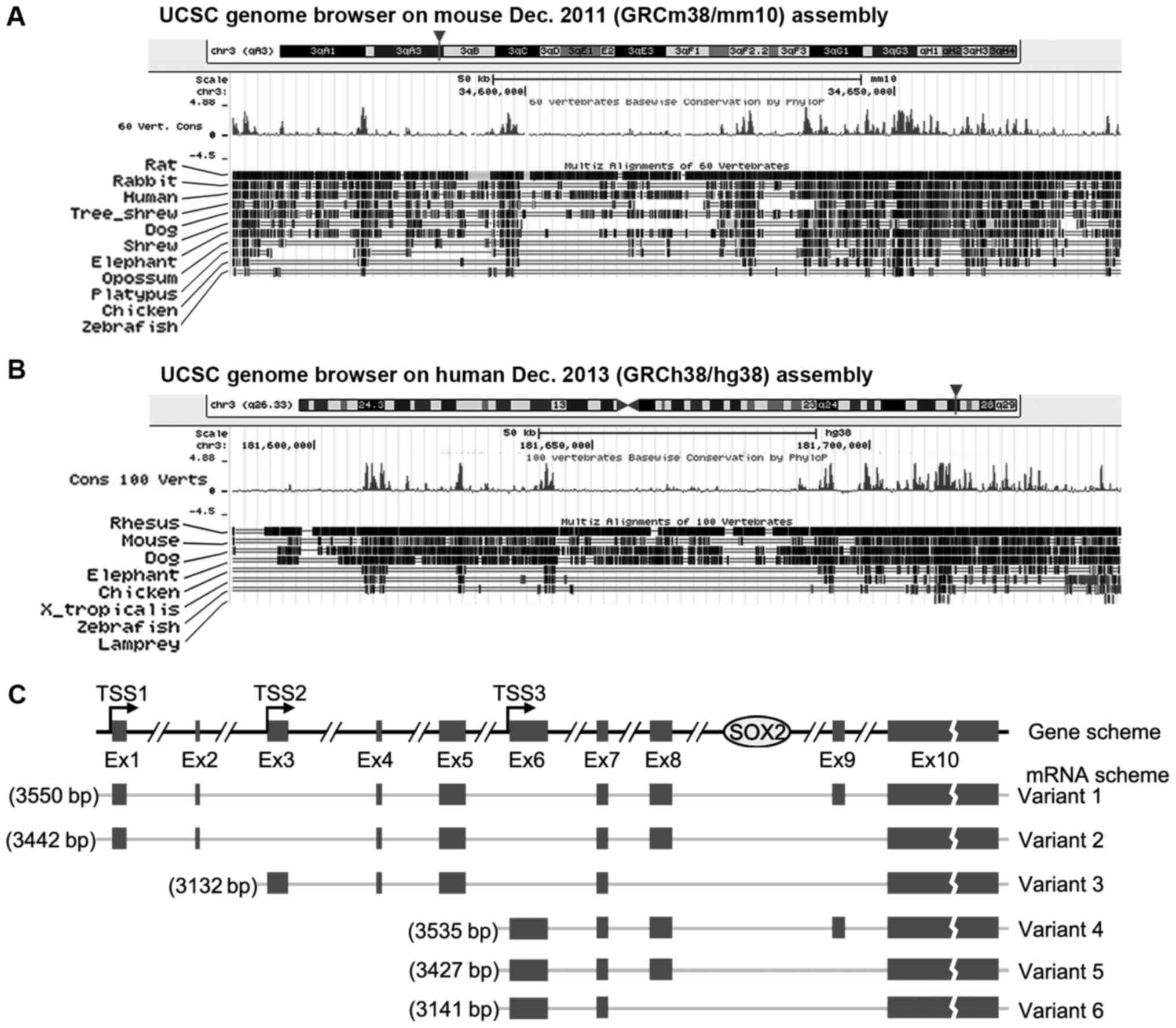
The gene structure of SOX2OT in mouse and human from UCSC is
presented in Fig. 5A and B. SOX2OT
is located on chromosome 3 in the two species and it overlaps with
mRNA encoding gene SOX2. Conservation tracking demonstrates that
SOX2OT is highly conserved between different vertebrates,
especially in mice and humans. SOX2OT has a number of transcript
variants. Human variants 1–6 have been documented in the NCBI
database and the structure diagrams are presented in Fig. 5C. A total of three transcription
starting sites (TSSs) are demonstrated and between Ex (exon) 8 and
9, which is where SOX2 resides.
Expression pattern of SOX2OT in HPCs
and HMCs
Since podocytes and mesangial cells are the two main
types of cells functionally associated with the onset of DN, HPCs
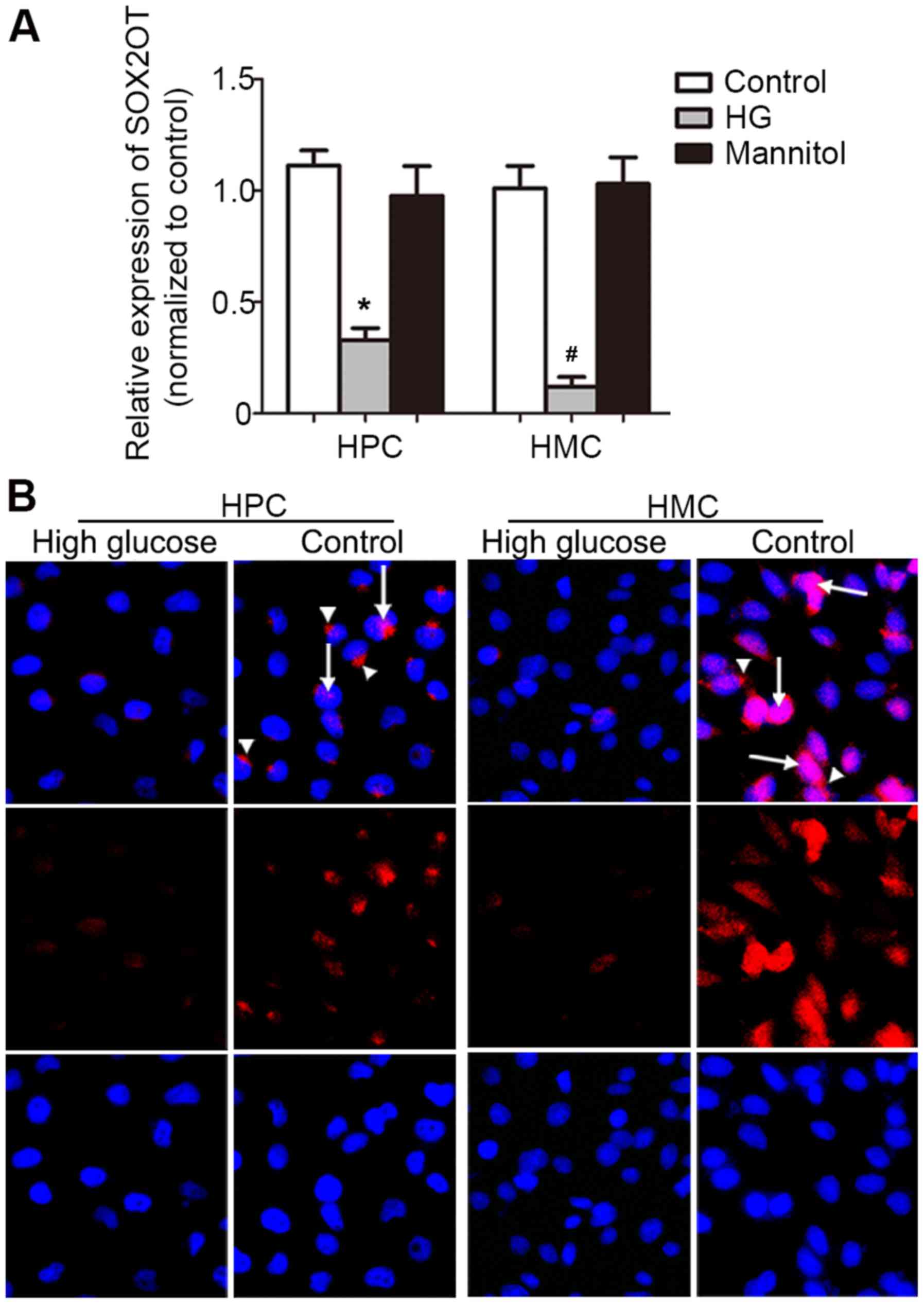
and HMCs were cultured to check SOX2OT expression. HG (30 mM for 24
h) could significantly downregulate SOX2OT expression compared with
the control group (P=0.0006 in HPC group and 0.0043 in HMC group,
Fig. 6A). This suggested that
SOX2OT may have the same expression pattern and function in human
podocytes and HMCs as that in mice. Then FISH was performed
(Fig. 6B). Results demonstrated
consistently weakened signal in HG-stimulated HPCs and HMCs. SOX2OT
expressed in the nucleus (arrow) and cytoplasm (arrow head).
However, in HPCs, SOX2OT was expressed in the cytoplasm, while in
HMCs, it was expressed in the nucleus. This very different
distribution indicated that SOX2OT may have different roles in HPCs
and HMCs under HG-stimulation.
Discussion
For decades, lncRNA has always been recognized as
byproduct of the gene transcription. Although recent studies
(23–25) have greatly improved current
knowledge about the function and associated mechanism of lncRNA, it
is mostly limited to the area of oncology. The role of lncRNA in
renal diseases is largely unknown.
In the present study, db/db mice were used as a
spontaneous diabetes model. Sustained high blood glucose ends in
the occurrence of protein urea and finally the onset of DN.
Microarray analysis using the renal cortex of db/db mice revealed
large number of dysregulated lncRNAs and mRNAs. To investigate what
biological processes and pathways were most altered GO and KEGG
pathway enrichment analysis was carried out. As expected, metabolic
associated biological processes were altered, including glucuronic
associated processes. The significant cellular component enrichment
of chylomicron was consistent with previous studies that
demonstrated lipid dysregulation served an important role in DN
(26,27). It is already well known that
oxidative stress is associated with the onset and progression of DN
(28). The present study's result
also added novel evidence to the role of oxidative stress by
describing alterations in certain known functional processes or
pathways, including flavonoid-associated processes (29), cytochrome P450 (30) and glutathione metabolism (31). Alterations in other DN-associated
pathways (32), including the
renin-angiotensin system, PPAR signaling pathway and arachidonic
acid metabolism were also demonstrated in the present study. These
discoveries, on one hand, proved the successful establishment of
the DN model at the molecular level and on the other hand they
provided a whole picture of disordered gene expression, which
indicates novel directions for future study. In addition, except
for the commonly enriched pathways, steroid-related alterations
were also demonstrated in GO and pathway enrichment maps. At
present, steroids are widely used for treatment of diverse
glomerular diseases. Although advanced stages of DN are often
accompanied by massive proteinuria, steroid hormones are rarely
used in these patients due to side effects or uncertain
effectiveness. However the results of the present study, together
with other microarray analysis (33) suggested that steroid hormone
disorders may also participate in the onset of DN, which requires
further study in the future.
As is currently known, the common way for lncRNA to
function is via interacting with mRNA. In the present study, the
lncRNA-associated mRNA alterations were also discussed which could
help prediction of the lncRNA-associated cellular functions. In the
present study, the correlation of differentially expressed lncRNAs
and mRNAs were presented as networks according to calculated PC
values.
RT-qPCR was introduced to validate microarray
results and a total of 10 lncRNAs and mRNAs were included. In these
lncRNAs, PVT1 and SOX2OT have already been reported and researched
in humans, especially in cancer (20,21).
PVT1 has also been studied intensively in DN (34). However, the function of SOX2OT in
DN has not been identified. Therefore, in the following studies,
the role of SOX2OT was focused on in DN. GO enrichment was carried
out again using co-expressed mRNAs of SOX2OT. As expected,
enrichment results were highly consistent with GO and KEGG results.
Flavonoid associated processes, oxidoreductase reaction,
glutathione metabolism and steroid metabolic processes were all
exhibited, indicating that SOX2OT is involved in the major
disorders of DN. Since the role of SOX2OT in DN has not been
reported previously, it is of interest to investigate the exact
function, which may provide a novel point of view on DN
treatment.
The location within the genome of SOX2OT is
chromosome 3 in humans and mice. Conservational analysis
demonstrated that SOX2OT is conserved in multiple vertebrates. This
conservation among species indicated that SOX2OT may have an
important role in normal physiology. SOX2OT has a number of
variants (35). At least 6
versions were documented in NCBI. A total of three TSSs were
demonstrated to start transcription of different variants. However,
the overlapping sequences prevented the design of specific and
highly efficient primers. As a result, only the total amount of
SOX2OT was measured. SOX2OT was first reported to be associated
with cell proliferation in breast cancer, lung cancer, colorectal
cancer and gastric cancer (36–38).
Su et al (39) proposed
that SOX2OT acted as a microRNA (miR)-194-5p or miR-122 sponge and
knockdown of SOX2OT inhibited the malignant biological behaviors of
glioblastoma stem cell. Recently, it is reported that, in
diabetes-induced retinal neurodegeneration, SOX2OT expression is
significantly downregulated in the retinas of STZ-induced diabetic
mice and in the Retinal Ganglion Cells upon HG or oxidative stress
(40). Diabetic-induced
nephropathy is also a micro-vascular complication and SOX2OT was
also downregulated in the mice renal cortex and cultured HPCs and
HMCs. FISH was performed to find out the cellular location. The
images exhibited downregulation of SOX2OT under HG stimulation.
Furthermore, it was demonstrated that the location of SOX2OT was
very different in these two cells, suggesting that SOX2OT may serve
different functions in different cells. Considering the
conservation of SOX2OT between mice and humans, the consistent
result of SOX2OT suggested it also plays a role in human DN.
However, the role of SOX2OT in certain cell types needs further
investigation.
In conclusion, the present study confirmed that
db/db mice exhibited different gene expression compared with db/m
mice and these DEGs were involved in diverse biological processes
and pathways associated with DN. Out of these dysregulated lncRNAs,
the significantly different and species-conserved lncRNA SOX2OT
were identified to be the putative controller of DN onset and
progression. SOX2OT may serve a central role in orchestrating
scenarios of molecular disorder and be the putative target for DN
treatment. The detailed function and role of SOX2OT in DN requires
further study in the future.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National
Natural Science Foundation of China (grant nos. 81570690 and
81700633), the Excellent Youth Foundation of Henan Scientific
Committee (grant no. 154100510017), the Youth Foundation of the
First Affiliated Hospital of Zhengzhou University to Jin Shang, Key
Scientific Research Projects of Henan Colleges and Universities
(grant no. 17A320065), Foundation and Frontier Technology Research
Program of Henan Province (grant no. 142300410211).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
ZZ and JX conceived and designed the experiments; JS
and XW performed the experiments; GC and DL analyzed the data; XZ
and YJ analyzed and interpreted data and wrote the manuscript; ZZ
revised and gave final approval of the manuscript to be published.
All authors read and approved the final manuscript.
Ethics approval and consent to
participate
All protocols were approved by the Institutional
Animal Care and Use Committee of the First Affiliated Hospital of
Zhengzhou University and conducted in accordance with the National
Institutes of Health Guide for the Care and Use of Laboratory
Animals.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Vicente PC, Kim JY, Ha JJ, Song MY, Lee
HK, Kim DH, Choi JS and Park KS: Identification and
characterization of site-specific N-glycosylation in the potassium
channel Kv3.1b. J Cell Physiol. 233:549–558. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Badal SS and Danesh FR: New insights into
molecular mechanisms of diabetic kidney disease. Am J Kidney Dis.
63 2 Suppl 2:S63–S83. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hwang SH, Han BI and Lee M: Knockout of
ATG5 leads to malignant cell transformation and resistance to Src
family kinase inhibitor PP2. J Cell Physiol. 233:506–515. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Frenette-Cotton R, Marcoux AA, Garneau AP,
Noel M and Isenring P: Phosphoregulation of K(+) -Cl(−)
cotransporters during cell swelling: Novel insights. J Cell
Physiol. 233:396–408. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Shang J, Wan Q, Wang X, Duan Y, Wang Z,
Wei X, Zhang Y, Wang H, Wang R and Yi F: Identification of NOD2 as
a novel target of RNA-binding protein HuR: Evidence from NADPH
oxidase-mediated HuR signaling in diabetic nephropathy. Free Radic
Biol Med. 79:217–227. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Liu R, Liao X, Li X, Wei H, Liang Q, Zhang
Z, Yin M, Zeng X, Liang Z and Hu C: Expression profiles of long
noncoding RNAs and mRNAs in post-cardiac arrest rat brains. Mol Med
Rep. 17:6413–6424. 2018.PubMed/NCBI
|
|
7
|
Liao Z, Zhao J and Yang Y: Downregulation
of lncRNA H19 inhibits the migration and invasion of melanoma cells
by inactivating the NFkappaB and PI3K/Akt signaling pathways. Mol
Med Rep. 17:7313–7318. 2018.PubMed/NCBI
|
|
8
|
Xu JH, Chang WH, Fu HW, Yuan T and Chen P:
The mRNA, miRNA and lncRNA networks in hepatocellular carcinoma: An
integrative transcriptomic analysis from Gene Expression Omnibus.
Mol Med Rep. 17:6472–6482. 2018.PubMed/NCBI
|
|
9
|
Li Z, Xu Y, Liu X, Nie Y and Zhao Z:
Urinary heme oxygenase-1 as a potential biomarker for early
diabetic nephropathy. Nephrology (Carlton). 22:58–64. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhang Z, Cheng X, Yue L, Cui W, Zhou W,
Gao J and Yao H: Molecular pathogenesis in chronic obstructive
pulmonary disease and therapeutic potential by targeting
AMP-activated protein kinase. J Cell Physiol. 233:1999–2006. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Guo J, Xiao J, Gao H, Jin Y and Zhao Z,
Jiao W, Liu Z and Zhao Z: Cyclooxygenase-2 and vascular endothelial
growth factor expressions are involved in ultrafiltration failure.
J Surg Res. 188:527–536e2. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Tian F, Gu C, Zhao Z, Li L, Lu S and Li Z:
Urinary Emmprin, matrix metalloproteinase 9 and tissue inhibitor of
metalloproteinase 1 as potential biomarkers in children with
ureteropelvic junction narrowing on conservative treatment.
Nephrology (Carlton). 20:194–200. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kong XD, Shi HR, Liu N, Wu QH, Xu XJ, Zhao
ZH, Lu N, Li-Ling J and Luo D: Mutation analysis and prenatal
diagnosis for three families affected by isolated methylmalonic
aciduria. Genet Mol Res. 13:8234–8240. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zou Y, Li C, Shu F, Tian Z, Xu W, Xu H,
Tian H, Shi R and Mao X: lncRNA expression signatures in
periodontitis revealed by microarray: The potential role of lncRNAs
in periodontitis pathogenesis. J Cell Biochem. 116:640–647. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang S, Zhang Y, Wei X, Zhen J, Wang Z,
Li M, Miao W, Ding H, Du P, Zhang W, et al: Expression and
regulation of a novel identified TNFAIP8 family is associated with
diabetic nephropathy. Biochim Biophys Acta. 1802:1078–1086. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Shi X, Xu Y, Zhang C, Feng L, Sun Z, Han
J, Su F, Zhang Y, Li C and Li X: Subpathway-LNCE: Identify
dysfunctional subpathways competitively regulated by lncRNAs
through integrating lncRNA-mRNA expression profile and pathway
topologies. Oncotarget. 7:69857–69870. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chen X: KATZLDA: KATZ measure for the
lncRNA-disease association prediction. Sci Rep. 5:168402015.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hu M, Wang R, Li X, Fan M, Lin J, Zhen J,
Chen L and Lv Z: LncRNA MALAT1 is dysregulated in diabetic
nephropathy and involved in high glucose-induced podocyte injury
via its interplay with β-catenin. J Cell Mol Med. 21:2732–2747.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang C, Han C, Zhang Y and Liu F: LncRNA
PVT1 regulate expression of HIF1alpha via functioning as ceRNA for
miR199a5p in nonsmall cell lung cancer under hypoxia. Mol Med Rep.
17:1105–1110. 2018.PubMed/NCBI
|
|
21
|
Tang X, Gao Y, Yu L, Lu Y, Zhou G, Cheng
L, Sun K, Zhu B, Xu M and Liu J: Correlations between lncRNA-SOX2OT
polymorphism and susceptibility to breast cancer in a Chinese
population. Biomark Med. 11:277–284. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Shahryari A, Jazi MS, Samaei NM and Mowla
SJ: Long non-coding RNA SOX2OT: Expression signature, splicing
patterns, and emerging roles in pluripotency and tumorigenesis.
Front Genet. 6:1962015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Munschauer M, Nguyen CT, Sirokman K,
Hartigan CR, Hogstrom L, Engreitz JM, Ulirsch JC, Fulco CP,
Subramanian V, Chen J, et al: The NORAD lncRNA assembles a
topoisomerase complex critical for genome stability. Nature.
561:132–136. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang Y, Pitchiaya S, Cieslik M, Niknafs
YS, Tien JC, Hosono Y, Iyer MK, Yazdani S, Subramaniam S, Shukla
SK, et al: Analysis of the androgen receptor-regulated lncRNA
landscape identifies a role for ARLNC1 in prostate cancer
progression. Nat Genet. 50:814–824. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hua JT, Ahmed M, Guo H, Zhang Y, Chen S,
Soares F, Lu J, Zhou S, Wang M, Li H, et al: Risk SNP-mediated
promoter-enhancer switching drives prostate cancer through lncRNA
PCAT19. Cell. 174:564–575e18. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wong R, Chen W, Zhong X, Rutka JT, Feng ZP
and Sun HS: Swelling-induced chloride current in glioblastoma
proliferation, migration, and invasion. J Cell Physiol.
233:363–370. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Bagheri V, Memar B, Momtazi AA, Sahebkar
A, Gholamin M and Abbaszadegan MR: Cytokine networks and their
association with Helicobacter pylori infection in gastric
carcinoma. J Cell Physiol. 233:2791–2803. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Feng B, Yan XF, Xue JL, Xu L and Wang H:
The protective effects of alpha-lipoic acid on kidneys in type 2
diabetic Goto-Kakisaki rats via reducing oxidative stress. Int J
Mol Sci. 14:6746–6756. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Olli KE, Li K, Galileo DS and
Martin-DeLeon PA: Plasma membrane calcium ATPase 4 (PMCA4)
co-ordinates calcium and nitric oxide signaling in regulating
murine sperm functional activity. J Cell Physiol. 233:11–22. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Noda S, Sumita Y, Ohba S, Yamamoto H and
Asahina I: Soft tissue engineering with micronized-gingival
connective tissues. J Cell Physiol. 233:249–258. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lin W, Izu Y, Smriti A, Kawasaki M,
Pawaputanon C, Bottcher RT, Costell M, Moriyama K, Noda M and Ezura
Y: Profilin1 is expressed in osteocytes and regulates cell shape
and migration. J Cell Physiol. 233:259–268. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Xu HZ, Cheng YL, Wang WN, Wu H, Zhang YY,
Zang CS and Xu ZG: 12-Lipoxygenase inhibition on microalbuminuria
in Type-1 and Type-2 diabetes is associated with changes of
glomerular angiotensin II Type 1 receptor related to insulin
resistance. Int J Mol Sci. 17:pii: E684. 2016. View Article : Google Scholar
|
|
33
|
Chen S, Dong C, Qian X, Huang S, Feng Y,
Ye X, Miao H, You Q, Lu Y and Ding D: Microarray analysis of long
noncoding RNA expression patterns in diabetic nephropathy. J
Diabetes Complications. 31:569–576. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Alvarez ML and Distefano JK: The role of
non-coding RNAs in diabetic nephropathy: Potential applications as
biomarkers for disease development and progression. Diabetes Res
Clin Pract. 99:1–11. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Strippoli R, Loureiro J, Moreno V,
Benedicto I, Lozano ML, Barreiro O, Pellinen T, Minguet S, Foronda
M, Osteso MT, et al: Caveolin-1 deficiency induces a
MEK-ERK1/2-Snail-1-dependent epithelial-mesenchymal transition and
fibrosis during peritoneal dialysis. EMBO Mol Med. 7:3572015.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Askarian-Amiri ME, Seyfoddin V, Smart CE,
Wang J, Kim JE, Hansji H, Baguley BC, Finlay GJ and Leung EY:
Emerging role of long non-coding RNA SOX2OT in SOX2 regulation in
breast cancer. PLoS One. 9:e1021402014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Hou Z, Zhao W, Zhou J, Shen L, Zhan P, Xu
C, Chang C, Bi H, Zou J, Yao X, et al: A long noncoding RNA Sox2ot
regulates lung cancer cell proliferation and is a prognostic
indicator of poor survival. Int J Biochem Cell Biol. 53:380–388.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Liu S, Xu B and Yan D: Enhanced expression
of long non-coding RNA Sox2ot promoted cell proliferation and
motility in colorectal cancer. Minerva Med. 107:279–286.
2016.PubMed/NCBI
|
|
39
|
Su R, Cao S, Ma J, Liu Y, Liu X, Zheng J,
Chen J, Liu L, Cai H, et al: Knockdown of SOX2OT inhibits the
malignant biological behaviors of glioblastoma stem cells via
up-regulating the expression of miR-194-5p and miR-122. Molecular
cancer. 16:1712017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Li CP, Wang SH, Wang WQ, Song SG and Liu
XM: Long Noncoding RNA-Sox2OT knockdown alleviates diabetes
mellitus-induced retinal ganglion cell (RGC) injury. Cell Mol
Neurobiol. 37:361–369. 2017. View Article : Google Scholar : PubMed/NCBI
|