Introduction
As the third most common malignancy worldwide,
colorectal cancer (CRC) leads to numerous cases of
cancer-associated mortalities annually (1). Several factors, including genetic
mutations, inflammation, alcohol consumption and smoking may induce
the development of CRC (2).
Therapeutic strategies have been developed; however, the 5-year
survival rate of patients with CRC remains unsatisfactory (3). Tumor metastasis and recurrence are
the main factors that affect the treatment of patients with CRC
(4). Numerous signaling pathways
and molecules have been reported to be associated with the
development or progression of CRC, including the Notch and
adenomatous polyposis coli tumor suppressor signaling pathways
(5,6); however, the molecular mechanisms
underlying the carcinogenesis of CRC remain largely unknown.
Furthermore, it is necessary to determine effective biomarkers and
therapeutic targets for CRC intervention.
MicroRNAs (miRNAs/miRs) are a collection of short
noncoding RNAs that regulate gene expression by targeting the
3′-untranslated region (UTR) of target mRNAs via base-pairing
(7). The functions of miRNAs have
been widely reported, such as their involvement in various
physiological and pathological processes (8), including the cell-cycle,
proliferation, apoptosis and metastasis (9,10).
The dysregulation of miRNAs has been demonstrated to be associated
with the carcinogenesis of human cancers, including CRC (11). Increasing evidence has demonstrated
that miRNAs are aberrantly expressed in CRC and regulate cancer
progression (12–15). Some miRNAs have been reported as
potential biomarkers or targets for the diagnosis, prognosis and
therapy of CRC (16). Thus,
understanding the function and mechanism of miRNAs in CRC may aid
the development of effective therapeutic methods for the treatment
of CRC.
Previous studies have demonstrated that miR-3666 was
detected in non-small cell lung cancer, thyroid carcinoma and
cervical cancer (17–19). Whether miR-3666 regulates the
progression of CRC remains unknown. In the present study, it was
reported that miR-3666 was significantly downregulated in CRC
tissues and may have suppressed the proliferation, migration and
invasion of CRC cells by targeting special AT-rich sequence binding
protein 2 (SATB2). The findings of the present study suggested that
miR-3666 may be a promising therapeutic target for the treatment of
CRC.
Materials and methods
Patient samples
A total of 53 CRC and 53 adjacent non-cancerous
specimens (28 males and 25 females; aged 33–67 years old with a
median age of 54 years) were obtained from patients undergoing
surgical resection at the Wenzhou Central Hospital (Wenzhou, China)
between January 2014 and December 2016. Patients who had previously
been subjected to chemotherapy and radiotherapy were excluded from
the present study. miR expression was categorized into low and high
groups according to the mean value, which were then analyzed by
Kaplan Meier analysis. The dissected tissue specimens were
immediately frozen in liquid nitrogen and stored at −80°C. All
participants provided written informed consent for the collection
of tissue samples. The present study was reviewed and approved by
the Institutional Human Experiment and Ethics Committee of Wenzhou
Central Hospital. The experimental procedures were performed in
accordance with the Declaration of Helsinki.
Cell lines and cell culture
The human CRC cell lines, HT29, HCT116, SW480 and
SW620 were obtained from the Cell Bank of Type Culture Collection
of the Chinese Academy of Sciences (Shanghai, China). Human normal
colon mucosal epithelial cells (NCM460) were obtained from Incell
Corporation LLC (San Antonio, TX, USA). The cells were cultured in
Dulbecco's modified Eagle's medium (DMEM; GE Healthcare Sciences,
Logan, UT, USA), supplemented with 10% fetal bovine serum (FBS,
Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) and
1% penicillin/streptomycin (Thermo Fisher Scientific, Inc.) in a
humidified incubator at 37°C containing 5% CO2. HCT116
and SW480 cells were selected for subsequent functional
investigation, as these cells expressed the lowest miR-3666
expression levels.
Cell transfection
miR-3666 mimics (5′-CAGUGCAAGUGUAGAUGCCGA-3′),
inhibitors (5′-UCGGCAUCUACACUUGCACUG-3′) and negative control
(miR-NC; 5′-UCACAACCUCCUAGAAAGAGUAGA-3′) were obtained from
Shanghai GenePharma Co., Ltd. (Shanghai, China). A pcDNA3.1-SATB2
overexpressing plasmid (Shanghai GenePharma Co., Ltd.) was
synthesized to overexpress SATB2 by cloning the coding sequence
into pcDNA3.1 vector as previously reported (20); empty pcDNA3.1 vectors served as a
negative control. Transfection was performed using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol. To
restore SATB2 expression, miR-3666 mimics (100 nM) and either
SATB2-overexpressing plasmids (1 µg) or empty vectors (1 µg) were
co-transfected into 2×106 CRC cells. A total of 48 h
post-transfection, the efficiency was confirmed using RT-qPCR
according to the aforementioned protocol.
Cell proliferation
For the Cell Counting Kit-8 (CCK-8) assay, cells
were seeded into a 96-well plate at a density of 1×104
cells/well and cultured for 48 h at 37°C containing 5%
CO2 in complete DMEM; cells were transfected as
aforementioned with miR-3666 mimics, inhibitor or miR-NC for 48 h.
Then, 10 µl CCK-8 solution (Sigma-Aldrich; Merck KGaA, Darmstadt,
Germany) was added to each well. Following incubation for 1 h at
37°C, the optical density at 450 nm was detected using an
enzyme-linked immunoassay analyzer (BioTek Instruments, Inc.
Winooski, VT, USA).
Bioinformatics analysis
Putative target genes of miR-3666 were predicted
using TargetScan 7.0 tool (http://www.targetscan.org/vert_70/).
Transwell assay
A Transwell chamber with 8-µm pore size (Corning
Incorporated, Corning, NY, USA) was used for the analysis of cell
migration and invasion. Following transfection as aforementioned
with miR-3666 mimics, inhibitor or miR-NC for 36 h, CRC cells
(5×104 cells) suspended in serum-free DMEM were plated
into the upper chamber with (for invasion analysis) or without (for
migration analysis) Matrigel. DMEM medium and 10% FBS was added to
the lower chamber as a chemoattractant. Cells were incubated at
37°C for 24 h, and the remaining cells on the upper chambers were
removed with cotton swabs. The invaded or migrated cells on the
lower chamber surface were fixed with 100% methanol for 30 min at
25°C and stained with 0.5% crystal violet for 30 min at 25°C. The
number of invaded and migrated cells were observed and counted in 5
randomly selected fields from each chamber under an inverted
microscope (magnification, ×100; Olympus Corporation, Tokyo,
Japan).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA from CRC tissues and cultured cell lines
was extracted with TRIzol (Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocols. Then, cDNA was
synthesized with TaqMan MicroRNA Reverse Transcription Kit (Applied
Biosystems; Thermo Fisher Scientific, Inc.). Subsequently, qPCR was
performed using a TaqMan MicroRNA Assay kit (Applied Biosystems;
Thermo Fisher Scientific, Inc.) with an ABI 7300 qPCR system
(Applied Biosystems; Thermo Fisher Scientific, Inc.). The
thermocycling conditions used for qPCR were as follows:
Denaturation at 95°C for 10 min; followed by 40 cycles of
denaturation at 95°C for 15 sec and elongation at 60°C for 1 min.
The relative expression levels of SATB2 and miRs were calculated
and normalized to that of endogenous β-actin or U6, respectively.
The relative fold change in expression was calculated by the
2−ΔΔCq method (21).
The primer sequences used were as follows: U6 forward,
5′-AACGAGACGACGACAGAC-3′ and reverse,
5′-GCAAATTCGTGAAGCGTTCCATA-3′; β-actin forward,
5′-CGGCGCCCTATAAAACCCA-3′ and reverse, 5′-GAGGCGTACAGGGATAGCAC-3′;
miR-3666 forward, 5′-AACGAGACGACGACAGAC-3′ and reverse,
5′-CAGTGCAAGTGTAGATGCCGA-3′; SATB2 forward,
5′-GCCTGATGACTCAACGCAAC-3′ and reverse, 5′-ACACCAAGAGCCGAGAAGAC-3′.
All assays were performed in triplicate.
Western blot analysis
CRC cells were lysed using radioimmunoprecipitation
buffer (Thermo Fisher Scientific, Inc.) and total protein lysates
were obtained. Protein concentration was determined using a
bicinchoninic acid assay. Total protein (20 µg) was separated using
4–20% SDS-PAGE gels and then transferred to polyvinylidene fluoride
membranes at 4°C. Membranes were subsequently blocked at 25°C for 1
h with 5% non-fat milk/Tris-buffered saline containing 0.1%
Tween-20, and then incubated at 4°C overnight with primary
antibodies against the following proteins: SATB2 (1:2,000; cat. no.
39229; Cell Signaling Technology, Inc., Danvers, MA, USA) and GAPDH
(1:2,000; cat. no. 5174; Cell Signaling Technology, Inc.).
Following this, membranes were incubated with goat anti-rabbit
horseradish peroxidase (HRP)-conjugated secondary antibodies
(1:2,000; cat. no. 7074; Cell Signaling Technology, Inc.) for 2 h
at 25°C. Protein signals on membranes were detected using enhanced
chemiluminescence reagents (GE Healthcare, Chicago, IL, USA).
Immunohistochemistry
CRC tissues were fixed in 10% formaldehyde at 4°C
for 12 h and then embedded in paraffin. Following this, tissues
were cut into 6 µm sections, placed on silanized glass slides,
deparaffinized in xylene and then rehydrated using ethanol
gradients. The sections were then pretreated with antigen target
retrieval solution [10 mmol/l citric acid monohydrate adjusted with
2 nM sodium hydroxide to (pH 6.0)] at 90°C for 40 min in citrate
buffer. Endogenous peroxidase activity was blocked via incubation
with methanol with 0.3% hydrogen peroxide for 30 min at room
temperature. Sections were then incubated with anti-SATB2 (SATB2;
1:500; cat. no. 39229; Cell Signaling Technology, Inc.) at 4°C for
12 h. Following this, tumor tissues were incubated with goat
anti-rabbit HRP-conjugated secondary antibodies (1:2,000; cat. no.
7074; Cell Signaling Technology, Inc.) for 1 h at 37°C. Proteins
were then visualized using the DAKO EnVision system (Dako; Agilent
Technologies GmbH, Waldbronn, Germany) in accordance with the
manufacturer's instructions and images were captured using a light
microscope (magnification, ×100).
Luciferase reporter assay
The 3′-UTR region of SATB2 containing the binding
site of miR-3666 was constructed into the pmirGLO vector (Promega
Corporation, Madison, WI, USA). In order to perform luciferase
reporter assays, 2×104 HCT116 and SW480 cells per well
were plated in 96-well plates and then incubated at 37°C for 24 h.
Subsequently, the cells were transfected with miR-3666 mimics and
pmirGLO-SATB2-3′UTR or pmirGLO-3′UTR-MUT plasmids using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol. A total
of 24 h post-transfection, luciferase activity was measured using
the Dual-Luciferase Reporter Assay System (Promega Corporation).
Luciferase activity was normalized to Renilla luciferase
activity.
Statistical analysis
Data were expressed as the mean ± standard deviation
of 3 independent experiments. Statistical analysis was performed
using SPSS software version 20.0 (IBM Corp., Armonk, NY, USA). The
statistical significance of the differences between groups was
assessed using a Student's t-test or one-way analysis of variance
followed by a Tukey's post-hoc test for multiple comparisons. The
association between miR-3666 levels and the clinical features of
patients with CRC was investigated via Chi-square test. Pearson's
correlation analysis was used to determine correlations between
miR-3666 and SATB1 in expression levels. Survival curves were
calculated using the Kaplan-Meier method and were analyzed using
with a log-rank test. P<0.05 was considered to indicate a
statistically significant difference.
Results
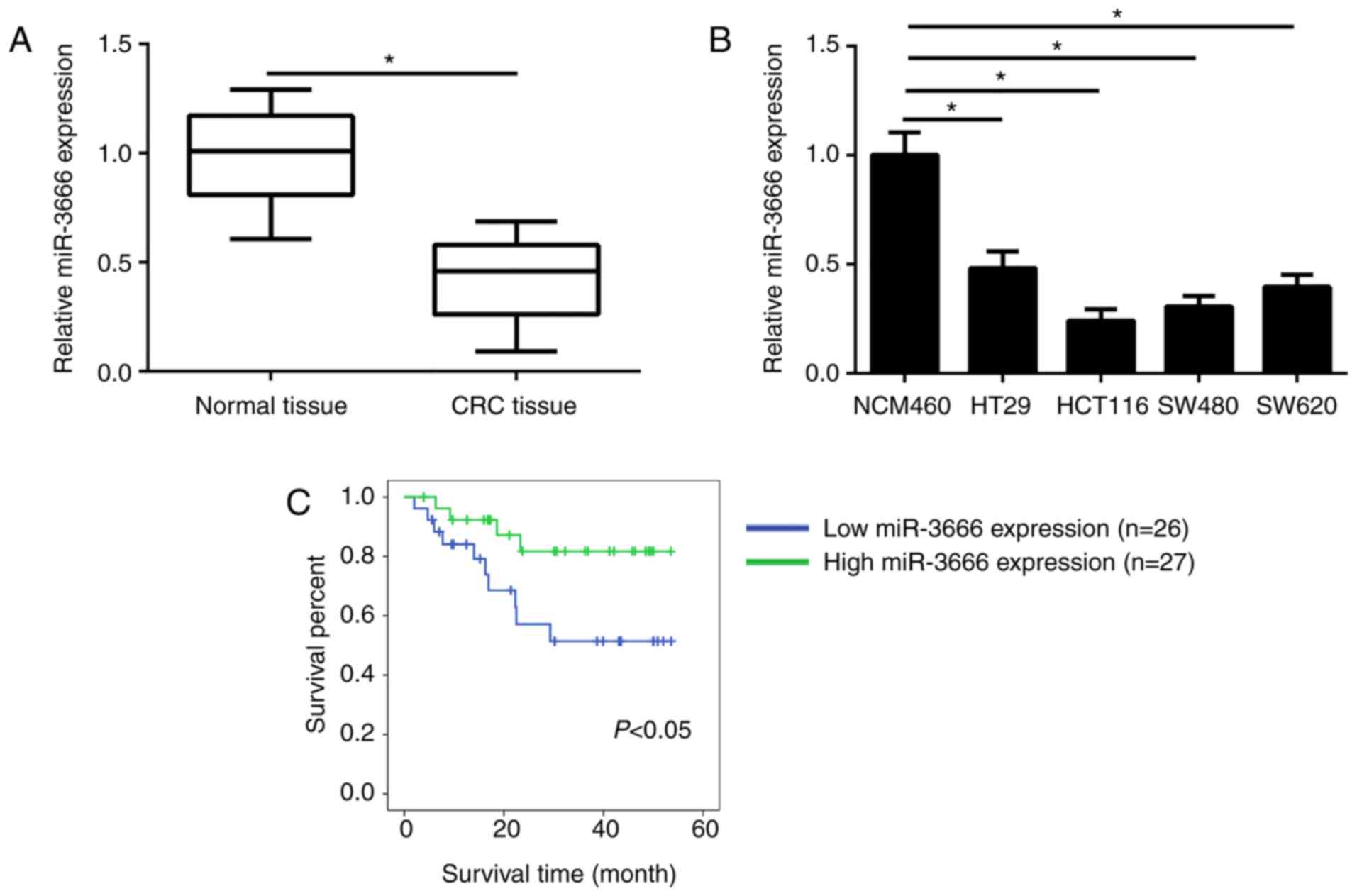
miR-3666 is downregulated in CRC
tissues
The expression levels of miR-3666 in 53 CRC tissues
and 53 adjacent normal tissues were analyzed by RT-qPCR. The
results revealed that the mean expression levels of miR-3666 were
significantly lower in CRC tissues compared with in adjacent normal
tissues (Fig. 1A). In addition,
compared with in the normal control cell line NCM470, the
expression levels of miR-3666 were significantly downregulated in
CRC cell lines (HT29, HCT116, SW480 and SW620 cells; Fig. 1B). Furthermore, the association
between miR-3666 expression and the clinical characteristics of
patients with CRC was determined. The results of the present study
revealed that miR-3666 expression levels were negatively correlated
with CRC tumor, node and metastasis (TNM) stage, tumor size, and
metastasis (Table I); however, no
association was observed between miR-3666 expression and other
clinicopathological characteristics, including age and gender. To
analyze the association between miR-3666 expression and prognosis,
Kaplan-Meier survival analysis was performed. The results revealed
that more patients with higher expression levels of miR-3666
exhibited longer survival times (P=0.037; Fig. 1C). These results indicated that
miR-3666 was aberrantly expressed in CRC tissues and may
participate in the progression of CRC.
 | Table I.Association between miR-3666
expression and clinicopathological features of patients with
CRC. |
Table I.
Association between miR-3666
expression and clinicopathological features of patients with
CRC.
| Features | Cases | Low expression | High expression | P-value |
|---|
| Sex |
|
|
| 0.414 |
| Male | 28 | 12 | 16 |
|
|
Female | 25 | 14 | 11 |
|
| Age (years) |
|
|
| 0.275 |
|
<60 | 30 | 16 | 15 |
|
| ≥60 | 23 | 10 | 12 |
|
| Tumor size (cm) |
|
|
| 0.028a |
|
<5 | 25 | 8 | 17 |
|
| ≥5 | 28 | 18 | 10 |
|
| Lymph node
metastasis |
|
|
| 0.028a |
|
Absent | 29 | 10 | 19 |
|
|
Present | 24 | 16 | 8 |
|
| TNM stage |
|
|
| 0.006a |
|
I–II | 25 | 7 | 18 |
|
III–IV | 28 | 19 | 9 |
miR-3666 suppresses the proliferation,
migration and invasion of CRC cells
To investigate the function of miR-3666 in CRC,
miR-3666 overexpressed or downregulated in HCT116 and SW480 cells
by transfection with miR-3666 mimics, inhibitors or miR-NC,
respectively. RT-qPCR analysis indicated that, compared with in the
control group, miR-3666 was significantly overexpressed and notably
downregulated following the transfection of HCT116 and SW480 cells
with miR-3666 mimics and inhibitors, respectively (Fig. 2A). Then, a CCK-8 assay was
conducted to determine the potential of cellular proliferation. The
results demonstrated that, compared with in the control group,
overexpression of miR-3666 significantly inhibited cell
proliferation and vice versa (Fig.
2B). Additionally, Transwell assays were used to evaluate cell
migration and invasion. The results revealed that overexpression of
miR-3666 significantly suppressed the number of migrated and
invaded cells, whereas inhibition of miR-3666 significantly
promoted cell migration and invasion compared with in the control
group (Fig. 2C and D). These data
indicated that miR-3666 suppressed the proliferation, migration and
invasion of CRC cells.
SATB2 is a target gene of
miR-3666
To further investigate the downstream mechanism, the
target gene of miR-3666 in CRC cells was determined. Analysis with
TargetScan 7.0 revealed that numerous genes were potential targets
of miR-3666. Among these targets, SATB2 ranked first and has been
reported to promote the development of CRC (22); the potential binding site of
miR-3666 in the 3′-UTR of SATB2 was presented in Fig. 3A. Therefore, SATB2 was selected for
investigation in the present study. Using luciferase reporter
assays, it was demonstrated that overexpression of miR-3666
significantly inhibited the luciferase activity of HCT116 and SW480
cells transfected with the wild type 3′-UTR, whereas knockdown of
miR-3666 significantly upregulated luciferase activity compared
with in the control (Fig. 3B).
Furthermore, the present study revealed that overexpression of
miR-3666 significantly enhanced the mRNA and notably increased the
protein expression levels of SATB2 in HCT116 and SW480 cells
compared with in the control (Fig. 3C
and D). In addition, RT-qPCR analysis indicated that the
expression of miR-3666 was negatively correlated with that of SATB2
in CRC tissues (Fig. 3E). Then,
the expression of SATB2 in CRC tissues was analyzed by RT-qPCR,
which demonstrated that the expression of SATB2 was significantly
upregulated in CRC tissues compared with in adjacent normal tissues
(Fig. 3F). Furthermore,
immunohistochemical and western blot analyses indicated that SATB2
was overexpressed in CRC tissues compared with in matched normal
tissues (Fig. 3G and H).
Collectively, the results of the study indicated that miR-3666
targeted SATB2, which was upregulated in CRC tissues.
 | Figure 3.SATB2 is a target gene of miR-3666.
(A) A diagram for the potential binding site of miR-3666 in the
3′-UTR of SATB2 mRNA. (B) A SATB2 3′-UTR fragment containing the Wt
or Mut miR-3666 binding site was cloned into the pmirGLO reporter
gene vector. Cells were cotransfected with the reporter gene
construct vector and miR-3666 mimics or inhibitor for 48 h. (C and
D) HCT116 and SW480 cells were transfected with miR-3666 mimics or
inhibitor for 48 h. Relative mRNA and protein levels of SATB2 were
detected by RT-qPCR and western blotting. (E) Correlation between
SATB2 mRNA and miR-3666 expression was assessed by Pearson's
correlation analysis. (F) Relative expression of SATB2 was measured
by RT-qPCR in CRC tissues and adjacent normal tissues. (G) SATB2
expression was determined by immunohistochemical analysis in paired
CRC tissues and normal tissues (magnification, ×100). (H) Western
blot analysis of SATB2 protein expression in 3 pairs of CRC tissues
and adjacent normal tissues. *P<0.05 vs. NC group or normal
tissues. miR, microRNA; Mut, mutant; N, normal tissue; NC, negative
control; RT-qPCR, reverse transcription-quantitative polymerase
chain reaction; SATB2, special AT-rich sequence binding protein 2;
T, tumor tissue; UTR, untranslated region; Wt, wild type. |
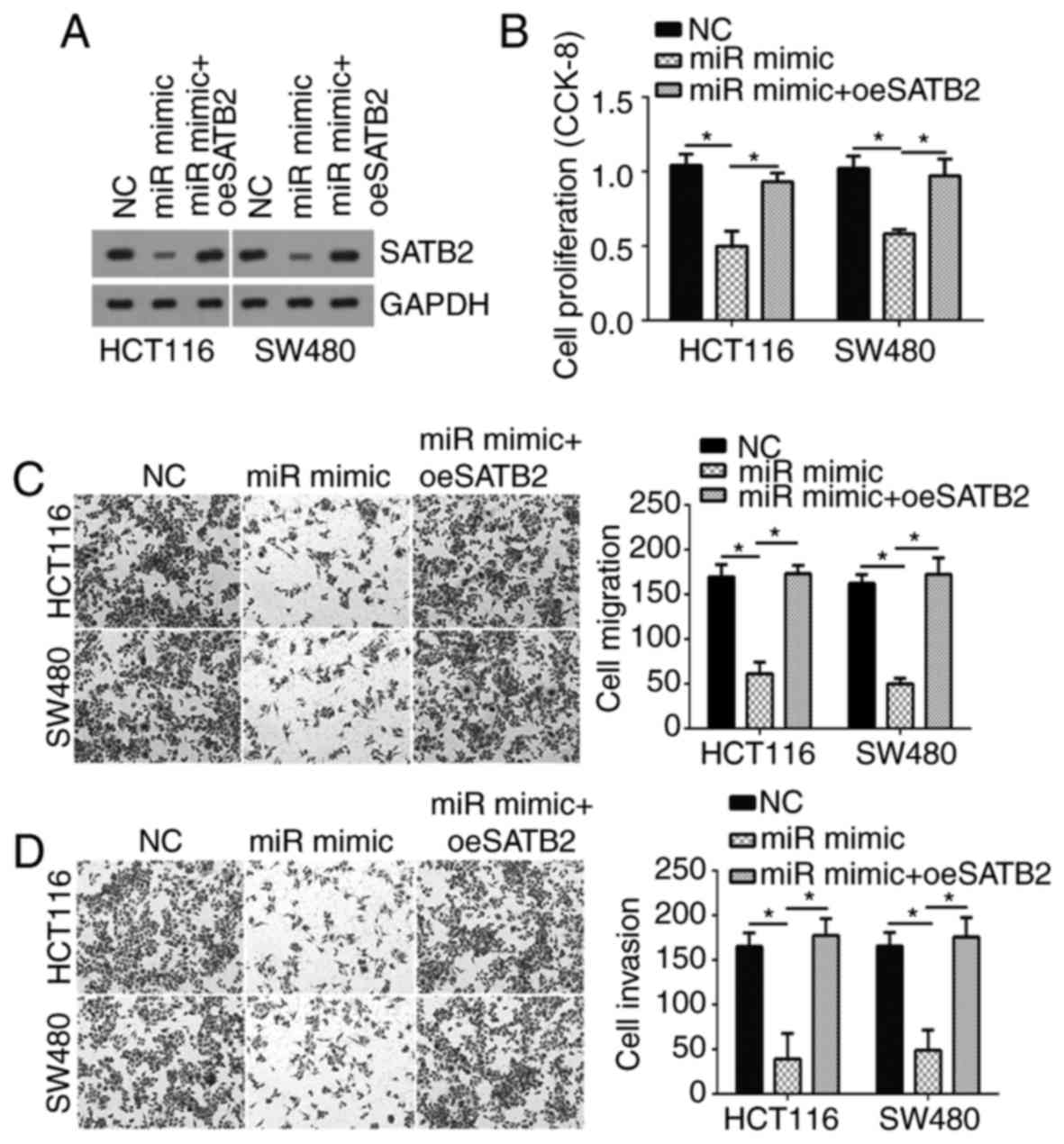
Restoration of SATB2 expression
abolishes the antitumor effects of miR-3666
To determine whether SATB2 is required for
miR-3666-mediated effects on CRC cells, the protein expression
levels of SATB2 were restored in HCT116 and SW480 cells transfected
with miR-3666. Western blotting indicated that the expression of
SATB2 protein was notably increased in HCT116 and SW480 cells
expressing the SATB2 overexpression vector compared with in cells
transfected with miR-3666 mimics (Fig.
4A). Then, CCK-8 and Transwell assays were conducted, which
revealed that cell proliferation, migration and invasion was
significantly inhibited in cells overexpressing miR-3666 compared
with in the control; however, restoration of SATB2 significantly
reversed these effects of miR-3666 (Fig. 4B-D). In conclusion, the data
suggested that miR-3666 suppressed CRC cell proliferation,
migration and invasion by targeting SATB2.
Discussion
CRC is one of the most malignant types of cancer and
one of the leading cause of cancer-associated mortality worldwide
(23). Improvements in the
treatment of CRC have been reported; however, the prognosis of
patients with CRC remains poor (1). Numerous patients with CRC are
diagnosed at advanced stages when tumor metastasis occurs (2). Therefore, in order to identify
biomarkers and therapeutic targets for the diagnosis, prognosis and
treatment of CRC, complete understanding of the underlying
molecular mechanisms of CRC progression is required (24). Increasing evidence has indicated
that miRNAs regulate cell proliferation, survival and metastasis
(25). Numerous miRNAs have been
reported as promising biomarkers for CRC (26). Thus, miRNAs may be considered as
important regulators in tumor progression. For example, miRNA-520c
was demonstrated to promote gastric cancer growth and metastasis
via targeting interferon regulatory factor 2 (27). In addition, miR-451 was observed to
enhance pancreatic cancer growth via inhibition of calcium-binding
protein 39 (28). miR-381
suppressed gastric cancer cell migration and invasion by inhibiting
transmembrane member 16A (29).
The function of miR-3666 has been recently studied
(17,18). Shi et al (17) reported that miR-3666 inhibited lung
cancer cell proliferation by targeting sirtuin 7. Wang et al
(18) revealed that miR-3666
upregulation suppressed the progression of thyroid carcinoma.
Additionally, Li et al (19) reported that miR-3666 overexpression
repressed cervical cancer growth and metastasis. Collectively,
these data indicated that miR-3666 serves as a tumor suppressor in
certain types of cancer; however, its role and clinical
significance in CRC remain unknown. In the present study, it was
reported that miR-3666 was downregulated in CRC tissues compared
with in adjacent normal tissues. In addition, miR-3666 expression
was associated with tumor size, TNM staging and metastasis in CRC,
and the downregulation of miR-3666 was associated with a poor
prognosis of CRC. Functional experiments demonstrated that miR-3666
suppressed the proliferation, migration and invasion of CRC cells.
The results of the present study indicated that miR-3666 serves as
a tumor suppressor in CRC and as a potential biomarker for the
prognosis of CRC.
To investigate the molecular mechanism underlying
miR-3666-suppressed CRC cell proliferation and metastasis, the
target gene of miR-3666 was determined; SATB2 was predicted as a
direct target gene of miR-3666 in CRC cells in the present study.
SATB2 is a transcription factor that has been associated with
tumorigenesis (30). For instance,
Wang et al (20) reported
that SATB2 promoted lung cancer metastasis. Luo et al
(31) demonstrated that SATB2
induced the progression of triple negative breast cancer. A recent
study also indicated that SATB2 promoted the progression of
colorectal cancer via the Wnt pathway (30). These studies demonstrated the role
of SATB2 as an oncogene in a variety of cancers; however, the
regulation of SATB2 expression in CRC requires further
investigation. In the present study, using a luciferase reporter
assay, miR-3666 was proposed to directly target SATB2 in CRC cells.
Additionally, the findings of the present study suggested that
miR-3666 significantly inhibited the expression of SATB2; miR-3666
expression was negatively correlated with that of SATB2 in CRC
tissues. In addition, SATB2 expression was significantly
upregulated in CRC tissues and cell lines. Furthermore, via a
series of functional experiments, the present study demonstrated
that restoration of SATB2 expression rescued the effects of
miR-3666 on CRC cell proliferation, migration and invasion.
In summary, miR-3666 was reported to be
downregulated in CRC tissues and overexpression of miR-3666
impaired the proliferation, migration and invasion of CRC cells by
targeting SATB2 in the present study. Collectively, these results
may provide novel insight to improve understanding of the mechanism
underlying miR-3666 and SATB2 in the regulation of CRC progression,
and may aid the identification of potential therapeutic targets for
the clinical treatment of CRC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
DY and HZ made substantial contributions to the
design of the present study, analyzed and interpreted the results,
and wrote the manuscript. RL, JX and WL performed some experiments.
All authors read and approved the final manuscript.
Ethics approval and consent to
participate
For the use of human samples, the protocol employed
in the present study was approved by the Institutional Ethics
Committee of Wenzhou Central Hospital (Wenzhou, China) and all
enrolled patients provided written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Vermeer NC, Snijders HS, Holman FA,
Liefers GJ, Bastiaannet E, van de Velde CJ and Peeters KC:
Colorectal cancer screening: Systematic review of screen-related
morbidity and mortality. Cancer Treat Rev. 54:87–98. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ballester V, Rashtak S and Boardman L:
Clinical and molecular features of young-onset colorectal cancer.
World J Gastroenterol. 22:1736–1744. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zafar SY, McNeil RB, Thomas CM, Lathan CS,
Ayanian JZ and Provenzale D: Population-based assessment of cancer
survivors' financial burden and quality of life: A prospective
cohort study. J Oncol Pract. 11:145–150. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Pan Y, Tong JHM, Lung RWM, Kang W, Kwan
JSH, Chak WP, Tin KY, Chung LY, Wu F, Ng SSM, et al: RASAL2
promotes tumor progression through LATS2/YAP1 axis of hippo
signaling pathway in colorectal cancer. Mol Cancer. 17:1022018.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yu W, Wang Y and Guo P: Notch signaling
pathway dampens tumor-infiltrating CD8+ T cells activity
in patients with colorectal carcinoma. Biomed Pharmacother.
97:535–542. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sakai E, Nakayama M, Oshima H, Kouyama Y,
Niida A, Fujii S, Ochiai A, Nakayama KI, Mimori K, Suzuki Y, et al:
Combined mutation of Apc, Kras and Tgfbr2 effectively drives
metastasis of intestinal cancer. Cancer Res. 78:1334–1346. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Esquela-Kerscher A and Slack FJ:
Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer.
6:259–269. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kloosterman WP and Plasterk RH: The
diverse functions of microRNAs in animal development and disease.
Dev Cell. 11:441–450. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Mei LL, Qiu YT, Wang WJ, Bai J and Shi ZZ:
Overexpression of microRNA-1470 promotes proliferation and
migration, and inhibits senescence of esophageal squamous carcinoma
cells. Oncol Lett. 14:7753–7758. 2017.PubMed/NCBI
|
|
10
|
Wang G, Fang X, Han M, Wang X and Huang Q:
MicroRNA-493-5p promotes apoptosis and suppresses proliferation and
invasion in liver cancer cells by targeting VAMP2. Int J Mol Med.
41:1740–1748. 2018.PubMed/NCBI
|
|
11
|
Slaby O, Svoboda M, Michalek J and Vyzula
R: MicroRNAs in colorectal cancer: Translation of molecular biology
into clinical application. Mol Cancer. 8:1022009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Koo KH and Kwon H: MicroRNA miR-4779
suppresses tumor growth by inducing apoptosis and cell cycle arrest
through direct targeting of PAK2 and CCND3. Cell Death Dis.
9:772018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Islam F, Gopalan V, Vider J, Lu CT and Lam
AK: miR-142-5p act as an oncogenic microRNA in colorectal cancer:
Clinicopathological and functional insights. Exp Mol Pathol.
104:98–107. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
He S, Wang G, Ni J, Zhuang J, Zhuang S,
Wang G, Ye Y and Xia W: MicroRNA-511 inhibits cellular
proliferation and invasion in colorectal cancer by directly
targeting hepatoma-derived growth factor. Oncol Res. Jan
10–2018.(Epub ahead of print). View Article : Google Scholar
|
|
15
|
Alipour A, Mojdehfarahbakhsh A, Tavakolian
A, Morshedzadeh T, Asadi M, Mehdizadeh A and Nami M: Neural
communication through theta-gamma cross-frequency coupling in a
bistable motion perception task. J Integr Neurosci. 15:539–551.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yuan Z, Baker K, Redman MW, Wang L, Adams
SV, Yu M, Dickinson B, Makar K, Ulrich N, Böhm J, et al: Dynamic
plasma microRNAs are biomarkers for prognosis and early detection
of recurrence in colorectal cancer. Br J Cancer. 117:1202–1210.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Shi H, Ji Y, Zhang D, Liu Y and Fang P:
MicroRNA-3666-induced suppression of SIRT7 inhibits the growth of
non-small cell lung cancer cells. Oncol Rep. 36:3051–3057. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang G, Cai C and Chen L: MicroRNA-3666
regulates thyroid carcinoma cell proliferation via MET. Cell
Physiol Biochem. 38:1030–1039. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li L, Han LY, Yu M, Zhou Q, Xu JC and Li
P: Pituitary tumor-transforming gene 1 enhances metastases of
cervical cancer cells through miR-3666-regulated ZEB1. Tumour Biol.
Sep 17–2015.(Epub ahead of print).
|
|
20
|
Wang J, Lu Y, Ding H, Gu T, Gong C, Sun J,
Zhang Z, Zhao Y and Ma C: The miR-875-5p inhibits SATB2 to promote
the invasion of lung cancer cells. Gene. 644:13–19. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sun X, Liu S, Chen P, Fu D, Hou Y, Hu J,
Liu Z, Jiang Y, Cao X, Cheng C, et al: miR-449a inhibits colorectal
cancer progression by targeting SATB2. Oncotarget. 8:100975–100988.
2017.PubMed/NCBI
|
|
23
|
Qu JJ, Qu XY and Zhou DZ: miR4262 inhibits
colon cancer cell proliferation via targeting of GALNT4. Mol Med
Rep. 16:3731–3736. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Dowling CM, Hayes SL, Phelan JJ, Cathcart
MC, Finn SP, Mehigan B, McCormick P, Coffey JC, O'sullivan J and
Kiely PA: Expression of protein kinase C gamma promotes cell
migration in colon cancer. Oncotarget. 8:72096–72107. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kok MG, Mandolini C, Moerland PD, de Ronde
MW, Sondermeijer BM, Halliani A, Nieuwland R, Cipollone F, Creemers
EE, Meijers JC and Pinto-Sietsma SJ: Low miR-19b-1-5p expression in
isolated platelets after aspirin use is related to aspirin
insensitivity. Int J Cardiol. 203:262–263. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hu WL and Zhou XH: Identification of
prognostic signature in cancer based on DNA methylation interaction
network. BMC Med Genomics. 10 Suppl 4:S632017. View Article : Google Scholar
|
|
27
|
Li YR, Wen LQ, Wang Y, Zhou TC, Ma N, Hou
ZH and Jiang ZP: MicroRNA-520c enhances cell proliferation,
migration, and invasion by suppressing IRF2 in gastric cancer. Febs
Open Bio. 6:1257–1266. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Guo R, Gu J, Zhang Z, Wang Y and Gu C:
miR-451 promotes cell proliferation and metastasis in pancreatic
cancer through targeting CAB39. Biomed Res Int. 2017:23814822017.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Cao Q, Liu F, Ji K, Liu N, He Y, Zhang W
and Wang L: MicroRNA-381 inhibits the metastasis of gastric cancer
by targeting TMEM16A expression. J Exp Clin Cancer Res. 36:292017.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yu W, Ma Y, Shankar S and Srivastava RK:
SATB2/β-catenin/TCF-LEF pathway induces cellular transformation by
generating cancer stem cells in colorectal cancer. Sci Rep.
7:109392017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Luo LJ, Yang F, Ding JJ, Yan DL, Wang DD,
Yang SJ, Ding L, Li J, Chen D, Ma R, et al: miR-31 inhibits
migration and invasion by targeting SATB2 in triple negative breast
cancer. Gene. 594:47–58. 2016. View Article : Google Scholar : PubMed/NCBI
|