Lung cancer is the leading cause of cancer-related
death worldwide. Despite improvements in treatment, the overall
5-year survival of lung cancer remains less than 18% in the United
States (1,2), and the age-standardised 5-year net
survival is in the range of 10–20% in most countries (2). Therefore, optimal therapies for lung
cancer are urgently needed, the development of which requires
extensive knowledge of the aetiology of lung cancer.
Interleukin (IL)-17 has been implicated in various
cancers and is regarded as a double-edged sword in lung cancer
(3). The greatest homogeneity is
between IL-17A and IL-17F, which, together with their producing
cells, have been implicated in multiple types of cancer. IL-17 is
generally referred to as IL-17A in the literature. Moreover, most
studies have reported that the expression of IL-17A is positively
correlated with tumour growth (4–7),
while various investigations have shown the opposite result
(8). For example, IL-17A produced
by tumour-infiltrating immune cells was found to promote cancer
cell growth through an IL-6/signal transducer and activator of
transcription 3 (STAT3) pathway (7), and transfection with IL-17A was found
to promote hepatocellular carcinoma (HCC) tumour growth via protein
kinase B (AKT)-dependent activation of IL-6/Janus kinase 2
(JAK2)/STAT3 signalling (9).
However, a low level of intratumoural IL-17A expression was found
to be indicative of a poor prognosis (10).
However, other members of the IL-17 family have not
been well investigated for their potential roles in cancer
development. The IL-17 family includes IL-17A through IL-17F and
their cognate receptors, IL-17RA to IL-17RE. IL-17A was first
cloned and initially named cytotoxic T lymphocyte-associated
antigen 8 (CTLA-8) in 1993. A decade later, IL-17A was found to be
a distinctive feature of Th17 cells, as it was not found in Th1 and
Th2 cells. The other five family members were discovered in quick
succession and designated as IL-17B, IL-17C, IL-17D, IL-25 (IL-17E)
and IL-17F.
IL-17B/IL-17RB signalling has been demonstrated to
be implicated in tumour malignancies, such as breast cancer
(11), pancreatic cancer (12) and gastric cancer (13). IL-17C is involved in the innate
immune response of human bronchial epithelial cells (14) and mediates the recruitment of
tumour-associated neutrophils with enhanced tumour growth (15). IL-17C has also been found to be
upregulated in intestinal cancers and is critical for the
microbiota-mediated contribution to tumour development (16). The function of IL-17D in cancer has
not been well described. Some studies have shown that IL-17D is
poorly expressed in cancer cells and induces tumour rejection
through the recruitment of NK cells (17). IL-17E (also known as IL-25) is the
most divergent member in the IL-17 family. The influence of IL-25
derived from tumour-associated fibroblasts (18), macrophages and T cells (19) on tumour progression has been
explored in recent years. Studies have shown that IL-25/IL-17RB
(IL-25R) signalling participates in inhibiting tumour metastasis
and growth (18–20). Moreover, IL-25 has been
demonstrated to bind IL-17RB through nuclear factor-κB (NF-κΒ) and
JAK/STAT3 pathways to promote proliferation and nourish cancer stem
cells in hepatocellular carcinoma (21). IL-17F, together with IL-17A, IL-25,
and their receptors, has been described to be elevated in benign
prostatic hyperplasia and prostate cancer (22). As mentioned above, IL-17F is the
most homologous with IL-17A but lacks the same contributions to
biological and physiological processes. Unlike the double-edged
sword role of IL-17A in cancer (3), there is little clinical evidence
illustrating the role of IL-17F in cancer, except that it has been
shown to be a negative modulator that suppressed tumour
angiogenesis in hepatocellular carcinoma in an animal model
(23) and is a potential
diagnostic biomarker when combined with vascular endothelial growth
factor (VEGF) in oral squamous cell carcinoma (24).
In summary, every member of the IL-17 family has
been identified to play critical roles in cancer, with the main
focus on IL-17A and the interstitial cells that produce IL-17A in
the tumour microenvironment. In this study, the expression and
genetic alterations in IL-17 family members were investigated
through the Catalogue of Somatic Mutations in Cancer (COSMIC),
Oncomine and cBioPortal databases. The prognostic value and
interaction network were then analysed by the Kaplan-Meier plotter
and FunRich software.
STRING database (Search Tool for the Retrieval of
Interacting Genes; available at http://string-db.org/) (30) provides uniquely comprehensive
coverage and easy access to protein-protein interaction
information. The common gene networks of the IL-17 family were
constructed independently by importing gene symbols. We selected
the interactions pertaining to Homo sapiens and showed
minimum interactions with a confidence score >0.9. Only the
interactions with a combined score >0.4 were considered
significant.
Functional enrichment analysis of the interaction
proteins was performed using FunRich (http://www.funrich.org), which is an open access,
standalone functional enrichment and network analysis tool
(31). The functional analysis of
related genes that interact with IL-17 family genes was conducted
with FunRich software. The STRING interaction proteins were
imported into FunRich, and significantly enriched pathways and site
of expression were analysed.
Student's t-test (two-tailed) was used to compare
the means between two groups. mRNA expression data are presented as
fold change, and P-values <0.05 were considered statistically
significant. Overall survival (OS) data are displayed as
Kaplan-Meier plots, with P-values calculated using the log-rank
test. P-values <0.05 were considered statistically
significant
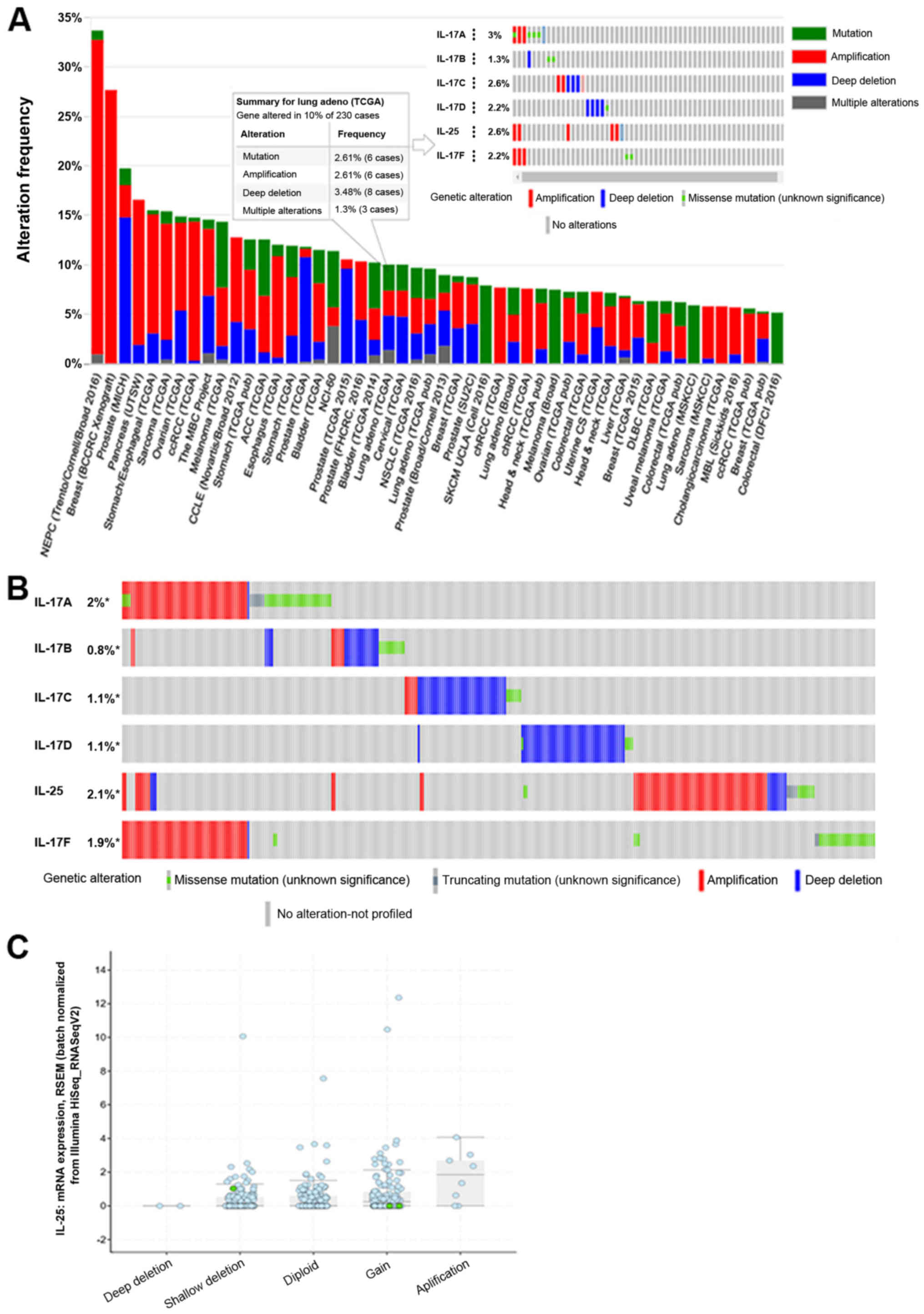
The expression levels of the six IL-17 family
members in cancers were investigated using Oncomine (26) and cBioPortal (29). We first measured the expression
levels of six IL-17 family members in 20 types of cancers and
compared the expression levels to those in normal individuals. Four
datasets showed significantly increased expression of IL-17A in
cancers such as brain cancer, leukaemia and sarcoma, and nine
datasets showed decreased expression of IL-17A in cancers such as
oesophageal carcinoma, cervical cancer and pancreatic cancer.
However, no dataset showed a significant difference in IL-17A
expression between lung cancer and controls. Likewise, two datasets
showed that IL-17B was highly expressed in brain cancer, and four
datasets showed decreased IL-17B expression in breast cancer. Two
datasets showed that IL-17C was overexpressed in other cancers, and
five datasets showed that IL-17E was decreased in four types of
cancer. Notably, five datasets showed that IL-17D was significantly
decreased in lung cancer. Data on IL-17F in cancer are rare, and no
results showed changes in its expression in cancer Fig. 2). Therefore, we assessed the mRNA
expression of IL-17 family members in different lung cancer
datasets. Table II shows that
IL-17A, IL-17B, IL-17C and IL-25 mRNA overexpression was not
significant in the Hou et al (35), Beer et al (36), Garber et al (37), Selamat et al (38) and Landi et al (39) lung datasets. According to the
Garber et al (37), Okayama
et al (40) and Hou lung
datasets, IL-17D downregulation was observed in lung adenocarcinoma
and squamous cell lung carcinoma compared with normal lung tissue,
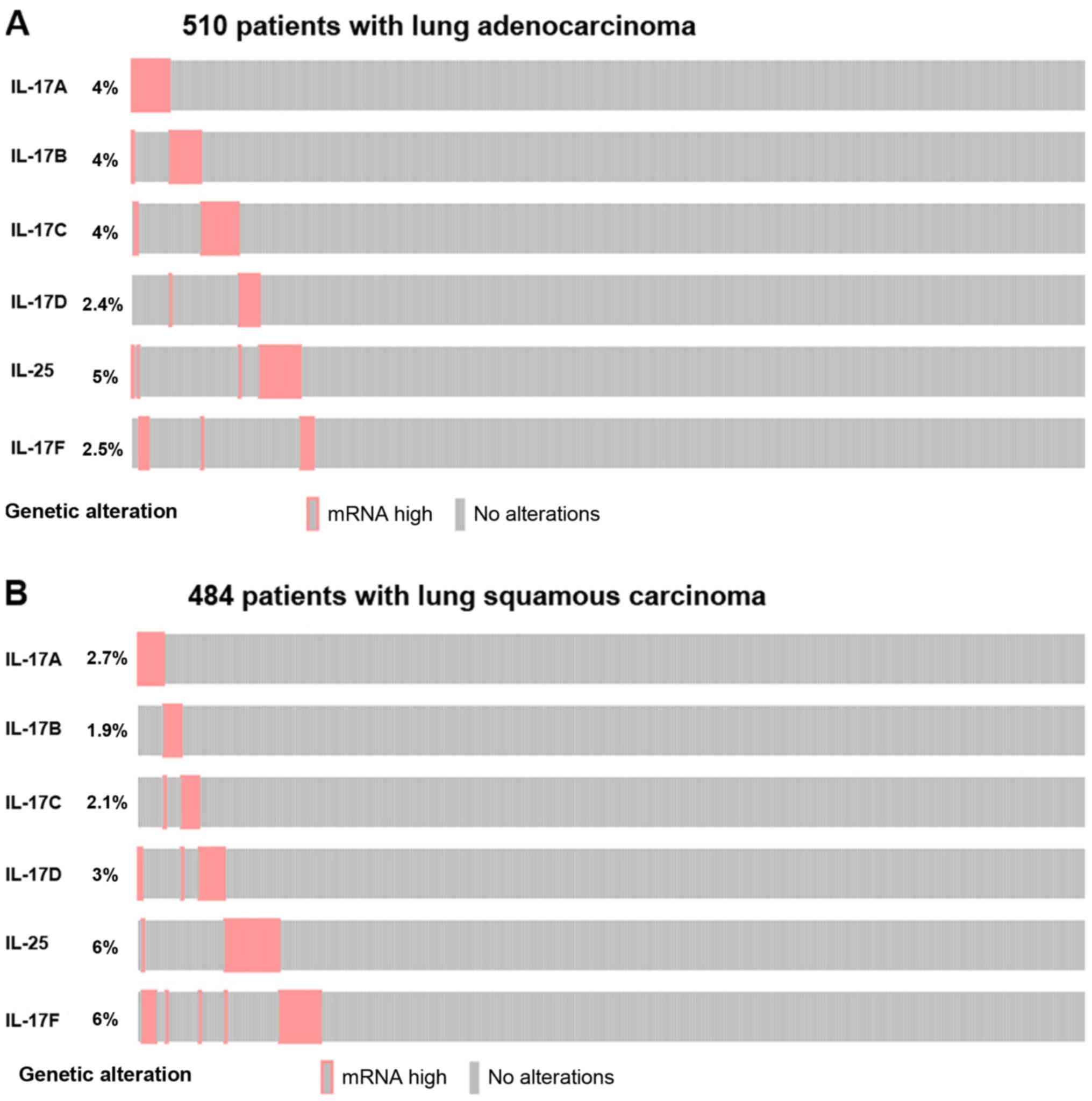
with a fold change >2.0. We also evaluated whether alterations
in IL-17 family gene mRNA are associated with a specific
histological type of non-small cell lung cancer (NSCLC) using
cBioPortal (Fig. 3). In the TCGA
PanCancer Atlas (https://gdc.cancer.gov/about-data/publications/pancanatlas)
(41), 510 samples of lung
adenocarcinoma and 484 samples of lung squamous cell carcinoma with
mRNA data (RNA Seq V2) were investigated. An mRNA expression
z-score ±2.0 was set as the threshold. The result showed that the
IL-17A-F mRNA upregulation frequencies in lung adenocarcinoma were
4, 4, 4, 2.4, 5 and 2.5%, respectively. The IL-17A, IL-17B and
IL-17C mRNA upregulation frequencies in lung squamous cell
carcinoma were lower than those in lung adenocarcinoma (2.7, 1.9
and 2.1%, respectively), whereas the IL-17D, IL-25 and IL-17F mRNA
upregulation rates were higher in lung squamous cell carcinoma (3,
6 and 6%, respectively).
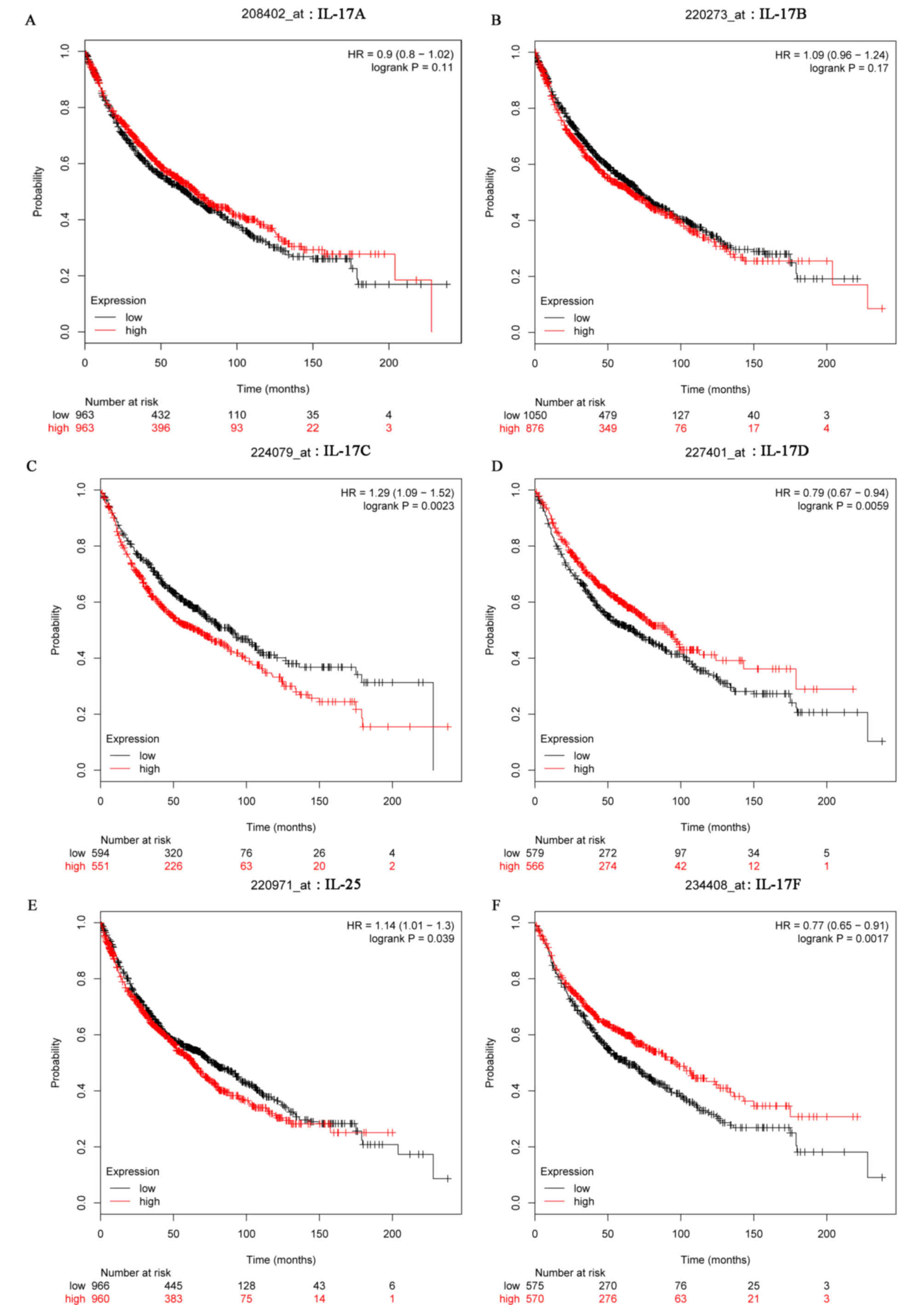
The prognostic information of IL-17 family genes in
lung cancer is freely available at http://kmplot.com/analysis (27). However, there were no significant
associations of IL-17A and IL-17B expression levels with the OS of
lung cancer patients (Fig. 4A and
B). High IL-17C (HR 1.29 (1.09–1.52); P=0.0023) and IL-25 (HR
1.14 (1.01–1.3); P=0.039) mRNA expression levels were associated
with poor OS in lung cancer patients. Conversely, higher mRNA
levels of IL-17D (HR 0.79 (0.67–0.94); P=0.0059) and IL-17F (HR
0.77 (0.65–0.91); P=0.0017) were associated with better long-term
OS (Fig. 4C-F).
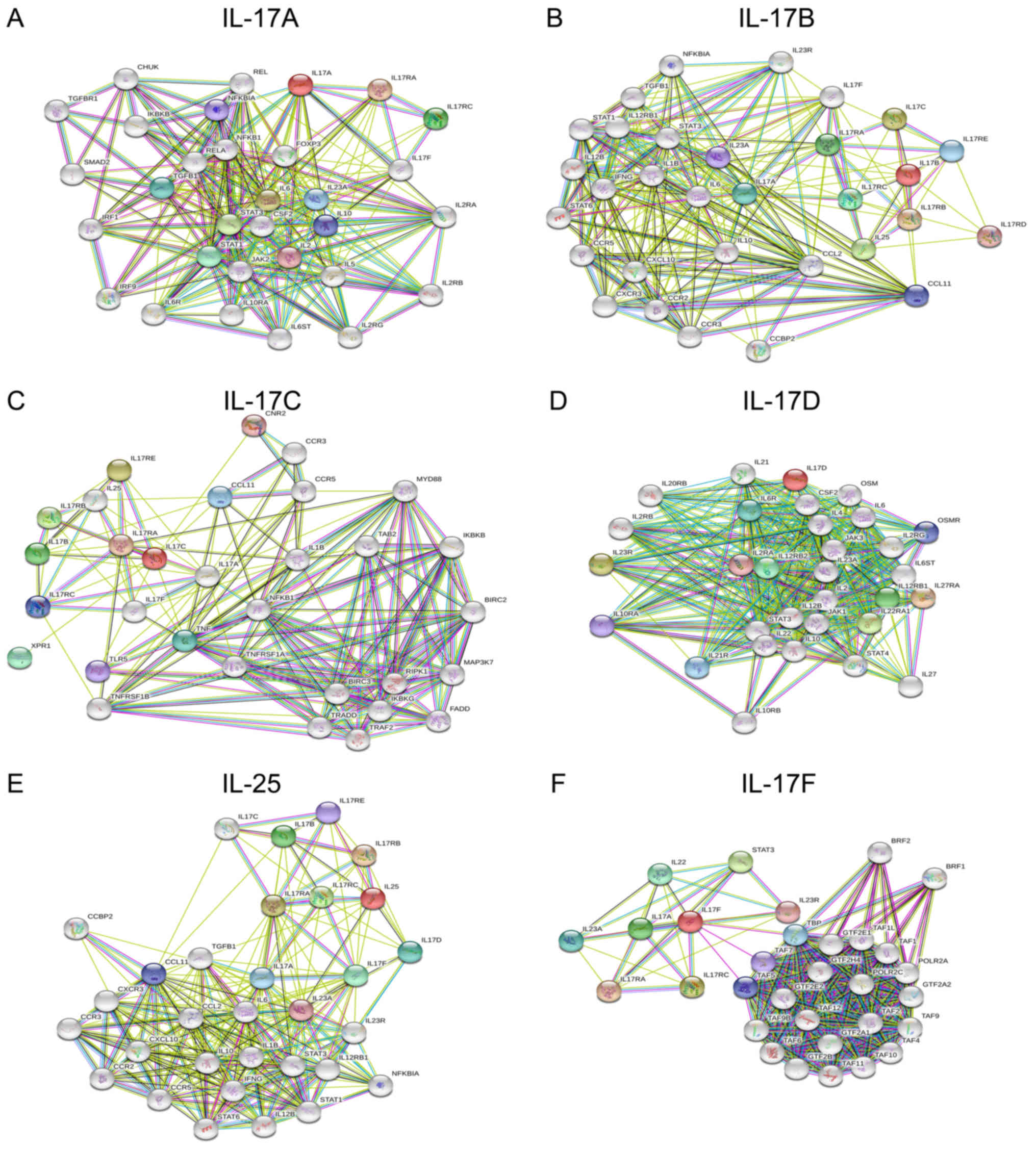
To explore the interactions of the IL-17 family and
regulation in biological processes, we constructed protein-protein
interaction (PPI) networks by submitting an IL-17 family gene list
to the STRING data and analysed the networks using FunRich software
(31). Each constructed common
gene network had 31 nodes, which were considered the most highly
connected protein interactions (Fig.
5). Many of the interacting proteins were also connected with
molecules in other networks, probably indicating that IL-17 family
members share functional associations with each other. The
potential targeted genes were used for the biological pathway and
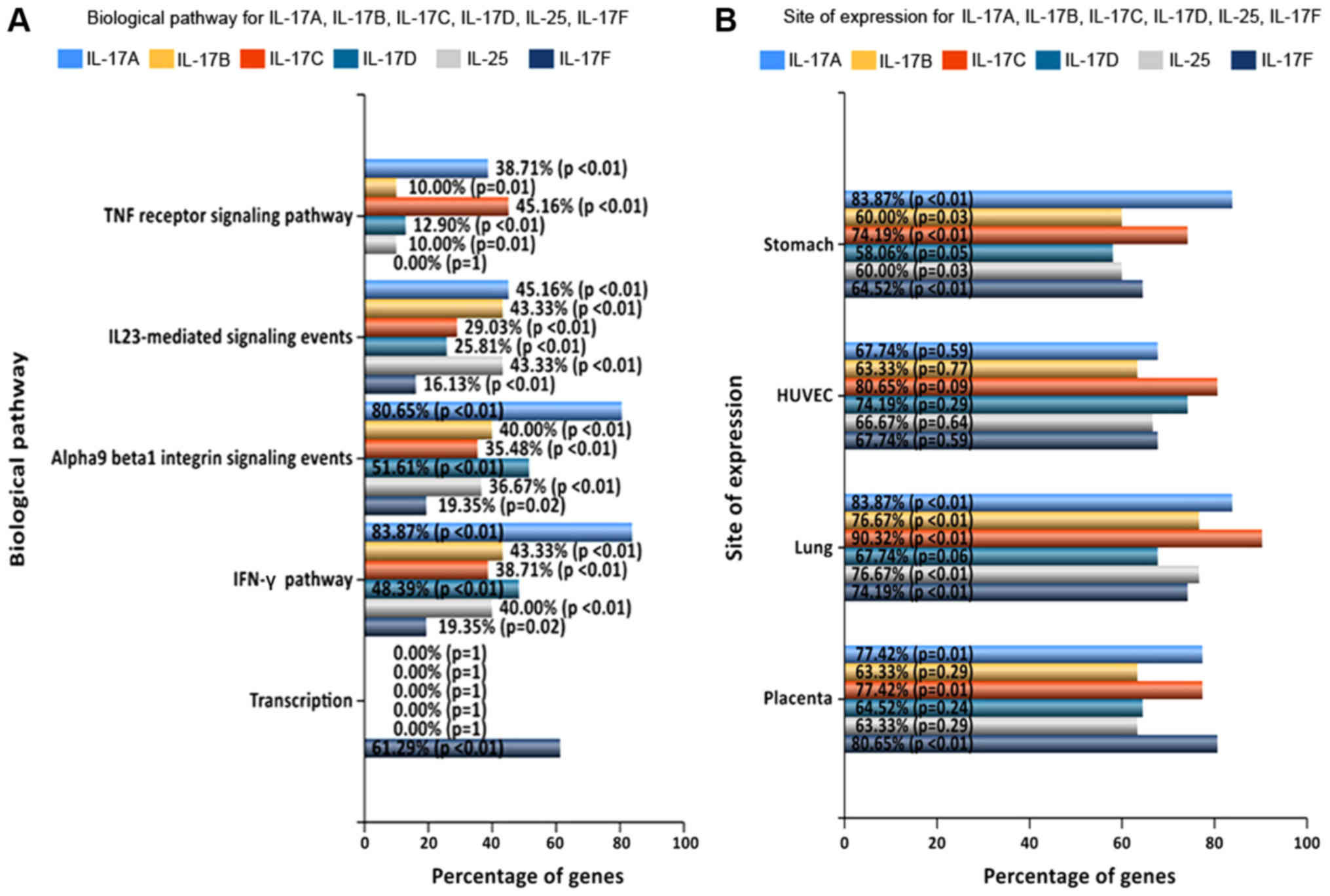
site expression analysis with FunRich software. The results showed
that IL-17A was significantly enriched in proteins involved in the
‘IFN-γ pathway’ (83.87%) and ‘α9β1 integrin signalling events’
(80.65%). IL-17B was significantly enriched in ‘IFN-γ’ (83.87%) and
‘IL-23-mediated signalling events’ (83.87%). IL-17C was enriched in
‘TNF receptor signalling pathway’ (38.71%) and ‘IFN-γ’ (45.16%).
IL-17D was enriched in ‘α9β1 integrin signalling events’ (51.61%)
and ‘IFN-γ’ (48.39%). IL-25 was enriched in ‘IL-23-mediated
signalling events’ (40%) and ‘IFN-γ’ (43.33%), and IL-17F was
mainly enriched in ‘transcription’ (61.29%) (Fig. 6A). Moreover, the analysis based on
the term ‘site of expression’ showed that IL-17C expression levels
were higher in the lung than in other family members in other sites
(Fig. 6B).
The IL-17 family, a subset of cytokines consisting
of IL-17A-F, plays crucial roles in inflammation, autoimmune
diseases and cancer. IL-17A and IL-17F are primarily secreted by
immune cells, whereas IL-17B, IL-17C, IL-17D, and IL-25 are derived
from a wide range of cells (42).
Among all members, the expression and biological function of IL-17A
have been widely studied in lung cancer (Table III). Initial studies have shown
that increased expression of IL-17A in cancer tissue and serum were
associated with poor survival of patients with lung cancer
(43–47). Overexpression or exogeneous IL-17A
can promote tumour growth (48),
angiogenesis (49,50) and metastasis (51). Other studies have observed that
high levels of intratumoral IL-17A expression may indicate good
prognosis in gastric adenocarcinoma (10), and similar results have been
obtained in oesophageal squamous cell carcinoma (52) and cervical adenocarcinoma (53). These previous studies with small
cohorts had limited statistical and clinical power. Furthermore,
very little is known about the expression of other IL-17 cytokines
in lung cancer. Large databases such as Oncomine and TCGA provide
large samples and datasets. Through the integration and analysis of
massive bioinformatic data, we can avoid errors associated with
small sample experimental research and increase the credibility of
the research results. In the present study, the gene mutations,
mRNA expression levels, prognostic values and network pathways of
different IL-17 family members were investigated, aiming to find
directions for further studies and potential therapeutic targets in
lung cancer.
A number of studies have reported that IL-17 genetic
family polymorphisms are associated with increased risk of cancer.
It was demonstrated that IL-17A and IL-17F gene polymorphisms were
associated with an increased risk of lung cancer (54–56),
gastric cancer (57), acute
myeloid leukaemia (58), and
colorectal cancer (59).
Furthermore, IL-17A polymorphisms may upregulate IL-17A expression
(60) and are associated with
clinicopathological features and tobacco smoking history in lung
cancer patients (54). The
relationships between the genetic polymorphisms of other IL-17
family members and lung cancer susceptibility are not fully
understood. In our study, the frequencies and types of alterations
of IL-17 family genes were analysed through the COSMIC and
cBioPortal databases. The present results revealed that IL-17
family gene mutation rates were in general low, and amplification
and deep deletion were the main mutation type. The incidence of CNV
was found to be higher than that of point mutations, and IL-17E
exhibited the highest CNV, which was positively related to mRNA
expression in lung cancer. IL-17E, which had the highest CNV in
lung cancer, represents a promising potential oncogene in lung
cancer that warrants further clinical and experimental
investigation in the future.
Among the IL-17 family, IL-17A, IL-17B, IL17C and
IL-17F were reported to have promoting effects on lung cancer
development. In the Oncomine database, our results revealed that
five datasets showed significantly decreased IL-17D expression in
lung cancer, and no dataset showed a significant difference in the
expression of IL-17A, IL-17B, IL-17C, IL-25 or IL17-F between lung
cancer and controls. In other lung cancer datasets, IL-17A, IL-17B,
IL-17C and IL-25 mRNA levels were upregulated, but the increase was
small. IL-17D mRNA levels were downregulated with a fold change
>2.0 in NSCLC. Analysis of the cBioPortal database revealed that
IL-17 family mRNA alteration frequencies may be associated with
specific histological types of NSCLC. IL-17A, IL-17B and IL-17C
mRNA upregulation frequencies were lower in lung squamous cell
carcinoma than in lung adenocarcinoma. IL-17D, IL-25 and IL-17F
mRNA upregulation rates were higher in lung squamous cell carcinoma
than in lung adenocarcinoma. However, the prognostic information
indicated that IL-17A and IL-17B showed no effect on the OS of
patients with lung cancer. High mRNA expression levels of IL-17C
and IL-25 were associated with poor OS in lung cancer patients;
conversely, high mRNA levels of IL-17D and IL-17F were correlated
with better OS.
Analysis of the interaction network and regulation
of IL-17 family genes revealed that the biological pathways of
IL-17D and IL-17A overlapped. Moreover, all IL-17 family genes,
except IL-17F, mainly participated in the ‘IFN-γ pathway’.
Th1-associated cytokines (IFN-γ) have been suggested to augment
anti-tumour responses by positively modulating cancer-directed
immune effectors, such as dendritic cells (DCs), T cells and NK
cells (61). This information
suggests that the relationship between IL-17 family genes and the
lung cancer immune microenvironment should be studied in-depth.
Based on our results, we can conclude that IL-17 family gene
mutation rates were in general low and that amplification and deep
deletion were the main mutation type. The expression and function
of IL-17A and IL-17B in lung cancer are still not fully elucidated
and require studies with larger sample sizes. The survival results
revealed that IL-17C, IL-25 and IL-17F have prognostic roles in
lung cancer. IL-17D was significantly decreased in lung cancer and
was correlated with better OS. The interaction network and
biological pathway analyses of the IL-17 family indicated that
IL-17 family genes share functional associations with each other.
These findings can provide a reference for further experimental
studies to identify the molecular mechanism of IL-17D in lung
cancer progression.
Nevertheless, some limitations exist in the present
study. First, the correlation between the IL-17 gene family and OS
within each subclass of clinical parameters, namely, pathologic N
stage, pathologic T stage, and pathologic M stage, was not analysed
due to data limitations. Second, the mutations and expression
levels of IL-17 genes in lung cancer cells need to be validated in
a future experimental study. In addition, future functional
investigations are required to explore the underlying mechanisms of
the IL-17 gene family in lung cancer development. In conclusion, in
the present study, we performed the first comprehensive
investigation of the IL-17 gene family in lung cancer, including
gene mutations, mRNA expression levels, prognostic values and
network pathway analysis. Although gene mutations and mRNA
expression levels were abnormal in lung cancer patients, IL-17
family gene mutation rates were low in general, and only IL-17D was
significantly decreased in lung cancer and was correlated with
better OS. The expression and function of IL-17A and IL-17B in lung
cancer are still not fully elucidated and require studies with
larger sample sizes. Studies of IL-17C-F in lung cancer are
limited. Therefore, more research attention should be given to the
association between IL-17D and lung cancer progression to identify
more effective therapeutic targets for lung cancer.
Not applicable.
The present work was supported by The Natural
Science Foundation of Hubei Province (grant no. 2014CFA057) and The
Health and Planning Commission Fund of Hubei Province (grant no.
WJ2017M098).
YJ and TTL conceived and designed the experiments.
TTL, JSF and ZLL prepared the figures and tables, and drafted and
revised the manuscript. JJX, FW, GHY, QH, GRH, MFG, MZ, LMD and SFW
prepared the figures and interpreted the data. All authors read and
approved the manuscript and agreed to be accountable for all
aspects of the research in ensuring that the accuracy or integrity
of any part of the work were appropriately investigated and
resolved.
Not applicable.
Not applicable.
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2018. CA Cancer J Clin. 68:7–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Allemani C, Matsuda T, Di Carlo V,
Harewood R, Matz M, Nikšić M, Bonaventure A, Valkov M, Johnson CJ,
Estève J, et al: Global surveillance of trends in cancer survival
2000–14 (CONCORD-3): Analysis of individual records for 37 513 025
patients diagnosed with one of 18 cancers from 322 population-based
registries in 71 countries. Lancet. 391:1023–1075. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wu F, Xu J, Huang Q, Han J, Duan L, Fan J,
Lv Z, Guo M, Hu G, Chen L, et al: The role of interleukin-17 in
lung cancer. Mediators Inflamm. 2016:84940792016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhang Q, Liu S, Ge D, Zhang Q, Xue Y,
Xiong Z, Abdel-Mageed AB, Myers L, Hill SM, Rowan BG, et al:
Interleukin-17 promotes formation and growth of prostate
adenocarcinoma in mouse models. Cancer Res. 72:2589–2599. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Akbay EA, Koyama S, Liu Y, Dries R, Bufe
LE, Silkes M, Alam MM, Magee DM, Jones R, Jinushi M, et al:
Interleukin-17A promotes lung tumor progression through neutrophil
attraction to tumor sites and mediating resistance to PD-1
blockade. J Thorac Oncol. 12:1268–1279. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Numasaki M, Fukushi J, Ono M, Narula SK,
Zavodny PJ, Kudo T, Robbins PD, Tahara H and Lotze MT:
Interleukin-17 promotes angiogenesis and tumor growth. Blood.
101:2620–2627. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang L, Yi T, Kortylewski M, Pardoll DM,
Zeng D and Yu H: IL-17 can promote tumor growth through an
IL-6-Stat3 signaling pathway. J Exp Med. 206:1457–1464. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kryczek I, Wei S, Szeliga W, Vatan L and
Zou W: Endogenous IL-17 contributes to reduced tumor growth and
metastasis. Blood. 114:357–359. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gu FM, Li QL, Gao Q, Jiang JH, Zhu K,
Huang XY, Pan JF, Yan J, Hu JH, Wang Z, et al: IL-17 induces
AKT-dependent IL-6/JAK2/STAT3 activation and tumor progression in
hepatocellular carcinoma. Mol Cancer. 10:1502011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chen JG, Xia JC, Liang XT, Pan K, Wang W,
Lv L, Zhao JJ, Wang QJ, Li YQ, Chen SP, et al: Intratumoral
expression of IL-17 and its prognostic role in gastric
adenocarcinoma patients. Int J Biol Sci. 7:53–60. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Huang CK, Yang CY, Jeng YM, Chen CL, Wu
HH, Chang YC, Ma C, Kuo WH, Chang KJ, Shew JY and Lee WH:
Autocrine/paracrine mechanism of interleukin-17B receptor promotes
breast tumorigenesis through NF-κB-mediated antiapoptotic pathway.
Oncogene. 33:2968–2977. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wu HH, Hwang-Verslues WW, Lee WH, Huang
CK, Wei PC, Chen CL, Shew JY, Lee EY, Jeng YM, Tien YW, et al:
Targeting IL-17B-IL-17RB signaling with an anti-IL-17RB antibody
blocks pancreatic cancer metastasis by silencing multiple
chemokines. J Exp Med. 212:333–349. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bie Q, Sun C, Gong A, Li C, Su Z, Zheng D,
Ji X, Wu Y, Guo Q, Wang S and Xu H: Non-tumor tissue derived
interleukin-17B activates IL-17RB/AKT/β-catenin pathway to enhance
the stemness of gastric cancer. Sci Rep. 6:254472016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Pfeifer P, Voss M, Wonnenberg B, Hellberg
J, Seiler F, Lepper PM, Bischoff M, Langer F, Schäfers HJ, Menger
MD, et al: IL-17C is a mediator of respiratory epithelial innate
immune response. Am J Respir Cell Mol Biol. 48:415–421. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Jungnickel C, Schmidt LH, Bittigkoffer L,
Wolf L, Wolf A, Ritzmann F, Kamyschnikow A, Herr C, Menger MD,
Spieker T, et al: IL-17C mediates the recruitment of
tumor-associated neutrophils and lung tumor growth. Oncogene.
36:4182–4190. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Song X, Gao H, Lin Y, Yao Y, Zhu S, Wang
J, Liu Y, Yao X, Meng G, Shen N, et al: Alterations in the
microbiota drive interleukin-17C production from intestinal
epithelial cells to promote tumorigenesis. Immunity. 40:140–152.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
O'Sullivan T, Saddawi-Konefka R, Gross E,
Tran M, Mayfield SP, Ikeda H and Bui JD: Interleukin-17D mediates
tumor rejection through recruitment of natural killer cells. Cell
Rep. 7:989–998. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yin SY, Jian FY, Chen YH, Chien SC, Hsieh
MC, Hsiao PW, Lee WH, Kuo YH and Yang NS: Induction of IL-25
secretion from tumour-associated fibroblasts suppresses mammary
tumour metastasis. Nat Commun. 7:113112016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Jiang Z, Chen J, Du X, Cheng H, Wang X and
Dong C: IL-25 blockade inhibits metastasis in breast cancer.
Protein Cell. 8:191–201. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Furuta S, Jeng YM, Zhou L, Huang L, Kuhn
I, Bissell MJ and Lee WH: IL-25 causes apoptosis of
IL-25R-expressing breast cancer cells without toxicity to
nonmalignant cells. Sci Transl Med. 3:78ra312011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Luo Y, Yang Z, Su L, Shan J, Xu H, Xu Y,
Liu L, Zhu W, Chen X, Liu C, et al: Non-CSCs nourish CSCs through
interleukin-17E-mediated activation of NF-κB and JAK/STAT3
signaling in human hepatocellular carcinoma. Cancer Lett.
375:390–399. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liu Y, Zhao X, Sun X, Li Y, Wang Z, Jiang
J, Han H, Shen W, Corrigan CJ and Sun Y: Expression of IL-17A, E,
and F and their receptors in human prostatic cancer: Comparison
with benign prostatic hyperplasia. Prostate. 75:1844–1856. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Xie Y, Sheng W, Xiang J, Ye Z and Yang J:
Interleukin-17F suppresses hepatocarcinoma cell growth via
inhibition of tumor angiogenesis. Cancer Invest. 28:598–607. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ding L, Hu EL, Xu YJ, Huang XF, Zhang DY,
Li B, Hu QG, Ni YH and Hou YY: Serum IL-17F combined with VEGF as
potential diagnostic biomarkers for oral squamous cell carcinoma.
Tumour Biol. 36:2523–2529. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Forbes SA, Beare D, Gunasekaran P, Leung
K, Bindal N, Boutselakis H, Ding M, Bamford S, Cole C, Ward S, et
al: COSMIC: Exploring the world's knowledge of somatic mutations in
human cancer. Nucleic Acids Res 43 (Database Issue). D805–D811.
2015. View Article : Google Scholar
|
|
26
|
Rhodes DR, Kalyana-Sundaram S, Mahavisno
V, Varambally R, Yu J, Briggs BB, Anstet MJ, Kincead-Beal C,
Kulkarni P, et al: Oncomine 3.0: Genes, pathways, and networks in a
collection of 18,000 cancer gene expression profiles. Neoplasia.
9:166–180. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Győrffy B, Surowiak P, Budczies J and
Lánczky A: Online survival analysis software to assess the
prognostic value of biomarkers using transcriptomic data in
non-small-cell lung cancer. PLoS One. 8:e822412013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yu T, Jia W, An Q, Cao X and Xiao G:
Bioinformatic analysis of GLI1 and related signaling pathways in
chemosensitivity of gastric cancer. Med Sci Monit. 24:1847–1855.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Cerami E, Gao J, Dogrusoz U, Gross BE,
Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et
al: The cBio cancer genomics portal: An open platform for exploring
multidimensional cancer genomics data. Cancer Discov. 2:401–404.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Szklarczyk D, Franceschini A, Kuhn M,
Simonovic M, Roth A, Minguez P, Doerks T, Stark M, Muller J, Bork
P, et al: The STRING database in 2011: Functional interaction
networks of proteins, globally integrated and scored. Nucleic Acids
Res 39 (Database Issue). D561–D568. 2011. View Article : Google Scholar
|
|
31
|
Pathan M, Keerthikumar S, Ang CS, Gangoda
L, Quek CY, Williamson NA, Mouradov D, Sieber OM, Simpson RJ, Salim
A, et al: FunRich: An open access standalone functional enrichment
and interaction network analysis tool. Proteomics. 15:2597–2601.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Cancer Genome Atlas Research Network, .
Comprehensive molecular profiling of lung adenocarcinoma. Nature.
511:543–550. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Beltran H, Prandi D, Mosquera JM, Benelli
M, Puca L, Cyrta J, Marotz C, Giannopoulou E, Chakravarthi BV,
Varambally S, et al: Divergent clonal evolution of
castration-resistant neuroendocrine prostate cancer. Nat Med.
22:298–305. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Eirew P, Steif A, Khattra J, Ha G, Yap D,
Farahani H, Gelmon K, Chia S, Mar C, Wan A, et al: Dynamics of
genomic clones in breast cancer patient xenografts at single-cell
resolution. Nature. 518:422–426. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Hou J, Aerts J, den Hamer B, van Ijcken W,
den Bakker M, Riegman P, van der Leest C, van der Spek P, Foekens
JA, Hoogsteden HC, et al: Gene expression-based classification of
non-small cell lung carcinomas and survival prediction. PLoS One.
5:e103122010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Beer DG, Kardia SL, Huang CC, Giordano TJ,
Levin AM, Misek DE, Lin L, Chen G, Gharib TG, Thomas DG, et al:
Gene-expression profiles predict survival of patients with lung
adenocarcinoma. Nat Med. 8:816–824. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Garber ME, Troyanskaya OG, Schluens K,
Petersen S, Thaesler Z, Pacyna-Gengelbach M, van de Rijn M, Rosen
GD, Perou CM, Whyte RI, et al: Diversity of gene expression in
adenocarcinoma of the lung. Proc Natl Acad Sci USA. 98:13784–13789.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Selamat SA, Chung BS, Girard L, Zhang W,
Zhang Y, Campan M, et al: Genome-scale analysis of DNA methylation
in lung adenocarcinoma and integration with mRNA expression. Genome
Res 22 (7). 1197–1211. 2012. View Article : Google Scholar
|
|
39
|
Landi MT, Dracheva T, Rotunno M, Figueroa
JD, Liu H, Dasgupta A, Mann FE, Fukuoka J, Hames M, Bergen AW, et
al: Gene expression signature of cigarette smoking and its role in
lung adenocarcinoma development and survival. PLoS One.
3:e16512008. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Okayama H, Kohno T, Ishii Y, Shimada Y,
Shiraishi K, Iwakawa R, Furuta K, Tsuta K, Shibata T, Yamamoto S,
et al: Identification of genes upregulated in ALK-positive and
EGFR/KRAS/ALK-negative lung adenocarcinomas. Cancer Res.
72:100–111. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Liu J, Lichtenberg T, Hoadley KA, Poisson
LM, Lazar AJ, Cherniack AD, Kovatich AJ, Benz CC, Levine DA, Lee
AV, et al: An integrated TCGA pan-cancer clinical data resource to
drive high-quality survival outcome analytics. Cell.
173:400–416.e11. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Gaffen SL: Recent advances in the IL-17
cytokine family. Curr Opin Immunol. 23:613–619. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Chen X, Wan J, Liu J, Xie W, Diao X, Xu J,
Zhu B and Chen Z: Increased IL-17-producing cells correlate with
poor survival and lymphangiogenesis in NSCLC patients. Lung Cancer.
69:348–354. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Liu J, Duan Y, Cheng X, Chen X, Xie W,
Long H, Lin Z and Zhu B: IL-17 is associated with poor prognosis
and promotes angiogenesis via stimulating VEGF production of cancer
cells in colorectal carcinoma. Biochem Biophys Res Commun.
407:348–354. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Lin Q, Xue L, Tian T, Zhang B, Guo L, Lin
G, Chen Z, Fan K and Gu X: Prognostic value of serum IL-17 and VEGF
levels in small cell lung cancer. Int J Biol Markers. 30:e359–e363.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Pan B, Che D, Cao J, Shen J, Jin S, Zhou
Y, Liu F, Gu K, Man Y, Shang L and Yu Y: Interleukin-17 levels
correlate with poor prognosis and vascular endothelial growth
factor concentration in the serum of patients with non-small cell
lung cancer. Biomarkers. 20:232–239. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Wang XF, Zhu YT, Wang JJ, Zeng DX, Mu CY,
Chen YB, Lei W, Zhu YH and Huang JA: The prognostic value of
interleukin-17 in lung cancer: A systematic review with
meta-analysis based on Chinese patients. PLoS One. 12:e01851682017.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Wei L, Wang H, Yang F, Ding Q and Zhao J:
Interleukin-17 potently increases non-small cell lung cancer
growth. Mol Med Rep. 13:1673–1680. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Pan B, Shen J, Cao J, Zhou Y, Shang L, Jin
S, Cao S, Che D, Liu F and Yu Y: Interleukin-17 promotes
angiogenesis by stimulating VEGF production of cancer cells via the
STAT3/GIV signaling pathway in non-small-cell lung cancer. Sci Rep.
5:160532015. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Huang Q, Duan L, Qian X, Fan J, Lv Z,
Zhang X, Han J, Wu F, Guo M, Hu G, et al: IL-17 promotes angiogenic
factors IL-6, IL-8, and Vegf production via Stat1 in lung
adenocarcinoma. Sci Rep. 6:365512016. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Li Q, Han Y, Fei G, Guo Z, Ren T and Liu
Z: IL-17 promoted metastasis of non-small-cell lung cancer cells.
Immunol Lett. 148:144–150. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Lu L, Pan K, Zheng HX, Li JJ, Qiu HJ, Zhao
JJ, Weng DS, Pan QZ, Wang DD, Jiang SS, et al: IL-17A promotes
immune cell recruitment in human esophageal cancers and the
infiltrating dendritic cells represent a positive prognostic marker
for patient survival. J Immunother. 36:451–458. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Punt S, van Vliet ME, Spaans VM, de Kroon
CD, Fleuren GJ, Gorter A and Jordanova ES: FoxP3(+) and IL-17(+)
cells are correlated with improved prognosis in cervical
adenocarcinoma. Cancer Immunol Immunother. 64:745–753. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
He Y, Du Y, Wei S, Shi J, Mei Z, Qian L,
Chen Z and Jie Z: IL-17A and IL-17F single nucleotide polymorphisms
associated with lung cancer in Chinese population. Clin Respir J.
11:230–242. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Ma QY, Chen J, Wang SH, Wu N, Hao ZH and
Chen XF: Interleukin 17A genetic variations and susceptibility to
non-small cell lung cancer. APMIS. 123:194–198. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Kaabachi W, ben Amor A, Kaabachi S,
Rafrafi A, Tizaoui K and Hamzaoui K: Interleukin-17A and −17F genes
polymorphisms in lung cancer. Cytokine. 66:23–29. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Li Z, Liu Y, Cao D, Jiang M and Luo F:
IL-17A and IL-17F polymorphisms and gastric cancer risk: A
meta-analysis. Genet Mol Res. 14:7008–7017. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Wróbel T, Gębura K, Wysoczańska B, Jaźwiec
B, Dobrzyńska O, Mazur G, Kuliczkowski K and Bogunia-Kubik K:
IL-17F gene polymorphism is associated with susceptibility to acute
myeloid leukemia. J Cancer Res Clin Oncol. 140:1551–1555. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Omrane I, Baroudi O, Bougatef K, Mezlini
A, Abidi A, Medimegh I, Stambouli N, Ayari H, Kourda N, Uhrhammer
N, et al: Significant association between IL23R and IL17F
polymorphisms and clinical features of colorectal cancer. Immunol
Lett. 158:189–194. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Cheng S, Shao Z, Liu X, Guo L, Zhang X, Na
Q, Chen X, Ma Y, Zheng J, Song B and Liu J: Interleukin 17A
polymorphism elevates gene expression and is associated with
increased risk of nonsmall cell lung cancer. DNA Cell Biol.
34:63–68. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Rožman P and Švajger U: The tolerogenic
role of IFN-γ. Cytokine Growth Factor Rev. 41:40–53. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Xu C, Hao K, Yu L and Zhang X: Serum
interleukin-17 as a diagnostic and prognostic marker for non-small
cell lung cancer. Biomarkers. 19:287–290. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Xu B, Guenther JF, Pociask DA, Wang Y,
Kolls JK, You Z, Chandrasekar B, Shan B, Sullivan DE and Morris GF:
Promotion of lung tumor growth by interleukin-17. Am J Physiol Lung
Cell Mol Physiol. 307:L497–L508. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Li Y, Cao ZY, Sun B, Wang GY, Fu Z, Liu
YM, Kong QF, Wang JH, Zhang Y, Xu XY and Li HL: Effects of IL-17A
on the occurrence of lung adenocarcinoma. Cancer Biol Ther.
12:610–616. 2014. View Article : Google Scholar
|
|
65
|
Numasaki M, Watanabe M, Suzuki T,
Takahashi H, Nakamura A, McAllister F, Hishinuma T, Goto J, Lotze
MT, Kolls JK and Sasaki H: IL-17 enhances the net angiogenic
activity and in vivo growth of human non-small cell lung cancer in
SCID mice through promoting CXCR-2-dependent angiogenesis. J
Immunol. 175:6177–6189. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Zhao C, Li Y, Zhang W, Zhao D, Ma L, Ma P,
Yang F, Wang Y, Shu Y and Qiu W: IL17 induces NSCLC A549 cell
proliferation via the upregulation of HMGA1, resulting in an
increased cyclin D1 expression. Int J Oncol. Mar 7–2018.DOI:
10.3892/ijo.2018.4307.
|
|
67
|
Chen X, Xie Q, Cheng X, Diao X, Cheng Y,
Liu J, Xie W, Chen Z and Zhu B: Role of interleukin-17 in
lymphangiogenesis in non-small-cell lung cancer: Enhanced
production of vascular endothelial growth factor C in
non-small-cell lung carcinoma cells. Cancer Sci. 101:2384–2390.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
You R, DeMayo FJ, Liu J, Cho SN, Burt BM,
Creighton CJ, Casal RF, Lazarus DR, Lu W, Tung HY, et al: IL17A
regulates tumor latency and metastasis in lung adeno and squamous
SQ.2b and AD.1 cancer. Cancer Immunol Res. 6:645–657. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Gu K, Li MM, Shen J, Liu F, Cao JY, Jin S
and Yu Y: Interleukin-17-induced EMT promotes lung cancer cell
migration and invasion via NF-κB/ZEB1 signal pathway. Am J Cancer
Res. 5:1169–1179. 2015.PubMed/NCBI
|
|
70
|
Kulig P, Burkhard S, Mikita-Geoffroy J,
Croxford AL, Hövelmeyer N, Gyülvészi G, Gorzelanny C, Waisman A,
Borsig L and Becher B: IL17A-mediated endothelial breach promotes
metastasis formation. Cancer Immunol Res. 4:26–32. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Chang SH, Mirabolfathinejad SG, Katta H,
Cumpian AM, Gong L, Caetano MS, Moghaddam SJ and Dong C: T helper
17 cells play a critical pathogenic role in lung cancer. Proc Natl
Acad Sci USA. 111:5664–5669. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Liu L, Ge D, Ma L, Mei J, Liu S, Zhang Q,
Ren F, Liao H, Pu Q, Wang T and You Z: Interleukin-17 and
prostaglandin E2 are involved in formation of an M2
macrophage-dominant microenvironment in lung cancer. J Thorac
Oncol. 7:1091–1100. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Yang YF, Lee YC, Lo S, Chung YN, Hsieh YC,
Chiu WC and Yuan SF: A positive feedback loop of IL-17B-IL-17RB
activates ERK/β-catenin to promote lung cancer metastasis. Cancer
Lett. 422:44–55. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Liao C, Yu ZB, Meng G, Wang L, Liu QY,
Chen LT, Feng SS, Tu HB, Li YF and Bai L: Association between
Th17-related cytokines and risk of non-small cell lung cancer among
patients with or without chronic obstructive pulmonary disease.
Cancer. 121 (Suppl 17):S3122–S3129. 2015. View Article : Google Scholar
|