Introduction
Atrial fibrillation (AF) is one of the most common
types of arrhythmia globally, affecting ~33 million individuals
worldwide. The incidence of AF increases with age (1–5). AF
induces palpitation, heart failure and thrombus formation (6,7),
which may impair quality of life (8) and increase the risk of mortality. In
fact, one study revealed that at 10 years of follow-up, 61.5% of
men with AF had died compared with 30% of men without AF, whereas
in women, 57.6% of those with AF had died compared with 20.9% women
without AF, which is an approximate doubling of the mortality rate
in both sexes (9,10). Since the etiology of AF is complex,
it is important to determine the underlying pathogenic
mechanisms.
Numerous studies have demonstrated that AF is
associated with unbalanced energy supply and consumption (11–13);
the downregulation of mitochondrial electron transport chain
activity has been found in AF (14,15).
Therefore, alterations in energy metabolism, particularly
mitochondrial dysfunction, may contribute to the pathogenesis of
AF. It has been reported that the development of AF is commonly
accompanied by alterations in gene expression levels, such as
potassium voltage-gated channel subfamily Q member 1, potassium
inwardly rectifying channel subfamily J member 3, collagen type XV
α1 chain and matrix metalloproteinase, thus resulting in abnormal
protein expression levels (16,17).
Mitochondrial transcription factor A (TFAM) is essential for the
maintenance of mitochondrial DNA (mtDNA) and regulates mtDNA
transcription (18). A number of
reports have demonstrated that TFAM dysfunction leads to
mitochondrial impairment, which causes cardiovascular diseases,
such as heart failure and cardiomyopathy (19–21);
however, its role in AF is unknown. The aim of the present study
was to investigate the association between TFAM and AF and the
effect of TFAM on ATP content in cardiomyocytes.
Materials and methods
Ethics statement
The present study was approved by the Ethics
Committee of the First Affiliated Hospital of China Medical
University (Shenyang, China; approval no. 2017-69-2) and performed
in accordance with Declaration of Helsinki (22). Written informed consent was
obtained from all patients prior to tissue collection.
Specimen collection
Between June 2017 and May 2019, left atrial
appendage (LAA) tissues were collected from 20 patients with AF who
underwent mitral valve repair and maze IV procedure, and from 20
patients with sinus rhythm (SR) and left atrial thrombus who
underwent mitral valve repair, LAA resection and thrombectomy. The
samples were divided into two parts: One part was quickly frozen in
liquid nitrogen and then stored at −80°C; the other part was fixed
with 10% formalin at 4°C for 48 h for further use. AF was diagnosed
using a 12-lead electrocardiogram and 72 h-holter.
Primary culture of rat atrial
cardiomyocytes
A total of 60 male Sprague Dawley neonatal rats
(age, 1 day old; weight, 5.5±0.8 g) were procured from Liaoning
Changsheng Biotechnology Co., Ltd. The animal protocol was approved
by the Ethics Review Committee for Animal Experimentation of China
Medical University. All animal procedures were performed in
accordance with the Guide for the Care and Use of Laboratory
Animals published by the National Institutes of Health.
Neonatal rats were sterilized and sacrificed by
cervical dislocation. The whole hearts were excised from the body
and atria were isolated from the ventricles. Cardiomyocytes were
dissociated with 0.08% trypsin and 0.1% type II collagenase. The
cells were resuspended in DMEM (Invitrogen; Thermo Fisher
Scientific, Inc.) supplemented with 10% fetal bovine serum
(HyClone; Cytiva) and cultured in 6-well plate at a density of
1×106 cells/ml, then 5-BrdU was added at a final
concentration of 0.1 mmol/l. The medium was replenished after 24 h
and thereafter every 48 h for up to 2 weeks. Beating myocardial
cells were observed with an inverted light microscope (DFC295;
Leica Microsystems GmbH), and video graphs of beating cells were
recorded with a digital camera (Coolpix5400; Nikon Corporation).
Detailed procedures are presented in Data S1.
Tachypacing of cardiomyocytes
The spontaneous firing rate of primary cultured
cardiomyocytes was ~1 Hz, which was measured by counting the
beating frequency of cardiomyocytes with a light microscope
(magnification, ×200). The cardiomyocytes were cultured in 6-well
plates and subjected to electrical field stimulation using a YC-2
stimulator (Cheng Yi). The cardiomyocytes were stimulated at 6 Hz
for 24 h as previously described (23,24)
using the following parameters: 1.5 V/cm field strength,
square-wave, 5-ms pulses. Non-pacing cells were used as a
control.
Cell transfection
The TFAM overexpression plasmid pEXP-RB-Mam-TFAM,
pEXP-RB-Mam-negative control (NC), three small interfering (si)RNAs
for the inhibition of the expression levels of TFAM and siRNA-NC
(cat. no. siN0000001-1-5) were commercially constructed by
Guangzhou RiboBio Co., Ltd. The siRNA sequences were as follows:
siRNA 1, forward 5′-GGAAGAGCAAAUGGCUGAA-3′, reverse
5′-CCUUCUCGUUUACCGACUU-3′; siRNA 2, forward
5′-GGCAGAAACGCCUAAAGAA-3′, reverse 5′-CCGUCUUUGCGGAUUUCUU-3′; and
siRNA 3, forward 5′-CCUGUCAGCCUUAUCUGTA-3′, reverse
5′-GGACAGUCGGAAUAGACAU-3′.
Cells were seeded in a 6-well plate at a density of
1×105 cells/well and transfected (2,500 ng
pEXP-RB-Mam-TFAM and pEXP-RB-Mam-NC or 50 nM siRNA) with
Lipofectamine® 3000 (Invitrogen; Thermo Fisher
Scientific, Inc.) and serum-free Opti-MEM (Gibco; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol. After 6
h transfection, the medium was replaced with DMEM containing 10%
FBS. After 48 h, cells transfected with pEXP-RB-Mam-TFAM were used
for tachypacing, and cells transfected with siRNAs were cultured in
non-paced conditions.
Cell viability assay
Cell viability was assessed by Cell Counting Kit 8
(CCK-8) assay (Vazyme Biotech Co., Ltd.), according to the
manufacturer's protocol. Cells were seeded into a 96-well plate at
a density of 1×104 cells/well in a final volume of 100
µl. Cells were cultured in 5% CO2 at 37°C for 12 h and
then stimulated at 6 Hz for 8, 16 and 24 h. A total of 10 µl CCK-8
solution was added to each well and cultured for an additional 2 h
at 37°C. Absorbance was measured at 450 nm using an Infinite F200
PRO multimode reader (Tecan Group, Ltd.) and the viability was
calculated using the following equation: Absorbance8/16/24
h/Absorbance0 h. Each experiment was repeated at
least three times.
Measurement of ATP content
ATP content was assessed using an ATP Assay kit
(Beyotime Institute of Biotechnology) according to the
manufacturer's instructions. Luciferase/luciferin chemiluminescence
method was used for measurement (25,26).
Cells were homogenized in lysis buffer (provided in kit) on ice.
Lysates were centrifuged at 12,000 × g for 10 min at 4°C. The
supernatants were collected and used to test protein concentration
and ATP content, separately. Proteins were quantified using a
bicinchoninic acid protein assay kit (Beyotime Institute of
Biotechnology). Each experiment was repeated at least three
times.
Reverse transcription-quantitative
(RT-q)PCR
Total RNA was isolated from LAA tissues and primary
cultured cardiomyocytes using TRIzol® reagent
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. Total RNA was reverse transcribed into
cDNA using the PrimeScript™ RT reagent kit (Takara Bio, Inc.) at
37°C for 15 min, 85°C for 5 sec and 4°C for 1 min. RT-qPCR was
performed using a TB Green™ Premix Ex II reagent kit (Takara Bio,
Inc.). β-actin was used as the internal control. The data were
analyzed using a LightCycler® 480 Real-Time PCR System
(Roche Diagnostics) and the relative changes in mRNA expression
levels were calculated using the 2−ΔΔCq method (27). The following thermocycling
conditions were used for the qPCR: Initial denaturation at 95°C for
30 sec; followed by 50 cycles of 95°C for 5 sec and 60°C for 30
sec; 1 cycle of 95°C for 5 sec and 60°C for 1 min; and a final
cycle at 50°C for 30 sec. The primers were as follows: Hsa-TFAM,
forward 5′-TTCCAAGAAGCTAAGGGTGATT-3′, reverse
5′-AGAAGATCCTTTCGTCCAACTT-3′; hsa-β-actin, forward
5′-CCTGGCACCCAGCACAAT-3′, reverse 5′-GGGCCGGACTCGTCATAC-3′;
rno-TFAM, forward 5′-GTGATCTCATCCGTCGCAGTGTG-3′, reverse
5′-TGCCCAATCCCAATGACAACTCTG-3′; and rno-β-actin, forward,
5′-TGTCACCAACTGGGACGATA-3′, reverse 5′-GGGGTGTTGAAGGTCTCAAA-3′.
Western blotting
Western blotting analysis was performed as
previously described (28).
Tissues or cells were homogenized on ice in RIPA lysis buffer
supplemented with phenylmethylsulfonyl fluoride (Beijing Solarbio
Science & Technology Co. Ltd.). Lysates were centrifuged at
12,000 × g for 10 min at 4°C. The supernatants were collected, and
protein concentrations were quantified using an enhanced BCA
protein assay kit (Beyotime Institute of Biotechnology. Proteins
(40 µg/lane) were separated by 10% SDS-PAGE and transferred onto
polyvinylidene fluoride membranes by electroblotting. The membranes
were blocked in 5% skim milk dissolved in TBS-0.1% Tween-20 for 2 h
at room temperature. Membranes were incubated at 4°C overnight with
primary antibodies against TFAM (cat. no. ab131607; Abcam;
1:2,000), mitochondrially-encoded (MT)-NADH dehydrogenase 1 (ND1)
(cat. no. ab181848; Abcam; 1:10,000), MT-cytochrome c
oxidase 1 (CO1) (cat. no. ab203912; Abcam; 1:1,000), NADH
ubiquinone oxidoreductase core subunit 1 (NDUFS1; cat. no.
ab169540; Abcam; 1:20,000) and cytochrome c oxidase subunit
6C (COX6C) (cat. no. ab150422; Abcam; 1:5,000). β-actin
(ProteinTech Group, Inc.; 1:10,000) was used as a loading control
and for normalization. Membranes were subsequently incubated with
horseradish peroxidase-conjugated Affinipure goat anti-rabbit
secondary antibodies (ProteinTech Group, Inc.; cat. no. SA00001-2;
1:10,000) at room temperature for 2 h. Protein bands were
visualized by exposure to ECL buffer (Beyotime Institute of
Biotechnology) and the signals were captured by MicroChemi system
(DNR Bio-Imaging Systems, Ltd.). The expression levels were
analyzed using ImageJ software (version 1.52a, National Institutes
of Health).
Immunohistochemistry and
immunocytochemistry
Tissues were fixed with 10% formalin at 4°C for 48 h
and then dehydrated with an increasing series of alcohol at room
temperature, made transparent with dimethylbenzene at room
temperature and paraffin embedded. Sections were cut to a thickness
of 5-µm and blocked with 5% goat serum (Fuzhou Maixin Biotech Co.,
Ltd.) for 10 min at room temperature. Meanwhile, the cells were
fixed with 4% paraformaldehyde at 4°C for 15 min, permeated with
0.5% Triton X-100 and blocked with 5% goat serum for 30 min at room
temperature. UltraSensitive™ S-P kit (Fuzhou Maixin Biotech Co.,
Ltd.) was used for immunohistochemical staining. Slides were
incubated with the primary antibody to TFAM (cat. no. ab176558;
Abcam; 1:300) at 4°C overnight. Cells (106 cells/ml)
were incubated with primary antibody against α-sarcomeric actin
(cat. no. BM0001; Wuhan Boster Biological Technology, Ltd.; 1:500)
at 4°C overnight. PBS without primary antibodies was used as a
negative control. Following the primary antibody incubation, the
membranes were incubated with biotin-labeled goat anti-mouse/rabbit
IgG secondary antibodies (solution C in S-P kit) for 10 min at room
temperature, followed by a further incubation with
anti-biotin-peroxidase complex (solution D in S-P kit) for 10 min
at room temperature. 3,3-diaminobenzidine was used as a chromogen
(Fuzhou Maixin Biotech Co., Ltd.). Subsequently, the slides or
cells were counterstained with 4% hematoxylin for 2 min at room
temperature (Beijing Solarbio Science & Technology Co., Ltd.),
blued in 1% ammonium hydroxide for 3 min at room temperature,
dehydrated and mounted. All sections were observed, and images were
captured with an Olympus BX51 light microscope (magnification,
×400; Olympus Corporation). Average optical density (AOD) of TFAM
protein was calculated by ImageJ software (version 1.52a, National
Institutes of Health).
Statistical analysis
Data analysis was performed using SPSS software
(version 22.0; IBM Corp.). All continuous data are presented as the
mean ± standard deviation of at least three independent repeats.
Unpaired Student's t-test was used for two-group comparisons. ANOVA
was used for comparison of multiple groups followed by post hoc
Bonferroni's correction. P<0.05 was considered to indicate a
statistically significant difference.
Results
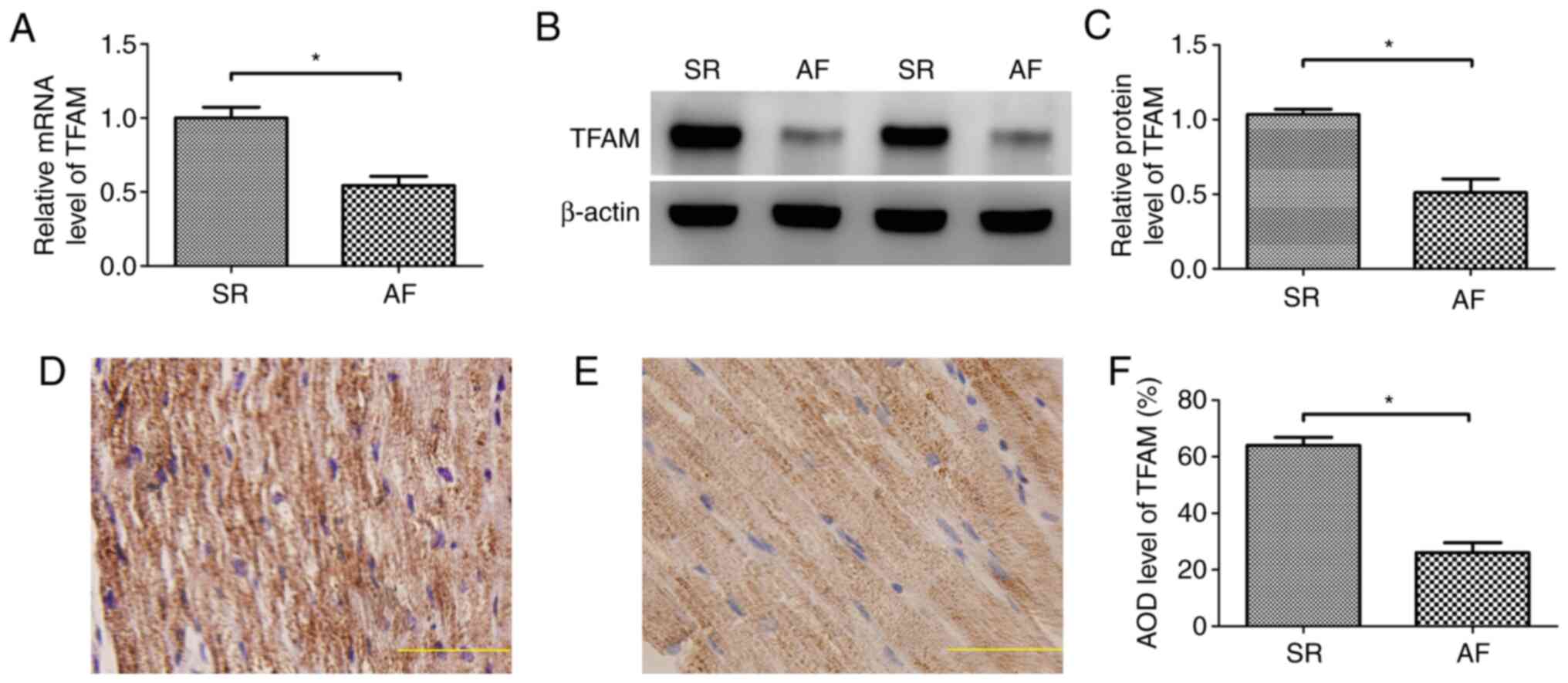
Expression levels of TFAM in SR and AF
tissues
RT-qPCR results indicated that the mRNA expression
levels of TFAM were decreased in AF compared with SR tissue
(P<0.05; Fig. 1A). Western
blotting demonstrated that the protein expression levels of TFAM
were also decreased in AF compared with SR tissue (P<0.05)
(Fig. 1B and C).
Immunohistochemical staining indicated that the TFAM protein was
expressed in the cytoplasm, and the AOD of TFAM protein in SR
tissue was 63.96±2.89%, whereas that in AF tissue was 26.04±3.52%
(P<0.05; Fig. 1D-F).
Identification of cardiomyocytes
The identification of cardiomyocytes was performed
by observation of beating cells and immunocytochemistry. The
myocardial cells observed under an inverted microscope are shown in
Fig. 2A, and video graphs of
beating cells are presented in Video
S1. Immunocytochemical staining of cells showed the presence of
myocardial cell-specific protein α-sarcomeric actin (Fig. 2B), which demonstrated that the
primary cultured cells were cardiomyocytes.
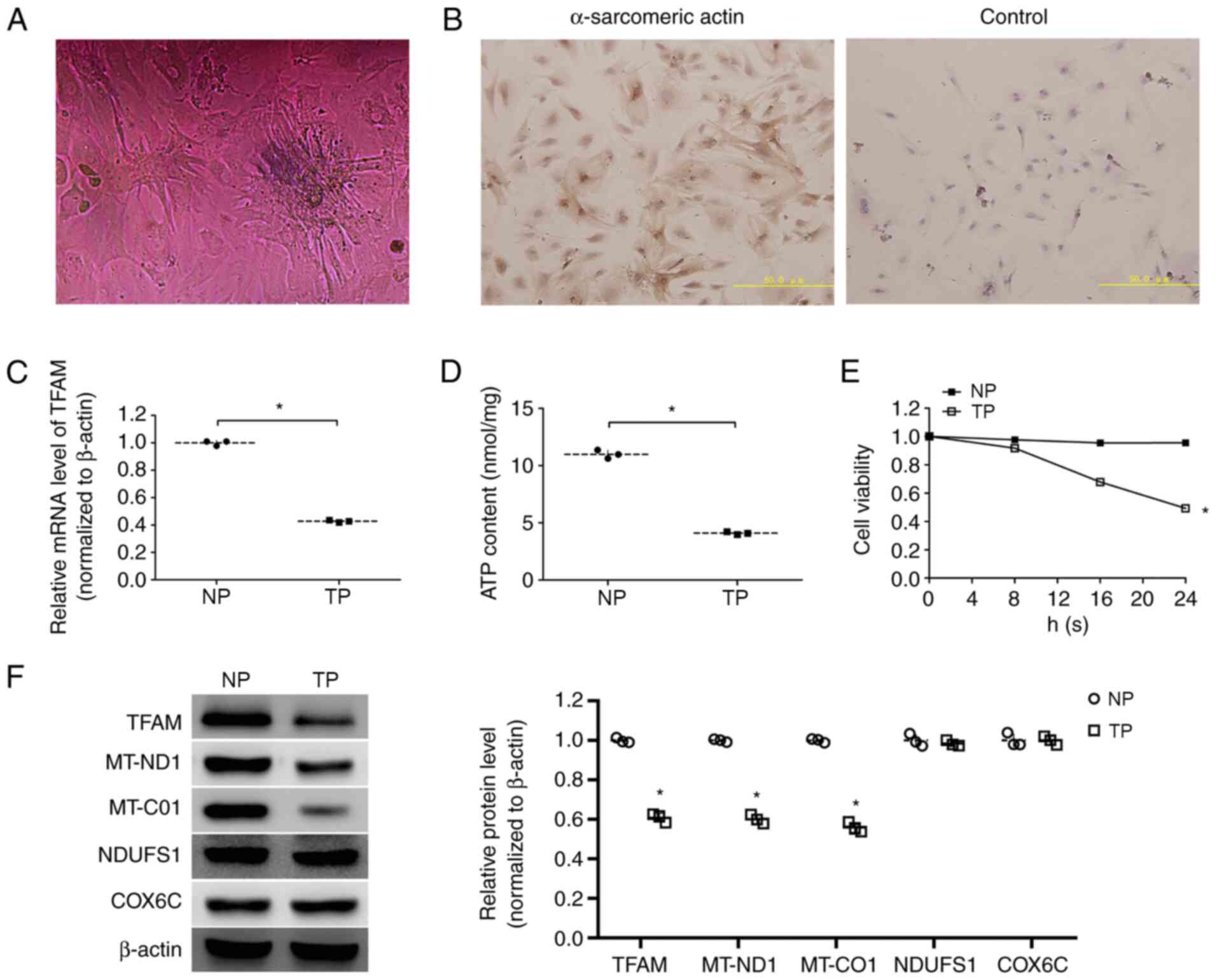 | Figure 2.Differential expression of mRNA,
proteins, ATP content and cell viability in NP and TP
cardiomyocytes. (A) Photomicrograph of primary cultured
cardiomyocytes; magnification, ×200. (B) Positive staining with
anti-α-sarcomeric actin antibody and negative control;
magnification, ×200; scale bar, 50 µm. (C) mRNA expression levels
of TFAM in NP and TP cardiomyocytes. (D) ATP content in NP and TP
cardiomyocytes. (E) Viability of primary cultured cardiomyocytes in
NP and TP groups. (F) Representative western blotting images and
semi-quantitative analysis of protein expression levels of TFAM,
MT-ND1, MT-CO1, NDUFS1 and COX6C in NP and TP cardiomyocytes. Data
are presented as the mean ± standard deviation. *P<0.05 vs. NP
cardiomyocytes. COX6C, cytochrome c oxidase subunit 6C; MT-CO1,
mitochondrially encoded-cytochrome c oxidase 1; MT-ND1,
mitochondrially encoded-NADH dehydrogenase 1; NDUFS1, NADH
ubiquinone oxidoreductase core subunit 1; NP, non-pacing; TFAM,
mitochondrial transcription factor A; TP, tachypacing. |
Expression levels of TFAM, ATP content
and cell viability of tachypacing cardiomyocytes
RT-qPCR results indicated that the mRNA expression
levels of TFAM were decreased in tachypacing cardiomyocytes
compared with non-pacing cardiomyocytes (P<0.05; Fig. 2C). Western blotting showed that the
protein expression levels of TFAM were also decreased in
tachypacing cardiomyocytes compared with non-pacing cardiomyocytes
(P<0.05) (Fig. 2F).
To explore the energy status, ATP content in
cardiomyocytes was evaluated. The results indicated that ATP
content was significantly lower in tachypacing cardiomyocytes
(4.09±0.13 nmol/mg) compared with that in non-pacing cardiomyocytes
(10.98±0.37 nmol/mg) (P<0.05; Fig.
2D).
The viability of cells in the non-pacing and the
tachypacing groups is presented in Fig. 2E. By measuring the viability at
different times, it was demonstrated that the cell viability in the
non-pacing group remained stable over 24 h; however, that in the
tachypacing group decreased gradually. There was a significant
difference between two groups following 24 h tachypacing
(P<0.05).
Expression levels of subunits of
oxidative respiratory chain complexes in tachypacing
cardiomyocytes
The protein expression levels of MT-ND1, MT-CO1,
NDUFS1 and COX6C were measured by western blotting, which
demonstrated that the protein expression levels of MT-ND1 and
MT-CO1 were lower in tachypacing cardiomyocytes compared with
expression in the non-pacing cardiomyocytes (P<0.05), whereas
there was no difference in the protein expression levels of NDUFS1
and COX6C between the two groups (Fig.
2F).
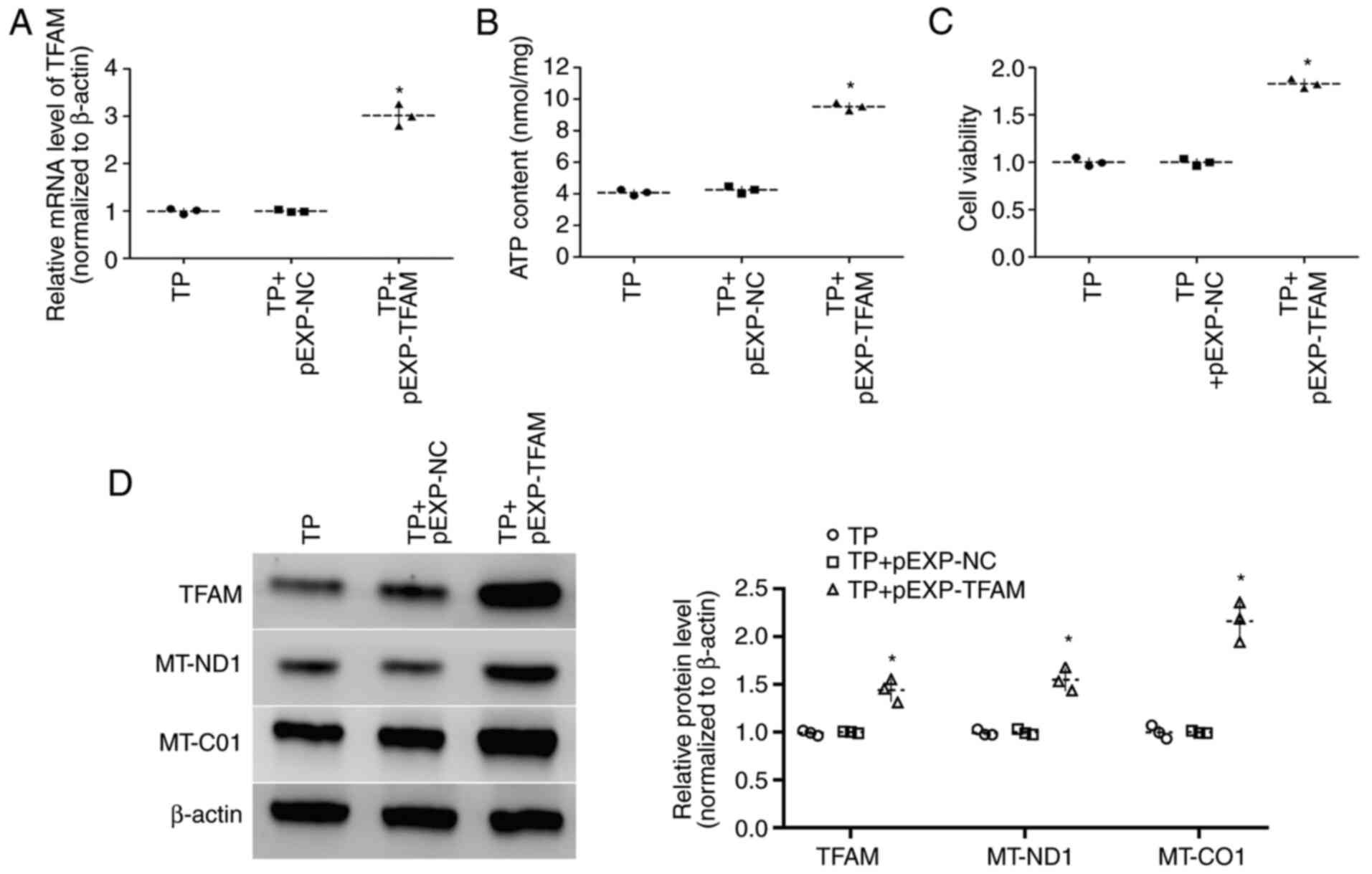
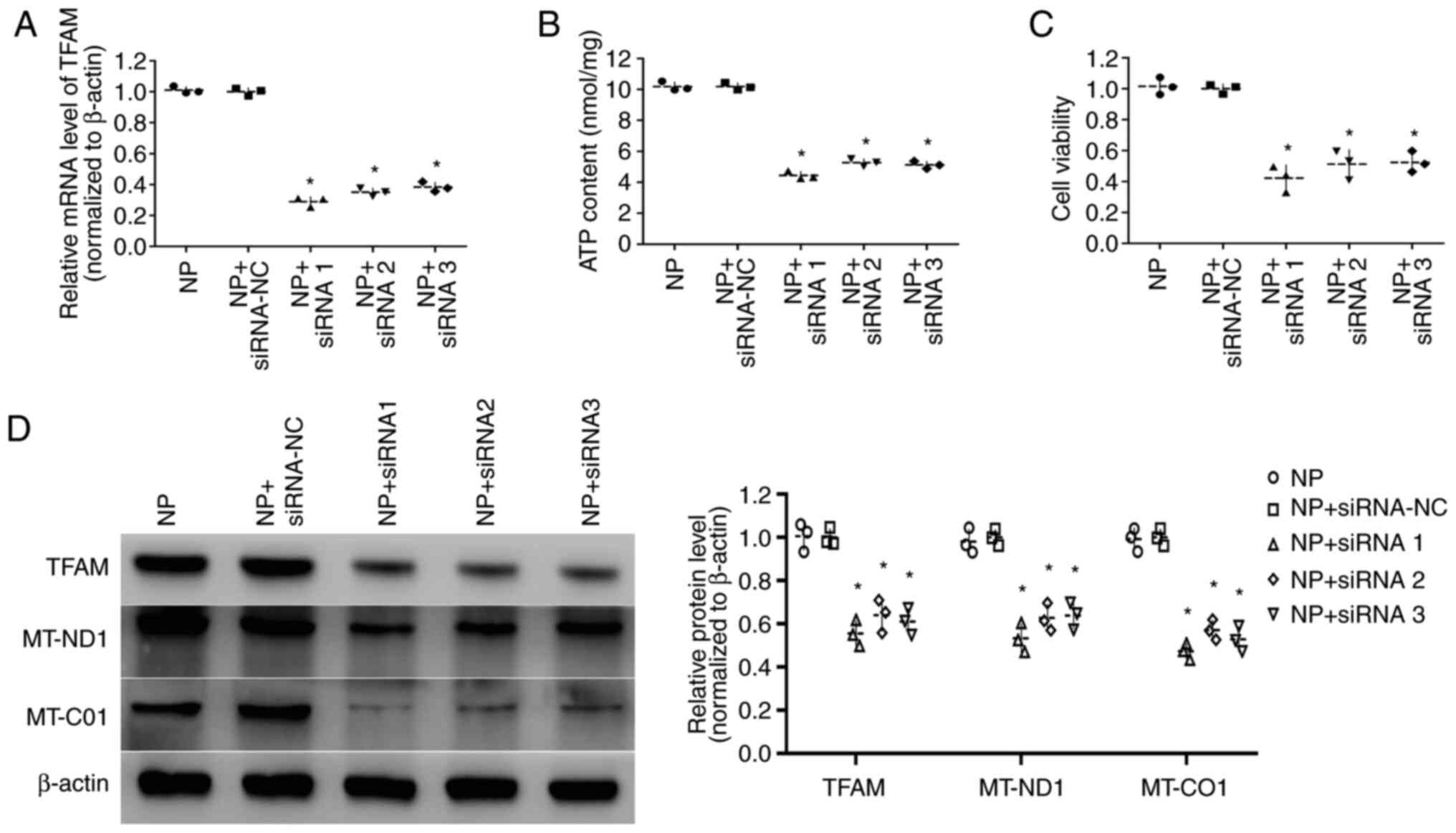
Effect of TFAM on ATP content and
viability in cardiomyocytes
To investigate the effect of TFAM on ATP content and
viability of cardiomyocytes, cells that were transfected with
pEXP-RB-Mam-TFAM were used for tachypacing and cells transfected
with TFAM siRNA were cultured in non-pacing conditions.
Transfection of pEXP-RB-Mam-TFAM upregulated TFAM expression levels
and increased viability and ATP content in tachypacing cells
compared with cells transfected with pEXP-RB-Mam-NC (9.52±0.25
nmol/mg vs. 4.25±0.23 nmol/mg, respectively) (all P<0.05;
Fig. 3A-D). By contrast, TFAM
siRNA transfection downregulated TFAM expression levels and
decreased the viability and ATP content of non-pacing cells
compared with siRNA-NC transfection (all P<0.05; Fig. 4A-D). These results indicated that
TFAM may be involved in the energy synthesis of cardiomyocytes.
Effect of TFAM on the expression
levels of MT-ND1 and MT-CO1 in cardiomyocytes
The expression levels of MT-ND1 and MT-CO1 were
measured by western blotting, which indicated that the expression
levels of MT-ND1 and MT-CO1 were increased in tachypacing cells
transfected with pEXP-RB-Mam-TFAM compared with
pEXP-RB-Mam-NC-transfected cells (P<0.05; Fig. 3D). However, compared with siRNA-NC
transfection, MT-ND1 and MT-CO1 levels were decreased in non-pacing
cells transfected with TFAM siRNAs (P<0.05; Fig. 4D).
Discussion
AF is a notable medical problem presenting a burden
to the individual and the society (29). However, current therapeutics and
radial frequency ablation used for the treatment of AF are unable
to provide a complete cure and may cause unexpected complications
(30–32). Thus, the treatment of AF is far
from satisfactory owing to insufficient knowledge of the mechanism
underlying AF; further research on the pathogenesis of AF and
molecular level of dysfunction may provide novel methods for
treatment. Combined metabolomic and proteomic analysis of AF has
demonstrated that energy metabolism-associated proteins are altered
in AF tissues (33,34), which indicated that energy
metabolism serves a role in the pathophysiology of AF.
The present study measured the expression levels of
TFAM in tissues and cells. TFAM is a transcription factor which is
produced in the nucleus and transported to the mitochondria
(35,36). TFAM is essential for mtDNA
maintenance; it serves a key role in mtDNA stability and modifies
mitochondrial gene expression levels (37,38).
Previous reports have demonstrated that TFAM dysfunction causes
cardiovascular diseases, such as heart failure and cardiomyopathy
(39,40). However, it remains unclear whether
TFAM is involved in the progression of AF. Results from the present
study demonstrated that expression levels of TFAM were decreased in
both AF tissue and tachypacing cardiomyocytes. Without the
protection of TFAM, mtDNA becomes unstable and degrades (35), resulting in decreased ATP
synthesis, which exacerbates the development of AF by decreasing
ion pump efficiency and cardiomyocyte contraction.
Primary cultured cardiomyocytes have been used to
study cardiac bioenergetics (41).
The whole heart contains a mixture of myocytes and non-myocytes;
however, primary cultured cardiomyocytes are pure, with minimal
contamination of fibroblasts and endothelial cells (42,43).
α-sarcomeric actin is specifically expressed in myocardial cells,
and was therefore selected to distinguish cardiomyocytes and
non-myocytes in primary cultured cells in the present study
(44). A tachypacing model of
cardiomyocytes was constructed to investigate the pathological
changes similar to those in patients with AF. Because the
spontaneous frequency of primary cultured cardiomyocytes was 1 Hz,
6 Hz was used to produce a similar frequency increment (6-fold
increase) to that which occurs in humans with AF (23,24).
The present study measured the ATP content in
cardiomyocytes because in order to sustain cardiac function,
cardiomyocytes need a constant supply of energy, primarily in the
form of ATP (45). Emelyanova
et al (14) reported that
the overall functional activity of the electron transport chain
(ETC) was reduced by 30% in AF tissues compared with non-AF
tissues, which was accompanied by reduced ATP production (14). In addition, Schild et al
(46) concluded that the rapid
pacing of cardiomyocytes decreased mitochondrial ATP synthesis
(46). In the study, ATP content
was decreased in tachypacing cardiomyocytes compared with
non-pacing cardiomyocytes. Furthermore, the effect of TFAM on ATP
content was investigated. The overexpression of TFAM in tachypacing
cardiomyocytes increased the ATP content, whereas inhibition of
TFAM in non-pacing cardiomyocytes decreased the ATP content. These
findings indicated that the expression levels of TFAM may regulate
ATP content in cardiomyocytes. Viability of cardiomyocytes was also
assessed; the cell viability decreased gradually in tachypacing
cardiomyocytes and was restored by overexpression of TFAM.
The majority (>90%) of the cellular ATP used in
the heart is produced through oxidative phosphorylation by the
mitochondria (47). To investigate
the downstream factors of TFAM that affect ATP synthesis,
expression levels of certain subunits [encoded by both nuclear DNA
(NDUFS1 and COX6C) and mtDNA (MT-ND1 and MT-CO1)] of oxidative
respiratory chain complexes were measured in the present study.
Previous studies revealed that the activities of complex I and II
were selectively reduced in AF and the function of ETC was impaired
in tachypacing cardiomyocytes (14,46).
In the present study, the protein levels of MT-ND1 and MT-CO1 were
decreased in tachypacing cardiomyocytes whereas there was no
difference in expression levels of NDUFS1 and COX6C between the two
groups. Furthermore, the effect of TFAM on the expression of MT-ND1
and MT-CO1 was investigated. The overexpression of TFAM in
tachypacing cardiomyocytes increased the expression levels of
MT-ND1 and MT-CO1, whereas inhibition of TFAM in non-pacing
cardiomyocytes decreased the expression levels of MT-ND1 and
MT-CO1. These results indicated that the decrease in ATP content
was induced by decreased MT-ND1 and MT-CO1 in tachypacing
cardiomyocytes, and that TFAM may regulate ATP content through the
expression levels of MT-ND1 and MT-CO1 in cardiomyocytes.
In summary, the present results demonstrated that
the expression levels of TFAM were decreased in AF tissue and
tachypacing cardiomyocytes and that the restoration of TFAM
increased ATP content by upregulating the levels of MT-ND1 and
MT-CO1 in tachypacing cardiomyocytes. Thus, TFAM may be a novel
beneficial target for treatment of patients with AF. The expression
levels of TFAM have an effect on the content of mtDNA, that is, the
overexpression of TFAM increased the mtDNA content, whereas the
knockdown of TFAM decreased the content; however, further
investigation is required to determine if expression levels of TFAM
affect the content of mtDNA in tachypacing cardiomyocytes. The
present study only measured 2 of the 13 subunits encoded by mtDNA;
further research is required to assess the other subunits, and
animal models are needed to further elucidate the function of TFAM
in AF.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
The authors would like to thank Dr Lei Yu (First
Affiliated Hospital of China Medical University, Shenyang, China)
for the assistance with tissue collection, and Professor Zhongyan
Shan and Dr Chengjiang Lin (Endocrine Laboratory of First
Affiliated Hospital of China Medical University, Shenyang, China)
for teaching the techniques for the experimental procedures.
Funding
The present work was supported by The Department of
Education of Liaoning Province, China (grant no. XLYC1802066).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
YL and YZ wrote and edited the manuscript and
designed the study. RT and XJ performed reverse
transcription-quantitative PCR and western blotting. YW cultured
the cardiomyocytes and measured the ATP content. TG is the
guarantor of this work and analyzed the data. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
Human and animal studies were approved by the Ethics
Committee of The First Affiliated Hospital of China Medical
University (Shenyang, China; approval no. 2017-69-2) and were
performed in accordance with Declaration of Helsinki. Written
informed consent was obtained from all patients prior to tissue
collection.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Rahman F, Kwan GF and Benjamin EJ: Global
epidemiology of atrial fibrillation. Nat Rev Cardiol. 11:639–654.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Friberg L and Bergfeldt L: Atrial
fibrillation prevalence revisited. J Intern Med. 274:461–468. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Piccini JP, Hammill BG, Sinner MF, Jensen
PN, Hernandez AF, Heckbert SR, Benjamin EJ and Curtis LH: Incidence
and prevalence of atrial fibrillation and associated mortality
among medicare beneficiaries, 1993–2007. Circ Cardiovasc Qual
Outcomes. 5:85–93. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Staerk L, Wang B, Preis SR, Larson MG,
Lubitz SA, Ellinor PT, McManus DD, Ko D, Weng LC, Lunetta KL, et
al: Lifetime risk of atrial fibrillation according to optimal,
borderline, or elevated levels of risk factors: Cohort study based
on longitudinal data from the Framingham heart study. BMJ.
361:k14532018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chugh SS, Havmoeller R, Narayanan K, Singh
D, Rienstra M, Benjamin EJ, Gillum RF, Kim YH, McAnulty JH Jr,
Zheng ZJ, et al: Worldwide epidemiology of atrial fibrillation: A
global burden of disease 2010 study. Circulation. 129:837–847.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Stewart S, Hart CL, Hole DJ and McMurray
JJ: A population-based study of the long-term risks associated with
atrial fibrillation: 20-year follow-up of the Renfrew/Paisley
study. Am J Med. 113:359–364. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Freedman B, Potpara TS and Lip GYH: Stroke
prevention in atrial fibrillation. Lancet. 388:806–817. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Thrall G, Lane D, Carroll D and Lip GY:
Quality of life in patients with atrial fibrillation: A systematic
review. Am J Med. 119:448.e1–e19. 2006. View Article : Google Scholar
|
|
9
|
Benjamin EJ, Wolf PA, D'Agostino RB,
Silbershatz H, Kannel WB and Levy D: Impact of atrial fibrillation
on the risk of death: The Framingham heart study. Circulation.
98:946–952. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Friberg L, Hammar N, Pettersson H and
Rosenqvist M: Increased mortality in paroxysmal atrial
fibrillation: Report from the Stockholm Cohort-study of atrial
fibrillation (SCAF). Eur Heart J. 28:2346–2353. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tsuboi M, Hisatome I, Morisaki T, Tanaka
M, Tomikura Y, Takeda S, Shimoyama M, Ohtahara A, Ogino K, Igawa O,
et al: Mitochondrial DNA deletion associated with the reduction of
adenine nucleotides in human atrium and atrial fibrillation. Eur J
Clin Invest. 31:489–496. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Barbey O, Pierre S, Duran MJ, Sennoune S,
Lévy S and Maixent JM: Specific up-regulation of mitochondrial
F0F1-ATPase activity after short episodes of atrial fibrillation in
sheep. J Cardiovasc Electrophysiol. 11:432–438. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Cha YM, Dzeja PP, Shen WK, Jahangir A,
Hart CYT, Terzic A and Redfield MM: Failing atrial myocardium:
Energetic deficits accompany structural remodeling and electrical
instability. Am J Physiol Heart Circ Physiol. 284:H1313–H1320.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Emelyanova L, Ashary Z, Cosic M,
Negmadjanov U, Ross G, Rizvi F, Olet S, Kress D, Sra J, Tajik AJ,
et al: Selective downregulation of mitochondrial electron transport
chain activity and increased oxidative stress in human atrial
fibrillation. Am J Physiol Heart Circ Physiol. 311:H54–H63. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ausma J, Coumans WA, Duimel H, Van der
Vusse GJ, Allessie MA and Borgers M: Atrial high energy phosphate
content and mitochondrial enzyme activity during chronic atrial
fibrillation. Cardiovasc Res. 47:788–796. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ou F, Rao N, Jiang X, Qian M, Feng W, Yin
L and Chen X: Analysis on differential gene expression data for
prediction of new biological features in permanent atrial
fibrillation. PLoS One. 8:e761662013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yeh YH, Kuo CT, Lee YS, Lin YM, Nattel S,
Tsai FC and Chen WJ: Region-specific gene expression profiles in
the left atria of patients with valvular atrial fibrillation. Heart
Rhythm. 10:383–391. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kang D, Kim SH and Hamasaki N:
Mitochondrial transcription factor A (TFAM): Roles in maintenance
of mtDNA and cellular functions. Mitochondrion. 7:39–44. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kunkel GH, Kunkel CJ, Ozuna H, Miralda I
and Tyagi SC: TFAM overexpression reduces pathological cardiac
remodeling. Mol Cell Biochem. 454:139–152. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lebrecht D, Setzer B, Ketelsen UP,
Haberstroh J and Walker UA: Time-dependent and tissue-specific
accumulation of mtDNA and respiratory chain defects in chronic
doxorubicin cardiomyopathy. Circulation. 108:2423–2429. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ide T, Tsutsui H, Hayashidani S, Kang D,
Suematsu N, Nakamura K, Utsumi H, Hamasaki N and Takeshita A:
Mitochondrial DNA damage and dysfunction associated with oxidative
stress in failing hearts after myocardial infarction. Circ Res.
88:529–535. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
World Medical Association: World medical
association declaration of Helsinki: Ethical principles for medical
research involving human subjects. JAMA. 310:2191–2194. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wiersma M, Meijering RAM, Qi XY, Zhang D,
Liu T, Hoogstra-Berends F, Sibon OCM, Henning RH, Nattel S and
Brundel BJJ: Endoplasmic reticulum stress is associated with
autophagy and cardiomyocyte remodeling in experimental and human
atrial fibrillation. J Am Heart Assoc. 6:e0064582017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Brundel BJ, Kampinga HH and Henning RH:
Calpain inhibition prevents pacing-induced cellular remodeling in a
HL-1 myocyte model for atrial fibrillation. Cardiovasc Res.
62:521–528. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lu L, Han H, Tian Y, Li W, Zhang J, Feng M
and Li Y: Aurora kinase A mediates c-Myc's oncogenic effects in
hepatocellular carcinoma. Mol Carcinog. 54:1467–1479. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chen Y, Zhang Z, Yang K, Du J, Xu Y and
Liu S: Myeloid zinc-finger 1 (MZF-1) suppresses prostate tumor
growth through enforcing ferroportin-conducted iron egress.
Oncogene. 34:3839–3847. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhang D, Wu CT, Qi X, Meijering RAM,
Hoogstra-Berends F, Tadevosyan A, Cubukcuoglu Deniz G, Durdu S,
Akar AR, Sibon OCM, et al: Activation of histone deacetylase-6
induces contractile dysfunction through derailment of α-tubulin
proteostasis in experimental and human atrial fibrillation.
Circulation. 129:346–358. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Chugh SS, Roth GA, Gillum RF and Mensah
GA: Global burden of atrial fibrillation in developed and
developing nations. Glob Heart. 9:113–119. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zimetbaum P: Antiarrhythmic drug therapy
for atrial fibrillation. Circulation. 125:381–389. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Scherr D, Khairy P, Miyazaki S,
Aurillac-Lavignolle V, Pascale P, Wilton SB, Ramoul K, Komatsu Y,
Roten L, Jadidi A, et al: Five-year outcome of catheter ablation of
persistent atrial fibrillation using termination of atrial
fibrillation as a procedural endpoint. Circ Arrhythm
Electrophysiol. 8:18–24. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Gillinov AM, Gelijns AC, Parides MK,
DeRose JJ Jr, Moskowitz AJ, Voisine P, Ailawadi G, Bouchard D,
Smith PK, Mack MJ, et al: Surgical ablation of atrial fibrillation
during mitral-valve surgery. N Engl J Med. 372:1399–1409. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Tu T, Zhou S, Liu Z, Li X and Liu Q:
Quantitative proteomics of changes in energy metabolism-related
proteins in atrial tissue from valvular disease patients with
permanent atrial fibrillation. Circ J. 78:993–1001. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Mayr M, Yusuf S, Weir G, Chung YL, Mayr U,
Yin X, Ladroue C, Madhu B, Roberts N, Souza AD, et al: Combined
metabolomic and proteomic analysis of human atrial fibrillation. J
Am Coll Cardiol. 51:585–594. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kunkel GH, Chaturvedi P and Tyagi SC:
Mitochondrial pathways to cardiac recovery: TFAM. Heart Fail Rev.
21:499–517. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Picca A and Lezza AM: Regulation of
mitochondrial biogenesis through TFAM-mitochondrial DNA
interactions: Useful insights from aging and calorie restriction
studies. Mitochondrion. 25:67–75. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Alam TI, Kanki T, Muta T, Ukaji K, Abe Y,
Nakayama H, Takio K, Hamasaki N and Kang D: Human mitochondrial DNA
is packaged with TFAM. Nucleic Acids Res. 31:1640–1645. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Ekstrand MI, Falkenberg M, Rantanen A,
Park CB, Gaspari M, Hultenby K, Rustin P, Gustafsson CM and Larsson
NG: Mitochondrial transcription factor A regulates mtDNA copy
number in mammals. Hum Mol Genet. 13:935–944. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wang J, Wilhelmsson H, Graff C, Li H,
Oldfors A, Rustin P, Brüning JC, Kahn CR, Clayton DA, Barsh GS, et
al: Dilated cardiomyopathy and atrioventricular conduction blocks
induced by heart-specific inactivation of mitochondrial DNA gene
expression. Nat Genet. 21:133–137. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Garnier A, Fortin D, Delomenie C, Momken
I, Veksler V and Ventura-Clapier R: Depressed mitochondrial
transcription factors and oxidative capacity in rat failing cardiac
and skeletal muscles. J Physiol. 551:491–501. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Judd J, Lovas J and Huang GN: Isolation,
culture and transduction of adult mouse cardiomyocytes. J Vis Exp.
28:540122016.
|
|
42
|
Zhou P and Pu WT: Recounting cardiac
cellular composition. Circ Res. 118:368–370. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Parameswaran S, Kumar S, Verma RS and
Sharma RK: Cardiomyocyte culture-an update on the in vitro
cardiovascular model and future challenges. Can J Physiol
Pharmacol. 91:985–998. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Belostotskaya GB and Golovanova TA:
Characterization of contracting cardiomyocyte colonies in the
primary culture of neonatal rat myocardial cells: A model of in
vitro cardiomyogenesis. Cell Cycle. 13:910–918. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Stanley WC, Recchia FA and Lopaschuk GD:
Myocardial substrate metabolism in the normal and failing heart.
Physiol Rev. 85:1093–1129. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Schild L, Bukowska A, Gardemann A, Polczyk
P, Keilhoff G, Täger M, Dudley SC, Klein HU, Goette A and Lendeckel
U: Rapid pacing of embryoid bodies impairs mitochondrial ATP
synthesis by a calcium-dependent mechanism-a model of in vitro
differentiated cardiomyocytes to study molecular effects of
tachycardia. Biochim Biophys Acta. 1762:608–615. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Harris DA and Das AM: Control of
mitochondrial ATP synthesis in the heart. Biochem J. 280:561–573.
1991. View Article : Google Scholar : PubMed/NCBI
|