Introduction
Dendritic cells (DCs) are antigen-presenting cells
that develop directly from myeloid progenitors in the bone marrow
and circulating blood monocytes and initiate primary T-cell
responses, linking innate and adaptive immune responses (1). DCs have mainly been explored as potent
stimulators of adaptive immunity, but DCs also establish and
maintain immunological tolerance (2). The state of maturation or activation
determines their capacity for initiating tolerance or immunity.
Immature (im)DCs induce tolerance, whereas mature (m)DCs induce
immunity (3). Some reports have
revealed that DCs can prevent, inhibit, or modulate T-cell-mediated
effector responses through releasing small (~40–150 nm in diameter)
membrane-enclosed vesicles (exosomes). These DC-derived exosomes
have been proposed to be involved in antigen presentation, immune
regulation and signal transduction (4,5).
imDC-derived exosomes (imDex) and mDC-derived
exosomes (mDex) have opposite biological functions in regulating
the immune network the same as their parental cells (5,6).
DC-derived exosomes (Dex) also regulate the cellular behavior of
recipient cells following uptake transferring cargo molecules. The
majority of studies have focused on the function of transmembrane
proteins on the surface of imDex and mDex (6,7).
Despite the importance of these cargo molecules, only a few studies
have paid attention to their intraluminal composition (8,9).
The majority of transcripts transcribed from the
human or mouse genome are noncoding (nc)RNAs (10). Among these ncRNAs, long noncoding
RNAs (lncRNAs) are >200 nucleotides in length. Numerous lncRNAs
have been identified to date and have emerged as significant
regulators of various biological processes, such as cell growth,
cell activation and metabolic rewiring (11–13).
In the context of Dex, the functions of lncRNA remain poorly
understood.
In the present study, RNA-sequencing analysis was
performed between mDex and imDex. Subsequently, markedly altered
biological functions and pathways were predicted by Gene Ontology
(GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway
analyses. In addition, the physiological and pathological functions
of differentially expressed lncRNAs were identified via the
annotation of their associated genes. Finally, the differential
expression of representative lncRNAs was further validated by
reverse transcription-quantitative (RT-q) PCR. Probable novel
mechanisms were identified via a combination of bioinformatics
methods. The study of lncRNAs might shed light on the different
functions of two kinds of exosomes.
Materials and methods
Animals and reagents
A total of 50 five-week-old male C57 mice (Animal
Center of Southern Medical University, Guangzhou, China), weighing
25–30 g, were used in the present study. All mice were were
acclimatized for 1 week before experiments at room temperature
under a controlled 12/12 h light/dark cycle and received food and
water ad libitum. Antibodies for western blot analysis were
as follows: Rabbit monoclonal anti-mouse CD63 (cat. no. ab217345;
Abcam), CD9 (cat. no. ab92726; Abcam), CD81 (cat. no. ab109201;
Abcam) and TSG101 (cat. no. ab125011; Abcam). Antibodies for flow
cytometry were as follows: FITC anti-mouse CD11c (clone N418;
Biolegend), PE anti-mouse MHC-II (clone 10-3.6; Biolegend), PE
anti-mouse CD80 (clone 16-10A1; Biolegend) and APC anti-mouse CD86
(clone GL-1; Biolegend).
All animal-related experiments were performed
according to the guidelines of the Care and Use of Laboratory
Animals (Ministry of Health, China, 1998) (14). The experiments were approved by the
Animal Use Committee of Shenzhen Hospital, Southern Medical
University.
Cultivation of bone marrow dendritic
cells
Bone marrow dendritic cells (BMDCs) were obtained
from C57 mice as previously described (6,9).
Bovine EV-depleted medium was obtained by overnight
ultracentrifugation of medium (RPMI-1640; Gibco; Thermo Fisher
Scientific, Inc.) supplemented with 20% fetal bovine serum (FBS;
Gibo; Thermo Fisher Scientific, Inc.) at 100,000 × g (4°C for 8 h)
to eliminate the interference of exosomes from FBS (9). Briefly, the mice were sacrificed by
cervical dislocation. Bone marrow progenitors were washed out from
long bones (femur and tibia) and cultured (37°C, 48 h) in
EV-depleted medium (10% EV-depleted FBS final) containing 20 ng/ml
granulocyte-macrophage colony-stimulating factor (GM-CSF;
PeproTech, Inc.) and 10 ng/ml IL-4 (PeproTech, Inc.). Non-adherent
cells were gently washed out after 48 h. The remaining clusters,
which were loosely adhered to the Petri dish, were cultured (37°C,
4 days) and the medium was changed every other day. On day 7, cells
were directly observed under a light microscope (magnification, ×40
and ×100). A total of 6 plates (4 fields of view/plate) were
selected to observe cells. Then, cells were collected for treatment
and treated with different protocols depending on different studies
subsequently conducted.
For exosome isolation, DCs were treated with
lipopolysaccharide (LPS; 5 µg/ml; Enzo Life Sciences, Inc.) or PBS
for 24 h, washed twice with PBS and replaced with fresh medium.
After another 48 h of continuous culture, the culture medium was
collected for exosome isolation.
BMDC analysis by flow cytometry
BMDCs were analyzed by flow cytometry for surface
marker expression. The dendritic cells were stained with antibodies
against CD11c-FITC (1:200; cat. no. 117305), MHC-II-PE (1:1,000;
cat. no. 116407), CD80-PE (1:300; cat. no. 104707), CD86-PE (1:160;
cat. no. 159203) from BioLegend, Inc. for 45 min at 4°C. The cells
were stained using a Fortessa flow cytometer (BD Biosciences) and
data were analyzed with FlowJo V10 software (FlowJo, LLC).
Exosome isolation and analysis
Exosomes were isolated by differential
ultracentrifugation as previously described (6,9).
Briefly, the culture medium collected using the aforementioned
protocol was centrifuged at 300 × g for 10 min at 4°C to obtain the
pellet. The supernatant was centrifuged at 2,000 × g for 20 min,
transferred to new tubes and centrifuged for 40 min at 10,000 × g
and finally for 90 min at 100,000 × g at 4°C. All pellets were
washed in 50–60 ml of PBS and recentrifuged at 100,000 × g for 90
min at 4°C before being resuspended in 50–100 µl of sterile
PBS.
The ultrastructure and size distribution of exosomes
were analyzed by transmission electron microscopy (TEM; Hitachi,
Ltd.) and NanoSight NS300 (Malvern Panalytical), respectively.
Briefly, exosome samples were fixed with 1% glutaraldehyde in PBS
at an optimal concentration at room temperature for 5 min. The
mixture was then spotted onto 300-mesh carbon/formvar-coated grids
and dried at room temperature. Next, the grids were washed with PBS
and stained for contrast using uranyl acetate (50%) in water at
room temperature for 10 min. Then, exosome size and morphology were
observed using a JEM-1011 electron microscope (magnification,
×25,000; JEOL Ltd.).
Western blotting
Protein markers, CD63, CD9, TSG101 and CD81 were
detected using western blot analysis. The collected exosomes were
dissected to extract total protein using RIPA buffer (Applygen
Technologies, Inc.) and quantified using a BCA assay. Following
quantification, equal amount of proteins (5 µg) were separated by
10% SDS-PAGE and transferred onto PVDF membranes (EMD
Millipore).
Following blocking with 5% skimmed milk at room
temperature for 1 h, the membranes were incubated with the
following primary antibodies at 4°C overnight (all Abcam): CD63
(1:1,000; cat. no. ab217345), CD9 (1:2,000; cat. no. ab92726), CD81
(1:3,000; cat. no. ab109201) and TSG101 (1:2,000; cat. no.
ab125011). The antibodies were detected using horseradish
peroxidase-conjugated IgG (goat anti-rabbit; 1:10,000; cat. no.
ab205718; Abcam) at room temperature for 2 h and visualized using
enhanced chemiluminescence (Bio-Rad Laboratories, Inc.).
High-throughput sequencing and
analysis of lncRNA
Exosomes were extracted from mDCs and imDCs as
described earlier and the exosomal RNA was extracted using
TRIzol® (Thermo Fisher Scientific, Inc.). Extracted
lncRNA was enriched with oligo(dT) magnetic beads. RNA-sequencing
(RNA-seq) libraries were produced and subjected to quality
inspection using an Agilent 2100 (Agilent Technologies GmbH). The
libraries were quantified by RT-qPCR. The samples were sequenced
using an Illumina HiSeq 4000 (Illumina, Inc.). All lncRNAs were
annotated based on the Ensembl. RNA-seq and subsequent
bioinformatics analysis were performed by ShuPu Biotechnology LLC
as previously reported (15).
RNA isolation and RT-qPCR validation
of lncRNAs
RT-qPCR was performed as reported previously further
to verify the differentially expressed identified lncRNAs (16). RNA from exosome samples (5 µg) was
extracted using TRIzol® (Thermo Fisher Scientific, Inc.)
following the manufacturer's protocols.
RNA quantity and quality were measured using a
NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies; Thermo
Fisher Scientific, Inc.). The RNA (1 µg) was reverse-transcribed
using a reverse transcript kit (TransGen Biotech Co., Ltd.)
following the manufacturer's protocol. RT-qPCR was performed with a
PerfectStart Green qPCR SuperMix kit (TransGen Biotech Co., Ltd.)
following the manufacturer's protocol. The primers for validated
lncRNAs are listed in Table SI.
PCR was performed using a final reaction volume of 20 µl and the
following thermocycling conditions: 5 min at 95°C for denaturation,
40 cycles of 10 sec at 95°C for denaturation, 30 sec at 60°C for
annealing and elongation, and final extension for 10 min at 72°C.
Amplification was performed with an ABI-7500 machine (Applied
Biosystems; Thermo Fisher Scientific, Inc.). The data were analyzed
with the SDS relative quantification software (version 2.2.2;
Thermo Fisher Scientific, Inc.). The relative fold change was
calculated using the 2−∆∆Cq method (17). All reactions were performed in
triplicate and normalized to the internal control products of
GAPDH.
GO and KEGG pathway analysis of
selected lncRNAs
GO (geneontology.org) and KEGG (genome.jp/kegg) analyses
of differentially expressed lncRNA-associated genes were performed
using the online Database for Annotation, Visualization and
Integrated Discovery tool (18,19).
The top 10 enriched GO terms among the two groups were identified.
GO analysis results consisted of ‘biological process’ (BP), ‘cell
composition’ (CC) and ‘molecular function’ (MF). The adjusted
P-value was also obtained using the Benjamini & Hochberg method
(20) and an adjusted P<0.05 was
considered to indicate a statistically significant difference. The
pathways associated with lncRNA-targeted mRNAs were identified by
KEGG pathway analysis.
Gene set enrichment analysis
Gene set enrichment analysis (GSEA) is a
computational method used to determine whether a given gene set has
significant differences among different groups, as previously
described (21). Briefly speaking,
the Subramanian method, using Java 11 software, was used to first
calculate the enrichment score (ES), estimate the importance of ES
and finally evaluate their importance by adjusting multiple
hypothesis tests.
Prediction of lncRNA-miRNA-mRNA
interactions
lncRNA-miRNA interaction was predicted using miRanda
(score >150; energy <-30) (22). The target genes of selected miRNAs
were predicted and analyzed using miRWalk2.0 (23). The results from six different
databases, namely, miRWalk, miRanda, miRDB (mirdb.org/),
miRNAMap (mirnamap.mbc.nctu.edu.tw/), RNA22 (cm.jefferson.edu/rna22/Precomputed/) and
TargetScan (targetscan.org/mamm_31/), were used for analysis. If a
gene was predicted to be a target of miRNA in >3 databases, the
gene was considered as a target of miRNA. Cytoscape 3.7.1 software
(24) was used to construct the
network.
Statistical analysis
All experimental data were expressed as the mean ±
standard deviation. Statistical analysis of the data was performed
with the two-tailed independent-samples Student's t test
using GraphPad Prism 5.0 (GraphPad Software, Inc.). One-way
analysis of variance followed by Tukey's honestly significant
difference test was used to compare differences among groups.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Characterization of BMDCs and
BMDC-derived exosomes
BMDCs were cultured in complete medium with
recombinant mouse GM-CSF (rmGM-CSF) and recombinant mouse IL-4
(rmIL-4), supplemented with 10% endotoxin-free and EV-depleted FBS.
They were directly observed under a microscope on day 7 and the
results demonstrated typical aggregate formation (Fig. 1A). Mature markers were also studied
by flow cytometry. Following stimulation with LPS (5 µg/ml) for 24
h, mDCs expressed high levels of MHC class II and co-stimulatory
CD80 and CD86 compared with imDCs (Fig.
1B; the original data are given in Fig. S1). These data indicated a
successful culture of BMDCs with EV-depleted complete medium.
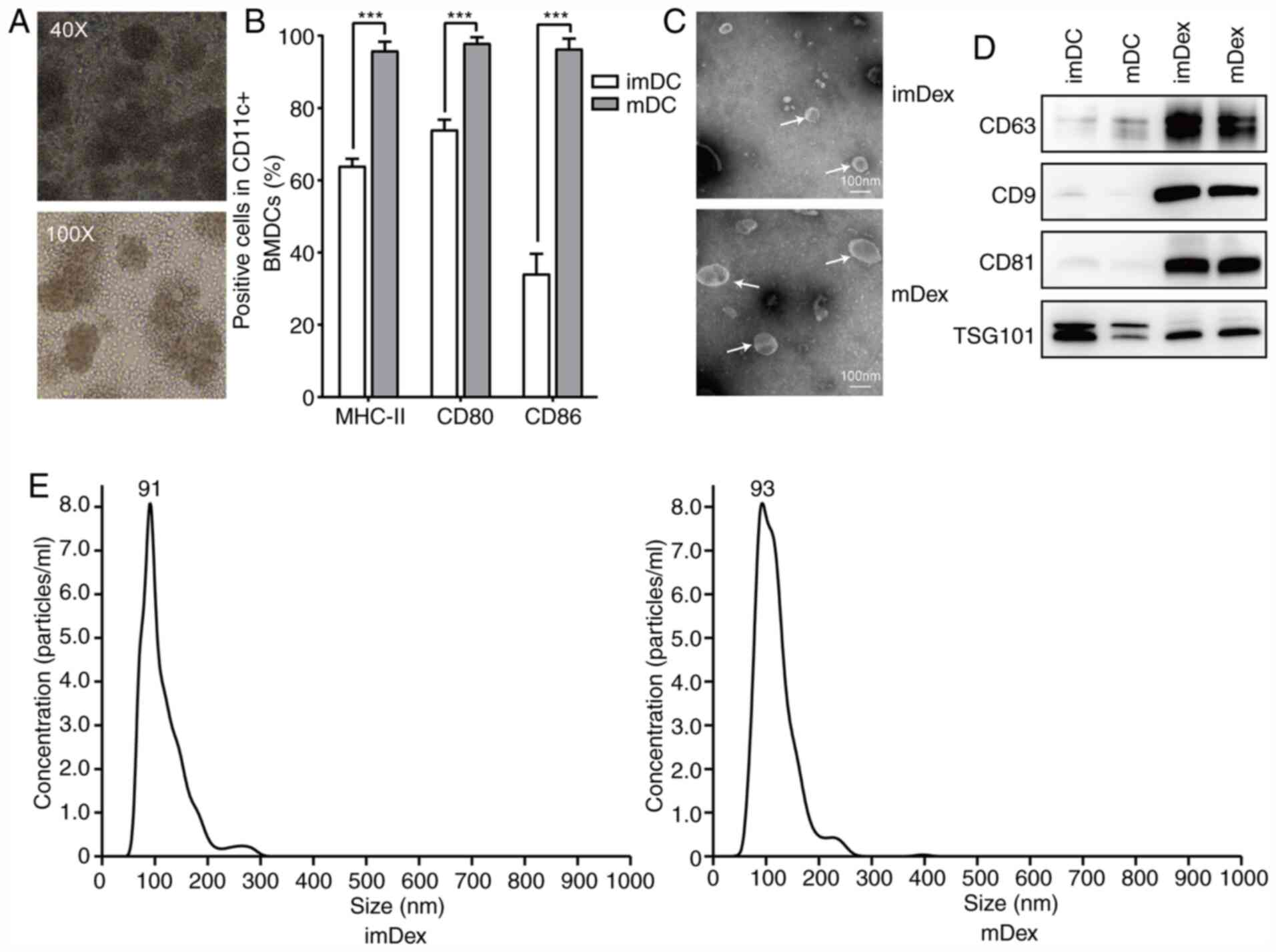 | Figure 1.Successful isolation of exosomes from
DC culture medium. (A) Morphological structure of BMDCs
(magnification, ×40 and ×100) cultured with previously
ultracentrifuged FBS on day 5. (B) Mature markers detected by flow
cytometry in imDCs and mDCs cultured with previously
ultracentrifuged FBS. ***P<0.001. (C) Ultrastructure of mDex and
imDex by transmission electron microscopy (white arrows); scale
bar, 100 nm. (D) Expression of exosome markers, CD63, CD9, CD81 and
TSG101 confirmed by western blot analysis. A total of 5 µg protein
from DC lysis and 5 µg protein from exosomes lysis were loaded onto
each lane (representative image of n=3). (E) Size distribution
profile of mDex and imDex using NanoSight NS300. DC, dendritic
cells; BMDCs, bone marrow dendritic cells; im, immature; m, mature;
Dex, dendritic cell-derived exosomes. |
On day 7 of DC culture, PBS or LPS was added to the
medium to generate immature or mature DCs. After 24 h of activation
by PBS or LPS, the culture medium was replaced completely. After
continuously culturing DCs for another 48 h, the exosomes were
isolated from the culture supernatants of mDCs and imDCs by
differential ultracentrifugation. TEM and nanoparticle tracking
analysis were used to analyze the ultrastructure and size
distribution of exosomes, respectively. TEM results of exosomes
revealed the characteristic saucer shape of exosomes (Fig. 1C). Nanoparticle Tracking Analysis
demonstrated that the imDex had a narrow size distribution with a
mean particle diameter of 91 nm, which was not significantly
different in size compared with mDex (Fig. 1E). Additionally, the expression
levels of CD9, CD63, CD81 and TSG101 were analyzed by western blot
analysis (Fig. 1D). As expected,
tetraspanins (CD9, CD63 and CD81) and TSG101 were more abundant in
the exosome protein lysis compared with their parental cell protein
lysis. These data indicated the successful isolation of exosomes
from the culture medium.
Identification of differentially
expressed lncRNAs between mDex and imDex
The expression pattern of lncRNAs was detected in
three mDex and three imDex samples. A total of 437 lncRNAs were
obtained through RNA-seq. Based on the Ensembl (25), ~153 lncRNAs were already annotated,
whereas ~284 lncRNAs were first identified in the present study.
Differentially expressed lncRNAs were identified by comparing the
differences in the expression levels of these RNAs between mDex and
imDex. A total of 108 differentially expressed lncRNAs were
analyzed by comparing the differential expression levels.
Upregulation was observed in 87 lncRNAs of mDex, whereas
downregulation was observed in 21 lncRNAs compared with imDex
(Table SII). The heat maps
(Fig. 2A), volcano plots at
different P-values and fold change (Fig. 2B) and scatter plots (Fig. 2C) were used to show the expression
ratios (log2 scale) of lncRNAs in mDex and imDex.
Prediction of lncRNA function
GO and KEGG function enrichment analyses for the
target genes of differentially expressed lncRNAs were applied to
thoroughly understand the functions of the lncRNAs listed in
Table SI.
In the GO analysis, prediction terms with the
P-value <0.05 were selected. The GO analysis was divided into
three parts: BP, CC and MF. The top 10 GO terms of three parts
associated with the research background were listed. GO analysis
clearly revealed that some important biological functions, such as
the immune system process and innate immune response, were
associated with upregulated lncRNAs (Fig. 3A), whereas the downregulated lncRNAs
were mainly involved in the BPs, such as poly(A) RNA binding
(Fig. 3B).
In addition, the analysis of the KEGG pathway of
lncRNAs of differentially expressed genes demonstrated the
relationship of TNF signaling pathway and Toll-like receptor
signaling pathway with the upregulated lncRNAs (Fig. 3C), while downregulated lncRNAs were
involved in the ribosome pathway (Fig.
3D).
Next, attempts were made to identify features of the
top 20 differentially expressed lncRNAs using GSEA. Differential
lncRNAs with the same BP and the same pathway were clustered. The
biological functions detected using GO analysis, such as T helper
cell differentiation, T-cell activation and Janus kinase (JAK)-STAT
cascade (Fig. 4A), revealed that
some immunoregulatory processes were associated with upregulated
lncRNA-associated genes (Procr-203, Clec4e-202 and Traf1-203). The
KEGG pathway analysis of the aforementioned three genes revealed
JAK-STAT signaling pathway, Th1/Th2 cell differentiation and TNF
signaling pathway (Fig. 4C).
Cellular components (Fig. S2A) and
molecular functions (Fig. S2B) of
GO analysis also provided some clues for lncRNA function
annotation.
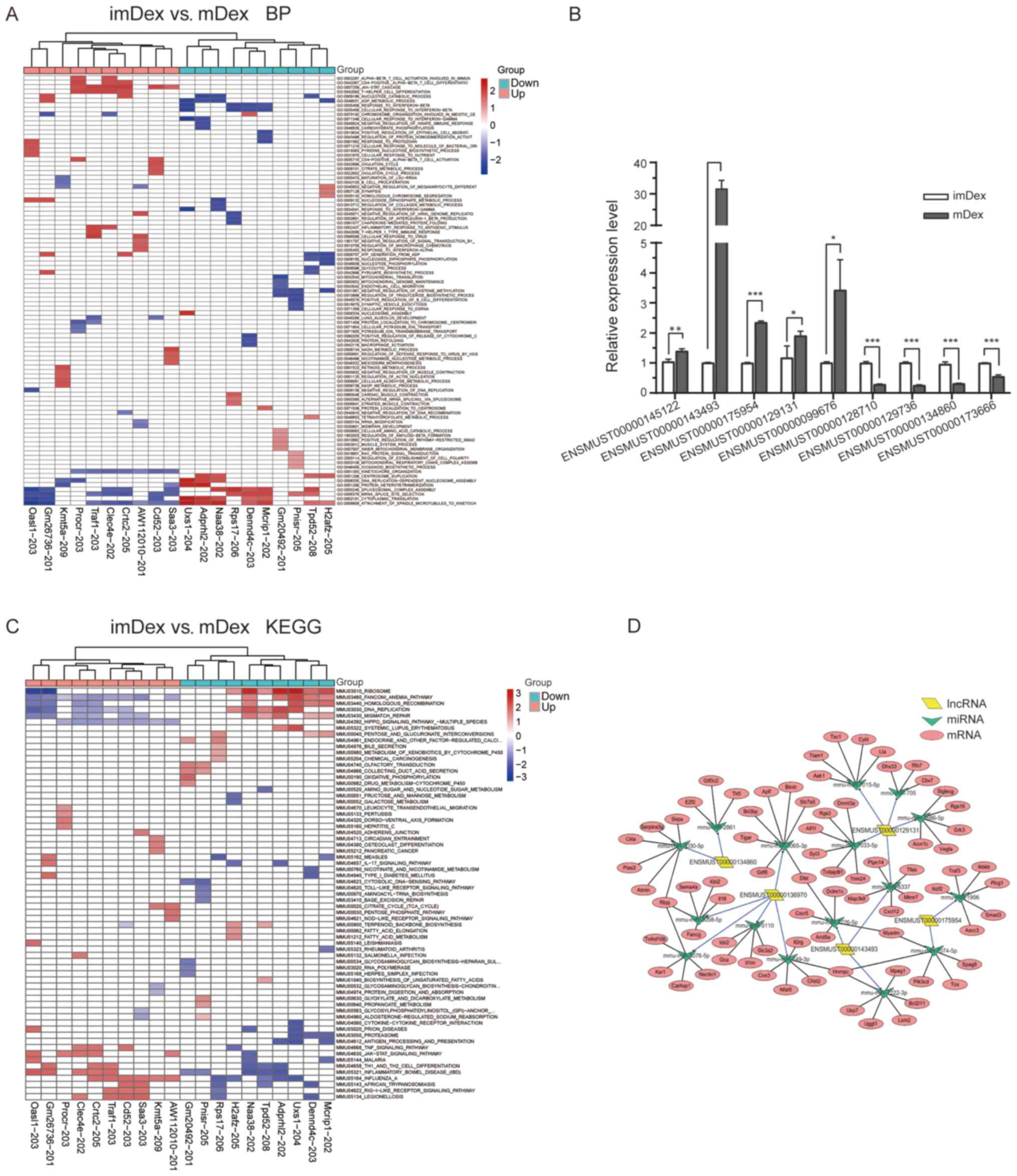 | Figure 4.Different lncRNAs GSEA prerank. (A)
BP. (B) KEGG pathway; each row represents a functional entry and
each column represents an lncRNA. (C) Changes in lncRNA expression
were confirmed using reverse transcription-quantitative PCR for
selected circRNAs in mDex and imDex groups. Bars represent mean ±
SEM (n=3; *P<0.05; **P<0.01; ***P<0.001). (D) lncRNA
candidates (n=5) were annotated in detail according to the
lncRNA/miRNA interaction information using Cytoscape. Based on the
miRNA prediction and bioinformatics analyses, mRNAs were found to
be regulated by selected miRNAs. Parallelogram, triangles and
circles indicate lncRNAs, miRNAs and mRNAs, respectively. lncRNA,
long noncoding RNA; GSEA, Gene set enrichment analysis; BP,
biological process; KEGG, Kyoto Encyclopedia of Genes and Genomes;
circRNA, circular RNA; m, mature; im, immature; Dex, dendritic
cell-derived exosomes; |
RT-qPCR validation of the
differentially expressed lncRNAs
A total of nine differentially expressed lncRNAs
were selected for validation by RT-qPCR to further validate the key
differentially expressed lncRNAs identified by RNA-seq. The nine
lncRNAs included five upregulated lncRNAs (ENSMUST00000145122,
ENSMUST00000143493, ENSMUST00000175954, ENSMUST00000129131 and
ENSMUST00000099676) and four downregulated lncRNAs
(ENSMUST00000128710, ENSMUST00000129736, ENSMUST00000134860 and
ENSMUST00000173666). They were chosen based on the GSEA of their
associated genes. These lncRNAs were involved in immunoregulation.
The expression levels of verified lncRNAs were consistent with the
sequencing results (Fig. 4B).
Identification of lncRNA-targeting
miRNAs and construction of the lncRNA-miRNA-mRNA competing
endogenous (ce)RNA regulatory network
Based on the GSEA results, five candidates were
selected (ENSMUST00000143493, ENSMUST00000175954,
ENSMUST00000129131, ENSMUST00000136970 and ENSMUST00000134860)
whose functions were most relevant to immunoregulation. An
lncRNA-miRNA-mRNA network of the five lncRNAs was constructed
(Fig. 4D). First, the target miRNAs
of lncRNAs were predicted by choosing the significant correlation
pairs according to the score and energy in miRanda. Then, the
miRNA-mRNA pairs were predicted using miRWalk2.0 and the top five
mRNAs involved in the immunoregulation function were selected and
shown in the networks. By integrating the miRNA-mRNA and
miRNA-lncRNA regulatory relationships, the miRNA-lncRNA-mRNA
network was finally constructed, providing key data for subsequent
works.
The resultant network consisted of 14 nodes,
including 5 lncRNAs, 16 miRNAs and 75 mRNAs. In total, 98 edges
were formed, including 17 lncRNA-miRNA regulation relationships and
81miRNA-mRNA regulation relationships.
Discussion
Exosomes derived from cell-culture supernatants of
dendritic cells are the choice to modulate immune response further
in antigen presentation, cancer therapy and a number of other
fields in immunology (26,27). In the context of transplantation,
allogeneic exosomes from immature DCs can modulate the rejection of
heart allografts (5,28). Notably, exosomes derived from tumor
peptide-stimulated mature DCs are able to prime tumor-specific
cytotoxic T lymphocyte responses in vivo, resulting in tumor
growth delay or the eradication of established murine tumors
(29). Dex can serve as cargoes to
transfer functional components to T cells to stimulate their
functions (8). Therefore, it was
hypothesized in the present study that Dex had different roles in
immune networks depending on the state of parental DCs and in part
via its intraluminal molecules. lncRNA in exosomes is an important
topic, thoroughly studied in types of cancer and a number of other
diseases (30,31). However, few studies have explored
the physiological or pathological function of lncRNAs in Dex. It
was hypothesized that upregulated lncRNAs in different-state
DC-derived exosomes may exert different immunoregulatory functions
in their recipient cells via vesicle transport through cell-to-cell
communication. Thus, the present study hypothesized that immune
activation or suppression could, to some extent, be induced by
differentially expressed lncRNA in Dex.
The present study investigated the differential
expression patterns of lncRNAs in imDex and mDex and predicted the
potential functions of selected lncRNAs. A total of 108
differentially expressed lncRNAs (87 upregulated and 21
downregulated) were identified in both imDex and mDex. Some novel
findings were deduced by bioinformatics analyses of genes
associated with differentially expressed lncRNAs, including
identification of the most significantly altered GO categories and
KEGG pathways. The KEGG analysis via GSEA demonstrated that the
JAK-STAT signaling pathway, reported to be involved in the
differentiation of T helper cells, was also associated with
upregulated lncRNAs (32). These
finding suggested that the alternation of lncRNA expression could
regulate the function of T cells through the JAK-STAT signaling
pathway, something which requires further investigation. In
addition, nine differentially expressed lncRNAs were selected for
further validation using RT-qPCR and the results demonstrated that
they were all significantly different.
Based on the GSEA results of the top 10
differentially expressed lncRNAs, the present study focused on five
lncRNAs (ENSMUST00000143493, ENSMUST00000175954,
ENSMUST00000129131, ENSMUST00000136970 and ENSMUST00000134860)
mostly associated with immune-modulating function (33–36).
That is also the reason why these five lncRNAs were chosen for
further ceRNA network analysis. For instance, protein C receptor,
an associated gene of ENSMUST00000143493, is identified to regulate
Th17 pathogenicity via several key molecules of the proinflammatory
module in Th17 cells (34). Also,
Clec4e, an associated gene of ENSMUST00000175954, is an endocytic
receptor expressed in DCs and implicated in corpse scavenging,
degradation, or antigen salvage pathways in DC (37). Astudy indicated that tumor necrosis
factor receptor-associated factor 1, an associated gene of
ENSMUST00000129131, is involved in multiple signaling pathways,
including NF-κB and MAPK pathways (38) and thus influences inflammatory and
apoptotic responses to tightly regulate the development of
rheumatoid arthritis and chronic infection (39,40).
These three lncRNAs, upregulated in mDex, may regulate immune cells
via their potential signaling pathways when mDex are taken up by
recipient cells. However, this hypothesis needs further
validation.
Finally, their lncRNA-miRNA-mRNA networks were also
constructed using bioinformatics tools for further investigation.
All of these miRNAs predicted using miRWalk were associated with
the T-cell-receptor signaling pathway. In addition, all the
selected targeted genes of miRNAs are reported to regulate the
functions of lymphocytes and are enriched in the spleen according
to the annotation of PubMed Gene (data not shown) (41,42).
On this basis, the present study proposed that the analyzed lncRNAs
enriched in different exosomes might participate in various BPs,
such as cell activation, differentiation, immune system regulation
and some other cellular functions of recipient cells.
In summary, the present study was unveiled lncRNA
expression patterns in exosomes derived from dendritic cells in
different states, which might help in understanding the role of
lncRNAs of exosomes. The present study also indicated the
importance of lncRNAs in the imDex- or mDex-modulated
immunoregulation.
However, the current study possessed several
limitations. First, RNA-seq is an important method to screen
possible lncRNAs associated with specific functions and pathways,
but the results of big-data analyses may be false positives.
Therefore, RT-qPCR should be performed to further verify the
differential expression. However, only 9 of these lncRNAs were
verified in the present study. Second, the functions of targeted
lncRNAs were predicted only indirectly by bioinformatics analysis.
Therefore, further functional studies on the mechanism of Dex are
warranted to clarify the role of lncRNAs. Finally, due to limited
conditions, it was not possible to obtain human cells for the
present study, but mouse model can also be helpful for the study of
human immune response. For instance, the role of BMDC in modulation
of immune response is the same in both human and mice as well as
crucial regulatory molecules found in BMDC (43).
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by grants from the
National Natural Science Foundation of China (grant no. 81672180)
and the Sanming Project of Medicine in Shenzhen (grant no.
SZSM201612019).
Availability of data and materials
The data used and/or analyzed in the present study
are available from the corresponding author on reasonable request.
The raw data (fastq) file has been uploaded to GEO database
(GSE156976).
Authors' contributions
JW and RZ performed the experiments, analyzed the
data and drafted the manuscript. JD and GL helped perform the
experiments and prepare the materials. XY and SW designed the
analytic strategy of the study. AUK, HS and JO designed experiments
and edited the manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
All animal-related experiments were performed
according to the guidelines of the Care and Use of Laboratory
Animals (Ministry of Health, China, 1998) and the experiments were
approved by the Animal Use Committee of Shenzhen Hospital, Southern
Medical University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Lu C, Huang X, Zhang X, Roensch K, Cao Q,
Nakayama KI, Blazar BR, Zeng Y and Zhou X: miR-221 and miR-155
regulate human dendritic cell development, apoptosis, and IL-12
production through targeting of p27kip1, KPC1, and SOCS-1. Blood.
117:4293–4303. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Takenaka MC and Quintana FJ: Tolerogenic
dendritic cells. Semin Immunopathol. 39:113–120. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Dhodapkar MV, Steinman RM, Krasovsky J,
Munz C and Bhardwaj N: Antigen-specific inhibition of effector T
cell function in humans after injection of immature dendritic
cells. J Exp Med. 193:233–238. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ma B, Yang JY, Song WJ, Ding R, Zhang ZC,
Ji HC, Zhang X, Wang JL, Yang XS, Tao KS, et al: Combining exosomes
derived from immature DCs with donor antigen-specific treg cells
induces tolerance in a rat liver allograft model. Sci Rep.
6:329712016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Pêche H, Heslan M, Usal C, Amigorena S and
Cuturi MC: Presentation of donor major histocompatibility complex
antigens by bone marrow dendritic cell-derived exosomes modulates
allograft rejection. Transplantation. 76:1503–1510. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gao W, Liu H, Yuan J, Wu C, Huang D, Ma Y,
Zhu J, Ma L, Guo J, Shi H, et al: Exosomes derived from mature
dendritic cells increase endothelial inflammation and
atherosclerosis via membrane TNF-α mediated NF-κB pathway. J Cell
Mol Med. 20:2318–2327. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Thery C, Ostrowski M and Segura E:
Membrane vesicles as conveyors of immune responses. Nat Rev
Immunol. 9:581–593. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hao S, Yuan J and Xiang J: Nonspecific
CD4(+) T cells with uptake of antigen-specific dendritic
cell-released exosomes stimulate antigen-specific CD8(+) CTL
responses and long-term T cell memory. J Leukoc Biol. 82:829–838.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kowal J, Arras G, Colombo M, Jouve M,
Morath JP, Primdal-Bengtson B, Dingli F, Loew D, Tkach M and Théry
C: Proteomic comparison defines novel markers to characterize
heterogeneous populations of extracellular vesicle subtypes. Proc
Natl Acad Sci USA. 113:E968–E977. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Rinn JL and Chang HY: Genome regulation by
long noncoding RNAs. Annu Rev Biochem. 81:145–166. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Militello G, Weirick T, John D, Doring C,
Dimmeler S and Uchida S: Screening and validation of lncRNAs and
circRNAs as miRNA sponges. Brief Bioinform. 18:780–788.
2017.PubMed/NCBI
|
|
12
|
Fatica A and Bozzoni I: Long non-coding
RNAs: New players in cell differentiation and development. Nat Rev
Genet. 15:7–21. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long noncoding RNAs. Cell. 136:629–641.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bao X, Zhang Q, Liu N, Zhuang S, Li Z,
Meng Q, Sun H, Bai J, Zhou X and Tang L: Characteristics of
circular RNA expression of pulmonary macrophages in mice with
sepsis-induced acute lung injury. J Cell Mol Med. 23:7111–7115.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Luo Z, Mao X and Cui W: Circular RNA
expression and circPTPRM promotes proliferation and migration in
hepatocellular carcinoma. Med Oncol. 36:862019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sheng F, Sun N, Ji Y, Ma Y, Ding H, Zhang
Q, Yang F and Li W: Aberrant expression of imprinted lncRNA MEG8
causes trophoblast dysfunction and abortion. J Cell Biochem.
120:17378–17390. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Huang da W, Sherman BT and Lempicki RA:
Bioinformatics enrichment tools: Paths toward the comprehensive
functional analysis of large gene lists. Nucleic Acids Res.
37:1–13. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Xiao B, Zhang W, Chen L, Hang J, Wang L,
Zhang R, Liao Y, Chen J, Ma Q, Sun Z and Li L: Analysis of the
miRNA-mRNA-lncRNA network in human estrogen receptor-positive and
estrogen receptor-negative breast cancer based on TCGA data. Gene.
658:28–35. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
An T, Zhang J, Ma Y, Lian J, Wu YX, Lv BH,
Ma MH, Meng JH, Zhou YT, Zhang ZY, et al: Relationships of
Non-coding RNA with diabetes and depression. Sci Rep. 9:107072019.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Enright AJ, John B, Gaul U, Tuschl T,
Sander C and Marks DS: MicroRNA targets in drosophila. Genome Biol.
5:R12003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Dweep H and Gretz N: miRWalk2.0: A
comprehensive atlas of microRNA-target interactions. Nat Methods.
12:6972015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang K, Li Q, Kang X, Wang Y and Wang S:
Identification and functional characterization of lncRNAs acting as
ceRNA involved in the malignant progression of glioblastoma
multiforme. Oncol Rep. 36:2911–2925. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Pitt JM, Andre F, Amigorena S, Soria JC,
Eggermont A, Kroemer G and Zitvogel L: Dendritic cell-derived
exosomes for cancer therapy. J Clin Invest. 126:1224–1232. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Monguio-Tortajada M, Lauzurica-Valdemoros
R and Borras FE: Tolerance in organ transplantation: From
conventional immunosuppression to extracellular vesicles. Front
Immunol. 5:4162014.PubMed/NCBI
|
|
28
|
Peche H, Renaudin K, Beriou G, Merieau E,
Amigorena S and Cuturi MC: Induction of tolerance by exosomes and
short-term immunosuppression in a fully MHC-mismatched rat cardiac
allograft model. Am J Transplant. 6:1541–1550. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zitvogel L, Regnault A, Lozier A, Wolfers
J, Flament C, Tenza D, Ricciardi-Castagnoli P, Raposo G and
Amigorena S: Eradication of established murine tumors using a novel
cell-free vaccine: Dendritic cell-derived exosomes. Nat Med.
4:594–600. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhu B, Zhang L, Liang C, Liu B, Pan X,
Wang Y, Zhang Y, Zhang Y, Xie W, Yan B, et al: Stem cell-derived
exosomes prevent aging-induced cardiac dysfunction through a novel
exosome/lncRNA MALAT1/NF-κB/TNF-α signaling pathway. Oxid Med Cell
Longev. 2019:97392582019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Dong H, Wang W, Chen R, Zhang Y, Zou K, Ye
M, He X, Zhang F and Han J: Exosome-mediated transfer of
lncRNA-SNHG14 promotes trastuzumab chemoresistance in breast
cancer. Int J Oncol. 53:1013–1026. 2018.PubMed/NCBI
|
|
32
|
Seif F, Khoshmirsafa M, Aazami H,
Mohsenzadegan M, Sedighi G and Bahar M: The role of JAK-STAT
signaling pathway and its regulators in the fate of T helper cells.
Cell Commun Signal. 15:232017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Pedros C, Altman A and Kong KF: Role of
TRAFs in signaling pathways controlling T follicular helper cell
differentiation and T cell-dependent antibody responses. Front
Immunol. 9:24122018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kishi Y, Kondo T, Xiao S, Yosef N,
Gaublomme J, Wu C, Wang C, Chihara N, Regev A, Joller N and Kuchroo
VK: Protein C receptor (PROCR) is a negative regulator of Th17
pathogenicity. J Exp Med. 213:2489–2501. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Iborra S, Martínez-López M, Cueto FJ,
Conde-Garrosa R, Del Fresno C, Izquierdo HM, Abram CL, Mori D,
Campos-Martín Y, Reguera RM, et al: Leishmania uses mincle to
target an inhibitory ITAM signaling pathway in dendritic cells that
dampens adaptive immunity to infection. Immunity. 45:788–801. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Hernandez JB, Chang C, LeBlanc M, Grimm D,
Le Lay J, Kaestner KH, Zheng Y and Montminy M: The CREB/CRTC2
pathway modulates autoimmune disease by promoting Th17
differentiation. Nat Commun. 6:72162015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Yamasaki S, Ishikawa E, Sakuma M, Hara H,
Ogata K and Saito T: Mincle is an ITAM-coupled activating receptor
that senses damaged cells. Nat Immunol. 9:1179–1188. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Ha H, Han D and Choi Y: TRAF-mediated
TNFR-family signaling. Curr Protoc Immunol. 11:Unit11 19D.
2009.PubMed/NCBI
|
|
39
|
Wang C, McPherson AJ, Jones RB, Kawamura
KS, Lin GHY, Lang PA, Ambagala T, Pellegrini M, Calzascia T,
Aidarus N, et al: Loss of the signaling adaptor TRAF1 causes
CD8+ T cell dysregulation during human and murine
chronic infection. J Exp Med. 209:77–91. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Abdul-Sater AA, Edilova MI, Clouthier DL,
Mbanwi A, Kremmer E and Watts TH: The signaling adaptor TRAF1
negatively regulates Toll-like receptor signaling and this
underlies its role in rheumatic disease. Nat Immunol. 18:26–35.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Chen Y, Yu M, Zheng Y, Fu G, Xin G, Zhu W,
Luo L, Burns R, Li QZ, Dent AL, et al: CXCR5(+)PD-1(+) follicular
helper CD8 T cells control B cell tolerance. Nat Commun.
10:44152019. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Bishop GA, Stunz LL and Hostager BS: TRAF3
as a multifaceted regulator of B lymphocyte survival and
activation. Front Immunol. 9:21612018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Mellor AL and Munn DH: IDO expression by
dendritic cells: Tolerance and tryptophan catabolism. Nat Rev
Immunol. 4:762–774. 2004. View Article : Google Scholar : PubMed/NCBI
|


















