Introduction
Intrauterine growth restriction (IUGR) (or fetal
growth restriction-FGR) implies fetal incapability to achieve its
genetically determined growth potential. The American College of
Obstetricians and Gynecologists (ACOG) and Society for
Maternal-Fetal Medicine (SMFM) define IUGR based on a sonographic
finding of estimated fetal weight (EFW) or abdominal circumference
(AC) below the 10th centile for gestation age (1) and is diagnosed in about 10% of
pregnancies (2). IUGR is a
heterogeneous entity since the growth restriction could be caused
by fetal, maternal, or placental pathology with possible common
overlapping. Abnormal placentation and placental vascular disease
lead to chronic uteroplacental hypoxia and IUGR (2). Our interest is IUGR due to
uteroplacental insufficiency and dysfunction since the placental
origin is the most common cause of late-onset IUGR (3). Adequate diagnosis and appropriate
pregnancy monitoring and delivery timing of growth-restricted
fetuses are essential because of the increased risk of fetal and
neonatal morbidity and mortality. That said, perinatal mortality
with IUGR is 6- to 10- fold increased (4). Also, numerous studies found that
infants born with IUGR had an increased risk for neurodevelopmental
abnormalities and lower cognitive performance (5–7).
The specific pattern of balanced differentiation of
various trophoblastic cell types is crucial for normal placentation
(8,9), and numerous studies reported a
decisive role of appropriate Wingless (Wnt) signaling in normal
placental development (10–12).
Wnt canonical pathway is an evolutionary conserved
cell-signaling system essential for the regulation of cell
differentiation, migration, invasion, and apoptosis (13), thus contributing to multiple organ
system development (14–17). Consequently, abnormal Wnt
signaling has been reported in various diseases and abnormalities
such as pregnancy-related diseases-preeclampsia or IUGR (18), birth defects, different
malignancies (19–22) and the pathophysiology of various
neuropsychiatric disorders (23).
β-catenin is a cytoplasmic protein with an essential role in Wnt
canonical signaling pathway. Wnt ligands activate Wnt signaling.
WNT5A is one of these ligands that plays a critical role in
convergent extension (CE), epithelial-mesenchymal transition (EMT),
and planar cell polarity (PCP) regulation during the embryonic
period (24). Once Wnt ligand
binds to the transmembrane Frizzled (Fz) receptor and LRP5 and LRP6
co-receptors (lipoprotein receptor-related protein 5 and 6),
cytoplasmic Dishevelled (DVL) protein activates and degrades the
APC (adenomatous polyposis coli)/Axin/CK1a (casein kinase 1a)/GSK3ß
(glycogen synthase kinase 3b) destruction complex causing the
stabilization and accumulation of β-catenin in the cytoplasm, in an
active, non-phosphorylated form. Accumulated β-catenin then
migrates to the nucleus. It associates to TCF/LEF (T-cell
factor/lymphoid-enhanced binding factor) family of transcription
factors that subsequently activate the target genes' transcription.
In the absence of Wnt ligands, when Wnt canonical pathway is not
activated, the APC/Axin/CK1a/GSK3ß destruction complex binds to
β-catenin, causing the phosphorylation (i.e., inactivation) of
β-catenin and its proteasomal degradation.
The Hh signaling pathway also has a decisive role
during embryonic development due to its function in trophoblast EMT
(25), cell growth and patterning
(26), angiogenesis and
vasculogenesis, as well as various tissue and organ systems
development (27,28).
Signaling begins when one of three Hedgehog
homologs, Indian (IHH), Desert (DHH) or Sonic (SHH), binds to the
membrane Patched receptor (PTCH), and then seven transmembrane
spanning protein Smoothened (SMO) initiates a signaling cascade and
activates GLI (glioma-associated oncogene) transcription factors
and target genes. Suppressor of fused (SUFU) exerts its function as
an essential negative regulator of the Hh signaling pathway by
binding to GLI, thus causing phosphorylation and inactivation of
GLI and further signaling. Min et al (29) demonstrated that SUFU could be a
negative regulator of the Wnt and Hh signaling pathways, showing a
potentially strong linkage between these pathways. This could be
very significant since SUFU might be an essential link between the
two pathways and their crosstalk.
DNA methylation is one of the most important and
well-studied DNA epigenetic modifications that have a decisive role
in placental development. Altered placental methylation patterns
have been associated with the disruption of placental morphology
and linked to pregnancy pathology (30). It has been shown that treating
pregnant rats with DNA methyltransferase inhibitor (5azaC) at
different stages of pregnancy results in altered trophoblast
proliferative activity, disruption of placental structure, and
reduced placental weight (31).
The altered DNA methylation patterns have also been reported in
placentas from pregnancies with underlying placental pathologies
such as preeclampsia or IUGR (32). Also, another study found
growth-related WNT2 gene promoter methylation to be
associated with low birth weight (33). Based on the knowledge of the
importance of DNA methylation in regulating gene expression in the
placenta and the fact that it is the best known and studied
epigenetic modification, we wanted to elucidate if this mechanism
also regulates the SUFU gene expression.
MicroRNAs (miRNAs), a class of small (19–25
nucleotides) evolutionary conserved endogenous single-stranded
non-coding RNAs, also play an important role in epigenetic
regulation of gene expression. They prevent protein production in a
sequence-specific manner by participating in either cleavage and
degradation or translation inhibition of target mRNAs (34–36). Up to now, aberrant expression of
regulatory miRNAs has been associated with the initiation and
progression of various pathological processes (37–39). Recent evidence also highlights
their regulatory roles in human fetoplacental growth (40–43). Some of them, such as miR-214 and
miR-378, have also regulated the Hh signaling pathway (44–46).
Based on these assumptions and since the role of the
Wnt and Hh signaling pathways is still insufficiently explored in
the placenta and IUGR, we wanted to investigate the expression of
WNT5A, β-catenin, and SUFU in placentas from IUGR and gather more
information regarding the role of these pathways in placental
pathology related to IUGR. Also, our study aimed to explore if DNA
methylation and miR-214-3p and miR-378-5p targeting could be
epigenetic mechanisms involved in regulating SUFU gene
expression in placentas.
To our knowledge, up to date, SUFU gene
methylation status and the status of miR-214-3p and miR-378-5p in
the term IUGR placentas have not been reported.
Materials and methods
Tissue samples
The samples used in the study were a part of a
collection of placental tissue samples belonging to the University
of Zagreb School of Medicine. They were collected in collaboration
with the University Hospital ‘Merkur’ Zagreb. Both institutions are
parts of the Scientific Center of Excellence for Reproductive and
Regenerative Medicine (CERRM).
In the examination of placentation, a control group
consisted of eighteen formalin-fixed paraffin-embedded (FFPE)
placentas, obtained from physiological singleton complication-free
pregnancies, delivered at term (between 38 and 42 weeks of
gestation) of a newborn with normal body weight (between 10th and
90th percentile for gestational age, newborn sex, and mother's
parity). The experimental group consisted of 34 term placentas from
pathological pregnancies with IUGR based on serial ultrasound
measurements of fetal biparietal diameter (BPD), head and abdominal
circumference (HC and AC, respectively), and femur length (FL),
with the assessment of the bodyweight below 10th percentile for the
duration of pregnancy, fetal sex, mother's parity, and confirmed at
birth by measuring newborn body weight. The only pregnancy
pathology that was included in the study was IUGR. Exclusion
criteria for pathological pregnancies and controls were as follows:
Multiple pregnancies, tobacco and drug use, intrauterine viral
infections (TORCH and Parvovirus B19), chorioamnionitis,
hypertension, preeclampsia, fetal malformations, and genetic
abnormalities as well as autoimmune diseases or eating disorders of
the mother. The board-certified pathologist (A.S.) examined each
placenta and rendered the diagnosis. A disc-shaped tissue sample
comprising an entire thickness of the placenta from the fetal to
maternal side, about 5 cm from the umbilical cord, was taken from
each placenta.
Immunohistochemistry (IHC)
IHC was performed on 34 IUGR placentas and 18
control placentas. FFPE tissue sections (4 µm thickness) were
placed on silanized glass slides (DakoCytomation) and analyzed by
immunohistochemistry as previously described (47). Antigen retrieval was performed by
heating the sections in Dako Target Retrieval Solution (Dako
Corporation) in a steamer for 20 min. Sections were incubated with
primary antibody overnight at 4°C. Dako REAL Envision detection
system (cat. no. K0679; Dako; Agilent Technologies, Inc.) was
utilized for visualization as suggested by the manufacturer, and
the sections were counterstained with hematoxylin at room
temperature (RT) for 1 min. The following primary antibodies were
used: anti-WNT5A (mouse monoclonal anti-human; Cat. No. ab86720,
Abcam, dilution 1:1,000), anti-β-catenin (rabbit polyclonal
anti-human; Cat. No. ab16051, Abcam, dilution 1:500) and anti-SUFU
(rabbit polyclonal anti-human; Cat. No. 26759-1-AP, Proteintech,
dilution 1:500). Tonsils (WNT5A), colon tissue (β-catenin), and
kidney (SUFU) were used as positive controls. Negative control was
treated the same way with the omission of incubation with primary
antibodies.
The expression pattern of WNT5A, β-catenin, and SUFU
in placentas was interpreted independently by two pathologists
(S.V., A.S.) as follows: 0 if no staining was observed; 1 if
<10% cells were stained; 2 if 10–50% cells were stained; and 3
if >50% cells were stained (48). Protein expression was observed in
trophoblasts, stromal cells, and endothelial cells. In discordant
interpretations, the pathologists reviewed cases together to obtain
an agreement.
Methylation-specific PCR (MSP)
DNA was isolated from two 10 µm sections of FFPE
control placental tissue (n=14) and IUGR placental tissue (n=10) as
previously described (49) and
treated with bisulfite using the MethylEdge Bisulfite Conversion
System (Promega, Madison, Wisconsin, USA) according to the
instructions from the manufacturer. Bisulfite-treated DNA was used
for methylation-specific PCR reaction (MSP). Primers for
SUFU promoter region (Table
I) were synthesized according to Paluszczak et al
(50). All PCRs were performed
using TaKaRa EpiTaq HS (for bisulfite-treated DNA) (TaKaRa Bio):
1XEpiTaq PCR Buffer (Mg2+ free), 2.5 mM
MgCl2, 0.3 mM dNTPs, 10 pmol of each primer
(Sigma-Aldrich), 25 ng of DNA, and 0.75 Units of TaKaRa EpiTaq HS
DNA Polymerase in a 25 µl final reaction volume. PCR cycling
conditions were as follows: initial denaturation at 95°C for 30
sec, followed by 40 cycles consisting of three steps: 95°C for 30
sec, the respective annealing temperature for 30 sec, 72°C for 30
sec, followed by a final extension at 72°C for 7 min. For the
amplification of the methylated SUFU promoter region, the
annealing temperature was 58°C, while for the unmethylated
SUFU promoter region was 55°C. PCR products were separated
on 2% agarose gel stained with GelStar nucleic acid stain (Lonza
Rockland, Inc.) and visualized on a UV transilluminator. Methylated
Human Control (Promega) was used as a positive control for
methylated reaction, and unmethylated human EpiTect Control DNA
(Qiagen) was used as a positive control for unmethylated reaction,
and nuclease-free water was used as a negative control.
 | Table I.Primer sequences for MSP and
RT-qPCR. |
Table I.
Primer sequences for MSP and
RT-qPCR.
| Targeted gene | Accession
number | Forward primer
(5′-3′) | Reverse primer
(5′-3′) | Product length,
bp |
|---|
| MSP primers |
|
|
|
|
|
SUFU | NC_000010.11 |
GTTTCGGGGAGTTTTATTTATC |
GAAAACCGAAAAAACAATCG | 180 |
|
methylated |
|
|
|
|
|
SUFU | NC_000010.11 |
GTTTTGGGGAGTTTTATTTATTGA |
AAACAAAAACCAAAAAAACAATCA | 183 |
|
unmethylated |
|
|
|
|
| RT-qPCR |
|
|
|
|
| primers |
|
|
|
|
|
WNT5A | NM_003392.7 |
GCACCAGAGCAGACAACC |
TCACAACACGGAGGAATCAG | 89 |
|
SUFU | NM_016169.4 |
GCTGCTGACAGAGGACCCACA |
GTGCAGACACCAACGATCTGGA | 84 |
|
CTNNB1 | NM_001904.4 |
TGCGTACTGTCCTTCGGGCT |
ATGGCAGGCTCAGTGATGTCT | 52 |
|
GAPDH | NM_002046.7 |
TGCACCACCAACTGCTTAGC |
GGCATGGACTGTGGTCATGAG | 87 |
RNA extraction, reverse transcription,
and RT-qPCR
For mRNA analysis, total RNA was isolated from five
consecutive 5-µm thick sections of FFPE control placental tissue
(n=14) and IUGR placental tissue (n=14). Shortly, all tissue
sections were deparaffinized by incubation in 1.0 ml xylene
(Invitrogen; Thermo Fisher Scientific, Inc.) for 3 min at 50°C,
followed by centrifugation (three times for 5 min at RT at 12,000 ×
g each). The supernatant was then discarded, and the obtained
tissue pellet was washed three times with 1.0 ml absolute ethanol
and subsequently incubated overnight at 55°C in 350 µl of protease
K digestion buffer (20 mM Tris-HCl pH 8.0; 1 mM CaCl2;
0.5% sodium dodecyl sulfate and 500 µg/ml protease K; all reagents
were obtained from Sigma-Aldrich; Merck KGaA). Total RNA was
isolated using TRIzol® reagent (Invitrogen; Thermo
Fisher Scientific, Inc.) following the manufacturer's recommended
procedure. The purity and quantity of total RNA were evaluated
using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific,
Inc.). Subsequently, one µg of total RNA from each sample was
reverse transcribed using the high-capacity cDNA Reverse
Transcription kit (Applied Biosystems; Thermo Fisher Scientific,
Inc.) according to the manufacturer's protocol. The mRNA expression
levels of targeted genes (CTNNB1, WNT5A, SUFU) were
determined using a CFX-96 real-time qPCR detection system (Bio-Rad
Laboratories, Inc.). All qPCR reactions were performed in
triplicate using TB Green™ Premix Ex Taq™ II (Tli RNaseH Plus PCR
master mix; Takara Biotechnology Co., Ltd.). The following
thermocycling conditions were used: Initial denaturation at 95°C
for 30 sec, followed by 40 cycles of 95°C for 5 sec and 60°C for 30
sec. The CFX96 manager software version 3.1 (Bio-Rad Laboratories,
Inc.) was used to generate the cycle threshold values, and the data
were analyzed using the 2−ΔΔCq method (51). The relative mRNA expression levels
of CTNNB1, WNT5A, and SUFU were normalized against
the mRNA expression levels of the glyceraldehyde-3-phosphate
dehydrogenase (GAPDH) gene as an endogenous control. The
specificity of qPCR amplification was confirmed using melting curve
analysis. The primer sequences used for mRNA analysis are presented
in Table I.
For microRNA analysis, total RNA was isolated from
five consecutive 5-µm thick sections of FFPE control placental
tissue (n=14) and IUGR placental tissue (n=14) as described above
and purified with the ‘NucleoSpin miRNA Plasma̛ kit
(Macherey-Nagel, Germany). Reverse transcription was performed
using the ‘TaqMan MicroRNA Reverse Transcription Kit’ (Applied
Biosystems) and specific loop primers for Hsa-miR-214-3p (assay ID
002306, Applied Biosystems, USA) and Has-miR-378a-5p (assay ID
000567, Applied Biosystems, USA) following the manufacturer's
procedure. The expression levels of targeted miRNAs were determined
using the ‘TaqMan microRNA Assay’ and ‘TaqMan™ Universal Master Mix
II, no UNG’ (Thermo Fisher Scientific, Inc.) following the
manufacturer's instructions. Small nuclear RNA U6 (U6 snRNA assay
ID 001973, Applied Biosystems, USA) was used as an endogenous
control. The following thermocycling conditions (CFX-96 real-time
qPCR detection system; Bio-Rad Laboratories, Inc.) were used:
Initial denaturation at 95°C for 10 min, followed by 40 cycles of
95°C for 15 sec and 60°C for 60 sec. All qPCR reactions were
performed in triplicate, and data were analyzed using the
2−ΔΔCq method as described above (51).
Statistical analysis
The normality of data distribution was tested using
the Shapiro-Wilk test. Continuous variables are shown as mean ±
standard deviation (SD) or median with interquartile range (Q1,
Q3). Categorical variables were presented as frequencies (n) and
percentages (%). Based on the normality of data distribution, the
group differences for quantitative variables were analyzed using
the unpaired Student's t-test (normally distributed data) or the
Mann-Whitney U test (non-normally distributed data),
correspondingly. Categorical variables were compared using the
Chi-square or Fisher's exact test, as appropriate. After Chi-square
or Fisher's exact test, and to correct for multiple comparisons,
where necessary, Bonferroni correction was used and corrective
t-values stated, as appropriate. The two-tailed P < 0.05 was
considered statistically significant. All analyses were performed
using the SPSS Statistics software version 27.0 (IBM Corp.).
Results
Clinical data - physiological and IUGR
pregnancies
The following clinical variables were analyzed:
maternal age, maternal body weight and height, maternal weight gain
and body mass index (BMI) before pregnancy and at time of delivery,
fetal body weight and height, placental weight and fetal/ placental
weight ratio. Moreover, the gender of the newborns and mode of
delivery was also analyzed. As expected, a statistically smaller
fetal weight, height and placental weight were found in newborns
from IUGR pregnancies (P<0.001). IUGR was more frequent in
female newborns (P=0.033), and pregnancies complicated with IUGR
were significantly more often completed with cesarean section
delivery than physiological pregnancies (P=0.001), which was also
expected. The mean maternal age in the IUGR group was 31.6 and was
significantly higher compared with healthy controls with a mean age
of 28.5 years (P=0.035). Median gestational age at delivery was
significantly lower in the IUGR group compared with the control
group, 38+3/7 (38+2/7, 39+4/7) and 39+3/7 weeks (39+3/7, 40+5/7),
respectively (P=0.001) (Table
II).
 | Table II.Clinical parameters of mothers and
newborns from IUGR and physiological pregnancies. |
Table II.
Clinical parameters of mothers and
newborns from IUGR and physiological pregnancies.
| Variable | Control (n=18) | IUGR (n=34) | P-value |
|---|
| Maternal age,
years | 28.5±3.6 | 31.6±5.5 | 0.035a |
| Gestational age at
delivery, weeks+days | 39+3/7 (39+2/7,
40+5/7) | 38+3/7 (38+2/7,
39+4/7) | 0.001a |
| Pre-pregnancy BMI,
kg/m2 | 21.9 (19.3,
22.9) | 20.2 (19.0,
21.7) | 0.532 |
| BMI at delivery,
kg/m2 | 26.6±2.7 | 26.0±3.0 | 0.532 |
| Maternal body
height, cm | 168.4±5.5 | 166.2±7.0 | 0.253 |
| Maternal body
weight at delivery, kg | 75.4±8.0 | 72.0±9.6 | 0.210 |
| Total weight gain
during pregnancy, kg | 16.0 (13.0,
18.5) | 14.0 (11.0,
20.0) | 0.159 |
| Fetal birth weight,
g | 3,516.7±336.9 | 2435.9±195.9 |
<0.001a |
| Fetal height,
cm | 51.0 (50.0,
51.2) | 46.0 (45.7,
48.0) |
<0.001a |
| Placental weight,
g | 554.6±76.6 | 369.7±80.2 |
<0.001a |
| Fetal/placental
weight ratio | 6.4±0.9 | 6.9±1.5 | 0.253 |
| Sex, n (%) |
|
|
|
|
Male | 13 (72.2) | 14 (41.2) | 0.033a |
|
Female | 5 (27.8) | 20 (58.8) |
|
| Mode of delivery, n
(%) |
|
|
|
|
Cesarean section | 0 (0.0) | 14 (41.2) | 0.001a |
| Vaginal
delivery | 18 (100.0) | 20 (58.8) |
|
Expression levels of WNT5A, SUFU, and
CTNNB1 mRNA
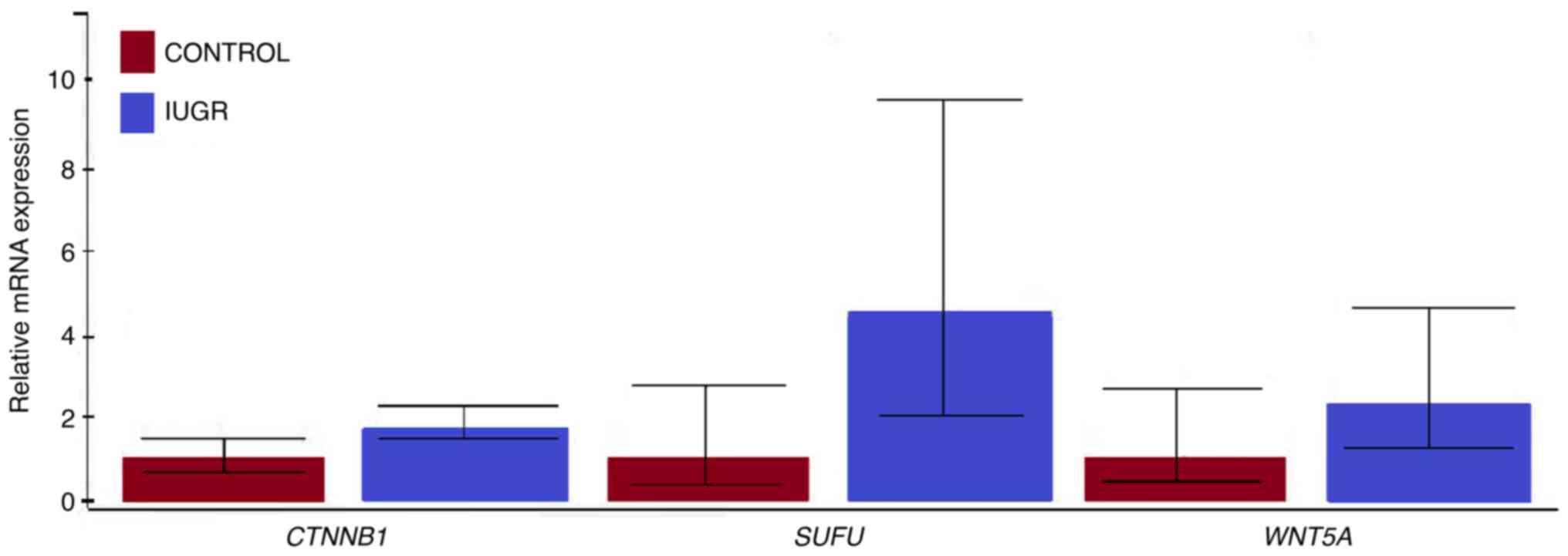
The RT-qPCR analysis showed that all targeted genes
(WNT5A, SUFU, and CTNNB1) were transcriptionally
active in both IUGR and control placenta tissue samples. The
relative mRNA expression levels of WNT5A, SUFU, and
CTNNB1 genes were higher in the IUGR than in the control
tissue (Fig. 1). However, none of
the observed differences in mRNA expression levels was
statistically significant. Regarding the mRNA expression levels of
targeted genes in IUGR tissue samples analyzed as a separate group,
the CTNNB1 gene showed the highest transcriptional activity,
followed by the SUFU and WNT5A genes. In control
tissue samples, the CTNNB1 gene also showed the highest
expression levels among the targeted genes analyzed. In contrast,
the expression levels of the SUFU gene were lower than those
observed for the WNT5A gene (data not shown). No
statistically significant gene expression was detected in the last
analysis as well.
WNT5A, β-catenin, and SUFU protein
expression in IUGR placentas
In IUGR placentas, WNT5A and β-catenin were
expressed in >10% of endothelial cells in 70.5 and 79.4%
samples, respectively. In contrast, these proteins were expressed
in <10% of endothelial cells in physiological placentas in 94.4
and 55.6% of samples. In IUGR placentas, β-catenin was expressed in
>10% of trophoblast cells in 94.1% of the samples, compared with
66.7% of the samples from placentas with uncomplicated pregnancies.
On the other hand, in both placental groups, WNT5A and β-catenin
expression in >10% of stromal cells were observed in 100% of the
samples (Table III).
 | Table III.WNT5A, β-catenin and SUFU protein
expression in IUGR and normal (control) placentas. |
Table III.
WNT5A, β-catenin and SUFU protein
expression in IUGR and normal (control) placentas.
|
| Throphoblasts | Stromal cells | Endothelial
cells |
|---|
|
|
|
|
|
|---|
| Protein | Normal placentas, n
(%) (n=18) | IUGR placentas, n
(%) (n=34) | P-value | Normal placentas, n
(%) (n=18) | IUGR placentas, n
(%) (n=34) | P-value | Normal placentas, n
(%) (n=18) | IUGR placentas, n
(%) (n=34) | P-value |
|---|
| WNT5A, % |
|
| 0.718 |
|
| 0.873 |
|
|
<0.001a |
|
>50 | 16 (88.9) | 29 (85.3) |
| 11 (61.1) | 20 (58.8) |
| 0 (0.0) | 6 (17.6) |
|
|
10-50 | 2 (11.1) | 5 (14.7) |
| 7 (38.9) | 14 (41.2) |
| 1 (5.6) | 18 (52.9) |
|
|
<10 | 0 (0.0) | 0 (0.0) |
| 0 (0.0) | 0 (0.0) |
| 9 (50.0) | 3 (8.8) |
|
| 0 | 0 (0.0) | 0 (0.0) |
| 0 (0.0) | 0 (0.0) |
| 8 (44.4) | 7 (20.7) |
|
| β-catenin, % |
|
| 0.059 |
|
| 0.602 |
|
| 0.002a |
|
>50 | 3 (16.7) | 11 (32.4) |
| 16 (88.8) | 32 (94.1) |
| 0 (0.0) | 16 (47.1) |
|
|
10-50 | 9 (50.0) | 21 (61.7) |
| 2 (11.2) | 2 (5.9) |
| 8 (44.4) | 11 (32.3) |
|
|
<10 | 5 (27.8) | 2 (5.9) |
| 0 (0.0) | 0 (0.0) |
| 2 (11.2) | 3 (8.8) |
|
| 0 | 1 (5.5) | 0 (0.0) |
| 0 (0.0) | 0 (0.0) |
| 8 (44.4) | 4 (11.8) |
|
| SUFU, % |
|
| 0.030b |
|
| 0.105 |
|
| 0.440 |
|
>50 | 5 (27.8) | 17 (50.0) |
| 8 (44.4) | 20 (58.8) |
| 0 (0.0) | 2 (5.9) |
|
|
10-50 | 9 (50.0) | 17 (50.0) |
| 7 (38.9) | 14 (41.2) |
| 10 (55.5) | 18 (52.9) |
|
|
<10 | 2 (11.1) | 0 (0.0) |
| 2 (11.2) | 0 (0.0) |
| 5 (27.8) | 12 (35.3) |
|
| 0 | 2 (11.1) | 0 (0.0) |
| 1 (5.5) | 0 (0.0) |
| 3 (16.7) | 2 (5.9) |
|
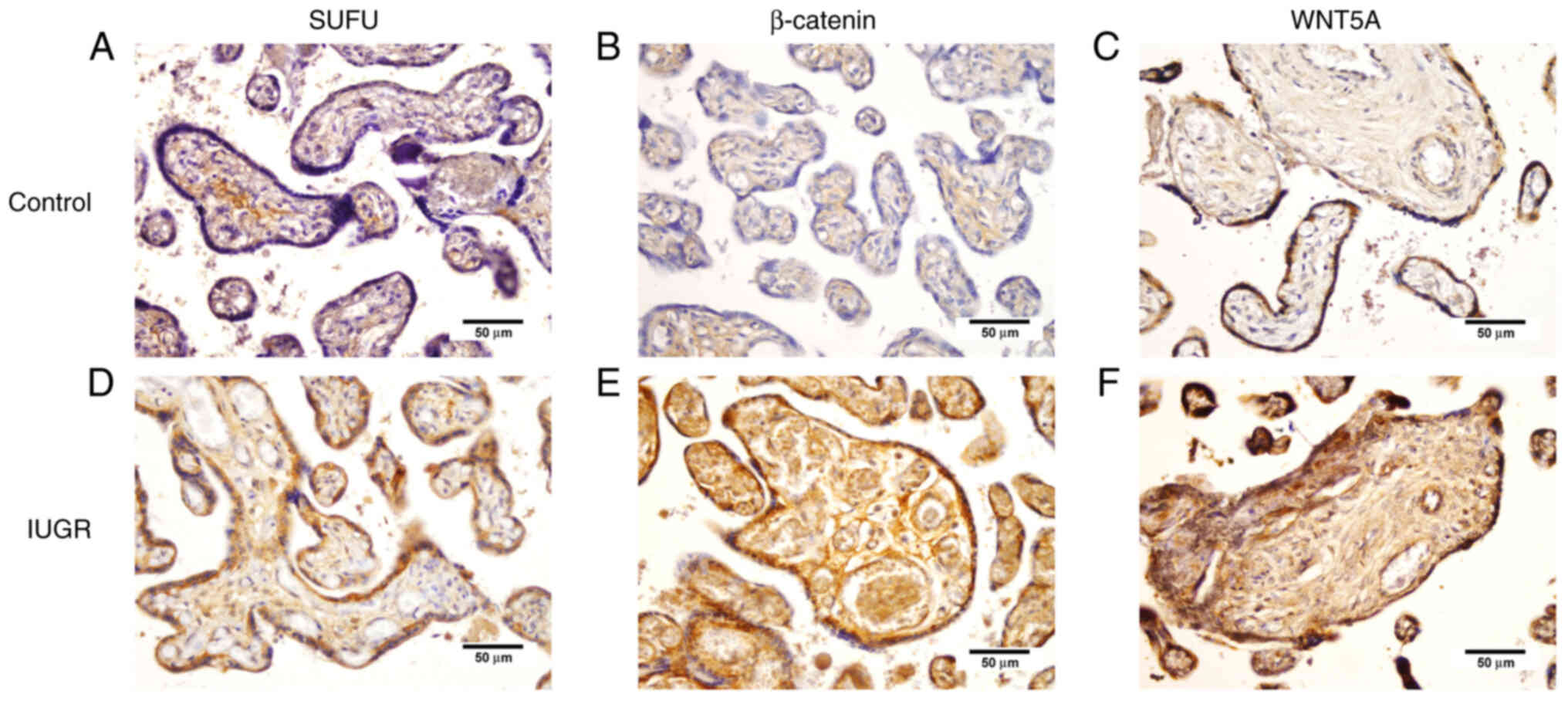
The expression of WNT5A, the positive regulator of
the Wnt signaling pathway, was significantly higher in endothelial
cells of placental villi (P<0.001) in placentas with IUGR
compared with term placentas from physiologic pregnancies.
β-catenin also exhibited a significantly higher expression in
trophoblasts (P=0.026) and endothelial cells of placental villi
(P<0.001) in IUGR placentas compared with the physiologic
placentas (Figs. 2 and 3).
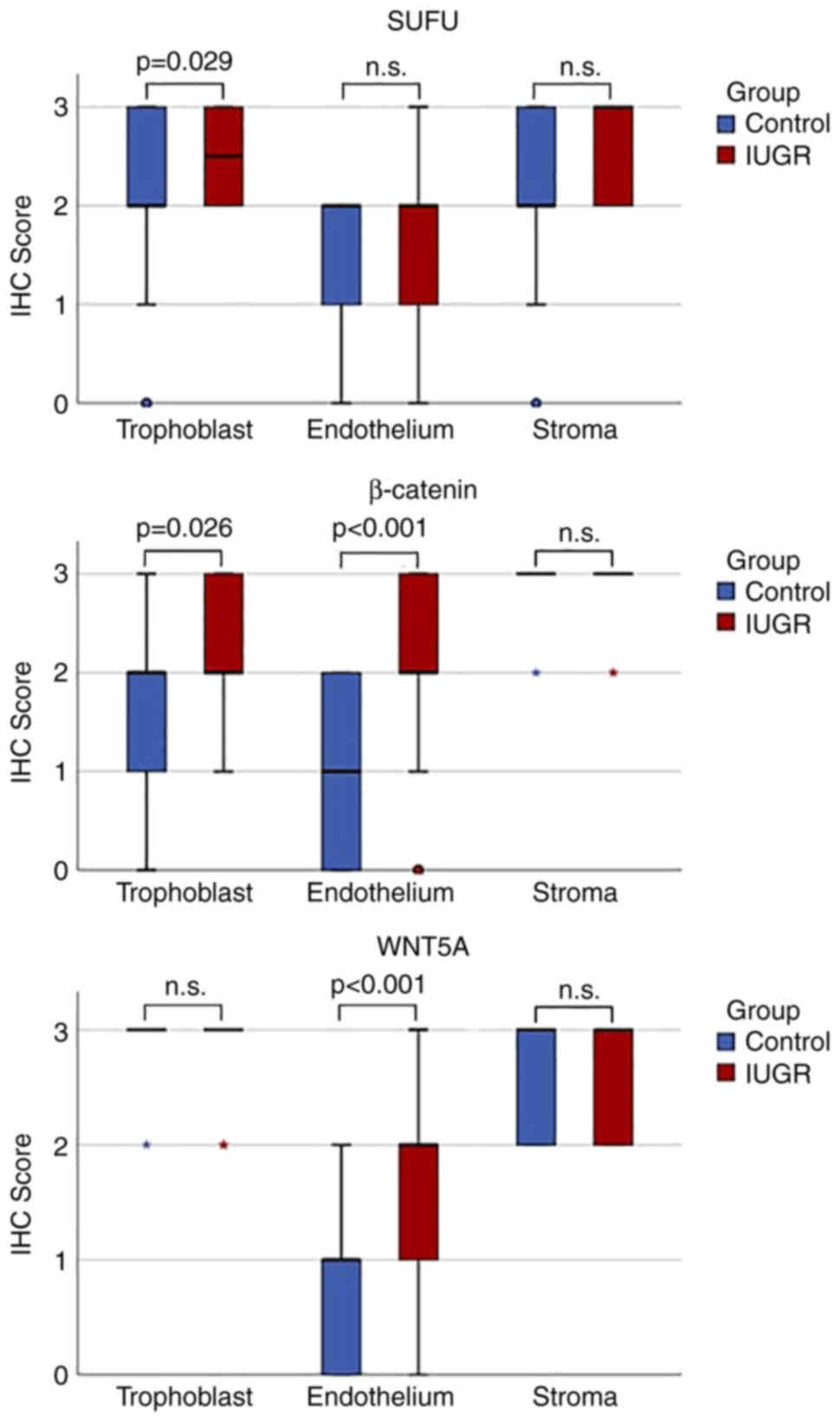 | Figure 3.Boxplots of WNT5A, β-catenin and SUFU
protein expression in IUGR and normal (control) placentas in
trophoblasts, endothelium and stroma. 0, no staining observed; 1,
<10% cells were stained; 2, 10–50% cells were stained; and 3,
>50% cells were stained. Asterisks denote extreme outliers,
while small circles denote outliers. IHC, immunohistochemistry;
IUGR, intrauterine growth restriction; n.s., not statistically
significant; SUFU, suppressor of fused. |
SUFU protein expression was observed in >10% of
trophoblast cells in 100% samples of IUGR placentas and 77.6%
samples of physiological placentas. The protein expression in
>10% of stromal cells of IUGR placentas was observed in 100% of
samples in the IUGR group and 83.3% in the control group. SUFU
protein expression was significantly higher in trophoblasts in IUGR
placentas (P=0.029) than in physiologic ones (Table III; Figs. 2 and 3).
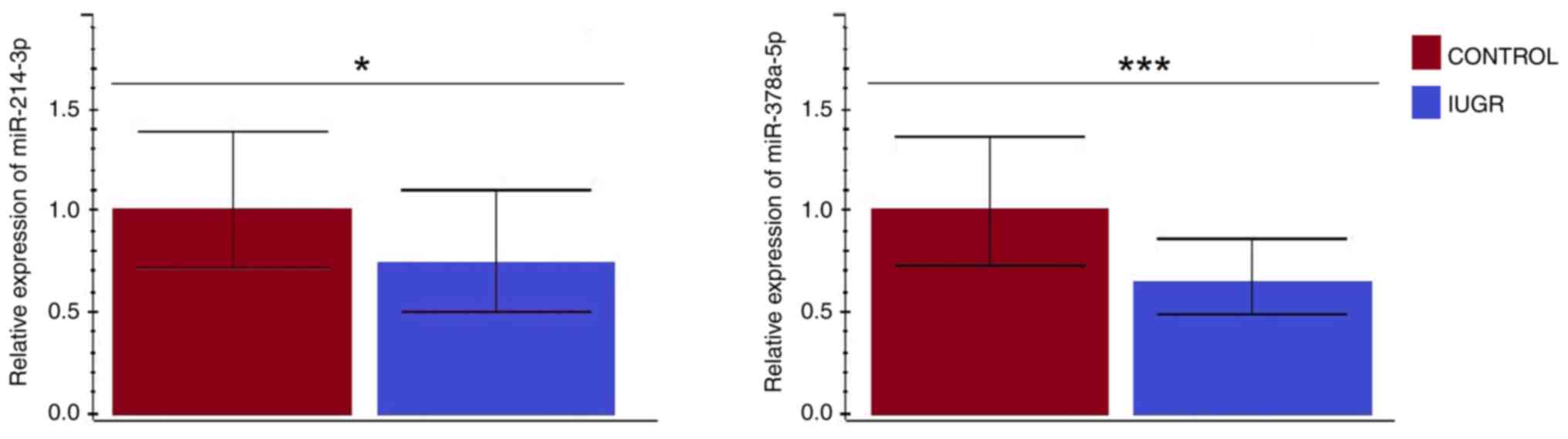
Expression levels of miR-214-3p and
miR-378a-5p
The RT-qPCR analysis results showed that targeted
miRNAs (miR-214-3p and miR-378a-5p) were expressed in all tissue
samples. Furthermore, both miR-214-3p (P=0.040) and miR-378a-5p
(P<0.001) showed a significantly lower expression in the IUGR
compared to control placental tissue (Fig. 4). Also, in control tissue samples
analyzed as a separate group, the miR-378a-5p showed higher
transcriptional activity. However, the observed difference was
insignificant (data not shown). Contrary to the IUGR tissue group,
both targeted miRNAs showed almost the same expression levels that
were lower than their expression values in the control tissue group
(data not shown).
DNA promoter methylation status of
SUFU gene in IUGR placentas
DNA promoter methylation of the SUFU gene was
analyzed by the methylation-specific PCR assay in 10 IUGR and 14
physiologic placentas. SUFU gene promoter was unmethylated
in all physiologic placentas, while in the IUGR group, one placenta
showed weak methylation of the SUFU gene promoter. In other
IUGR placentas, the SUFU gene promoter was unmethylated
(Fig. 5).
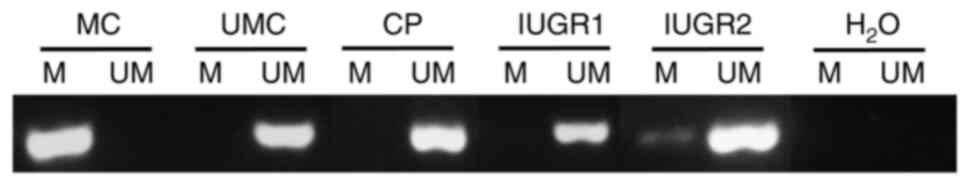 | Figure 5.A representative example of
methylation-specific PCR analysis for the suppressor of fused gene
promoter in term placentas from physiologic pregnancies (CP) and
term placentas from pregnancies complicated with IUGR. IUGR1 and
IUGR2 are samples from different patients. IUGR1 is a
representative example of the unmethylated promoter of the
SUFU gene in IUGR placentas. IUGR2 is one sample of IUGR
placenta with the presence of methylated promoter of the
SUFU gene. CP, control placenta; H2O, water,
negative control; IUGR, intrauterine growth restriction; M,
methylated reaction; MC, methylated human control, positive control
for methylated reaction; UM, unmethylated reaction; UMC,
unmethylated human control, positive control for unmethylated
reaction. |
Discussion
Since WNT5A and β-catenin are positive regulators of
the Wnt pathway, their significantly lower expression in placentas
from IUGR compared to the placentas from physiologic, uncomplicated
pregnancies could have been expected. Our results showed
significantly higher protein expression of WNT5A and β-catenin in
IUGR placentas compared to placentas from uncomplicated
pregnancies. These results align with our previous study that
revealed significantly higher expression of all three Dishevelled
proteins (DVL1-3) in IUGR placentas, indicating their potential
effects on placental hypoxia and angiogenesis in IUGR (52).
Uteroplacental insufficiency causes placental
hypoperfusion and chronic hypoxia and is one of the leading causes
of IUGR. Numerous studies report the association between oxidative
stress and IUGR (53–55) and the contribution of oxidative
stress to the IUGR metabolic sequelae (56). Zhang et al (57) reported that oxidative stress
upregulates Wnt signaling in a concentration-dependent manner and
induces angiogenic activity, thus contributing to
neovascularization. Funato et al (58) also found that reactive oxygen
species (ROS) promoted β-catenin stabilization. Vikram et al
(59) reported that suppressing
oxidative stress by antioxidants prevents β-catenin
dephosphorylation in endothelial cells. At the same time, the
β-catenin expression also increased ROS in endothelial cells and
whole blood vessels, suggesting that ROS could be both upstream
mediators and downstream effectors of Wnt signaling (59). Moreover, it has been reported that
active WNT3A and β-catenin could upregulate tumor necrosis
factor-alpha (TNF-α) in endothelial cells, thus promoting
endothelial dysfunction (60).
Increased expression of β-catenin also diminished vascular nitrogen
oxide (NO) bioavailability and impaired endothelium-dependent
vasorelaxation (59). Moreover,
endothelial cells from patients suffering from type 2 diabetes
mellitus had a 1.3-fold higher WNT5A expression. Furthermore,
inhibition of WNT5A restored endothelial NO synthase activity,
improved nitric oxide production and abrogated endothelial
dysfunction (61).
This data suggests that placental dysfunction was
triggered by hypoxia and oxidative stress. This may be partially
explained by the higher β-catenin expression in endothelial and
trophoblast cells and higher WNT5A expression in endothelial cells
in IUGR placentas obtained in our research.
Various studies emphasized the importance of Wnt
signaling in regulating apoptosis and suggested its antiapoptotic
activity (62,63). That said, Wnt signaling
activation, as showed in our IUGR placentas, could be a protective
mechanism that, besides inducing angiogenesis and supporting
vasorelaxation, could improve uteroplacental blood flow and fetal
oxygenation and be protective by reducing apoptotic activity and
negative consequences of oxidative stress.
On the other hand, other studies are not in
accordance with our results. Fan et al (64) reported active, dephosphorylated
β-catenin and Matrix Metallopeptidase 9 (MMP-9) levels to be
significantly lower in preeclampsia, especially a severe form,
compared with placentas from normal pregnancies, thus suggesting
the shallow invasion that is associated with preeclampsia (65) to be regulated by β-catenin via
Snail and MMP-9 (64). Other
studies also confirmed lower β-catenin expression in trophoblast
cells affected by hypoxia and inhibited proliferation, weakened
migration, invasion, and excessive apoptosis (66). Trophoblast invasion is vital for
normal embryonic development. The extravillous trophoblast is
formed in epithelium-mesenchymal transition (EMT) (67). Its capacity to invade the spiral
arteries, mediate the destruction of the arterial wall and replace
the endothelium is essential for pregnancy progress (68). There is evidence that Hh signaling
plays an essential role in EMT and invasion. Tang et al
(69) found higher expression of
Hh ligands in the villous core as Wnt-producing tissues and higher
expression of Hh receptors PTCH1 and SMO in trophoblast layers as
Wnt-responding tissues. They also found that the Hh ligand
stimulates the EMT of human cytotrophoblast cells.
In contrast, knockdown of Gli1 and Gli2 attenuated
SHH-induced EMT (indicated by lower vimentin and higher E-cadherin
expressions) and colony formation (69). Zhang and Zhang found that Forkhead
Box C2 (FOXC2) facilitates trophoblast invasion through the
activation of the Hh pathway-trophoblast cells with overexpression
of FOXC2 also experienced higher expression levels of SHH, Gli and
Snail and were more invasive (70). The effect was reverted when the
samples were treated with siRNA targeting FOXC2 (70). All these data emphasize the
importance of Hh signaling in human trophoblast physiology. To the
best of our knowledge, the role of SUFU in placentation and IUGR
has not been addressed by any previous study. Our results showed
that its expression is significantly higher in the trophoblast
cells of the IUGR placentas, whereas there were no differences in
endothelium and stromal cells. As a negative regulator of the Hh
pathway, higher expression of SUFU in IUGR trophoblast cells may
contribute to lower activity of Hh signaling and subsequently to
impaired trophoblast function in IUGR placentas.
Since SUFU is a negative regulator of the Hh pathway
and possibly the Wnt pathway, its greater protein expression in
IUGR placentas could be expected. Our results align with that, but
higher protein expression of positive Wnt pathway regulators, WNT5A
and β-catenin, was also found in IUGR placentas than normal ones.
Higher expression of SUFU protein in that context could reveal
another role of SUFU. Liu et al (71) demonstrated that SUFU could also be
a positive regulator of the Hh pathway, thus maximizing Hh pathway
activation. Moreover, it has been reported that RIO kinase 3
(RIOK3) acts as a SUFU-dependent positive regulator of Hh signaling
(72), suggesting that SUFU could
exert its double function as a positive regulator through other
compounds.
In contrast to their protein expressions, the
RT-qPCR analysis revealed no significant difference in WNT5A,
SUFU, and CTNNB1 mRNA expressions between IUGR placentas
and controls. This could be explained by the fact that IHC analysis
enabled protein expression analysis and quantification in different
placental compartments (trophoblasts, stromal cells and endothelial
cells). In contrast, RT-qPCR analysis was performed in whole
placental tissue sections.
In our study, other than the SUFU gene and
protein expression, we also wanted to perceive the SUFU gene
promoter methylation status. Our results show that the SUFU
gene promoter is unmethylated in all but one IUGR placenta. It is
also unmethylated in all the placentas from uncomplicated
pregnancies. We conclude that other epigenetic mechanisms might
regulate SUFU gene expression based on these results.
Although it has been demonstrated that DNA methylation is an
essential epigenetic mechanism in the human placenta and that IUGR
is significantly associated with altered DNA methylation patterns
in the placenta (30,73), growing evidence, including our
results, suggests other epigenetic mechanisms could also be
fundamental in placental gene expression regulation. Kimura et
al (74) demonstrated that
histone post-translational modifications could be an essential
mechanism of placental gene expression regulation. Moreover, Chuang
et al (75) reported that
histone modification is linked with the expression of genes that
are decisive for mediating trophoblastic fusion and, therefore,
proper placental structure and function.
MiRNAs appear to be actively involved in placental
gene regulation and development (76). It has been reported that the
expression of several placenta-specific miRNAs has been reduced in
placentas from pregnancies with IUGR than in placentas from
uncomplicated pregnancies (77).
Pineles et al (78)
reported specific miRNA expression patterns associated with
preeclampsia, which was also confirmed by Zhu et al
(79). Another study also
confirmed functional miRNAs in the trophoblast. The specific miRNAs
in the placenta can be up or down-regulated by the varying oxygen
levels, primarily in a hypoxic environment (80). This is an interesting finding that
could be important in IUGR since IUGR is associated with chronic
hypoxia, as we already stressed earlier. Peng et al
(81) reported that SUFU
was regulated by miRNA-20b and induced cell proliferation,
migration and EMT by negatively regulating both Wnt and Hh
signalling pathways.
Several studies reported SUFU gene expression
to be epigenetically downregulated by miRNAs and thus silenced in
various tumors such as breast, gastric, basal cell, or non-small
cell lung carcinomas (82–85).
Alimirah et al (82)
reported that miR-214 targets the SUFU gene, which then
inhibits its expression in breast cancer. This finding was also
confirmed in another study that found SUFU gene expression
negatively correlated with miR-214 expression, indicating that
miR-214 directly targeted SUFU expression and Hh signaling
in promoting liver fibrosis (86). Moreover, He et al (44) reported precisely the
miR-214-3p-SUFU-GLI1 axis as the critical signaling pathway
responsible for smooth muscle cell (SMC) differentiation and
generation from adventitial stem/progenitor cells (AdSPCs)
important for controlling neointimal hyperplasia. MiR-214-3p
controls vascular SMC proliferation and migration, while
SUFU is identified as its true target gene that operates as
a transcriptional repressor of SMC contractile genes, which is
essential in the context of vascular remodeling after injury
(44).
MiR-378a-5p was reported to negatively regulate the
expression of SUFU as a target gene in melanomas. That is
important since miR-378a-5p was found to increase cell migration
and invasion and to have proangiogenic activity by significantly
inducing angiogenic growth factor VEGF secretion, which then
increases in vivo and in vitro angiogenesis (87). Earlier studies also reported
miR-378a-5p as a promoter of angiogenesis by upregulating VEGF,
thus inducing neovascularization in hypoxia by targeting the
SUFU gene (45,88).
Based on these studies on other tissues showing
miR-214-3p and miR-378a-5p to modulate SUFU expression, we
wanted to identify their potential involvement in the regulation of
SUFU expression in placentas and, especially, in IUGR
placentas. This is particularly interesting due to the role of
these miRNAs in vascular remodeling and angiogenesis. Our results
showed that miR-214-3p and miR-378a-5p expressions were increased
in control placentas compared with IUGR placentas. This aligns with
protein expression results where SUFU protein expression was
increased in IUGR term placentas compared with physiological ones.
Our results also suggest that miR-214-3p and miR-378a-5p targeting
could be involved in epigenetic regulation of SUFU gene
expression in normal and IUGR placental tissue.
We found mean maternal age to be significantly
higher in the IUGR group compared with the control group. This is
interesting since there are conflicting reports regarding the
association between maternal age and risk for IUGR. Some studies
found increased maternal age to be an independent risk factor for
IUGR, which is in line with our findings (89,90). Other studies did not find any
association between maternal age and IUGR risk (91–93), while Yu et al (94) reported younger maternal age as a
risk factor for IUGR.
There are several limitations of the present study.
First, the sample size was moderate. Second, the study did not
explore the functional impact of the targeted mRNA and miRNA
expression on trophoblast cells. It would also be interesting to
focus on other epigenetic mechanisms besides the reported
miR-214-3p, miR378a-5p and DNA methylation in placentas with IUGR.
Our future studies will focus on the shortcomings of the current
study.
Our study provides new insights on the involvement
of the Wnt and Hh signaling pathways and epigenetic regulation of
SUFU gene expression in the placenta and IUGR. However, our
results should be further explored in a larger cohort to specify
more closely their exact functional roles in placental
insufficiency, detection or surveillance of IUGR, and optimal
delivery planning. The precise functional impact of miR-214-3p and
miR-378a-5p on SUFU gene expression in these tissue and
pathological settings should be scrutinized as well.
Acknowledgements
Not applicable.
Funding
This research was co-financed by the European Union through the
Europe Regional Development Fund, Operational Programme
Competitiveness and Cohesion, under grant agreement no.
KK.01.1.1.01.0008, Reproductive and Regenerative Medicine-Exploring
New Platforms and Potentials.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
IMS contributed to conceptualization, data
interpretation, data acquisition and analysis, performed
experimental work, wrote and edited the manuscript, and revised the
manuscript for important intellectual content. VKK contributed to
conceptualization, interpretation and design of experiments, edited
the manuscript, and revised the manuscript for important
intellectual content. FP contributed to data analysis and
interpretation, performed experimental work, and revised the
manuscript for important intellectual content. LL contributed to
data analysis and interpretation, and revised the manuscript for
important intellectual content. MG contributed to data
interpretation, performed experimental work and revised the
manuscript for important intellectual content. NS contributed to
data interpretation, and revised the manuscript for important
intellectual content. TD contributed to data interpretation and
performed experimental work. AS contributed to data analysis and
interpretation, and revised the manuscript for important
intellectual content. KK contributed to data interpretation, and
revised the manuscript for important intellectual content. SV
contributed to data analysis and interpretation, and revised the
manuscript for important intellectual content. LS conceived the
idea, contributed to conceptualization, data collection and
analysis and interpretation of the results, and revised the
manuscript for important intellectual content. LS and IMS confirm
the authenticity of all the raw data. All authors read and approved
the final manuscript.
Ethics approval and consent to
participate
The study was conducted according to the guidelines
of the Declaration of Helsinki and approved by the Ethics Committee
of the School of Medicine, University of Zagreb, Zagreb, Croatia
(641-01/22-02/01; 30th October 2018) and the Ethics Committee of
the University Hospital Merkur Zagreb, Zagreb, Croatia (03/1-1341;
14th February 2018). Written informed consent was obtained from all
participants involved in the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
ACOG Practice bulletin no. 134, . Fetal
growth restriction. Obstet Gynecol. 121:1122–1133. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hutter D, Kingdom J and Jaeggi E: Causes
and mechanisms of intrauterine hypoxia and its impact on the fetal
cardiovascular system: A review. Int J Pediatr. 2010:4013232010.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Krishna U and Bhalerao S: Placental
insufficiency and fetal growth restriction. J Obstet Gynaecol
India. 61:505–511. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Pollack RN and Divon MY: Intrauterine
growth retardation: Definition, classification, and etiology. Clin
Obstet Gynecol. 35:99–107. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Guellec I, Lapillonne A, Renolleau S,
Charlaluk ML, Roze JC, Marret S, Vieux R, Monique K and Ancel PY;
EPIPAGE Study Group, : Neurologic outcomes at school age in very
preterm infants born with severe or mild growth restriction.
Pediatrics. 127:e883–e891. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sacchi C, Marino C, Nosarti C, Vieno A,
Visentin S and Simonelli A: Association of intrauterine growth
restriction and small for gestational age status with childhood
cognitive outcomes: A systematic review and meta-analysis. JAMA
Pediatr. 174:772–781. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Levine TA, Grunau RE, McAuliffe FM,
Pinnamaneni R, Foran A and Alderdice FA: Early childhood
neurodevelopment after intrauterine growth restriction: A
systematic review. Pediatrics. 135:126–141. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Latos PA and Hemberger M: From the stem of
the placental tree: Trophoblast stem cells and their progeny.
Development. 143:3650–3660. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hemberger M, Hanna CW and Dean W:
Mechanisms of early placental development in mouse and humans. Nat
Rev Genet. 21:27–43. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Knöfler M and Pollheimer J: Human
placental trophoblast invasion and differentiation: A particular
focus on Wnt signaling. Front Genet. 4:1902013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Matsuura K, Jigami T, Taniue K, Morishita
Y, Adachi S, Senda T, Nonaka A, Aburatani H, Nakamura T and Akiyama
T: Identification of a link between Wnt/β-catenin signalling and
the cell fusion pathway. Nat Commun. 2:5482011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Aoki M, Mieda M, Ikeda T, Hamada Y,
Nakamura H and Okamoto H: R-spondin3 is required for mouse
placental development. Dev Biol. 301:218–226. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Miller JR: The Wnts. Genome Biol.
3:Reviews30012002.PubMed/NCBI
|
|
14
|
Logan CY and Nusse R: The Wnt signaling
pathway in development and disease. Annu Rev Cell Dev Biol.
20:781–810. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Cadigan KM and Peifer M: Wnt signaling
from development to disease: Insights from model systems. Cold
Spring Harb Perspect Biol. 1:a0028812009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
van Amerongen R and Nusse R: Towards an
integrated view of Wnt signaling in development. Development.
136:3205–3214. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
MacDonald BT, Tamai K and He X:
Wnt/beta-catenin signaling: Components, mechanisms, and diseases.
Dev Cell. 17:9–26. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kaufmann P, Black S and Huppertz B:
Endovascular trophoblast invasion: Implications for the
pathogenesis of intrauterine growth retardation and preeclampsia.
Biol Reprod. 69:1–7. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sonderegger S, Pollheimer J and Knöfler M:
Wnt signalling in implantation, decidualisation and placental
differentiation-review. Placenta. 31:839–847. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ma XR, Edmund Sim UH, Pauline B, Patricia
L and Rahman J: Overexpression of WNT2 and TSG101 genes in
colorectal carcinoma. Trop Biomed. 25:46–57. 2008.PubMed/NCBI
|
|
21
|
Geng M, Cao YC, Chen YJ, Jiang H, Bi LQ
and Liu XH: Loss of Wnt5a and Ror2 protein in hepatocellular
carcinoma associated with poor prognosis. World J Gastroenterol.
18:1328–1338. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bui TD, Zhang L, Rees MC, Bicknell R and
Harris AL: Expression and hormone regulation of Wnt2, 3, 4, 5a, 7a,
7b and 10b in normal human endometrium and endometrial carcinoma.
Br J Cancer. 75:1131–1136. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ge JF, Xu YY, Qin G, Cheng JQ and Chen FH:
Resveratrol Ameliorates the anxiety- and depression-like behavior
of subclinical hypothyroidism rat: Possible involvement of the HPT
Axis, HPA Axis, and Wnt/β-Catenin Pathway. Front Endocrinol
(Lausanne). 7:442016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Oishi I, Suzuki H, Onishi N, Takada R,
Kani S, Ohkawara B, Koshida I, Suzuki K, Yamada G, Schwabe GC, et
al: The receptor tyrosine kinase Ror2 is involved in non-canonical
Wnt5a/JNK signalling pathway. Genes Cells. 8:645–654. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Tang Y, Gholamin S, Schubert S, Willardson
MI, Lee A, Bandopadhayay P, Bergthold G, Masoud S, Nguyen B, Vue N,
et al: Epigenetic targeting of Hedgehog pathway transcriptional
output through BET bromodomain inhibition. Nat Med. 20:732–740.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Rubin LL and de Sauvage FJ: Targeting the
Hedgehog pathway in cancer. Nat Rev Drug Discov. 5:1026–1033. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Jia Y, Wang Y and Xie J: The Hedgehog
pathway: Role in cell differentiation, polarity and proliferation.
Arch Toxicol. 89:179–191. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Jeng KS, Chang CF and Lin SS: Sonic
Hedgehog signaling in organogenesis, tumors, and tumor
microenvironments. Int J Mol Sci. 21:7582020. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Min TH, Kriebel M, Hou S and Pera EM: The
dual regulator Sufu integrates Hedgehog and Wnt signals in the
early Xenopus embryo. Dev Biol. 358:262–276. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Koukoura O, Sifakis S and Spandidos DA:
DNA methylation in the human placenta and fetal growth (review).
Mol Med Rep. 5:883–889. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Serman L, Vlahović M, Sijan M, Bulić-Jakus
F, Serman A, Sincić N, Matijević R, Jurić-Lekić G and Katusić A:
The impact of 5-azacytidine on placental weight, glycoprotein
pattern and proliferating cell nuclear antigen expression in rat
placenta. Placenta. 28:803–811. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chelbi ST, Mondon F, Jammes H, Buffat C,
Mignot TM, Tost J, Busato F, Gut I, Rebourcet R, Laissue P, et al:
Expressional and epigenetic alterations of placental serine
protease inhibitors: SERPINA3 is a potential marker of
preeclampsia. Hypertension. 49:76–83. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ferreira JC, Choufani S, Grafodatskaya D,
Butcher DT, Zhao C, Chitayat D, Shuman C, Kingdom J, Keating S and
Weksberg R: WNT2 promoter methylation in human placenta is
associated with low birthweight percentile in the neonate.
Epigenetics. 6:440–449. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Dexheimer PJ and Cochella L: MicroRNAs:
From mechanism to organism. Front Cell Dev Biol. 8:4092020.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Duchaine TF and Fabian MR: Mechanistic
insights into MicroRNA-Mediated gene silencing. Cold Spring Harb
Perspect Biol. 11:a0327712019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Gebert LFR and MacRae IJ: Regulation of
microRNA function in animals. Nat Rev Mol Cell Biol. 20:21–37.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Paul P, Chakraborty A, Sarkar D, Langthasa
M, Rahman M, Bari M, Singha RS, Malakar AK and Chakraborty S:
Interplay between miRNAs and human diseases. J Cell Physiol.
233:2007–2018. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Ciesla M, Skrzypek K, Kozakowska M, Loboda
A, Jozkowicz A and Dulak J: MicroRNAs as biomarkers of disease
onset. Anal Bioanal Chem. 401:2051–2061. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Huang W: MicroRNAs: Biomarkers,
diagnostics, and therapeutics. Methods Mol Biol. 1617:57–67. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Kochhar P, Dwarkanath P, Ravikumar G,
Thomas A, Crasta J, Thomas T, Kurpad AV and Mukhopadhyay A:
Placental expression of miR-21-5p, miR-210-3p and miR-141-3p:
Relation to human fetoplacental growth. Eur J Clin Nutr.
76:730–738. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zarkovic M, Hufsky F, Markert UR and Marz
M: The Role of Non-Coding RNAs in the Human Placenta. Cells.
11:15882022. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Xu P, Ma Y, Wu H and Wang YL:
Placenta-Derived MicroRNAs in the pathophysiology of Human
pregnancy. Front Cell Dev Biol. 9:6463262021. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Awamleh Z, Gloor GB and Han VKM: Placental
microRNAs in pregnancies with early onset intrauterine growth
restriction and preeclampsia: Potential impact on gene expression
and pathophysiology. BMC Med Genomics. 12:912019. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
He S, Yang F, Yang M, An W, Maguire EM,
Chen Q, Xiao R, Wu W, Zhang L, Wang W and Xiao Q:
miR-214-3p-Sufu-GLI1 is a novel regulatory axis controlling
inflammatory smooth muscle cell differentiation from stem cells and
neointimal hyperplasia. Stem Cell Res Ther. 11:4652020. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Lee DY, Deng Z, Wang CH and Yang BB:
MicroRNA-378 promotes cell survival, tumor growth, and angiogenesis
by targeting SuFu and Fus-1 expression. Proc Natl Acad Sci USA.
104:20350–20355. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Hyun J, Wang S, Kim J, Rao KM, Park SY,
Chung I, Ha CS, Kim SW, Yun YH and Jung Y: MicroRNA-378 limits
activation of hepatic stellate cells and liver fibrosis by
suppressing Gli3 expression. Nat Commun. 7:109932016. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Kardum V, Karin V, Glibo M, Skrtic A,
Martic TN, Ibisevic N, Skenderi F, Vranic S and Serman L:
Methylation-associated silencing of SFRP1 gene in high-grade serous
ovarian carcinomas. Ann Diagn Pathol. 31:45–49. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Rizzardi AE, Johnson AT, Vogel RI,
Pambuccian SE, Henriksen J, Skubitz AP, Metzger GJ and Schmechel
SC: Quantitative comparison of immunohistochemical staining
measured by digital image analysis versus pathologist visual
scoring. Diagn Pathol. 7:422012. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Vrsalovic MM, Korac P, Dominis M, Ostojic
S, Mannhalter C and Kusec R: T- and B-cell clonality and frequency
of human herpes viruses-6, −8 and Epstein Barr virus in
angioimmunoblastic T-cell lymphoma. Hematol Oncol. 22:169–177.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Paluszczak J, Wiśniewska D,
Kostrzewska-Poczekaj M, Kiwerska K, Grénman R, Mielcarek-Kuchta D
and Jarmuż-Szymczak M: Prognostic significance of the methylation
of Wnt pathway antagonists-CXXC4, DACT2, and the inhibitors of
sonic hedgehog signaling-ZIC1, ZIC4, and HHIP in head and neck
squamous cell carcinomas. Clin Oral Investig. 21:1777–1788. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Sola IM, Serman A, Karin-Kujundzic V, Paic
F, Skrtic A, Slatina P, Kakarigi L, Vranic S and Serman L:
Dishevelled family proteins (DVL1-3) expression in intrauterine
growth restriction (IUGR) placentas. Bosn J Basic Med Sci.
21:447–453. 2021.PubMed/NCBI
|
|
53
|
Julian CG, Wilson MJ, Lopez M, Yamashiro
H, Tellez W, Rodriguez A, Bigham AW, Shriver MD, Rodriguez C,
Vargas E and Moore LG: Augmented uterine artery blood flow and
oxygen delivery protect Andeans from altitude-associated reductions
in fetal growth. Am J Physiol Regul Integr Comp Physiol.
296:R1564–R1575. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Williams LA, Evans SF and Newnham JP:
Prospective cohort study of factors influencing the relative
weights of the placenta and the newborn infant. BMJ. 314:1864–1868.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Thompson LP: Effects of chronic hypoxia on
fetal coronary responses. High Alt Med Biol. 4:215–224. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Rashid CS, Bansal A and Simmons RA:
Oxidative stress, intrauterine growth restriction, and
developmental programming of type 2 diabetes. Physiology
(Bethesda). 33:348–359. 2018.PubMed/NCBI
|
|
57
|
Zhang C, Tannous E and Zheng JJ: Oxidative
stress upregulates Wnt signaling in human retinal microvascular
endothelial cells through activation of disheveled. J Cell Biochem.
120:14044–14054. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Funato Y, Michiue T, Asashima M and Miki
H: The thioredoxin-related redox-regulating protein nucleoredoxin
inhibits Wnt-beta-catenin signalling through dishevelled. Nat Cell
Biol. 8:501–508. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Vikram A, Kim YR, Kumar S, Naqvi A,
Hoffman TA, Kumar A, Miller FJ Jr, Kim CS and Irani K: Canonical
Wnt signaling induces vascular endothelial dysfunction via
p66Shc-regulated reactive oxygen species. Arterioscler Thromb Vasc
Biol. 34:2301–2309. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Spillmann F, Van Linthout S, Miteva K,
Lorenz M, Stangl V, Schultheiss HP and Tschöpe C: LXR agonism
improves TNF-α-induced endothelial dysfunction in the absence of
its cholesterol-modulating effects. Atherosclerosis. 232:1–9. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Bretón-Romero R, Feng B, Holbrook M, Farb
MG, Fetterman JL, Linder EA, Berk BD, Masaki N, Weisbrod RM,
Inagaki E, et al: Endothelial dysfunction in human diabetes is
mediated by Wnt5a-JNK signaling. Arterioscler Thromb Vasc Biol.
36:561–569. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Caricasole A, Copani A, Caraci F, Aronica
E, Rozemuller AJ, Caruso A, Storto M, Gaviraghi G, Terstappen GC
and Nicoletti F: Induction of Dickkopf-1, a negative modulator of
the Wnt pathway, is associated with neuronal degeneration in
Alzheimer's brain. J Neurosci. 24:6021–6027. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Alvarez AR, Godoy JA, Mullendorff K,
Olivares GH, Bronfman M and Inestrosa NC: Wnt-3a overcomes
beta-amyloid toxicity in rat hippocampal neurons. Exp Cell Res.
297:186–196. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Fan M, Xu Y, Hong F, Gao X, Xin G, Hong H,
Dong L and Zhao X: Rac1/β-Catenin signalling pathway contributes to
trophoblast cell invasion by targeting Snail and MMP9. Cell Physiol
Biochem. 38:1319–1332. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Pennington KA, Schlitt JM, Jackson DL,
Schulz LC and Schust DJ: Preeclampsia: Multiple approaches for a
multifactorial disease. Dis Model Mech. 5:9–18. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Wu Q, Wu G and Li JX: Effect of hypoxia on
expression of placental trophoblast cells SATB1 and β-catenin and
its correlation with the pathogenesis of preeclampsia. Asian Pac J
Trop Med. 9:567–571. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Bischof P and Campana A: Molecular
mediators of implantation. Baillieres Best Pract Res Clin Obstet
Gynaecol. 14:801–814. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Nadeem L, Munir S, Fu G, Dunk C, Baczyk D,
Caniggia I, Lye S and Peng C: Nodal signals through activin
receptor-like kinase 7 to inhibit trophoblast migration and
invasion: Implication in the pathogenesis of preeclampsia. Am J
Pathol. 178:1177–1189. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Tang C, Mei L, Pan L, Xiong W, Zhu H, Ruan
H, Zou C, Tang L, Iguchi T and Wu X: Hedgehog signaling through
GLI1 and GLI2 is required for epithelial-mesenchymal transition in
human trophoblasts. Biochim Biophys Acta. 1850:1438–1448. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Zhang Y and Zhang Y: Forkhead box C2
promotes the invasion ability of human trophoblast cells through
Hedgehog (Hh) signaling pathway. Cell Biol Int. 42:859–866. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Liu J, Heydeck W, Zeng H and Liu A: Dual
function of suppressor of fused in Hh pathway activation and mouse
spinal cord patterning. Dev Biol. 362:141–153. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Tariki M, Wieczorek SA, Schneider P,
Bänfer S, Veitinger S, Jacob R, Fendrich V and Lauth M: RIO kinase
3 acts as a SUFU-dependent positive regulator of Hedgehog
signaling. Cell Signal. 25:2668–2675. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Banister CE, Koestler DC, Maccani MA,
Padbury JF, Houseman EA and Marsit CJ: Infant growth restriction is
associated with distinct patterns of DNA methylation in human
placentas. Epigenetics. 6:920–927. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Kimura AP, Liebhaber SA and Cooke NE:
Epigenetic modifications at the human growth hormone locus predict
distinct roles for histone acetylation and methylation in placental
gene activation. Mol Endocrinol. 18:1018–1032. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Chuang HC, Chang CW, Chang GD, Yao TP and
Chen H: Histone deacetylase 3 binds to and regulates the GCMa
transcription factor. Nucleic Acids Res. 34:1459–1469. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Fu G, Brkić J, Hayder H and Peng C:
MicroRNAs in Human placental development and pregnancy
complications. Int J Mol Sci. 14:5519–5544. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Higashijima A, Miura K, Mishima H,
Kinoshita A, Jo O, Abe S, Hasegawa Y, Miura S, Yamasaki K, Yoshida
A, et al: Characterization of placenta-specific microRNAs in fetal
growth restriction pregnancy. Prenat Diagn. 33:214–222. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Pineles BL, Romero R, Montenegro D, Tarca
AL, Han YM, Kim YM, Draghici S, Espinoza J, Kusanovic JP, Mittal P,
et al: Distinct subsets of microRNAs are expressed differentially
in the human placentas of patients with preeclampsia. Am J Obstet
Gynecol. 196:261.e1–e6. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Zhu XM, Han T, Sargent IL, Yin GW and Yao
YQ: Differential expression profile of microRNAs in human placentas
from preeclamptic pregnancies vs normal pregnancies. Am J Obstet
Gynecol. 200:661.e1–e7. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Donker RB, Mouillet JF, Nelson DM and
Sadovsky Y: The expression of Argonaute2 and related microRNA
biogenesis proteins in normal and hypoxic trophoblasts. Mol Hum
Reprod. 13:273–279. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Peng Y, Qin Y, Zhang X, Deng S, Yuan Y,
Feng X, Chen W, Hu F, Gao Y, He J, et al: MiRNA-20b/SUFU/Wnt axis
accelerates gastric cancer cell proliferation, migration and EMT.
Heliyon. 7:e066952021. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Alimirah F, Peng X, Gupta A, Yuan L, Welsh
J, Cleary M and Mehta RG: Crosstalk between the vitamin D receptor
(VDR) and miR-214 in regulating SuFu, a hedgehog pathway inhibitor
in breast cancer cells. Exp Cell Res. 349:15–22. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Peng Y, Zhang X, Ma Q, Yan R, Qin Y, Zhao
Y, Cheng Y, Yang M, Wang Q, Feng X, et al: MiRNA-194 activates the
Wnt/β-catenin signaling pathway in gastric cancer by targeting the
negative Wnt regulator, SUFU. Cancer Lett. 385:117–127. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Park M, Kim M, Hwang D, Park M, Kim WK,
Kim SK, Shin J, Park ES, Kang CM, Paik YK and Kim H:
Characterization of gene expression and activated signaling
pathways in solid-pseudopapillary neoplasm of pancreas. Mod Pathol.
27:580–593. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Long H, Wang Z, Chen J, Xiang T, Li Q,
Diao X and Zhu B: microRNA-214 promotes epithelial-mesenchymal
transition and metastasis in lung adenocarcinoma by targeting the
suppressor-of-fused protein (Sufu). Oncotarget. 6:38705–38718.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Ma L, Yang X, Wei R, Ye T, Zhou JK, Wen M,
Men R, Li P, Dong B, Liu L, et al: MicroRNA-214 promotes hepatic
stellate cell activation and liver fibrosis by suppressing Sufu
expression. Cell Death Dis. 9:7182018. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Tupone MG, D'Aguanno S, Di Martile M,
Valentini E, Desideri M, Trisciuoglio D, Donzelli S, Sacconi A,
Buglioni S, Ercolani C, et al: microRNA-378a-5p iS a novel positive
regulator of melanoma progression. Oncogenesis. 9:222020.
View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Hua Z, Lv Q, Ye W, Wong CK, Cai G, Gu D,
Ji Y, Zhao C, Wang J, Yang BB and Zhang Y: MiRNA-directed
regulation of VEGF and other angiogenic factors under hypoxia. PLoS
One. 1:e1162006. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Odibo AO, Nelson D, Stamilio DM, Sehdev HM
and Macones GA: Advanced maternal age is an independent risk factor
for intrauterine growth restriction. Am J Perinatol. 23:325–328.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Palatnik A, De Cicco S, Zhang L, Simpson
P, Hibbard J and Egede LE: The association between advanced
maternal age and diagnosis of small for gestational age. Am J
Perinatol. 37:37–43. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Vega J, Sáez G, Smith M, Agurto M and
Morris NM: Risk factors for low birth weight and intrauterine
growth retardation in Santiago, Chile. Rev Med Chil. 121:1210–1219.
1993.(In Spanish). PubMed/NCBI
|
|
92
|
Kalinka J, Hanke W and Szymczak W: Risk
factors of intrauterine growth retardation: A study of an urban
population in Poland. Cent Eur J Public Health. 4:192–196.
1996.PubMed/NCBI
|
|
93
|
Tierney-Gumaer R and Reifsnider E: Risk
factors for low birth weight infants of Hispanic, African American,
and White women in Bexar County, Texas. Public Health Nurs.
25:390–400. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Yu SH, Mason J, Crum J, Cappa C and
Hotchkiss DR: Differential effects of young maternal age on child
growth. Glob Health Action. 9:311712016. View Article : Google Scholar : PubMed/NCBI
|