Introduction
Hepatitis B virus X (HBX) protein has a long peptide
chain composed of 154 amino acids. The N-terminus of HBX is the
negative regulatory domain and the C-terminus is the deactivation
domain (1). In 1998, a protein
that interacts with the C-terminus of HBX was first identified and
named HBX-interacting protein (IP), also known as Lamtor5. HBXIP is
encoded by four exons located on human chromosome 1p13.3, and is a
conserved protein with a molecular weight of 18 kDa. HBXIP
localizes subcellularly to the surface of lysosomes and forms a
pentameric regulatory complex with the other four proteins of the
Lamtor family to activate mammalian target of rapamycin complex
(mTORC1) (2,3). mTORC1 is the major growth regulator
in humans and serves a role in cell growth (4,5).
HBXIP is highly expressed in numerous types of
cancer, such as breast cancer, gastric cancer and esophageal
squamous cell carcinoma, and its expression is associated with
certain clinicopathological characteristics (6–14).
Furthermore, high levels of HBXIP expression are associated with a
poor prognosis.
Research on HBXIP has revealed multiple carcinogenic
mechanisms. For example, in cervical cancer, HBXIP can interact
with LIM domain 2 and activate the Wnt signaling pathway to promote
malignant progression (15). In
ovarian cancer, HBXIP can coactivate S-phase kinase-associated
protein 2 via transcription factor SP1 to accelerate malignant
progression (16). HBXIP can also
promote the metastasis of tongue squamous cell carcinoma and
regulate the malignant progression of gastric cancer cells through
the PI3K/AKT signaling pathway (17,18).
Therefore, targeting HBXIP has been suggested as a potential cancer
treatment approach. Germacone, a monocyclic sesquiterpene, has been
reported to regulate the cell cycle and apoptosis in renal cell
carcinoma by inhibiting HBXIP expression (19). Reduction or loss of HBXIP
expression increases the number of cells containing single-phase
spindles, which impedes the ability of cells to divide (20). Inhibition of HBXIP expression was
reported to reduce tumor drug resistance and knockdown of HBXIP
expression sensitized patients with osteosarcoma and liver cancer
cell lines to chemotherapy (21).
The carcinogenic mechanism of HBXIP in most tumors
has been established, but its role in normal cells has been less
well studied. At present, the role of HBXIP in maintaining normal
glucose tolerance phenotype and normal embryonic tissue development
in mice has been preliminarily studied (22,23).
Likewise, HBXIP is involved in DNA damage repair (21). In summary, HBXIP serves roles in
both normal and cancer cells. The present review summarizes the
current knowledge of HBXIP, including its normal physiological
function and role in tumor cells, and provides a reference for
subsequent research on HBXIP.
Normal physiological role of HBXIP
mTORC1 is a major growth regulator in humans and is
activated by translocation to the lysosomal surface in response to
numerous cell signals (2). HBXIP
is a membrane protein on the surface of lysosomes and forms a
pentameric regulatory complex with P18, P14, MEK partner 1 (MP1)
and chromosome 7 open reading frame 59 (C7orf59). This complex
serves a key role in activating mTORC1. HBXIP and C7orf59 form the
core of this pentameric regulatory complex, which is responsible
for the initial nucleation of the complex as well as the
stabilization of the P18 conformation, which in turn allows the
subsequent binding of MP1 and P14 (2,24).
Therefore, loss of HBXIP leads to the formation and dysfunction of
this complex, which then inhibits mTORC1 activation and impedes the
downstream signaling pathways. For example, the self-renewal and
differentiation of embryonic stem cells are regulated by mTORC1
signaling. In mice, knockout of HBXIP causes mTORC1 inactivation,
which further affects the differentiation of embryonic stem cells
and eventually leads to embryonic lethality (23).
HBXIP has also been reported to act as a
transcription coactivator of oncoproteins in tumors and has a
similar function in normal cells (22,25,26).
HBXIP knockout mice have impaired glucose tolerance and reduced
insulin production (22).
Furthermore, HBXIP is highly expressed in pancreatic islets, acting
as a coactivator of pancreatic and duodenal homeobox transcription
factor and forming a complex with neurogenic differentiation 1 to
upregulate insulin transcription genes and promote insulin
secretion (22). Furthermore, the
DNA damage response is an intrinsic signaling network in cells that
recognizes and repairs DNA damage (27). Ataxia telangiectasia mutated (ATM)
is a potential first-step sensor of DNA damage response, which can
regulate downstream phosphorylation of p53, murine double minute 2
and checkpoint kinase (Chk) (28).
HBXIP delays or blocks cell cycle progression by activating ATM,
phosphorylating Chk and activating the G2/M phase DNA damage
checkpoint (21). Therefore,
patients with high HBXIP expression may be resistant to
chemotherapy by regulating DAN damage repair.
In summary, HBXIP may serve a role at several points
in the physiological regulation of cells (Fig. 1). When HBXIP expression is
dysregulated, it affects the malignant progression of cancer cells.
Therefore, an improved understanding of the physiological functions
of HXBIP in normal cells will help to elucidate the mechanisms
affected by its dysregulation in a more systematic manner.
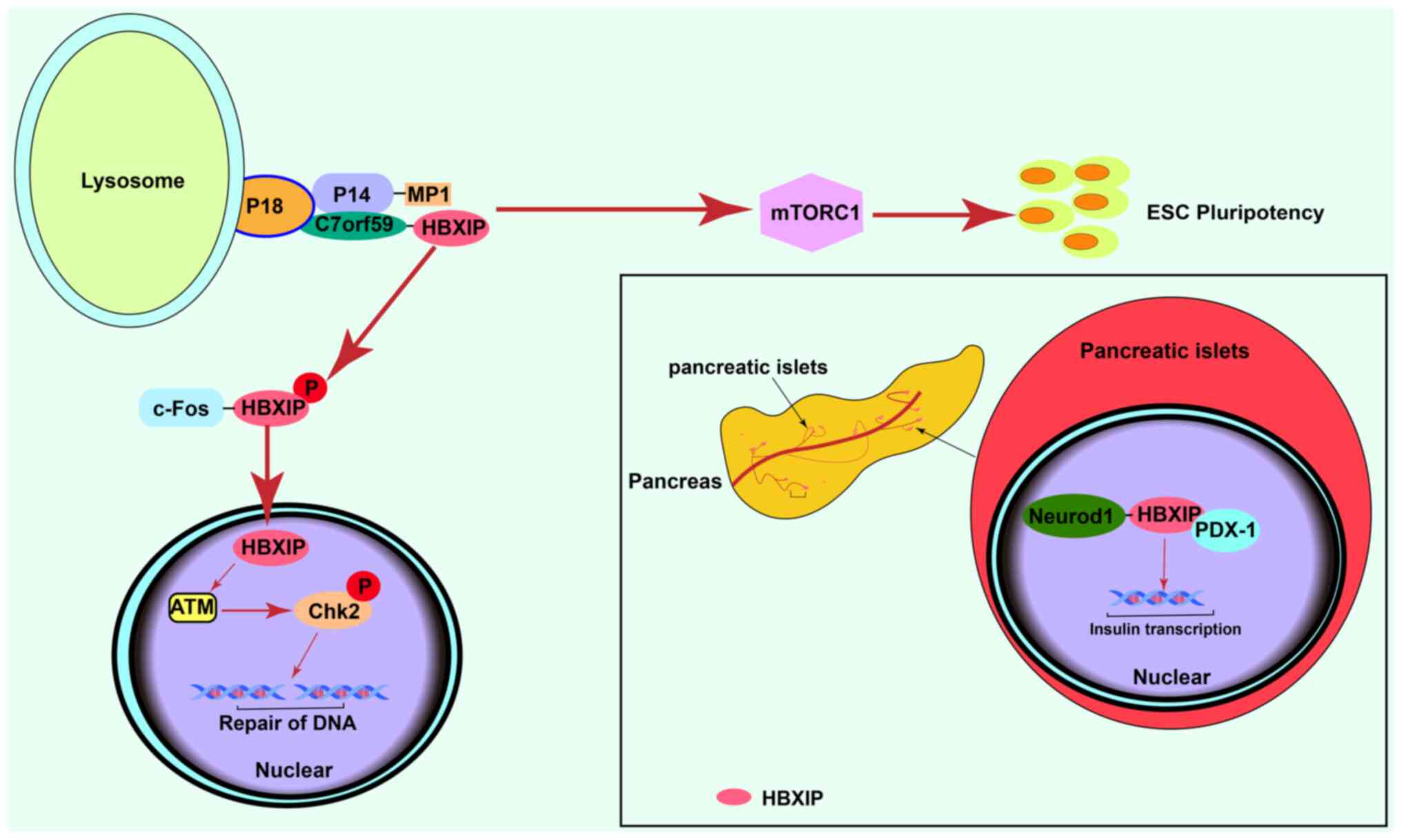 | Figure 1.Normal physiological effects of
HBXIP. Activation of mTORC1 signaling promotes normal
differentiation of ESCs. HBXIP is involved in DNA damage repair by
activating ATM. In the normal adult pancreas, HBXIP is highly
expressed in the pancreatic islets in comparison with other
pancreatic tissue and promotes the transcription of insulin. HBXIP,
hepatitis B X-interacting protein; mTORC1, mammalian target of
rapamycin complex 1; ESCs, embryonic stem cells; ATM, ataxia
telangiectasia mutated; Chk2, checkpoint kinase; c-Fos, c-Fos
proto-oncogene protein; PDX-1, programmed cell death 1 ligand 1;
Neurod1, neuronal differentiation 1Gene; P, phosphorylated. |
HBXIP and tumors
Prediction of worse survival in
patients with tumors and high expression of HBXIP
In the past decade, studies have reported that HBXIP
is highly expressed in breast cancer, esophageal squamous cell
carcinoma, colorectal, gastric cancer, renal cancer, liver cancer,
tongue squamous cell carcinoma and pancreatic cancer, and its
expression associated with the clinicopathological characteristics,
prognosis and survival of patients (Table I) (6–14).
Furthermore, HBXIP expression is associated with tumor metastasis,
stage and survival of different tumor types (6–14).
Therefore, high expression of HBXIP often predicts a higher degree
of malignancy, and promotes tumor cell proliferation and invasion
through different mechanisms.
 | Table I.Association between cancer type and
clinicopathological features with high expression of hepatitis B
X-interacting protein. |
Table I.
Association between cancer type and
clinicopathological features with high expression of hepatitis B
X-interacting protein.
| First author/s,
year |
| Clinicopathological
feature |
|
|
|---|
|
|
|
|
|
|---|
| Tumor type |
Differentiation | Invasion | Stage | Survival | Age | Mechanism | (Refs.) |
|---|
| Zhou et
al, | Pancreatic | Association | Association | Association | Association | Negative | Unknown. | (6) |
| 2019 | ductal |
|
|
|
|
| Cancer cell |
|
|
| carcinoma |
|
|
|
|
| metastasis is |
|
|
|
|
|
|
|
|
| promoted by |
|
| Wang et
al, | Colorectal | Not shown | Association | Association | Association | Not shown | epithelial- | (7) |
| 2020 | cancer |
|
|
|
|
| mesenchymal |
|
|
|
|
|
|
|
|
| transition |
|
| Piao et
al, | Gastric | Not shown | Association | Association | Association | Not shown | Cancer cell | (8) |
| 2017 | cancer |
|
|
|
|
| metastasis is |
|
|
|
|
|
|
|
|
| promoted by |
|
|
|
|
|
|
|
|
| regulating |
|
|
|
|
|
|
|
|
| metabolism and |
|
|
|
|
|
|
|
|
|
post-transcriptional |
|
|
|
|
|
|
|
|
| translation |
|
| Li et
al, | Cervical | Association | Association | Association | Association | Not shown | Tumor | (9) |
| 2017 | cancer |
|
|
|
|
| progression is |
|
|
|
|
|
|
|
|
| promoted via
the |
|
|
|
|
|
|
|
|
| Wnt signaling |
|
|
|
|
|
|
|
|
| pathway |
|
| Wang et
al, | Ovarian | Not shown | Association | Association | Association | Not shown | Co-activation | (10) |
| 2017 | cancer |
|
|
|
|
| of
transcription |
|
|
|
|
|
|
|
|
| factors
promotes |
|
|
|
|
|
|
|
|
| cancer
progression |
|
| Xia et
al, | Esophageal | Not shown | Association | Association | Association | Not shown | Cancer | (11) |
| 2017 | squamous |
|
|
|
|
| progression is |
|
|
| cell |
|
|
|
|
| influenced
through |
|
|
| carcinoma |
|
|
|
|
| translational
level |
|
|
|
|
|
|
|
|
| regulation |
|
| Cheng et
al, | Breast | Not shown | Association | Association | Association | Negative | Promotes
breast | (12) |
| 2014 | cancer |
|
|
|
|
| cancer via |
|
|
|
|
|
|
|
|
| metabolism and |
|
|
|
|
|
|
|
|
| co-activation
of |
|
|
|
|
|
|
|
|
| transcription |
|
|
|
|
|
|
|
|
| factors |
|
| Guo et
al, | Liver | Association | Association | Association | Association | Association | Promotes liver | (13) |
| 2021 | cancer |
|
|
|
|
| cancer |
|
|
|
|
|
|
|
|
| by regulating |
|
|
|
|
|
|
|
|
| level of RNA |
|
|
|
|
|
|
|
|
| modification |
|
|
|
|
|
|
|
|
| and
translation |
|
| Wang et
al, | Non-small | Not shown | Association | Association | Association | Not shown | Promotes non- | (14) |
| 2017 | cell lung |
|
|
|
|
| small cell
cancer |
|
|
| cancer |
|
|
|
|
| cells via MEPK |
|
|
|
|
|
|
|
|
| signaling
pathway |
|
Tumor-promoting mechanism of
HBXIP
HBXIP regulates expression of key proteins in
tumors at multiple stages
The regulation process from DNA to protein is called
gene expression regulation (29),
which is ordered in time and space. Gene expression is regulated at
multiple stages, including at the chromatin, transcription,
post-transcription, translation and post-translation stage.
Dysregulation of any of these stages can lead to changes in the
protein levels in the cell and affect related physiological
processes. HBXIP has been reported to influence the malignant
progression of tumor cells at multiple levels (Fig. 2).
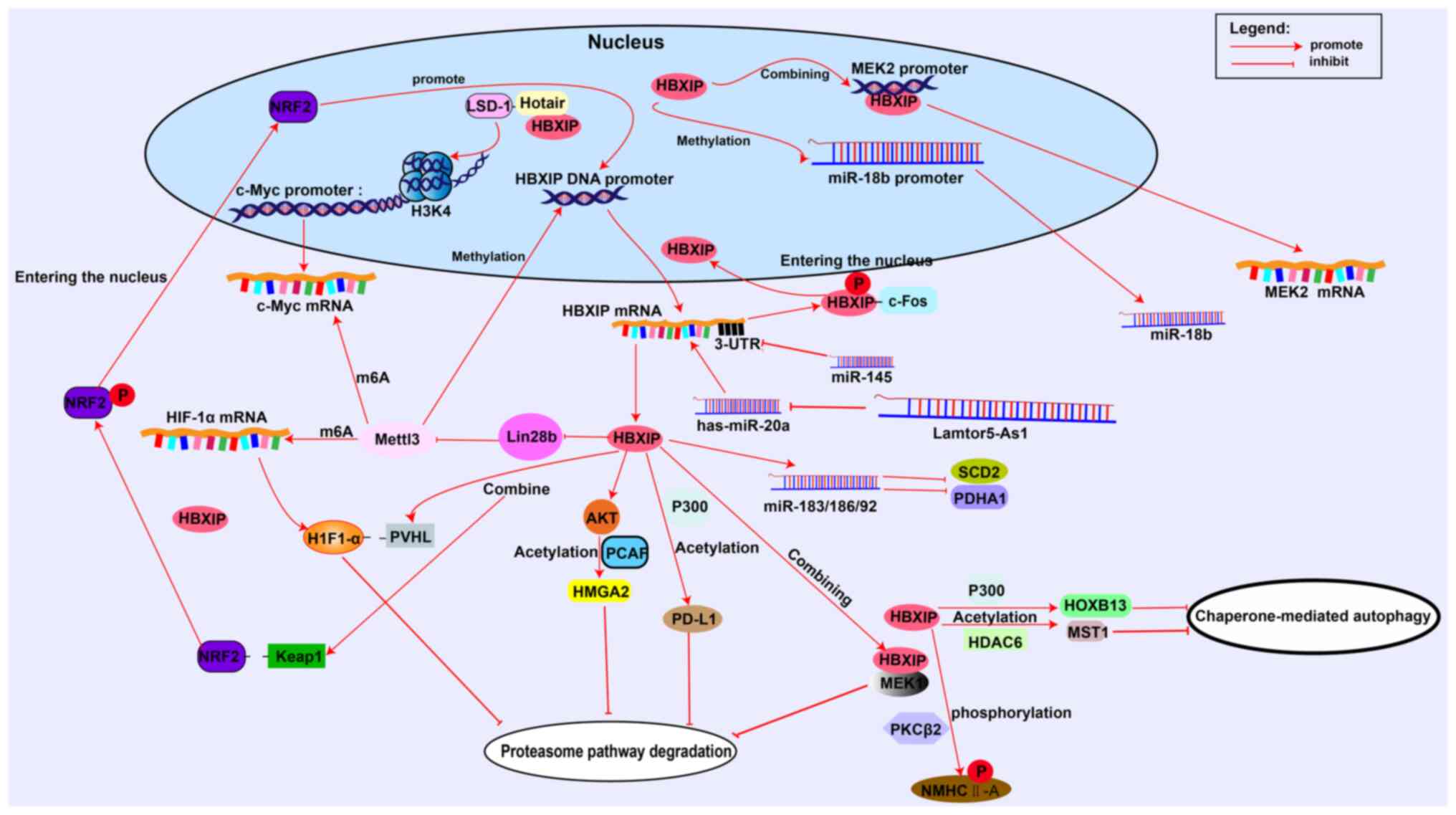 | Figure 2.HBXIP regulates tumor cells at
multiple levels. HBXIP promotes c-Myc histone demethylation through
LSD-1 and binds to the MEK2 promoter to promote transcription.
HBXIP directly promotes transcription via the miR-18b promoter.
HBXIP modifies HIF-1α and c-Myc mRNA methylation through Mettl3.
Micro RNAs and long non-coding RNAs can target or be targeted by
HBXIP. HBXIP can regulate the malignant progression of tumors by
acetylase, phosphorylase or promoting the separation of target
proteins from ubiquitin recognition receptors. LSD-1,
lysine-specific demethylase 1; HIF-1α, hypoxia-inducible factor-1α;
Mettl3, methyltransferase-like 3; HBXIP, hepatitis B X-interacting
protein; Keap1, Kelch-like ECH associated protein 1; PVHL, The
von-Hippel Lindau tumor suppressor; MEK2, MAP Kinase Kinase2; MEK1,
MAP Kinase Kinase1; PD-L1, programmed death ligand 1; PCAF,
P300/CBP-associated factor; m6A, N6-methyladenosine; c-Myc,
Cellular-myelocytomatosis viral oncogene; c-Fos, c-Fos
proto-oncogene protein; HMGA2, The high mobility group protein 2;
Hotair, HOX transcript antisense RNA; HDAC6, Histone deacetylase 6;
HOXB13, Homeobox B13; MST, STE20-like kinase 1; NMHC-IIA,
non-muscle heavy chain myosin IIA; SCD2, synthetic cytochrome c
oxidase 2; PDHA1, pyruvate dehydrogenase A1; H3K4, histone H3
Lysine 4 trimethylation; PKCβII, protein kinase βC. |
i) HBXIP regulates chromatin and transcriptional
processes. Nucleosomes are composed of histones and DNA and are the
basic units of chromatin. When transcription occurs, the structure
of chromatin is altered in such a way as to allow transcription
factors to bind to DNA. Thus, transcriptional activation is closely
related to the chromatin environment, and histone H3 modification
is the primary mode of chromatin remodeling (30). c-Myc is an oncoprotein that binds
to the E-box on the DNA sequence and affects the transcription of
thousands of human genes (31).
Studies have reported that HBXIP mediates demethylation of the
c-Myc histone H3K4 by lysine-specific demethylase 1, with the
support of long noncoding RNA (lncRNA) HOX transcript antisense
RNA. This causes local DNA oxidation and drives the assembly of the
c-Myc transcription initiation complex (32). Promoters are DNA sequences with
transcriptional initiation specificity and their methylation is
associated with transcriptional inhibition (33). HBXIP induces methylation of the
microRNA (miRNA/miR)-18b promoter, thereby inhibiting its
transcription and then affecting the expression of subsequent
proteins, such as up-regulation of mouse double-minute 2 (MDM)
expression, which ultimately degrades P53 and promotes the
development of breast cancer (34). HBXIP also directly interacts with
promoters to affect the expression of related proteins.
Mitogen-activated protein kinase kinase kinase (MEKK) 2 is a member
of the MEKK family, which activates extracellular signal-regulated
kinase (ERK) 1/2 (35). Chromatin
immunoprecipitation has shown that HBXIP directly binds to the
promoter of MEKK2 to upregulate its expression and activate
downstream ERK 1/2 to regulate the invasion and proliferation of
breast cancer cells (36).
ii) HBXIP affects post-transcriptional regulation of
related proteins. Eukaryotic transcription products undergo a
series of processing and modification steps. Common modifications
include N1 methyladenosine, 5-methylcytosine, pseudouracil and
N6-methyladenosine (m6A) (37,38).
Of these, m6A is the most common in mammals (39) and is mediated by
methyltransferase-like 3 (Mettl3). The negative regulation of
Mettl3 by miR-let-7g is attenuated by HBXIP through the oncogene
Lin-28 Homolog B, thereby increasing the level of Mettl3 in cells.
There is also an m6A site in the promoter sequence of HBXIP, which
can be modified by Mettl3 to promote transcription of HBXIP and
thus produce positive feedback (40). Furthermore, HBXIP mediates the mRNA
methylation of several proteins involved in the malignant
progression of tumors through the regulation of Mettl3. For
example, HBXIP mediates the level of m6A modifications in
hypoxia-inducible factor (HIF)-1α mRNA via Mettl3 to promote the
metabolic reprogramming of hepatocellular carcinoma (41). Likewise, in gastric cancer, HBXIP
modifies the level of methylation of c-Myc mRNA via Mettl3 and
promotes expression of c-Myc (42).
Most genes in the eukaryotic genome do not encode
proteins, and their transcript products are called noncoding
(nc)RNAs, which serve a role in normal physiological and
pathological states. For example, miRNAs are members of the ncRNA
family, which regulate the expression of target proteins by
targeting and interacting with the 3′ untranslated regions of mRNA.
This induces the degradation or inhibition of translation of mRNA
and affects many processes such as cell proliferation and
differentiation, and serves a role in promoting or suppressing
cancer (43,44). There is a close relationship
between miRNAs and HBXIP levels in tumors. For example, both
miR-let-7g and miR-18b are downregulated by HBXIP to promote tumor
development (34,40). HBXIP can also inhibit the synthesis
of human synthetic cytochrome c oxidase 2 and pyruvate
dehydrogenase A1 by upregulating the miR-183/182/96 cluster,
thereby reprogramming the glucose metabolism of tumor cells to
promote malignant progression (45). Certain miRNAs directly affect the
mRNA of HBXIP and serve a role in cancer inhibition. For example,
miR-145 targets the noncoding region of HBXIP mRNA, inhibiting its
translation and preventing malignant progression of breast cancer
(46). Similarly, both miR-520b
and miR-548p inhibit HBXIP-induced breast and liver cancer
(47,48). lncRNAs are another member of the
ncRNA family. LncRNAs are >200 nucleotides in length and serve a
role in the degradation and regulation of mRNA or protein stability
through numerous mechanisms (49).
LAMTOR5-AS1 is a lncRNA located next to the HBXIP gene. LAMTOR5-AS1
expression is positively associated with the age of patients with
colorectal cancer. LAMTOR5-AS1 may also affect the expression of
HBXIP mRNA by inhibiting the miRNAs hsa-let-7b and hsa-miR-20a
(50).
iii) HBXIP regulates the expression of related
proteins through post-translational modifications (PTMs). Protein
PTMs are the addition of moieties, catalyzed by relevant enzymes,
to ≥1 amino acid residues to alter the biochemical properties of a
protein (51). Common PTMs include
acetylation, ubiquitination and phosphorylation, which can change
the stability and activity of proteins (52–54).
HBXIP affects the malignant progression of tumor cells by altering
the acetylation or ubiquitination of target proteins through
several mechanisms, thereby regulating their intracellular
abundance and activity (Table
II). The ubiquitination-proteasome pathway is one of the major
pathways of protein degradation (55). HBXIP can prevent this pathway in
three ways: i) The first mechanism is though the activation of
acetylases. In esophageal squamous cell carcinoma, HBXIP activates
the acetylase PCAF via AKT to acetylate a specific lysine residue
of HMGA2, preventing ubiquitination at this site and thereby
enhancing its stability (56).
HBXIP also maintains the stability of programmed death ligand 1
(PD-L1) by increasing the level of acetylation through the
acetylase P300. Notably, HBXIP inhibits the degradation of PD-L1 to
increase its intracellular levels. HBXIP also acts as a coactivator
of the transcription factor ETS2 which upregulates the expression
of PD-L1 (57). However, the
mechanism by which HBXIP activates P300 remains unclear; ii) HBXIP
can directly bind to the target protein to prevent
proteasome-mediated degradation. In non-small cell carcinoma, HBXIP
binds to MEK1 preventing its degradation by the proteasome
(58); and iii) HBXIP prevents the
binding of target proteins to ubiquitin ligase. The von-Hippel
Lindau tumor suppressor (pVHL) is a tumor suppressor protein which
is part of the E3-ubiquitin ligase complex. PVH2 binds the target
protein allowing it to be ubiquitinated (59). A study reported that HBXIP
abrogates the interaction between pVHL and H1F-1α inhibiting the
ubiquitination and therefore degradation of HIF-1α (45). Similarly, HBXIP reduces binding of
E3 ubiquitin ligase adaptor protein Kelch-like ECH associated
protein 1 (KEAP1) to nuclear factor erythroid 2-related factor 2
(NRF2) and inhibits ubiquitin regulated degradation of NRF2
(60).
 | Table II.Hepatitis B X-interacting protein
regulates protein post-translational modifications through
different mechanisms. |
Table II.
Hepatitis B X-interacting protein
regulates protein post-translational modifications through
different mechanisms.
| First author/s,
year | Type of tumor | Enzyme | Target protein | Residue | Effect | (Refs.) |
|---|
| Ye et al,
2020 | Esophageal squamous
cell carcinoma | PCAF | HMGA2 | K26 | Blocking
ubiquitination-proteasome pathway degradation | (56) |
| Zhang et al,
2021 | Non-small cell
carcinoma | None | MEK1 | Binding |
| (58) |
| Xu et al,
2021 | Breast cancer | P300 | PD-L1 | K270 |
| (57) |
| Liu et al,
2015 |
| None | PVHL and
H1F-1α | Promote
dissociation |
| (45) |
| Zhou et al,
2019 |
| None | Keap1 and Nrf2 | Promote
dissociation |
| (60) |
| Liu et al,
2018 |
| P300 | HOXB13 | K277 | Prevent degradation
by the | (62) |
| Li et al,
2015 |
| HDAC6 | MST1 | L35 | lysosomal
pathway | (63) |
| Zhang et al,
2023 |
| PKCβII | NMHC-IIA | S1916 | Enhanced
aggressiveness of cancer cells | (64) |
Another major pathway of protein degradation is
chaperon-mediated autophagy. This selectively transports proteins
to the lysosomes for degradation (61). HBXIP is also involved in this
pathway. Unlike the ubiquitin-proteasome pathway, HBXIP mainly
regulates the acetylation of target proteins by modulating
acetylases. For example, tissue chip technology showed that HBXIP
expression in tumor cells was related to HOXB13 and STEM20 like
kinase1 (MST1) (62,63); further research into the mechanism
found that HBXIP can change the acetylation levels of HOXB13 and
tumor suppressor proteinMST1 via the acetylase P300 and Histone
deacetylase 6 (HDAC6) to regulate chaperone-mediated autophagy
HBXIP also recruits protein kinase CβII to stimulate
phosphorylation of non-muscle heavy chain myosin IIA, thereby
enhancing breast cancer invasion (64).
HBXIP acts as an oxidative regulator
to promote tumor development
Mitochondria are among the main sources of
intracellular reactive oxygen species (ROS), which are continuously
produced as a byproduct of aerobic metabolism and simultaneously
scavenged by the cellular antioxidant mechanisms, thus maintaining
a non-toxic levels (65). Under
normal physiological conditions, ROS can act as specific molecular
regulators of cell signaling and function. For example, one of the
more typical modes of ROS regulation is to reversibly oxidize the
sulfhydryl group of a target protein to cystine thereby mediating
its biological effects. ROS-induced changes in the intracellular
oxidation-reduction (redox) status can affect cellular activities,
including signaling, metabolism, growth and apoptosis (66,67).
However, in pathological conditions, such as when tumors or
inflammation occurs, excessive accumulation of ROS usually leads to
redox imbalance, causing oxidative stress. Tumor cells increase
their antioxidant capacity to adapt to the elevated ROS levels
(68,69). HBXIP reduces ROS levels in
tamoxifen-resistant breast cancer cells, demonstrating its
potential antioxidant capacity (70). The KEAP1-NRF2 signaling pathway is
commonly dysregulated in tumor cells to maintain oxidative balance
(71). In breast cancer, HBXIP
competitively binds to KEAP1 and NRF2, with a significantly higher
affinity for KEAP1 than NRF2 (60). Therefore, when HBXIP is highly
expressed in tumors, it binds to KEAP1, activates the nuclear
displacement of NRF2, and binds to the related antioxidant response
element (ARE), thereby improving the antioxidant capacity of tumor
cells. Furthermore, there is an ARE sequence in the HBXIP promoter.
Therefore, nuclear translocation of NRF2 allows NRF2 to bind to the
HBXIP promoter sequence to upregulate HBXIP. The positive feedback
between NRF2 and HBXIP increases the antioxidant capacity of tumor
cells (72).
HBXIP regulates ferroptosis and
promotes tumor development
Ferroptosis induces programmed cell death by
catalyzing the lipid peroxidation of unsaturated fatty acids highly
expressed on the cell membrane, under the action of divalent iron
or ester oxygenase (73). Lipid
metabolism disorder is closely related to ferroptosis. Liver X
receptor (LXR) is a gene that induces and controls cholesterol
homeostasis and adipogenesis. When the LXR receptor binds to its
ligands, it recruits nuclear coactivators containing NR motifs to
activate target gene transcription. In the absence of ligands,
nuclear co-repressors containing (CoRNR) motifs are recruited to
repress transcription (74,75).
HBXIP, which contains a CoRNR motif in the side chain of the amino
acid sequence, activates LXR independently in a ligand-independent
manner, thereby upregulating sterol regulatory element-binding
protein-1c expression (76). The
altered LXR/Sterol regulatory element-binding transcription factor
1 (SREBP-1c) axis can disrupt fatty acid regulation in normal
cells, indicating that HBXIP has the potential to regulate cell
ferroptosis. Stearyl-CoA desaturase (SCD) catalyzes the conversion
of saturated fatty acids to unsaturated fatty acids and its
expression prevents ferroptosis (75). HBXIP acts as a coactivator of
transcription factor zinc finger protein 263, which binds to the
−315/-165 region of the SCD promoter to upregulate SCD expression,
prevent ferroptosis and enhance the drug resistance of tumor cells
(77). In summary, the
relationship between HBXIP and ferroptosis has not been widely
assessed, but we hypothesize there is an association between the
two and this is worth further exploration.
Other mechanisms of HBXIP tumor
regulation
i) Involvement in the regulation of metabolic
reprogramming of tumor cells. HBXIP regulation of glucose
metabolism reprogramming in tumor cells has been previously
elucidated and its mechanisms of action are as follows: First,
HBXIP targets the PI3K/AKT axis and its downstream molecule mTOR to
regulate glucose metabolism reprogramming in gastric and bladder
cancer (18,78); second, HBXIP, as a coactivator of
the transcription factor E2F1, directly induces the transcription
of pyruvate kinase, which promotes glycolysis (26); third, HBXIP mediates the m6A levels
of HIF-1α mRNA to promote transcription and reduces ubiquitination
and degradation of HIF-1α, which increases HIF-1α protein levels in
cells to modulate glucose metabolism in tumor cells (41,45);
and fourth, HBXIP regulates lipid metabolism in tumor cells through
the LXR/SREBP-1c/FAS signaling cascade (76).
ii) HBXIP is a co-activator of multiple transcribed
genes. Following phosphorylation at Ser26, HBXIP interacts with the
leucine zipper domain of c-Fos and translocates from the cytoplasm
to the nucleus (79). The leucine
zipper domain of HBXIP binds to transcription factors, allowing
HBXIP to act as a coactivator to regulate the transcription of
downstream related proteins and signaling pathways (79). For example, transcription factors
such as E2F1, SP1, c-Myc, STAT4 and CREB have been reported to bind
to HBXIP and act on downstream-related genes and proteins to
promote tumor development (25,26,80,81).
iii) Involvement in endoplasmic reticulum (ER)
stress. When the demand for protein folding exceeds the limit of
the ER, ER stress is triggered and the unfolded protein response
(UPR) is activated to maintain ER homeostasis (82). Appropriate ER stress can promote
the survival of tumor cells (83).
HBXIP is a novel UPR factor that can bind to the ER stress protein
sensor inositol-requiring transmembrane kinase/endoribonuclease 1α,
thereby regulating UPR in the appropriate range to promote tumor
cell survival (70).
iv) High expression of HBXIP can cause abnormal
activation of multiple signaling pathways: Abnormal activation of
the PI3K/AKT signaling pathway in liver, stomach cancer, breast
cancer and tongue squamous cell carcinoma promotes the malignant
progression of tumor cells (17,18,80,84);
the MAPK/ERK signaling pathway can be aberrantly activated by HBXIP
in non-small cell lung cancer and breast cancer, thereby promoting
tumor cell invasion inhibiting targeting by the immune complement
system (36,58); and knockdown of HBXIP can block the
Wnt signaling pathway and inhibit the malignant progression of
cervical cancer (15).
v) Positive feedback signaling in tumors. HBXIP has
positive feedback signaling in multiple types of tumors (Fig. 3). For example, HBXIP upregulates
the expression of Mettl3, which methylates HBXIP promoter to
upregulate HBXIP protein expression. HBXIP also upregulates the
expression of tumor necrosis factor receptor 1, which in turn
increases the expression of HBXIP by activating the NF-kB/STAT3
signaling pathway (40,85). In summary, such a positive feedback
mechanism could enhance the role of HBXIP in promoting tumor cell
proliferation and invasion, or other molecular mechanisms of
malignant progression.
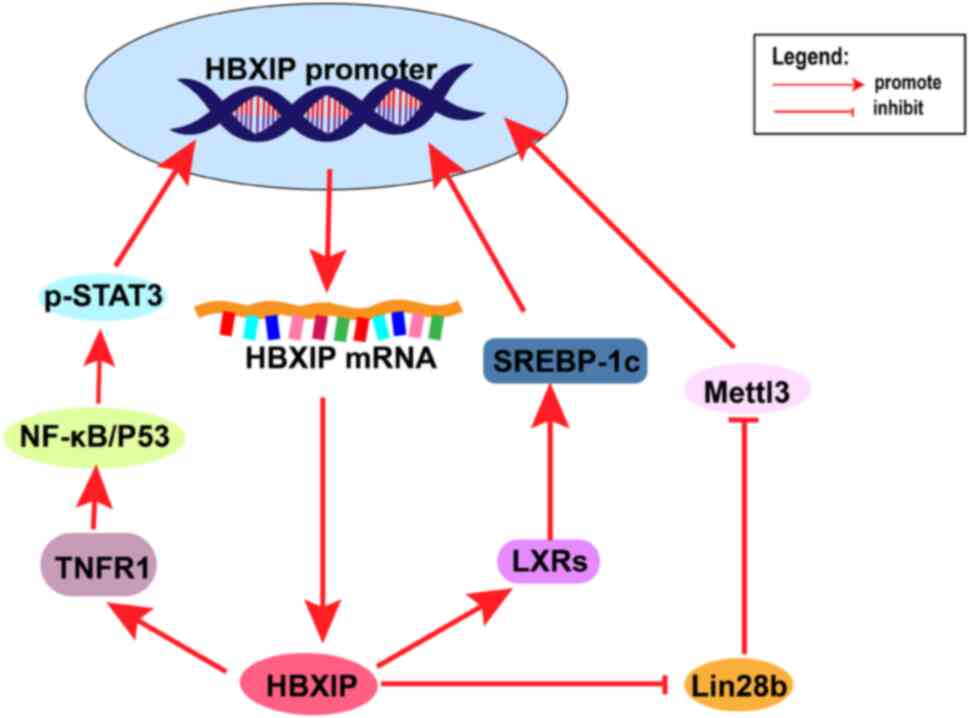 | Figure 3.Multiple positive feedback processes
involving HBXIP. After HBXIP upregulates TNFR1, this activates the
NF-κB pathway, which activates STAT3 in the nucleus and activates
the HBXIP promoter. After HBXIP upregulates LXRs, activated
SREBP-1c stimulates the HBXIP promoter. HBXIP inhibits the
inhibitory effect of Lin28b on Mettl3, which methylates the HBXIP
promoter to promote HBXIP expression. HBXIP, hepatitis B
X-interacting protein; Lin28b, Lin-28 homolog B; TNFR1, tumor
necrosis factor receptor 1; NF-κB, nuclear factor-kB; STAT3, signal
transducer and activator of transcription 3; LXRs, liver X
receptor; Mettl3, methyltransferase-like 3; SREBP-1c, sterol
regulatory element-binding transcription factor 1. |
Discussion
HBXIP is a membrane protein localized on the surface
of lysosomes and is encoded by the Lamtor gene. Under normal
physiological conditions, the pentameric complex that includes
HBXIP serves an important role in the regulation of mTORC. HBXIP is
essential for the normal development of mouse embryos and HBXIP
promotes the transcription of insulin transcription factors,
thereby maintaining normal cellular glucose metabolism. As an
oncoprotein, HBXIP is highly expressed in numerous tumor types and
is associated with poor clinicopathological features. Moreover,
drug resistance is associated with a high expression of HBXIP
protein in different tumors. For example, in
estrogen-receptor-positive breast cancer, HBXIP enhances
interleukin-6 transcription and maintains appropriate estrogen
receptor activation to confer tamoxifen resistance. Furthermore,
doxorubicin acts as a tumor suppressor by regulating DNA damage
response and inducing apoptosis. However, high expression of HBXIP
increase of DNA damage repair and thus serves a role in resistance
to doxorubicin-induced DNA damage. In summary, the carcinogenic
mechanism of HBXIP should be further studied.
In the present review, three types of mechanisms by
which high expression of HBXIP promotes tumor progression were
discussed: i) HXBIP promotes tumor development by regulating
transcriptional, post-transcriptional and translational processes;
ii) HBXIP promotes tumor development as a regulator of oxidative
stress; and iii) HBXIP prevents ferroptosis of tumor cells and
promotes tumor development. The reprogramming of tumor cell
metabolism by HBXIP was also discussed, as well as its role as a
coactivator of transcription factors and as a novel effector of
UPRs.
Over the past decade, HBXIP has been studied;
however, further research is still required to elucidate the
relationship between HBXIP and ferroptosis to determine whether
HBXIP regulates ferroptosis-related proteins, such as glutathione
peroxidase 4 and solute carrier family 7 member 11 to inhibit
ferroptosis of tumor cells. Furthermore, the protein function of
HBXIP may be involved in interference of the Keap1-NRF2 signaling
pathway. In previous studies, direct targeting of the
Keap1-Nrf2-ARE signaling pathway was the main strategy to inhibit
or activate oxidative stress. For example, the natural compound
curcumin enhances cellular antioxidant capacity by directly
disrupting Keap1 and thereby activating NRF2 in the nucleus
(86). However, the fact that
HBXIP competes with NRF2 to bind Keap1 brings new considerations
for modulating with this pathway. Future studies should assess a
mechanism to inhibit the binding of HBXIP and Keap1, to regulate
the level of NRF2 in the nucleus to indirectly regulate the ROS
levels to kill tumor cells. Moreover, further research on the
interaction between HBXIP and PTM is required.
HBXIP can affect the expression of related proteins
through acetylation, ubiquitination and phosphorylation. The
diversity and universality of HBXIP, through the different
modifications demonstrated in several types of tumors are worthy of
further evaluation. The present review demonstrated that the
expression of HBXIP and different PTM enzymes verified by tissue
chip technology has a good preliminary screening effect, and
subsequent testing in different tumor types should be considered to
expand knowledge of the relationship between HBXIP and PTM. For the
development of drugs targeting HBXIP. Although the mechanism of
action of HBXIP in numerous malignant tumors has been established,
research on drugs targeting HBXIP is scarce. At present, Germacrone
extracted from the traditional herbal medicine Curcuma
zedoaria, was reported by Fang et al (19) to inhibit the expression of HBXIP.
Therefore, considering traditional Chinese herbal medicine may be a
potential direction for the selection of HBXIP targeted drugs in
the future. Further research should develop natural or synthetic
compounds for the clinical treatment of HBXIP-related tumors.
In conclusion, HBXIP can promote the proliferation,
invasion and migration of tumor cells through numerous mechanisms.
Inhibition of HBXIP expression has potential therapeutic
significance for cancers. Finally, HBXIP has therapeutic potential
in tumors, and more in-depth studies are required to further
explore its carcinogenic mechanisms.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
Not applicable.
Authors' contributions
The original draft of the manuscript was written by
LC, manuscript editing and review was performed by LG, TZ and YY.
SL and RT revised and edited the final version of the manuscript.
All authors have read and approved the final version of the
manuscript. Data authentication is not applicable.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Chaturvedi VK, Singh A, Dubey SK, Hetta
HF, John J and Singh MP: Molecular mechanistic insight of hepatitis
B virus mediated hepatocellular carcinoma. Microb Pathog.
128:184–194. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bar-Peled L, Schweitzer LD, Zoncu R and
Sabatini DM: Ragulator Is a GEF for the Rag GTPases that signal
amino acid levels to mTORC1. Cell. 150:1196–1208. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Xiu M, Zeng X, Shan R, Wen W, Li J and Wan
R: The oncogenic role of HBXIP. Biomed Pharmacother.
133:1110452021. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Giguère V: Canonical signaling and nuclear
activity of mTOR-a teamwork effort to regulate metabolism and cell
growth. FEBS J. 285:1572–1588. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Villa E, Sahu U, O'Hara BP, Ali ES, Helmin
KA, Asara JM, Gao P, Singer BD and Ben-Sahra I: mTORC1 stimulates
cell growth through SAM synthesis and m(6)A mRNA-dependent control
of protein synthesis. Mol Cell. 81:2076–2093.e9. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhou X, Wang X, Duan J, Sun W, Chen Z, Li
Q, Ou Z, Jiang G, Ren X and Liu S: HBXIP protein overexpression
predicts the poor prognosis of pancreatic ductal adenocarcinomas.
Pathol Res Pract. 215:343–346. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang X, Feng Q, Yu H, Zhou X, Shan C,
Zhang Q and Liu S: HBXIP: A potential prognosis biomarker of
colorectal cancer which promotes invasion and migration via
epithelial-mesenchymal transition. Life Sci. 245:1173542020.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Piao JJ, Li N, Wang YX, Lin ZH and Liu SP:
HBXIP expression in gastric adenocarcinoma predicts poor prognosis.
Zhonghua Bing Li Xue Za Zhi. 46:88–92. 2017.(In Chinese).
PubMed/NCBI
|
|
9
|
Li N, Wang Y, Che S, Yang Y, Piao J, Liu S
and Lin Z: HBXIP over expression as an independent biomarker for
cervical cancer. Exp Mol Pathol. 102:133–137. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang Y, Sun J, Li N, Che S, Jin T, Liu S
and Lin Z: HBXIP overexpression is correlated with the clinical
features and survival outcome of ovarian cancer. J Ovarian Res.
10:262017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Xia H, Ma L, Li J, Bai H and Wang D:
Elevated HBXIP expression is associated with aggressive phenotype
and poor prognosis in esophageal squamous cell carcinoma. Am J
Cancer Res. 7:2190–2198. 2017.PubMed/NCBI
|
|
12
|
Cheng D, Liang B and Li Y: HBXIP
expression predicts patient prognosis in breast cancer. Med Oncol.
31:2102014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Guo ZY, Jiang LP and Zhu ZT: High HBXIP
expression is related to poor prognosis in HCC by extensive
database interrogation. Eur Rev Med Pharmacol Sci. 25:6196–6207.
2021.PubMed/NCBI
|
|
14
|
Wang Y, Li N, Che S, Jin T, Piao J, Liu S
and Lin Z: HBXIP suppression reduces cell proliferation and
migration and its overexpression predicts poor prognosis in
non-small-cell lung cancer. Tumour Biol. 39:10104283177096752017.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gao X and Yang L: HBXIP knockdown inhibits
FHL2 to promote cycle arrest and suppress cervical cancer cell
proliferation, invasion and migration. Oncol Lett. 25:1862023.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Xu F, Zhu X, Han TAO, You X, Liu F, Ye L,
Zhang X, Wang X and Yao Y: The oncoprotein hepatitis B
X-interacting protein promotes the migration of ovarian cancer
cells through the upregulation of S-phase kinase-associated protein
2 by Sp1. Int J Oncol. 45:255–263. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Meng X and Liu W: The effects of HBXIP on
the biological functions of tongue squamous cell carcinoma cells
and correlation with PI3K/Akt. Transl Cancer Res. 9:3375–3384.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Qiu L, Lu F, Zhang L, Wang G, Geng R and
Miao Y: HBXIP regulates gastric cancer glucose metabolism and
malignancy through PI3K/AKT and p53 signaling. Onco Targets Ther.
13:3359–3374. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Fang X, Tan T, Gao B, Zhao Y, Liu T and
Xia Q: Germacrone regulates HBXIP-Mediated cell cycle, apoptosis
and promotes the formation of autophagosomes to inhibit the
proliferation of gastric cancer cells. Front Oncol. 10:5373222020.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Fujii R, Zhu C, Wen Y, Marusawa H,
Bailly-Maitre B, Matsuzawa S, Zhang H, Kim Y, Bennett CF, Jiang W
and Reed JC: HBXIP, cellular target of hepatitis B virus
oncoprotein, is a regulator of centrosome dynamics and cytokinesis.
Cancer Res. 66:9099–9107. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Fei H, Zhou Y, Li R, Yang M, Ma J and Wang
F: HBXIP, a binding protein of HBx, regulates maintenance of the
G2/M phase checkpoint induced by DNA damage and enhances
sensitivity to doxorubicin-induced cytotoxicity. Cell Cycle.
16:468–476. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Li H, Wang Z, Li Y, Fang R, Wang H, Shi H,
Zhang X, Zhang W and Ye L: Hepatitis B X-interacting protein
promotes the formation of the insulin gene-transcribing protein
complex Pdx-1/Neurod1 in animal pancreatic β-cells. J Biol Chem.
293:2053–2065. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Qin Y, Ni P, Zhang Q, Wang X, Du X, Yin Z,
Wang L, Ye L and Chen L: Hbxip is essential for murine
embryogenesis and regulates embryonic stem cell differentiation
through activating mTORC1. Development. 149:dev2005272022.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yonehara R, Nada S, Nakai T, Nakai M,
Kitamura A, Ogawa A, Nakatsumi H, Nakayama KI, Li S, Standley DM,
et al: Structural basis for the assembly of the Ragulator-Rag
GTPase complex. Nat Commun. 8:16252017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Jiang Y, Wang D, Ren H, Shi Y and Gao Y:
Oncogenic HBXIP enhances ZEB1 through Sp1 to accelerate breast
cancer growth. Thorac Cancer. 9:1664–1670. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Liu BW, Wang TJ, Li LL, Zhang L, Liu YX,
Feng JY, Wu Y, Xu FF, Zhang QS, Bao MZ, et al: Oncoprotein HBXIP
induces PKM2 via transcription factor E2F1 to promote cell
proliferation in ER-positive breast cancer. Acta Pharmacol Sin.
40:530–538. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Haradhvala NJ, Polak P, Stojanov P,
Covington KR, Shinbrot E, Hess JM, Rheinbay E, Kim J, Maruvka YE,
Braunstein LZ, et al: Mutational strand asymmetries in cancer
genomes reveal mechanisms of DNA damage and repair. Cell.
164:538–549. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Smith J, Tho LM, Xu N and Gillespie DA:
The ATM-Chk2 and ATR-Chk1 pathways in DNA damage signaling and
cancer. Adv Cancer Res. 108:73–112. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Holoch D and Moazed D: RNA-mediated
epigenetic regulation of gene expression. Nat Rev Genet. 16:71–84.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Easwaran H, Tsai HC and Baylin SB: Cancer
epigenetics: Tumor heterogeneity, plasticity of stem-like states,
and drug resistance. Mol Cell. 54:716–727. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Pourdehnad M, Truitt M, Siddiqi I, Ducker
G, Shokat K and Ruggero D: Myc and mTOR converge on a common node
in protein synthesis control that confers synthetic lethality in
Myc-driven cancers. Proc Natl Acad Sci USA. 110:11988–11993. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Li Y, Wang Z, Shi H, Li H, Li L, Fang R,
Cai X, Liu B, Zhang X and Ye L: HBXIP and LSD1 Scaffolded by lncRNA
hotair mediate transcriptional activation by c-Myc. Cancer Res.
76:293–304. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Smith J, Sen S, Weeks RJ, Eccles MR and
Chatterjee A: Promoter DNA hypermethylation and paradoxical gene
activation. Trends Cancer. 6:392–406. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Li H, Wang Z, Jiang M, Fang RP, Shi H,
Shen Y, Cai XL, Liu Q, Ye K, Fan SJ, et al: The oncoprotein HBXIP
promotes human breast cancer growth through down-regulating p53 via
miR-18b/MDM2 and pAKT/MDM2 pathways. Acta Pharmacol Sin.
39:1787–1796. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Maruyama T, Kadowaki H, Okamoto N, Nagai
A, Naguro I, Matsuzawa A, Shibuya H, Tanaka K, Murata S, Takeda K,
et al: CHIP-dependent termination of MEKK2 regulates temporal ERK
activation required for proper hyperosmotic response. EMBO J.
29:2501–2514. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Li Y, Zhang Z, Zhou X, Li L, Liu Q, Wang
Z, Bai X, Zhao Y, Shi H, Zhang X and Ye L: The oncoprotein HBXIP
enhances migration of breast cancer cells through increasing
filopodia formation involving MEKK2/ERK1/2/Capn4 signaling. Cancer
Lett. 355:288–296. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Halbeisen RE, Galgano A, Scherrer T and
Gerber AP: Post-transcriptional gene regulation: From genome-wide
studies to principles. Cell Mol Life Sci. 65:798–813. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Sánchez-Vásquez E, Alata Jimenez N,
Vázquez NA and Strobl-Mazzulla PH: Emerging role of dynamic RNA
modifications during animal development. Mech Dev. 154:24–32. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhou Y, Yin Z, Hou B, Yu M, Chen R, Jin H
and Jian Z: Expression profiles and prognostic significance of RNA
N6-methyladenosine-related genes in patients with hepatocellular
carcinoma: Evidence from independent datasets. Cancer Manag Res.
11:3921–3931. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Cai X, Wang X, Cao C, Gao Y, Zhang S, Yang
Z, Liu Y, Zhang X, Zhang W and Ye L: HBXIP-elevated
methyltransferase METTL3 promotes the progression of breast cancer
via inhibiting tumor suppressor let-7g. Cancer Lett. 415:11–19.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Yang N, Wang T, Li Q, Han F, Wang Z, Zhu R
and Zhou J: HBXIP drives metabolic reprogramming in hepatocellular
carcinoma cells via METTL3-mediated m6A modification of HIF-1α. J
Cell Physiol. 236:3863–3880. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Yang Z and Jiang X, Li D and Jiang X:
viaHBXIP promotes gastric cancer METTL3-mediated MYC mRNA m6A
modification. Aging (Albany NY). 12:24967–24982. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Fabian MR, Sonenberg N and Filipowicz W:
Regulation of mRNA translation and stability by microRNAs. Annu Rev
Biochem. 79:351–379. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Shukla GC, Singh J and Barik S: MicroRNAs:
Processing, maturation, target recognition and regulatory
functions. Mol Cell Pharmacol. 3:83–92. 2011.PubMed/NCBI
|
|
45
|
Liu F, Zhang W, You X, Liu Y, Li Y, Wang
Z, Wang Y, Zhang X and Ye L: The oncoprotein HBXIP promotes glucose
metabolism reprogramming via downregulating SCO2 and PDHA1 in
breast cancer. Oncotarget. 6:27199–27213. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Jiang Y, Wang D, Ren H, Shi Y and Gao Y:
MiR-145-targeted HBXIP modulates human breast cancer cell
proliferation. Thorac Cancer. 10:71–77. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Zhang W, Lu Z, Kong G, Gao Y, Wang T, Wang
Q, Cai N, Wang H, Liu F, Ye L and Zhang X: Hepatitis B virus X
protein accelerates hepatocarcinogenesis with partner survivin
through modulating miR-520b and HBXIP. Mol Cancer. 13:1282014.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Hu XM, Yan XH, Hu YW, Huang JL, Cao SW,
Ren TY, Tang YT, Lin L, Zheng L and Wang Q: miRNA-548p suppresses
hepatitis B virus X protein associated hepatocellular carcinoma by
downregulating oncoprotein hepatitis B x-interacting protein.
Hepatol Res. 46:804–815. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Sebastian-delaCruz M, Gonzalez-Moro I,
Olazagoitia-Garmendia A, Castellanos-Rubio A and Santin I: The role
of lncRNAs in gene expression regulation through mRNA
Stabilization. Noncoding RNA. 7:32021.PubMed/NCBI
|
|
50
|
Zaniani NR, Oroujalian A, Valipour A and
Peymani M: LAMTOR5 expression level is a biomarker for colorectal
cancer and lncRNA LAMTOR5-AS1 predicting miRNA sponging effect. Mol
Biol Rep. 48:6093–6101. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Balasooriya ER, Madhusanka D,
Lopez-Palacios TP, Eastmond RJ, Jayatunge D, Owen JJ, Gashler JS,
Egbert CM, Bulathsinghalage C, Liu L, et al: Integrating clinical
cancer and PTM proteomics data identifies a mechanism of ACK1
kinase activation. Mol Cancer Res. 22:137–151. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Huang L, Wen X, Jin L, Han H and Guo H:
HOOKLESS1 acetylates AUTOPHAGY-RELATED PROTEIN18a to promote
autophagy during nutrient starvation in Arabidopsis. Plant Cell.
36:136–157. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Yin X, Wang X and Komatsu S:
Phosphoproteomics: Protein phosphorylation in regulation of seed
germination and plant growth. Curr Protein Pept Sci. 19:401–412.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Cockram PE, Kist M, Prakash S, Chen SH,
Wertz IE and Vucic D: Ubiquitination in the regulation of
inflammatory cell death and cancer. Cell Death Differ. 28:591–605.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Pispa J, Mikkonen E, Arpalahti L, Jin C,
Martínez-Fernández C, Cerón J and Holmberg CI: AKIR-1 regulates
proteasome subcellular function in Caenorhabditis elegans.
iScience. 26:1078862023. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Ye L, Zhang W, Jin T, Zhang L, Wang T, Fu
X, Jin T, Zhang W and Ye L: The regulation of acetylation and
stability of HMGA2 via the HBXIP-activated Akt-PCAF pathway in
promotion of esophageal squamous cell carcinoma growth. Nucleic
Acids Res. 48:4858–4876. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Xu FF, Sun HM, Fang RP, Zhang L, Shi H,
Wang X, Fu XL, Li XM, Shi XH, Wu Y, et al: The modulation of PD-L1
induced by the oncogenic HBXIP for breast cancer growth. Acta
Pharmacol Sin. 43:429–445. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Zhang J, Sun B, Ruan X, Hou X, Zhi J, Meng
X, Zheng X and Gao M: Oncoprotein HBXIP promotes tumorigenesis
through MAPK/ERK pathway activation in non-small cell lung cancer.
Cancer Biol Med. 18:105–119. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Min JH, Yang H, Ivan M, Gertler F, Kaelin
WG Jr and Pavletich NP: Structure of an HIF-1alpha-pVHL complex:
Hydroxyproline recognition in signaling. Science. 296:1886–1889.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Zhou XL, Zhu CY, Wu ZG, Guo X and Zou W:
The oncoprotein HBXIP competitively binds KEAP1 to activate NRF2
and enhance breast cancer cell growth and metastasis. Oncogene.
38:4028–4046. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Bopape M, Tiloke C and Ntsapi C: Moringa
oleifera and Autophagy: Evidence from in vivo studies on
chaperone-mediated autophagy in HepG2 cancer cells. Nutr
Cancer. 75:1822–1847. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Liu B, Wang T, Wang H, Zhang L, Xu F, Fang
R, Li L, Cai X, Wu Y, Zhang W and Ye L: Oncoprotein HBXIP enhances
HOXB13 acetylation and co-activates HOXB13 to confer tamoxifen
resistance in breast cancer. Hematol Oncol. 11:262018. View Article : Google Scholar
|
|
63
|
Li L, Fang R, Liu B, Shi H, Wang Y, Zhang
W, Zhang X and Ye L: Deacetylation of tumor-suppressor MST1 in
Hippo pathway induces its degradation through HBXIP-elevated HDAC6
in promotion of breast cancer growth. Oncogene. 35:4048–4057. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Zhang L, Zhou X, Liu B, Shi X, Li X, Xu F,
Fu X, Wang X, Ye K, Jin T, et al: HBXIP blocks myosin-IIA assembly
by phosphorylating and interacting with NMHC-IIA in breast cancer
metastasis. Acta Pharm Sin B. 13:1053–1070. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Yoneyama M, Kawada K, Gotoh Y, Shiba T and
Ogita K: Endogenous reactive oxygen species are essential for
proliferation of neural stem/progenitor cells. Neurochem Int.
56:740–746. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Freyre-Fonseca V, Delgado-Buenrostro NL,
Gutiérrez-Cirlos EB, Calderón-Torres CM, Cabellos-Avelar T,
Sánchez-Pérez Y, Pinzón E, Torres I, Molina-Jijón E, Zazueta C, et
al: Titanium dioxide nanoparticles impair lung mitochondrial
function. Toxicol Lett. 202:111–119. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Cremers CM and Jakob U: Oxidant sensing by
reversible disulfide bond formation. J Biol Chem. 288:26489–26496.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Galadari S, Rahman A, Pallichankandy S and
Thayyullathil F: Reactive oxygen species and cancer paradox: To
promote or to suppress? Free Radic Biol Med. 104:144–164. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Moldogazieva NT, Lutsenko SV and Terentiev
AA: Reactive oxygen and nitrogen species-induced protein
modifications: Implication in carcinogenesis and anticancer
therapy. Cancer Res. 78:6040–6047. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Zhang S, Wang R, Wang X, Guo X, Du Y, Guo
X, Zong X, Zhu C and Zhou X: HBXIP is a novel regulator of the
unfolded protein response that sustains tamoxifen resistance in ER+
breast cancer. J Biol Chem. 298:1016442022. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Baird L and Yamamoto M: Immunoediting of
KEAP1-NRF2 mutant tumours is required to circumvent NRF2-mediated
immune surveillance. Redox Biol. 67:1029042023. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Zhou X, Li L, Guo X, Zhang C, Du Y, Li T,
Tong K, Zhu C and Wang Z: HBXIP induces anoikis resistance by
forming a reciprocal feedback loop with Nrf2 to maintain redox
homeostasis and stabilize Prdx1 in breast cancer. NPJ Breast
Cancer. 8:72022. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Tang R, Luo J, Zhu X, Miao P, Tang H, Jian
Y, Ruan S, Ling F and Tang M: Recent progress in the effect of
ferroptosis of HSCs on the development of liver fibrosis. Front Mol
Biosci. 10:12588702023. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Zhao C and Dahlman-Wright K: Liver X
receptor in cholesterol metabolism. J Endocrinol. 204:233–240.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Cohen RN, Brzostek S, Kim B, Chorev M,
Wondisford FE and Hollenberg AN: The specificity of interactions
between nuclear hormone receptors and corepressors is mediated by
distinct amino acid sequences within the interacting domains. Mol
Endocrinol. 15:1049–1061. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Zhao Y, Li H, Zhang Y, Li L, Fang R, Li Y,
Liu Q, Zhang W, Qiu L, Liu F, et al: Oncoprotein HBXIP modulates
abnormal lipid metabolism and growth of breast cancer cells by
activating the LXRs/SREBP-1c/FAS signaling cascade. Cancer Res.
76:4696–4707. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Zhang L, Li XM, Shi XH, Ye K, Fu XL, Wang
X, Guo SM, Ma JQ, Xu FF, Sun HM, et al: Sorafenib triggers
ferroptosis via inhibition of HBXIP/SCD axis in hepatocellular
carcinoma. Acta Pharmacol Sin. 44:622–634. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Liu X, Li H, Che N, Zheng Y, Fan W, Li M,
Li X and Xuan Y: HBXIP accelerates glycolysis and promotes cancer
angiogenesis via AKT/mTOR pathway in bladder cancer. Exp Mol
Pathol. 121:1046652021. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Zhang Y, Zhao Y, Li H, Li Y, Cai X, Shen
Y, Shi H, Li L, Liu Q, Zhang X and Ye L: The nuclear import of
oncoprotein hepatitis B X-interacting protein depends on
interacting with c-Fos and phosphorylation of both proteins in
breast cancer cells. J Biol Chem. 288:18961–18974. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Liu S, Li L, Zhang Y, Zhang Y, Zhao Y, You
X, Lin Z, Zhang X and Ye L: The oncoprotein HBXIP uses two pathways
to up-regulate S100A4 in promotion of growth and migration of
breast cancer cells. J Biol Chem. 287:30228–30239. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Liu F, You X, Wang Y, Liu Q, Liu Y, Zhang
S, Chen L, Zhang X and Ye L: The oncoprotein HBXIP enhances
angiogenesis and growth of breast cancer through modulating FGF8
and VEGF. Carcinogenesis. 35:1144–1153. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Clarke HJ, Chambers JE, Liniker E and
Marciniak SJ: Endoplasmic reticulum stress in malignancy. Cancer
Cell. 25:563–573. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Clarke R, Shajahan AN, Wang Y, Tyson JJ,
Riggins RB, Weiner LM, Bauman WT, Xuan J, Zhang B, Facey C, et al:
Endoplasmic reticulum stress, the unfolded protein response, and
gene network modeling in antiestrogen resistant breast cancer. Horm
Mol Biol Clin Investig. 5:35–44. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Meng X, Qi XY, Wang QX and Liu WX: Effect
of HBXIP on biological function and PI3K/Akt signaling pathway of
adenoid cystic carcinoma cell line ACC-M. Shanghai Kou Qiang Yi
Xue. 26:389–394. 2017.(In Chinese). PubMed/NCBI
|
|
85
|
Cai X, Cao C, Li J, Chen F, Zhang S, Liu
B, Zhang W, Zhang X and Ye L: Inflammatory factor TNF-α promotes
the growth of breast cancer via the positive feedback loop of
TNFR1/NF-κB (and/or p38)/p-STAT3/HBXIP/TNFR1. Oncotarget.
8:58338–58352. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Zhuang C, Narayanapillai S, Zhang W, Sham
Y and Xing C: Rapid identification of Keap1-Nrf2 small-molecule
inhibitors through structure-based virtual screening and hit-based
substructure search. J Med Chem. 57:1121–1126. 2014. View Article : Google Scholar : PubMed/NCBI
|

















