Introduction
Human papillomavirus (HPV)-mediated transformation
of human cervical epithelial cells has been recognized as a
multi-step process in which not only viruses but also certain
additional unknown factors and (epi)genetic events are
required.
Our previous study indicated that one of the factors
that may be involved in cervical carcinogenesis is tumor
susceptibility gene 101 (TSG101) (1). The TSG101 gene was mapped to
chromosome 11.p15.1–p15.2., a region that is associated with the
loss of heterozygosity in several tumor types, including breast
cancer and cervical cancer (2,3). The
TSG101 protein is involved in a variety of important biological
functions, such as ubiquitination, transcriptional regulation,
endosomal trafficking, virus budding, proliferation and cell
survival (4–16). It has been suggested that TSG101 is
an important factor for maintaining cellular homeostasis and that
perturbation of TSG101 functions leads to transformation (17). To identify the mechanism responsible
for TSG101 downregulation during cervical cancer development, we
analyzed the TSG101 promoter using cis-element cluster
finder (Cister) software. One of the transcription factors, whose
binding site was detected in the TSG101 promoter, was LSF
(1).
LSF was initially identified due to its ability to
activate a major late Simian virus 40 (SV40) promoter (18). Late SV40 factor (LSF) is commonly
named as CP2 and is encoded by the TFCP2 gene located in
chromosome 12q13 (19,20). LSF belongs to an evolutionary
conserved family of transcription factors consisting of two
subfamilies: LSF/CP2 and grainyhead (21–24).
Human LSF is a 502-amino acid protein with a
molecular weight of ~57 kDa (24).
It consists of two functional domains. The N-terminal domain is a
DNA interaction region located between amino acids 67 and 260, and
is similar in structure to p53/p63/p73 DNA binding domain. The
C-terminal region is responsible for oligomerization and contains
tetramerization and dimerization domains. The tetramerization
domain is located between amino acids 326 and 89, and is
structurally similar to the sterile α-motif protein-protein
interaction domain (24,25). The dimerization domain contains
residues 448–502 and is structurally similar to the ubiquitin-like
fold domain (24–26). Amino acid residues 189–239 mediate
nuclear localization of LSF (24,27,28).
With the exception of its important role in
regulation of viral and cellular promoters, including SV40, and
HIV-1 promoters, including fibrinogen and α-globin, LSF is involved
in numerous other important biological functions, such as
regulation of the cell cycle, cell growth and development, DNA
synthesis, cell survival and Alzheimer’s disease. Moreover, LSF
also functions as an anti-apoptotic factor (24,29,30).
The aim of the present study was to analyze TSG101
and LSF protein expression during cervical cancer development.
Materials and methods
Clinical samples
Samples were collected from 116 patients (median
age, 49 years; range, 23–61 years) undergoing gynecological
surgical procedures at the Department of Obstetrics and Pathology
of Pregnancy, Medical University (Lublin, Poland). The study group
consisted of 29 HPV-positive cervical samples (carcinoma colli
uteri/squamous cell carcinoma), 30 HPV-positive high-grade squamous
intraepithelial lesion (HSIL) samples, 28 HPV-positive low-grade
squamous intraepithelial lesion (LSIL) samples and 29
histopathologically normal, HPV-negative cervical tissues obtained
from women undergoing treatment for reasons other than cervical
cancer. The study was approved by the Ethics Committee of the
Medical University of Lublin. For cancer and neoplastic
localization, all specimens initially underwent hematoxylin and
eosin staining followed by a pathological review. Cervical sections
comprising ≥70% cancer cells were used as cancer samples. The
tissue samples were frozen immediately in liquid nitrogen and
stored at −80°C until further analysis. Patients provided written
informed consent.
Isolation of DNA
Total DNA was isolated from study cells using a
QIAmp DNA Midi kit (Qiagen, Hilden, Germany) according to the
manufacturer’s instructions.
Identification of HPV DNA
HPV DNA was identified by polymerase chain reaction
(PCR) amplification of the HPV gene sequence using isolated DNA and
primers: MY09, MY11 (31) and LC1,
and LC2 (32), complementary to the
genome sequence of the majority of common types of HPV viruses, as
described previously (31,32).
RNA extraction/isolation
Total RNA was isolated from normal, dysplastic (LSIL
and HSIL) and cancer tissues using an RNeasy Mini kit (Qiagen)
following the manufuacturer’s instructions. DNA was removed by
DNase treatment (RNase-Free DNase set, Qiagen).
Quantitative PCR (qPCR) analysis
Total RNA (1 μg) was reverse transcribed to cDNA
using the QuantiTect Reverse Transcription kit (Qiagen). PCR
reactions were run under the following conditions: pre-denaturation
at 95°C for 5 min, then 40 cycles at 95°C for 10 sec, 63.8°C for 20
sec, 72°C for 20 sec and a final extension at 72°C for 6 min. qPCR
was performed on a Corbett Rotor-Gene 6000 (Corbett Life Science,
Concorde, NSW, Australia).
PCR was performed using SYBR Green PCR master mix
(Invitrogen Life Technologies, Carlsbad, CA, USA) and appropriate
primers (forward, 5′TGGCCGACGAAGTGATTGAA 3′ and reverse,
5′GGGCAATGCAAGGACATCAC 3′ for the LSF gene). Total cDNA (2 μl) and
primers were added to 8 μl Power SYBR Green PCR master mix.
Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was
used as reference gene (33). All
reactions for samples and housekeeping genes were run in
triplicate.
Immunohistochemical analysis of TSG101
and LSF expression
Immunohistochemical analysis was prepared using a
LSAB System-HRP visualization kit (K0679; Dako, Carpinteria, CA,
USA), mouse monoclonal anti-TSG101 antibody (Santa Cruz
Biotechnology, Inc., Santa Cruz, CA, USA) and mouse monoclonal
anti-LSF antibody (BD Transduction Laboratories™, San Jose, CA,
USA). Antibodies were diluted in Dako antibody diluent with
background-reducing component (S3022; Dako).
Immunohistochemical evaluations of TSG101 and LSF
expression were performed independently by two pathomorphologists.
The cells were counted on an Axiophote (Opton) fluorescence
microscope (Carl Zeiss, Oberkochen, Germany) with ×200
magnification on a field of 16 squares (4×4 squares), which
corresponded to an area of 0.25 mm2 (0.5×0.5 mm).
Positive staining was scored according to the
percentage of cells with positive staining and staining intensity.
Measurement of immunoreactive cells was performed using Cell-2
software, version 4.1 (University of Medical Sciences, Poznan,
Poland). The quantitative method is based on the analysis of the
color distribution and the optical density. The software identifies
cells with a greater optical density than the background and, on
the basis of the color ratio, classifies cells as immunoreactive.
To determine the percentage of positive cells in the sections, the
counts of immunopositive cells were divided by the total cell
count. For each case, a minimum of 5,000 total cells were counted
in a single section.
Statistical analysis
Data were analyzed with Statistica software, version
6.1. (Statsoft, Krakow, Poland). Statistical significance was
analyzed using Kruskal-Wallis with post-hoc Dunn’s test
(immunohistochemistry) and one-way analysis of variance with
Tukey’s post hoc test (qPCR). The difference was considered to be
statistically significant when P-values were <0.05.
Bioinformatics analysis
The localization and frequency of LSF binding sites
in the TSG101 promoter sequence were analyzed using Cister
software (Zlab gene regulation tools, Boston Univeristy, Boston,
MA, USA) as described previously (1).
Results
Bioinformatics analysis of LSF binding
site frequency in the TSG101 promoter sequence
Analysis using Cister software distinguished 14
binding sites for LSF transcription factor in the TSG101
promoter sequence (Table I).
 | Table IPosition and probability of LSF
transcription factor binding to TSG101 promoter sequence according
to Cister software. |
Table I
Position and probability of LSF
transcription factor binding to TSG101 promoter sequence according
to Cister software.
| Position | Strand | Sequence | Probablility |
|---|
| 3262–3276 | + |
agtggcttacgcctg | 0.56 |
| 5238–5252 | − |
ccggcccagccaagc | 0.49 |
| 1158–1172 | − |
ccactgcactccagc | 0.48 |
| 3398–3412 | + |
ggtggtgggcacctg | 0.31 |
| 3293–3307 | + |
gcaggctgaggcggg | 0.27 |
| 1333–1347 | − |
ctactgcactccagc | 0.24 |
| 5174–5188 | + |
gctgcgacgcgctcg | 0.21 |
| 1125–1139 | + |
ggtaggtggagcttg | 0.19 |
| 5064–5078 | − |
ctggggcagcccagc | 0.17 |
| 5233–5247 | − |
ccgtcccggcccagc | 0.14 |
| 5053–5067 | + |
tgtgggacggtctgg | 0.13 |
| 3493–3507 | − |
ctatcgcactccagc | 0.12 |
| 6654–6668 | − |
caggcgtgagccacc | 0.12 |
| 1063–1077 | + |
ggtggcaggtgcctg | 0.10 |
Detection of HPV viruses in clinical
samples
Prior to molecular analysis, clinical samples were
screened for the presence of HPV viruses. Only HPV-positive HSIL,
LSIL and cancer cells, and the HPV-negative control, were used in
these studies (Table II).
 | Table IICharacteristics of the studied
group. |
Table II
Characteristics of the studied
group.
| Clinical
characteristics | Control | HSIL | LSIL | Cervical
cancer |
|---|
| HPV detection | − | + | + | + |
| Number of
patients | 29 | 30 | 28 | 29 |
Immunohistochemical analysis of TSG101
expression
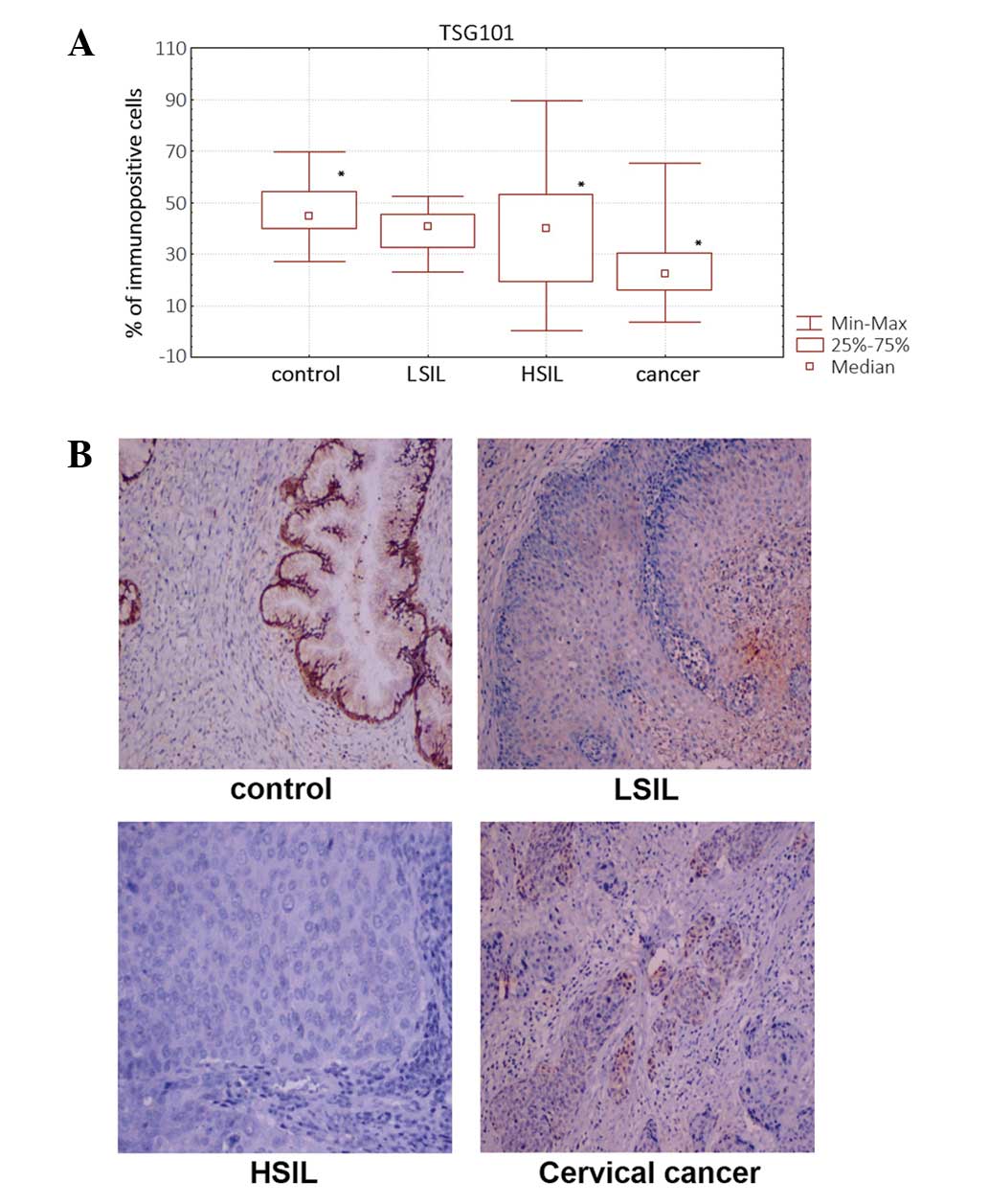
Quantitive immunohistochemical analysis based on the
percentage of TSG101-immunopositive cells revealed a significantly
(P<0.05) lower percentage of TSG101-immunopositive cells in
cervical cancer and HSIL samples compared with that in non-tumor
(HPV-negative) cells (Fig. 1A). The
TSG101 protein showed mainly nuclear localization (Fig. 1B).
Analysis of LSF mRNA and protein
level
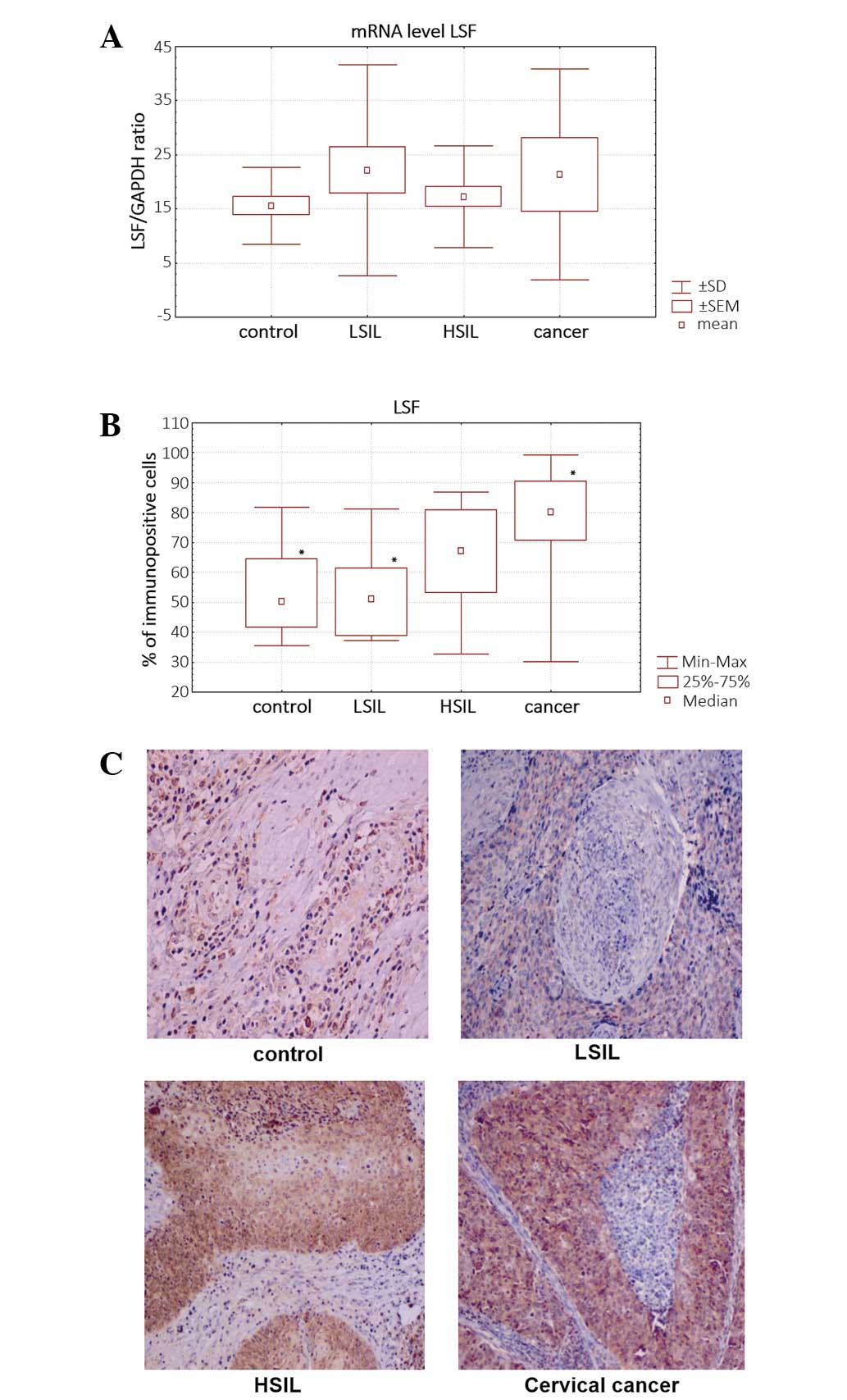
qPCR analysis showed an increased level of
LSF mRNA in dysplastic (LSIL and HSIL) and cervical cancer
cells. LSF mRNA level was upregulated to 42.43% in HSIL
samples, 30.6% in LSIL samples and 37.54% in cancer samples,
compared with that in non-tumor HPV-negative samples (Fig. 2A). The differences in LSF expression
were not statistically significant (P>0.05).
Quantitive immunohistochemical analysis based on the
percentage of LSF-immunopositive cells confirmed the increased
expression of LSF in cervical cancer cells.
The percentage of LSF-immunostained cells was
significantly higher in cervical cancer and LSIL samples compared
with that in HPV-negative non-tumor controls (P<0.05) (Fig. 2B). LSF was observed mainly in the
nucleus (Fig. 2C).
Discussion
In addition to HPV viruses, numerous factors, such
as oncogenes and tumor suppressor genes are involved in cervical
cancer development.
Our previous results demonstrated a decreased
expression of TSG101 in cervical cancer cells (1). In the present study, we confirmed
TSG101 protein downregulation during cervical cancer development
using immunohistochemical analysis.
TSG101 is constitutively expressed in a
number of human tissues (34).
However, upregulation of TSG101 was found in thyroid
papillary carcinomas, and breast, ovarian and gastrointestinal
tumors, while downregulation of TSG101 was observed in
endometrial and cervical cancers (1,35,36).
Our bioinformatics analysis showed that one of the
factors that may bind with the TSG101 promoter and regulate
its expression is LSF.
qPCR and immunohistochemical analysis revealed high
expression levels of LSF in cervical cancer cells compared with
non-cancer samples. These results suggest that LSF is important in
cervical tumorigenesis and that LSF mRNA levels generally do not
fluctuate. LSF mRNA has been suggested to be a normalization
control in gene expression profiling (37). LSF is also expressed ubiquitously in
cell lines and in the developing mouse, and protein levels are
unaltered during cell cycle progression (38–40).
LSF is a transcription factor involved in the
regulation of a variety of viral and cellular promoters. It acts as
a transcription activator and repressor (24). LSF stimulates transcription of the
SV40 late promoter (18). It also
binds to the sequence within the HIV-1 long terminal repeat (LTR)
initiation region and recruits YY1 and histone deacetylase 1 to the
LTR, inhibiting transcription and thereby contributing to HIV
persistence within resting CD4 T cells (41). The cellular factor YY1 also plays a
critical role in tumorigenesis and HPV infection, as a positive and
negative regulator of cellular and viral gene expression. The
YY1-mediated downregulation of HPV transcription, as well as other
promoters, act together with LSF (42).
High expression of LSF in cervical cancer
HPV-positive cells suggests that this protein may be involved in
downregulation of the TSG101 gene promoter and HPV-dependent
cervical carcinogenesis. Fan et al identified LSF as a
downstream mediator of Notch1 signaling and showed that LSF
mediates, at least in part, Notch-1-induced carcinogenesis
(43). Notch genes encode
heterodimeric transmembrane receptors, which play a critical role
in maintaining the balance between cell proliferation,
differentiation and apoptosis. Aberrant Notch signaling may
contribute to cervical carcinogenesis (44), head and neck cancer (45), lung cancer (46), colon cancer (47), acute myeloid leukemia (48) and diffuse large B-cell lymphoma
(30). LSF was also significantly
upregulated in hepatocellular carcinoma compared with non-cancer
samples (43). In liver cancer, its
expression is strongly correlated with tumor grade and
aggressiveness (49). Microarray
studies revealed that LSF modulated the expression of specific
genes involved in regulating invasion, angiogenesis,
chemoresistance and senescence (24). It has been suggested that LSF may
function as an oncogene in hepatocarcinogenesis (29,30,50).
Osteopontin, matrix metalloproteinase 9, c-Met and complement
factor H have been identified as a proteins directly regulated by
LSF and involved in hepatocarcinogenesis (24).
A major cellular target of LSF is the thymidylate
synthase gene, which encodes the enzyme involved in the production
of dTTP, required for DNA synthesis. Deregulated LSF expression may
facilitate entry into the G1/S phase of the cell cycle, promote DNA
synthesis, stimulate transformation and facilitate cancer cell
survival. Inhibition of LSF results in either apoptosis during S
phase or cell cycle arrest at the G1/S transition (29,30,50).
The role of LSF as a mediator in cervical cancer
development must be confirmed in future studies.
Acknowledgements
This study was supported by a grant (N N401 012136)
from the Ministry of Science and Higher Education in Poland.
References
|
1
|
Broniarczyk J, Olejnik-Schmidt AK, Luczak
MW, Schmidt MT, Dabrowski M, Józefiak A, Kedzia W, Kwaśniewska A
and Goździcka-Józefiak A: Analysis of expression and structure of
the TSG101 gene in cervical cancer cells. Int J Mol Med.
25:777–783. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Li L, Li X, Francke U and Cohen SN: The
TSG101 tumor susceptibility gene is located in chromosome 11 band
p15 and is mutated in human breast cancer. Cell. 88:143–154. 1997.
View Article : Google Scholar
|
|
3
|
Singh RK, Dasgupta S, Bhattacharya N,
Chunder N, Mondal R, Roy A, Mandal S, Roychowdhury S and Panda CK:
Deletion in chromosome 11 and Bcl-1/Cyclin D1 alterations are
independently associated with the development of uterine cervical
carcinoma. J Cancer Res Clin Oncol. 131:395–406. 2005. View Article : Google Scholar
|
|
4
|
Carstens MJ, Krempler A, Triplett AA, Van
Lohuizen M and Wagner KU: Cell cycle arrest and cell death are
controlled by p53-dependent and p53-independent mechanisms in
Tsg101-deficient cells. J Biol Chem. 279:35984–35994. 2004.
View Article : Google Scholar
|
|
5
|
Koonin EV and Abagyan RA: TSG101 may be
the prototype of a class of dominant negative ubiquitin regulators.
Nat Genet. 16:330–331. 1997. View Article : Google Scholar
|
|
6
|
Ponting CP, Cai YD and Bork P: The breast
cancer gene product TSG101: a regulator of ubiquitination? J Mol
Med. 75:467–469. 1997.PubMed/NCBI
|
|
7
|
Watanabe M, Yanagi Y, Masuhiro Y, Yano T,
Yoshikawa H, Yanagisawa J and Kato S: A putative tumor suppressor,
TSG101, acts as a transcriptional suppressor through its
coiled-coil domain. Biochem Biophys Res Commun. 245:900–905. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hittelman AB, Burakov D, Iñiguez-Lluhí JA,
Freedman LP and Garabedian MJ: Differential regulation of
glucocorticoid receptor transcriptional activation via
AF-1-associated proteins. EMBO J. 18:5380–5388. 1999. View Article : Google Scholar
|
|
9
|
Babst M, Odorizzi G, Estepa EJ and Emr SD:
Mammalian tumor susceptibility gene 101 (TSG101) and the yeast
homologue, Vps23p, both function in late endosomal trafficking.
Traffic. 1:248–258. 2000. View Article : Google Scholar
|
|
10
|
Garrus JE, von Schwedler UK, Pornillos OW,
et al: Tsg101 and the vacuolar protein sorting pathway are
essential for HIV-1 budding. Cell. 107:55–65. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lu Q, Hope LW, Brasch M, Reinhard C and
Cohen SN: TSG101 interaction with HRS mediates endosomal
trafficking and receptor down-regulation. Proc Natl Acad Sci USA.
100:7626–7631. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bache KG, Brech A, Mehlum A and Stenmark
H: Hrs regulates multivesicular body formation via ESCRT
recruitment to endosomes. J Cell Biol. 162:435–442. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhong Q, Chen Y, Jones D and Lee WH:
Perturbation of TSG101 protein affects cell cycle progression.
Cancer Res. 58:2699–2702. 1998.PubMed/NCBI
|
|
14
|
Xie W, Li L and Cohen SN: Cell
cycle-dependent subcellular localization of the TSG101 protein and
mitotic and nuclear abnormalities associated with TSG101
deficiency. Proc Natl Acad Sci USA. 95:1595–1600. 1998. View Article : Google Scholar
|
|
15
|
Ruland J, Sirard C, Elia A, MacPherson D,
Wakeham A, Li L, de la Pompa JL, Cohen SN and Mak TW: p53
accumulation, defective cell proliferation, and early embryonic
lethality in mice lacking tsg101. Proc Natl Acad Sci USA.
98:1859–1864. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Krempler A, Henry MD, Triplett AA and
Wagner KU: Targeted deletion of the Tsg101 gene results in cell
cycle arrest at G1/S and p53-independent cell death. J Biol Chem.
277:43216–43223. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wagner KU, Krempler A, Qi Y, Park K, Henry
MD, Triplett AA, Riedlinger G, Rucker EB and Hennighausen L: Tsg101
is essential for cell growth, proliferation, and cell survival of
embryonic and adult tissues. Mol Cell Biol. 23:150–162. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kim CH, Heath C, Bertuch A and Hansen U:
Specific stimulation of simian virus 40 late transcription in vitro
by a cellular factor binding the simian virus 40 21-base-pair
repeat promoter element. Proc Natl Acad Sci USA. 84:6025–6029.
1987. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Swendeman SL, Spielholz C, Jenkins NA,
Gilbert DJ, Copeland NG and Sheffery M: Characterization of the
genomic structure, chromosomal location, promoter, and development
expression of the alpha-globin transcription factor CP2. J Biol
Chem. 269:11663–11671. 1994.
|
|
20
|
Cunningham JM, Vanin EF, Tran N, Valentine
M and Jane SM: The human transcription factor CP2 (TFCP2), a
component of the human gamma-globin stage selector protein, maps to
chromosome region 12q13 and is within 250kb of the NF-E2 gene.
Genomics. 30:398–399. 1995.
|
|
21
|
Venkatesan K, McManus HR, Mello CC, Smith
TF and Hansen U: Functional conservation between members of an
ancient duplicated transcription factor family, LSF/Grainyhead.
Nucleic Acids Res. 31:4304–4316. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wilanowski T, Tuckfield A, Cerruti L,
O’Connell S, Saint R, Parekh V, Tao J, Cunningham JM and Jane SM: A
highly conserved novel family of mammalian developmental
transcription factors related to Drosophila grainyhead. Mech Dev.
114:37–50. 2002. View Article : Google Scholar
|
|
23
|
Traylor-Knowles N, Hansen U, Dubuc TQ,
Martindale MQ, Kaufman L and Finnerty JR: The evolutionary
diversification of LSF and Grainyhead transcription factors
preceded the radiation of basal animal lineages. BMC Evol Biol.
10:102010. View Article : Google Scholar
|
|
24
|
Santhekadur PK, Rajasekaran D, Siddiq A,
Gredler R, Chen D, Schaus SE, Hansen U, Fisher PB and Sarkar D: The
transcription factor LSF: a novel oncogene for hepatocellular
carcinoma. Am J Cancer Res. 2:269–285. 2012.PubMed/NCBI
|
|
25
|
Shirra MK and Hansen U: LSF and NTF-1
share a conserved DNA recognition motif yet require different
oligomerization states to form a stable protein-DNA complex. J Biol
Chem. 273:19260–19268. 1998. View Article : Google Scholar
|
|
26
|
Kokoszynska K, Ostrowski J, Rychlewski L
and Wyrwicz LS: The fold recognition of CP2 transcription factors
gives new insights into the function and evolution of tumor
suppressor protein p53. Cell Cycle. 7:2907–2915. 2008. View Article : Google Scholar
|
|
27
|
Drouin EE, Schrader CE, Stavnezer J and
Hansen U: The ubiquitously expressed DNA-binding protein late SV40
factor binds Ig switch regions and represses class switching to
IgA. J Immunol. 168:2847–2856. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zambrano N, Minopoli G, de Candia P and
Russo T: The Fe65 adaptor protein interacts through its PID1 domain
with the transcription factor CP2/LSF/LBP1. J Biol Chem.
273:20128–20133. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hansen U, Owens L and Saxena UH:
Transcription factors LSF and E2Fs: tandem cyclists driving G0 to
S? Cell Cycle. 8:2146–2151. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Powell CM, Rudge TL, Zhu Q, Johnson LF and
Hansen U: Inhibition of the mammalian transcription factor LSF
induces S-phase-dependent apoptosis by downregulating thymidylate
synthase expression. EMBO J. 19:4665–4675. 2000. View Article : Google Scholar
|
|
31
|
Manos MM, Ting Y, Wright DK, et al: The
use of polymerase chain reaction amplification for the detection of
genital human papillomaviruses. Cancer Cells. 7:209–214. 1989.
|
|
32
|
Yoshikawa H, Kawana T, Kitagawa K, Mizuno
M, Yoshikura H and Iwamoto A: Detection and typing of multiple
genital human papillomaviruses by DNA amplification with consensus
primers. J Cancer Res. 82:524–531. 1991.PubMed/NCBI
|
|
33
|
Daud II and Scott ME: Validation of
reference genes in cervical cell samples from human
papillomavirus-infected and -uninfected women for quantitative
reverse transcription-PCR assays. Clin Vaccine Immunol.
15:1369–1373. 2008. View Article : Google Scholar
|
|
34
|
Wagner KU, Dierisseau P, Rucker EB,
Robinson GW and Hennighausen L: Genomic architecture and
transcriptional activation of the mouse and human tumor
susceptibility gene TSG101: common types of shorter transcripts are
true alternative splice variants. Oncogene. 17:2761–2770. 1998.
View Article : Google Scholar
|
|
35
|
Liu RT, Huang CC, You HL, Chou FF, Hu CC,
Chao FP, Chen CM and Cheng JT: Overexpression of tumor
susceptibility gene TSG101 in human papillary thyroid carcinomas.
Oncogene. 21:4830–4837. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Young TW, Mei FC, Rosen DG, Yang G, Li N,
Liu J and Cheng X: Up-regulation of tumor susceptibility gene 101
protein in ovarian carcinomas revealed by proteomics analyses. Mol
Cell Proteomics. 6:294–304. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Kidd M, Nadler B, Mane S, Eick G,
Malfertheiner M, Champaneria M, et al: GeneChip, geNorm and
gastrointestinal tumors: novel reference genes for real-time PCR.
Physiol Genomics. 30:363–370. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Swendeman SL, Spielholz C, Jenkins NA,
Gilbert DJ, Copeland NG and Sheffery M: Characterization of the
genomic structure, chromosomal location, promoter, and
developmental expression of the B-globin transcription factor CP2.
J Biol Chem. 269:11663–11671. 1994.PubMed/NCBI
|
|
39
|
Ramamurthy L, Barbour V, Tuckfield A,
Clouston DR, Topham D, Cunningham JM, et al: Targeted disruption of
the CP2 gene, a member of the NTF family of transcription factors.
J Biol Chem. 276:7836–7842. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Saxena UH, Powell CMH, Fecko JK, Cacioppo
R, Chou HS, Cooper GM and Hansen U: Phosphorylation by cyclin
C/cyclin-dependent kinase 2 following mitogenic stimulation of
murine fibroblasts inhibits transcriptional activity of LSF during
G1 progression. Mol Cell Biol. 29:2335–2345. 2009. View Article : Google Scholar
|
|
41
|
Coull JJ, Romerio F, Sun JM, Volker JL,
Galvin KM, Davie JR, Shi Y, Hansen U and Margolis DM: The human
factors YY1 and LSF repress the human immunodeficiency virus type 1
long terminal repeat via recruitment of histone deacetylase 1. J
Virol. 74:6790–6799. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Lace MJ, Yamakawa Y, Ushikai M, Anson JR,
Hangen TH and Turek LP: Cellular factor YY1 downregulates the human
papillomavirus 16 E6/E7 promoter, P97, in vivo and in vitro from a
negative element overlapping the transcrition-initiation site. J
Gen Virol. 90:2402–2412. 2009. View Article : Google Scholar
|
|
43
|
Fan R, Chen P, Zhao D, Tonq JL, Li J and
Liu F: Cooperation of deregulated Notch signaling and Ras pathway
in human hepatocarcinogenesis. J Mol Histol. 42:473–481. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Zagouras P, Stifani S, Blaumeller CM,
Carcangiu ML and Artavanis-Tsakonas S: Alterations in Notch
signaling in neoplastic lesions of human cervix. Proc Natl Acad Sci
USA. 92:6414–6418. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Leethanakul C, Patel V, Gillespie J,
Pallente M, Ensley JF, Koontongkaew S, Liotta LA, Emmert-Buck M and
Gutkind JS: Distinct pattern of expression of differentiation and
growth-related genes in squamous cell carcinomas of the head and
neck revealed by use of laser capture microdissection and cDNA
arrays. Oncogene. 19:3220–3224. 2000. View Article : Google Scholar
|
|
46
|
Westhoff B: Colaluca IN, D’Ario G,
Donzelli M, Tosoni D, Volorio S, Pelosi G, Spaggiari L, Mazzarol G,
Viale G, Pece S and Di Fiore PP, Alterations of the Notch pathway
in lung cancer. Proc Natl Acad Sci USA. 106:22293–22298.
2009.PubMed/NCBI
|
|
47
|
Zhang Y, Li B, Ji ZZ and Zheng PS: Notch 1
regulates the growth of human colon cancer. Cancer. 116:5207–5218.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Okuhashi Y, Nara N and Tohoda S: Effects
of gamma-secretase inhibitors on the growth of leukemia cells.
Anticancer Res. 30:495–498. 2010.PubMed/NCBI
|
|
49
|
Shlomai A: Targeting late SV40 factor: is
the achilles heel of hepatocarcinogenesis revealed? World J
Gastroenterol. 18:6709–6711. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Veljkovic J and Hansen U: Lineage-specific
and ubiquitous biological roles of the mammalian transcription
factor LSF. Gene. 343:23–40. 2004. View Article : Google Scholar : PubMed/NCBI
|