Introduction
Hepatocellular carcinoma (HCC), as one of the most
malignant cancers worldwide, causes >500,000 mortalities
worldwide every year and has an extremely poor prognosis (1,2).
Currently, chemotherapy, liver transplantation, surgical resection,
and local ablation are used for the treatment of HCC, which is
dependent on the stage of HCC (3).
HCC leads to a high mortality due to the lack of reliable early
detection (4). Therefore, a reliable
target is urgently required for the development of early detection
and effective treatment techniques for HCC.
Ubiquitin-specific protease 9X (USP9X) is a X-linked
ubiquitin specific peptidase which belongs to the
ubiquitin-specific protease family (5). USP9X serves as a deubiquitinase and
effectively regulates the proliferation, adhesion and signal
transduction of cells, therefore is crucial in controlling
proteasome activity in organogenesis, transcriptional regulation
and tumorigenesis (6–9). Previous studies have demonstrated an
association between USP9X and lung, colon (5) and breast cancer (10), and lymphoma (11). Specifically, previous studies have
shown that high USP9X expression results in a poor prognosis in
lymphoma and lung cancer (10–14). Thus,
we hypothesize that high USP9X expression is associated with the
growth and metastasis of tumor cells.
However, to date, the role of USP9X in HCC has not
been reported. In the present study, the expression of USP9X was
blocked in SMMC7721 and HepG2 cells using specific small
interfering RNA (siRNA) to explore the potential influence of USP9X
in HCC cell lines.
Materials and methods
Cell culture
The human HCC cell lines SMMC7721 and HepG2, and
human liver cell line, L02 were purchased from American Type
Culture Collection (Manassas, VA, USA). Cells were cultured in
RPMI-1640 (Invitrogen Life Technologies, Carlsbad, CA, USA)
supplemented with 10% fetal bovine serum (FBS; Invitrogen Life
Technologies) at 37°C in a humidified atmosphere of 5%
CO2.
RNA interference
USP9X-specific siRNA (USP9X-siRNA) and a negative
control (NC group) with no significant homology with human gene
sequences were purchased from Gene Pharma (Shanghai, China). The
sequences of USP9X-siRNA and NC are shown in Table I. Cells were seeded into a 6-well
plate for 24 h at a density of 5×104 cells/well. In each
well, 20 µM USP9X-siRNA or negative control were transfected using
Lipofectamine 2000 (Invitrogen Life Technologies) according to the
manufacture's instruction.
 | Table I.Sequences of USP9X-siRNA and negative
control. |
Table I.
Sequences of USP9X-siRNA and negative
control.
| Name | Sequences |
|---|
| USP9X-siRNA | Sense (5′-3′)
AGAAAUCGCUGGUAUAAAUUU |
|
| Antisense (5′-3′)
AAAUUUAUACCAGCGAUUUCU |
| Negative control | Sense (5′-3′)
UUCUCCGAACGUGUCACGUTT |
|
| Antisense (5′-3′)
ACGUGACACGUUCGGAGAGTT |
Antibodies
The anti-USP9X (1:1,500; cat. no. 66026-1-Ig)
antibody was purchased from Proteintech (Chicago, IL, USA). The
anti β-tubulin antibodies (1:5,000; cat. no. KM9003) were purchased
from Tianjin Sungene Biotechnology Company (Tianjin, China).
Western blot analysis
Total protein was extracted using the Total Protein
Extraction Kit (Beyotime Institute of Biotechnology, Shanghai,
China); USP9X protein was separated by 6% SDS-PAGE and then
transferred to nitrocellulose membranes (Pierce, Rockford, USA) and
incubated at 37°C for 1 h with the indicated primary and
horseradish peroxidase-secondary antibodies (cat. no. LK2001;
Beyotime Institute of Biotechnology). The labeled proteins were
detected using chemiluminescent agents. Images were collected by
Image-Pro plus software 6.0 (Media Cybernetics, Silver Spring, MD,
USA). The relative levels of the target protein are represented as
the density ratio versus β-tubulin. Independent experiments were
repeated three times for each sample and the relative expression
levels of protein were normalized to the endogenous reference gene
β-tubulin and analyzed by using 2−∆∆CT method
(15).
Flow cytometry assay (FCM) for
apoptosis
Annexin-V/propidium iodide (PI) double assay was
performed using the Annexin V-FITC Apoptosis Detection kit (Nanjing
KeyGen Biotechnology, Nanjing, China). Cells were released from the
culture dish with trypsin and washed twice with phosphate buffered
saline (PBS; BoPei Biotech Co. Ltd., Chongqing, China). Cells
(1×106) were resuspended in 500 µl binding buffer and
stained with 5 µl FITC-labeled Annexin-V according to the
manufacturer's instructions. Following this, 5 µl PI was added and
allowed to incubate with the cells for 10 min at room temperature
in the dark. After one wash with PBS, cells were subjected to FCM
analysis using BD FACSCalibur (BD Biosciences, San Jose, CA, USA).
The data were analyzed using CellQuest data acquisition and
analysis software (version 5.1; BD Biosciences).
MTT assay
Cells were plated in 96-well plates at a density of
~4×103 cells/well. Following treatment, the plates were
incubated in a 37°C humidified incubator for the time periods
indicated below. To assess cell viability, the MTT assay was
performed according to the manufacturer's instructions
(Sigma-Aldrich, St Louis, MO, USA). In brief, 20 µl of MTT reagent
(5 mg/ml) was added to each well, and the cells were incubated for
a further 4 h at 37°C, followed by the addition of 150 µl DMSO
(Sigma-Aldrich). Absorbance (A) was determined by measuring the
absorbance at 490 nm using a Victor3 spectrofluorimeter (Perkin
Elmer, Foster City, USA) at 24, 48 and 72 h post-transfection. Each
assay was performed in triplicate and each experiment was repeated
at least three times. Cell-growth curves were calculated as A mean
values of triplicates per group.
Transwell assay
The migration assay was performed in a 6-well
transwell chamber (Corning, Cambridge, MA, USA), which contained an
8 µm pore size polycarbonate membrane filter for migration assay.
Cells were trypsinized and suspended in a serum-free medium
containing 1% bovine serum albumin at a concentration of
5×104 cells/insert. Medium supplemented with 10% fetal
calf serum was added to the lower chamber. After reculturing with
5% CO2 at 37°C for 24 h, the transwell chambers were
inverted and the cells were removed by swabbing and stained with
crystal violet (Beyotime Institute of Biotechnology, Shanghai,
China).
Statistical analysis
All statistical analyses were performed using SPSS
17.0 (SPSS, Chicago, USA). Data are presented as the mean ±
standard deviation. The differences of two groups and three groups
were statistically analyzed using the Student's t-test and
Analyze Compare Means One-Way ANOVA. The differences of ratios were
statistically analyzed using the χ2 test. P<0.05 was
considered to indicate a statistically significant difference.
Results
Expression of USP9X is upregulated in
hepatoma SMMC7721 and HepG2 cell lines
The protein expression of USP9X was detected in
SMMC7721, HepG2 and L02 by western blot analysis. The results
suggested that the protein expression of USP9X was upregulated in
SMMC7721 and HepG2 (Table II;
P<0.01).
 | Table II.Protein level of USP9X in three cell
groups (n=3, mean ± standard deviation). |
Table II.
Protein level of USP9X in three cell
groups (n=3, mean ± standard deviation).
| Groups | USP9X relative
protein expression |
|---|
| SMMC7721 |
0.53±0.03a |
| HepG2 |
0.47±0.05a |
| L02 |
0.18±0.03 |
USP9X expression was inhibited by
siRNA in SMMC7721 and HepG2 cells
To examine the off-target effect of RNAi,
USP9X-siRNA and NC were transfected into SMMC7721 and HepG2 cells
using Lipofectamine 2000. USP9X expression was evaluated by western
blot analysis. The results showed that USP9X-siRNA could
effectively inhibit the expression of USP9X in SMMC7721 and HepG2
cell lines (Table III;
P<0.01).
 | Table III.USP9X protein level in different
treated SMMC7721 and HepG2 cells after siRNA interference (n=3,
mean ± standard deviation). |
Table III.
USP9X protein level in different
treated SMMC7721 and HepG2 cells after siRNA interference (n=3,
mean ± standard deviation).
| Groups | SMMC7721 | HepG2 |
|---|
| USP9X-siRNA |
0.21±0.01a |
0.36±0.01a |
| NC |
0.62±0.02 |
0.94±0.01 |
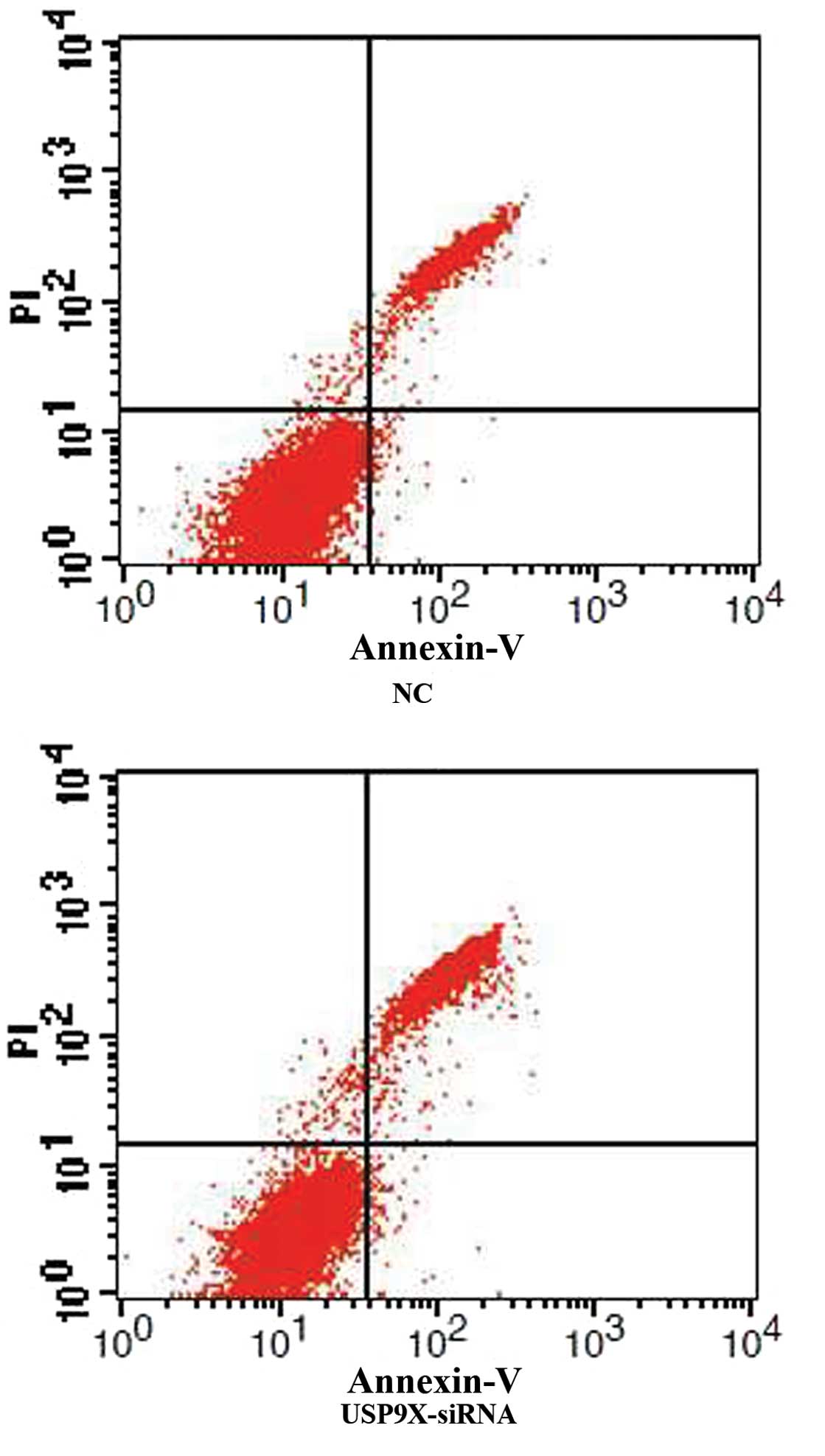
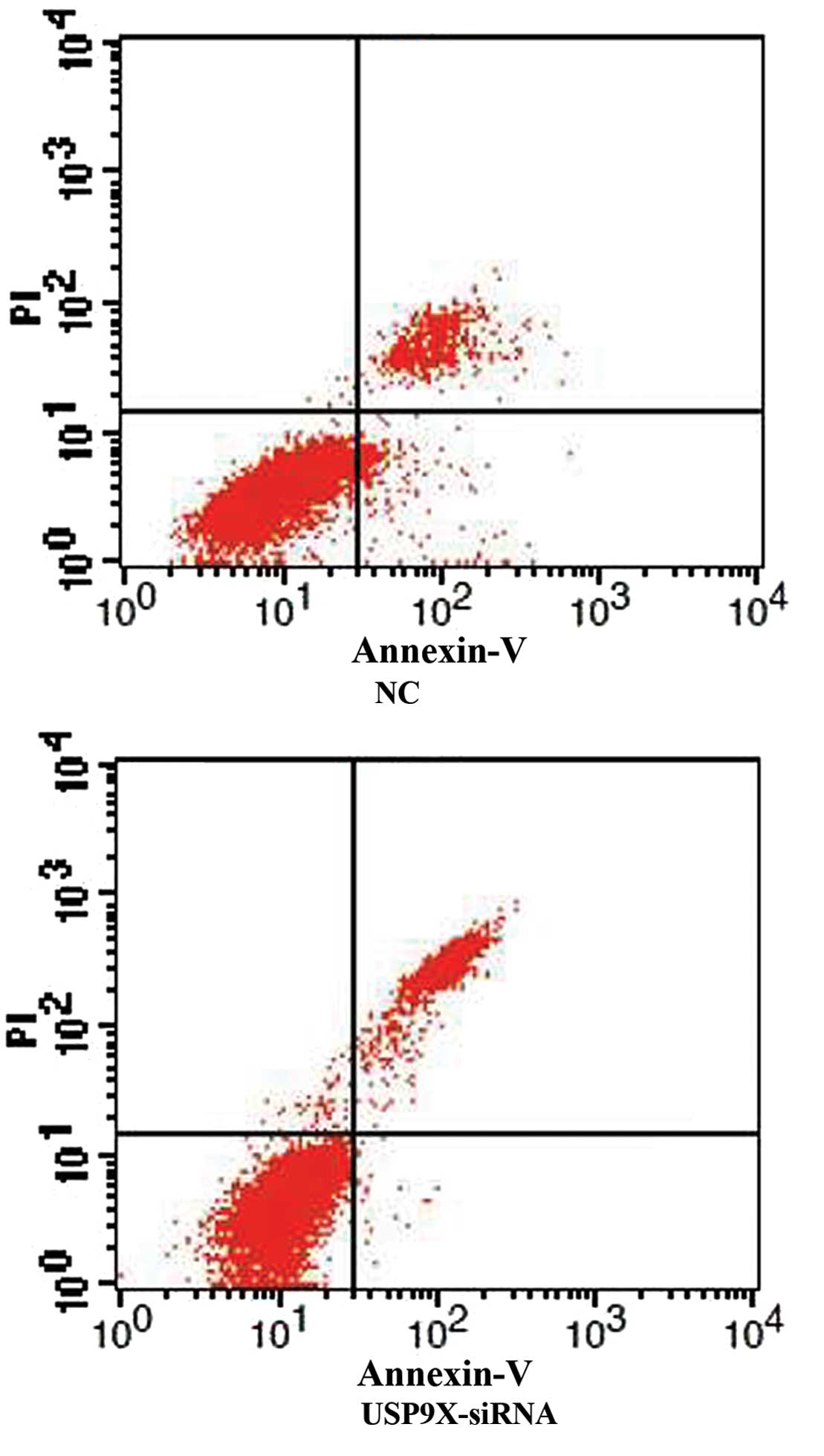
USP9X-siRNA enhances SMMC7721 and
HepG2 cell apoptosis
After SMMC7721 and HepG2 cells were transfected with
USP9X-siRNA, cellular apoptosis was first examined by Annexin V and
PI staining followed by FCM analysis. The percentage of apoptotic
cells in the USP9X-siRNA group was higher than the NC group in both
SMMC7721 and HepG2 (Figs. 1 and
2 and Table IV; P<0.01).
 | Table IV.The apoptosis of different treated
SMMC7721 and HepG2 cells (n=3; %, mean ± standard deviation). |
Table IV.
The apoptosis of different treated
SMMC7721 and HepG2 cells (n=3; %, mean ± standard deviation).
| Groups | SMMC7721 | HepG2 |
|---|
| USP9X-siRNA |
27.96±2.49a |
23.48±1.60a |
| NC |
15.02±3.03 |
9.56±2.33 |
USP9X-siRNA reduces SMMC7721 and HepG2
cell viability
As shown in Table V,
the MTT assay revealed that the USP9X-siRNA group exhibited
significantly reduced cell viability compared with the NC group
cells (P<0.01).
 | Table V.Comparison of cell absorbance in
different treated SMMC7721 and HepG2 cells (n=3, mean ± standard
deviation). |
Table V.
Comparison of cell absorbance in
different treated SMMC7721 and HepG2 cells (n=3, mean ± standard
deviation).
|
| SMMC7721 | HepG2 |
|---|
|
|
|
|
|---|
| Groups | 24 h | 48 h | 72 h | 24 h | 48 h | 72 h |
|---|
| USP9X-siRNA |
0.30±0.08a |
0.53±0.12a |
0.96±0.21a |
0.24±0.10a |
0.41±0.17a |
0.77±0.36a |
| NC |
0.40±0.08 |
0.72±0.12 |
1.11±0.22 |
0.32±0.10 |
0.56±0.16 |
0.88±0.34 |
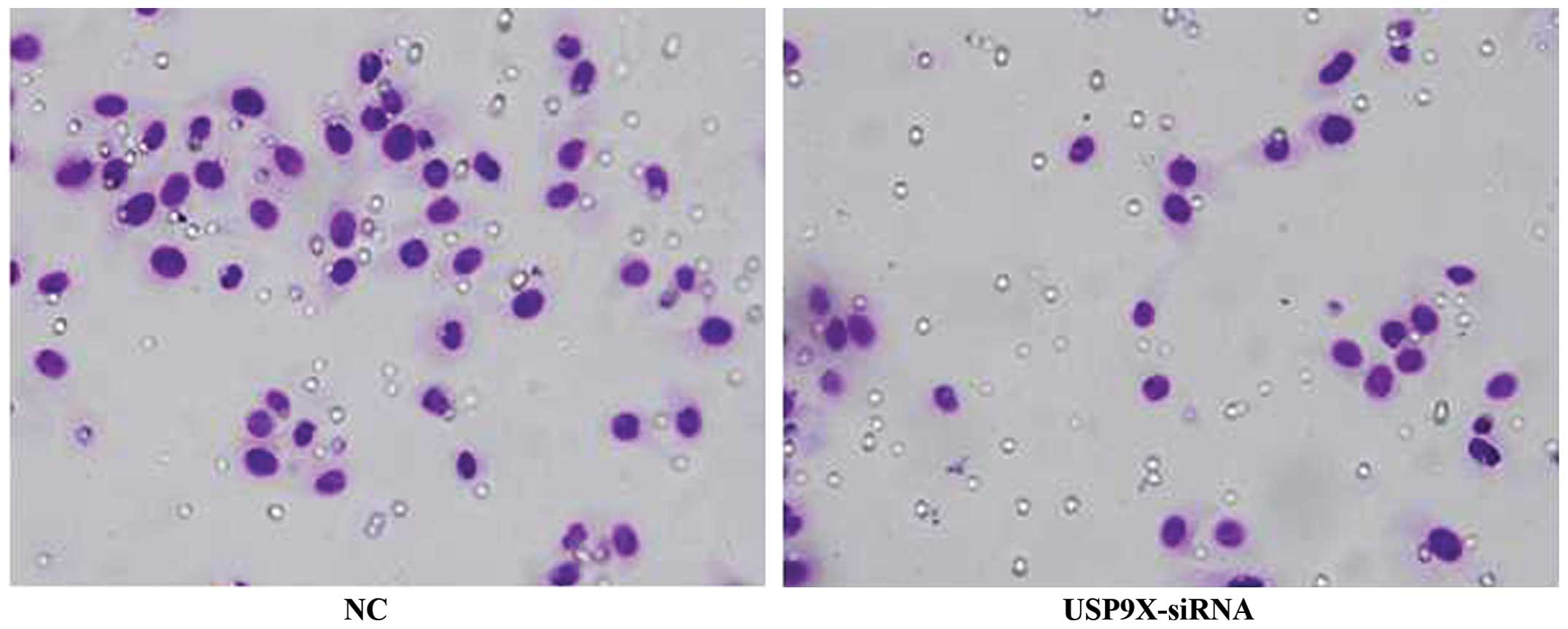
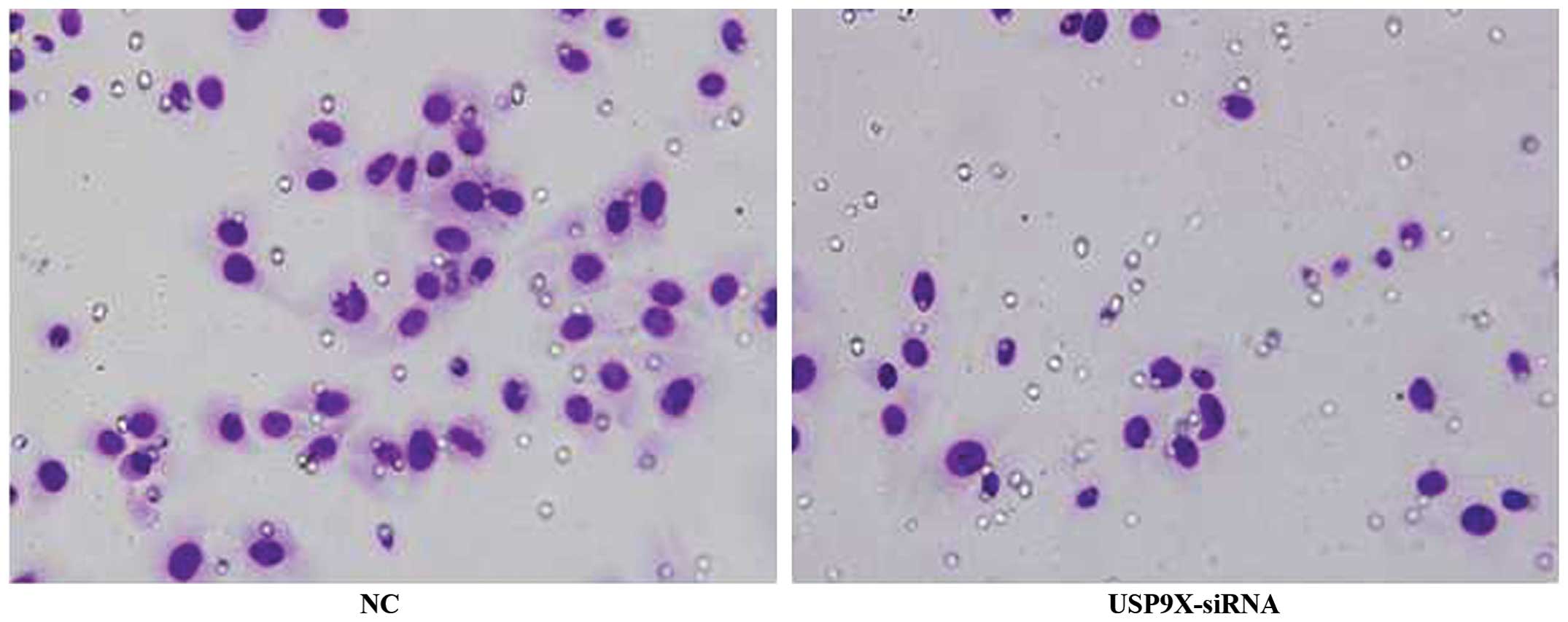
USP9X-siRNA inhibits the migration of
SMMC7721 and HepG2 cells
The results of the transwell assay revealed that the
migration of SMMC7721 and HepG2 cells transfected with USP9X-siRNA
was significantly reduced when compared with that of the NC groups
(P<0.01), indicating that suppression of USP9X inhibited the
migration of SMMC7721 and HepG2 cells (Figs. 3 and 4
and Table VI; P<0.01).
 | Table VI.Comparison of amounts of cell
migration in different treated SMMC7721 and HepG2 cells (n=3, mean
± standard deviation). |
Table VI.
Comparison of amounts of cell
migration in different treated SMMC7721 and HepG2 cells (n=3, mean
± standard deviation).
| Groups | SMMC7721 | HepG2 |
|---|
| USP9X-siRNA |
27.00±4.36a |
24.67±4.51a |
| NC |
55.70±4.04 |
56.00±2.65 |
Discussion
Recently, an increasing number of studies have
demonstrated that USP9X is involved with cancer (5–14). USP9X
expression was found to be higher in lung, colon and breast cancer,
when compared with normal tissues in vitro (5,10).
Furthermore, in lung and colon cancers, decreased USP9X expression
resulted in the promotion of cellular apoptosis in vivo
(5), which indicated that USP9X
expression may be a potential predictor of such cancers. However,
at present, the association between USP9X expression and HCC has
not been reported. Therefore, in the present study, the USP9X gene
was knocked down in SMMC7721 and HepG2 cells using siRNA to study
the potential effect of USP9X on HCC cells. In SMMC7721 and HepG2
cells, along with the effective silencing of USP9X by specific
siRNA, cell viability and migration were reduced while cell
apoptosis was increased.
Cell viability and apoptosis are important in the
oncogenesis and chemotherapy resistance of HCC cells. This study
demonstrated that the viability of SMMC7721 and HepG2 cells was
reduced by USP9X-siRNA. The apoptosis ratio in the USP9X-siRNA
group was significantly higher than in the NC group. This evidence
indicates that USP9X is a crucial factor in HCC tumor growth. The
current study also shows that cell migration was downregulated when
USP9X expression was inhibited, indicating that USP9X-siRNA alone
may inhibit cell migration. Additionally, USP9X-siRNA significantly
reduced cell viability, which consequently resulted in a relative
reduction in cell migration.
In conclusion, the results in the current
investigation revealed that RNAi-mediated downregulation of USP9X
effectively inhibits the growth and migration of SMMC7721 and HepG2
cells. However the mechanisms associated with USP9X regulating the
bioactivity of HCC remain unknown. USP9X has been previously shown
to bind to the Mcl-1 protein, which was reported to inhibit cell
apoptosis in HCC (16,17), and inhibit proteasomal degradation in
lymphoma (13). In other tumors, such
as lung cancer, colon cancer and lymphoma, USP9X inhibited cancer
cell apoptosis by influencing the degradation of specific protein
Mcl-1 (5). Therefore we speculate
that USP9X may inhibit cell apoptosis by influencing Mcl-1 in HCC
cells. USP9X may be a potential target for the treatment and early
detection of HCC, however, further studies are required to clarify
the mechanism by which USP9X is involved in the development and
progression of HCC, and its related pathway in HCC.
Acknowledgements
This study was supported by the National Natural
Science Foundation of China (grant no. 81171562), the Natural
Science Foundation of Chongqing, China (grant no. CSTC,
2013yykfA110010) and the Science Foundation of Yuzhong district,
Chongqing, China (grant no. 20130118).
References
|
1
|
Llovet JM, Burroughs A and Bruix J:
Hepatocellular carcinoma. Lancet. 362:1907–1917. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Marrero JA: Multidisciplinary management
of hepatocellular carcinoma: where are we today? Semin Liver Dis.
33(Suppl 1): 3–10. 2013.PubMed/NCBI
|
|
3
|
Yau T, Chan P, Epstein R and Poon RT:
Evolution of systemic therapy of advanced hepatocellular carcinoma.
World J Gastroenterol. 14:6437–6441. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
El-Serag HB and Rudolph KL: Hepatocellular
carcinoma: epidemiology and molecular carcinogenesis.
Gastroenterology. 132:2557–2576. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Peddaboina C, Jupiter D, Fletcher S, et
al: The downregulation of Mcl-1 via USP9X inhibition sensitizes
solid tumors to Bcl-xl inhibition. BMC Cancer. 12:5412012.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
DiAntonio A and Hicke L:
Ubiquitin-dependent regulation of the synapse. Annu Rev Neurosci.
27:223–246. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yang Bo and XIEcong Hua: Ubiquitin
specific peptidase 9X and tumor. J Int Oncol. 38:900–902. 2011.
|
|
8
|
Kitagawa K, Kotake Y and Kitagawa M:
Ubiquitin-mediated control of oncogene and tumor suppressor gene
products. Cancer Sci. 100:1374–1381. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Becuwe M, Herrador A, Haguenauer-Tsapis R,
Vincent O and Léon S: Ubiquitin-mediated regulation of endocytosis
by proteins of the arrestin family. Biochem Res Int.
2012:122012.
|
|
10
|
Deng S, Zhou H, Xiong R, et al:
Over-expression of genes and proteins of ubiquitin specific
peptidases (USPs) and proteasome subunits (PSs) in breast cancer
tissue observed by the methods of RFDD-PCR and proteomics. Breast
Cancer Res Treat. 104:21–30. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sun H, Kapuria V, Peterson LF, et al:
Bcr-Abl ubiquitination and Usp9X inhibition block kinase signaling
and promote CML cell apoptosis. Blood. 117:3151–3162. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kapuria V, Peterson LF, Fang D, Bornmann
WG, Talpaz M and Donato NJ: Deubiquitinase inhibition by
small-molecule WP1130 triggers aggresome formation and tumor cell
apoptosis. Cance Res. 70:9265–9276. 2010. View Article : Google Scholar
|
|
13
|
Schwickart M, Huang X, Lill JR, et al:
Deubiquitinase USP9X stabilizes MCL1 and promotes tumor cell
survival. Nature. 463:103–107. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Xie Y, Avello M, Schirle M, et al:
Deubiquitinase FAM/USP9X interacts with the E3 ubiquitin ligase
SMURF1 protein and protects it from ligase activity-dependent
self-degradation. J Biol Chem. 288:2976–2985. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2 (-Delta Delta C (T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Schulze-Bergkamen H, Fleischer B,
Schuchmann M, et al: Suppression of Mcl-1 via RNA interference
sensitizes human hepatocellular carcinoma cells towards apoptosis
induction. BMC Cancer. 6:2322006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sieghart W, Losert D, Strommer S, et al:
Mcl-1 overexpression in hepatocellular carcinoma: a potential
target for antisense therapy. J Hepatol. 44:151–157. 2006.
View Article : Google Scholar : PubMed/NCBI
|