Introduction
The incidence of breast cancer continues to rise
worldwide, particularly in the USA where approximately one in eight
females will be diagnosed with breast cancer during her lifetime
(1). While early stage breast cancer
is readily treatable, patients with metastatic disease pose a
greater therapeutic challenge and account for the majority of
breast cancer-related mortalities. This is largely due to the
paucity of knowledge with regard to the underlying molecular
mechanisms associated with malignant progression.
Regulated cellular proliferation and differentiation
are dependent upon functional tight junctions (TJs), and the loss
of TJ integrity may be important in cancer development and
progression. Located immediately beneath the apical surface of
adjacent endothelial and epithelial cells, TJs form an effective
barrier to the diffusion of solutes through the paracellular
pathway and exhibit ion-selective permeability in a cell
type-dependent manner. In association with adherens junctions, TJs
have been shown to establish and maintain epithelial cell polarity
by preventing the diffusion of membrane proteins and lipids between
the apical and basolateral regions of the plasma membrane. Recent
studies also suggest that the TJ plaque proteins, located on the
cytoplasmic side of the TJ, are important for the integration of
signaling molecules that regulate processes including gene
transcription, cellular proliferation, differentiation and
morphogenesis. This was reviewed by Turksen and Troy (2).
Claudins, the predominant integral membrane proteins
that form the backbone of TJs, are required for the assembly,
barrier and pore functions of vertebrate TJs (2). Deregulated expression of various claudin
proteins has been reported in breast cancer; overexpression of
claudin-3 and claudin-4 was demonstrated in 62 and 26% of primary
breast tumors, respectively (3).
Furthermore, Lanigan et al (4)
showed claudin-4 expression in 90.9% of primary breast cancers,
with the highest levels of claudin-4 associated with high grade
breast tumors. Notably, although immunohistochemical analysis
indicated a decrease in claudin-4 in 64% of grade I breast tumors,
its expression was found to be robust in grade II and III tumors
(5). In contrast to the general
upregulation of claudins-3 and −4 in breast cancer, two other
claudin proteins have been shown to be downregulated in breast
cancer. Specifically, Tokés et al (5) reported a decrease in claudin-1 protein
expression in 80% of invasive ductal breast carcinomas. In
addition, claudin-7 expression has been shown to decrease with
increasing breast tumor grade (6,7). These
studies indicate that deregulated levels of a number of claudin
proteins may contribute to breast tumorigenesis.
The present study aimed to investigate the potential
role of claudin-3 in tumor progression, by assessing the levels of
this protein in panels of normal tissues (to examine the basal
expression range) and in metastatic breast cancer cell lines MCF-7
and MDA-MB-415 (adenocarcinomas derived from pleural effusions) and
by evaluating the effects of siRNA-mediated suppression of
claudin-3 on cellular motility.
Materials and methods
Cell lines
The following human breast cancer cell lines were
obtained from the American Type Culture Collection (Manassas, VA,
USA): MCF-7 (catalog no. HTB-22), MDA-MB-415 (catalog no. HTB-128)
and MDA-MB-157 (catalog no. HTB-24). The three cell lines were
cultured in minimum essential media (MEM; catalog no. 11095-080)
supplemented with 10% fetal bovine serum (catalog no. 16140-071)
and 1% penicillin-streptomycin-L-glutamine (catalog no. 10378-016),
all of which were obtained from Life Technologies (Grand Island,
NY, USA). Human mammary epithelial cells (HMEC; catalog no.
CC-2551) were obtained from Lonza (Walkersville, MD, USA) and
cultured in mammary epithelial basal medium (catalog no. CC-3151;
Lonza) supplemented with the MEGM Bullet Kit (catalog no. CC-3150;
Lonza). All cells were maintained in a humidified atmosphere of 5%
CO2 at 37°C for 7–10 days prior to use.
Western blot analysis
Log phase cells were harvested with trypsin-EDTA
(0.25%/1 mM; Life Technologies) and subjected to centrifugation at
81.7 × g for 5 min. The cell pellets were then resuspended in 1X
sample buffer (10% v/v glycerol, 1% SDS, 0.125% w/v bromophenol
blue and 0.04M Tris pH 7.0) plus 10% β-mercaptoethanol, and
subjected to gel electrophoresis on precast 10% or 12%
SDS-polyacrylamide gels (catalog no. 456–1033 and 456–1043; BioRad,
Hercules, CA, USA). Following transfer to polyvinylidene fluoride
membranes (catalog no. IPVH304F0; Merck Millipore Ltd, Co. Cork,
Ireland), the membranes were incubated with the following
antibodies in 5% milk/phosphate buffered saline (PBS) for 2 h at
room temperature: 1 µg/ml Rabbit anti-claudin-1 (dilution, 1:1,000;
catalog no.49339; Cell Signaling Technologies, Inc, Beverly, MA,
USA); 2 µg/ml rabbit anti-claudin-3 (dilution, 1:250; catalog no.
34–1700; Life Technologies); 3 µg/ml mouse anti-claudin-4
(dilution, 1:167; catalog no. 32–9400; Life Technologies); and 1
µg/ml rabbit anti-actin (I-19; dilution, 1:200; catalog no.
SC1616-R; Santa Cruz Biotechnology, Santa Cruz, CA, USA). The
membranes were subsequently incubated in a 1:3,000 dilution of goat
anti-rabbit (catalog no. 172–1019; BioRad, Hercules, CA, USA) or
goat anti-mouse HRP-conjugated secondary antibodies (catalog no.
170–6516; BioRad) in 5% milk/PBS for 1 h at room temperature.
Signals were visualized using Amersham ECL Prime Western Blotting
Detection Reagent (catalog no. RPN2232; GE Healthcare,
Buckinghamshire, UK).
Differential detergent cell
fractionation
Using the ProteoExtract Protein Extraction Kit
(catalog no. 539791; Calbiochem/EMD Chemicals, Inc, San Diego, CA,
USA), ~1×106 log phase cells were incubated sequentially
in four extraction buffers to yield cytosolic, membranous, nuclear
and cytoskeletal fractions, following the manufacturer's
instructions.
Transient siRNA transfection
Approximately 1×105 cells were plated
into 100 mm wells and cultured in MEM supplemented with 10% fetal
bovine serum and 1% penicillin-streptomycin-L-glutamine. After
reaching ~30% confluency, cells were transfected with 20 pmol
Silencer Select siRNA targeted to claudin-3 (catalog no. s3444;
Life Technologies) using 6 µl/well DharmFect I cationic lipid
(catalog no. T-2001; Fisher Scientific, Pittsburgh, PA, USA) in
serum-free medium. For controls, cells were mock-transfected with
serum-free medium. Following incubation at 37°C for 24 h, the
medium was replaced with fresh medium containing serum.
Indirect immunofluorescence
analysis
Log phase cells were harvested, plated at a density
of 5×105 cells per chambered cover slide (catalog no.
177437; LabTek, Fisher Scientific) and incubated at 37°C until 80%
confluency was reached. Cells were rinsed three times with
pre-cooled PBS and fixed in pre-cooled 95% ethanol for 30 min on
ice. Following blocking with 1% bovine serum albumin, the cells
were incubated with 1 mg/ml rabbit-anti claudin-3 overnight at 4°C.
Cells were subsequently counterstained with goat
anti-rabbit-Alexa-488 (dilution, 1:400; catalog no. A-11034; Life
Technologies) for 1 h, rinsed with PBS and treated with Prolong
Gold antiFade Reagent (catalog no. I37156; Life Technologies).
Images were collected using a Nikon TE2000U eipfluorescent system
with a PlanFluor 40X objective and a Coolsnap HQ camera.
Wound-healing assay
Claudin-3 siRNA-transfected and mock transfected
control cells were plated at 2.5×105 cells per well into
Cytoselect wound-healing assay plates (catalog no. CBA-120; Cell
Biolabs San Diego, CA, USA) and grown until 95% confluent. Each
well contained an insert that created a defined, rectangular wound
field. Following removal of the inserts, the cells were stained
(stain included in Cytoselect kit) and photographed on days 0, 1, 2
and 3 to monitor migration of the cells into the wound field.
Protein extracts were prepared from the mock- and siRNA-transfected
cells (as described previously) on days 0 and 3, and subjected to
western blot analysis to assess the degree of claudin-3
suppression.
Results
Claudin-3 protein expression in normal
human tissues
To determine the level and variation of expression
of claudin-3 in normal tissues, a panel of protein extracts derived
from human bladder, breast, cervix, kidney, ovary, placenta,
prostate, testis and uterus were analyzed using a pre-made tissue
western blot (ProSci). The 23 kDa claudin-3 band was detectable in
all tissues with the exception of the bladder, cervix, placenta and
uterus (Fig. 1). The most intense
signal was observed in the lane containing normal breast
tissue.
Overexpression of claudin proteins in
metastatic breast cancer cell lines
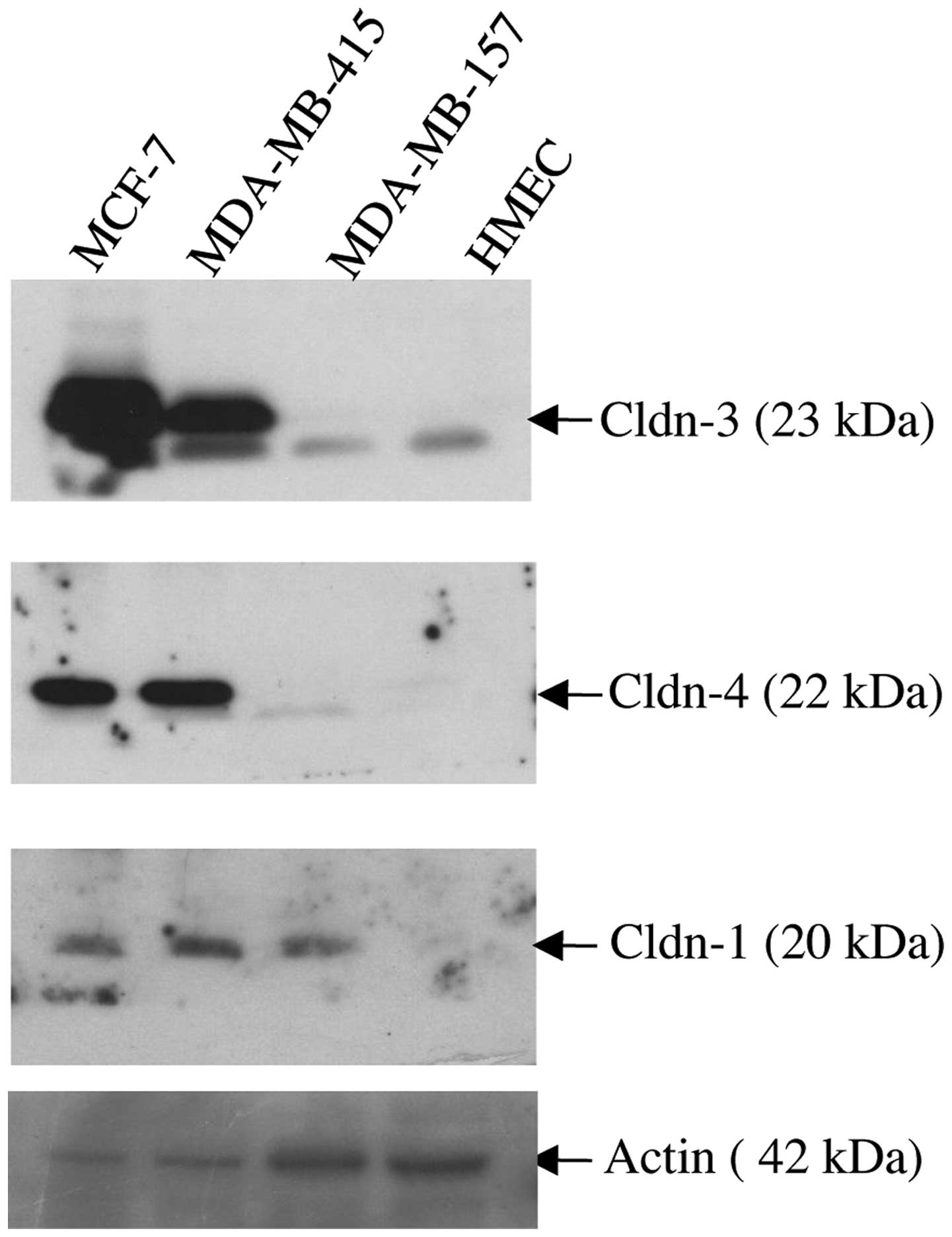
Immunoblot analysis was used to assess the
expression of claudin-1, −3 and −4 proteins in three breast cancer
cells lines (MCF-7, MDA-MB-415 and MDA-MB-157), all of which were
derived from patients with metastatic disease (Fig. 2). As a control, the expression of the
same three proteins was assessed in HMEC cells. Whereas the HMEC
cells expressed barely detectable levels of each of the three
claudin proteins, marked overexpression of claudins-3 and −4 was
observed in the MCF-7 and MDA-MB-415 cell lines. By contrast, the
MDA-MB-157 cell line expressed a similar level of expression of
claudins-3 and −4 as the HMEC cells in the form of a lower
molecular weight band (~23kDa). All three cancer cell lines
expressed readily detectable levels of claudin-1 protein relative
to the virtually undetectable expression of claudin-1 demonstrated
by the HMEC cells.
Delocalization of overexpressed
claudin-3 protein in the MCF-7 and MDA-MB-415 breast cancer cell
lines
Indirect immunofluorescence and differential
detergent cell fractionation were utilized to determine and compare
the effect of endogenous overexpression versus siRNA-mediated
suppression of claudin-3 protein on its sub-cellular localization
in the two claudin-3 overexpressing breast epithelial cell lines,
MCF-7 and MDA-MB-415.
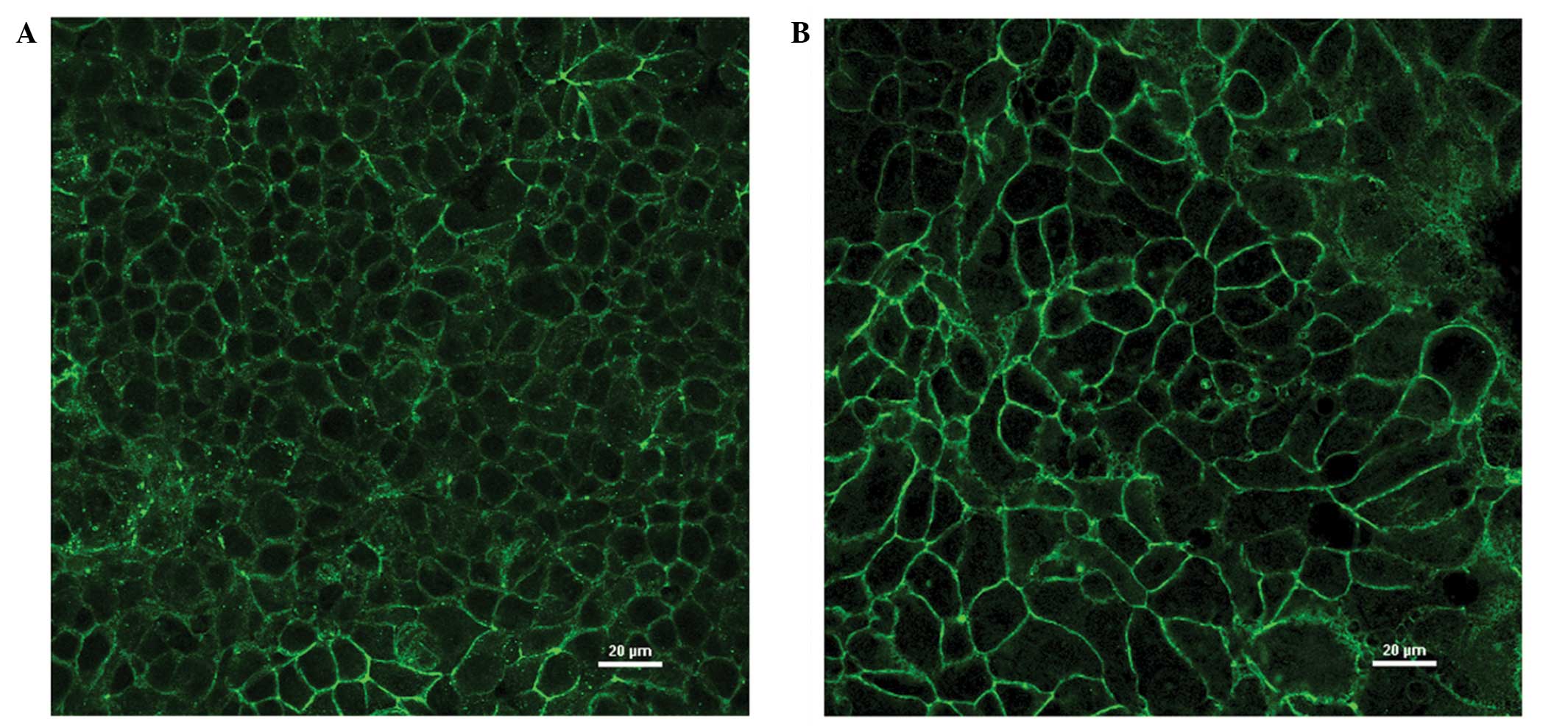
In the immunofluorescence analysis, MCF-7 and
MDA-MB-415 cells showed marked fluorescence at cell junctions,
consistent with the established role of claudin-3 in TJs (Fig. 3A and B). In addition, evidence of
intracellular claudin-3 staining was detected in the two cell
lines. To determine whether or not the intracellular fluorescence
was claudin-3-specific, signal intensities following siRNA-mediated
suppression of claudin-3 in the MCF-7 cells were calculated
following subtraction of secondary antibody controls. The cells
were visualized by differential interference contrast (DIC)
microscopy (Fig. 4, bottom panels)
and claudin-3 localization was visualized by immunofluorescence
(Fig. 4, top right panels). The
immunofluorescence images were then superposed onto the DIC images
(Fig. 4, top left panels), enabling
the comparison of claudin-3 signal intensities, at the cell
junctions and within the cells, in the mock- versus
siRNA-transfected conditions. Successful transfection of
claudin-3-specific siRNA resulted in the loss of claudin-3 signal
at cell junctions, however, a small fraction of the cells showed
junctional staining, which suggested a transfection efficiency of
<100% (Fig. 4B top right panel).
Notably, a significant (55%) reduction in intracellular
fluorescence was observed in siRNA-transfected cells that showed a
decrease in junctional fluorescence, and in those that demonstrated
claudin-3 signal at cell junctions (P<0.001) (determined by
analysis of variance with Tukey post-hoc comparison; Fig. 4B, top right panel). These data suggest
that the intracellular signal observed in the breast cancer cell
lines was claudin-3 specific and not the result of non-specific
background staining.
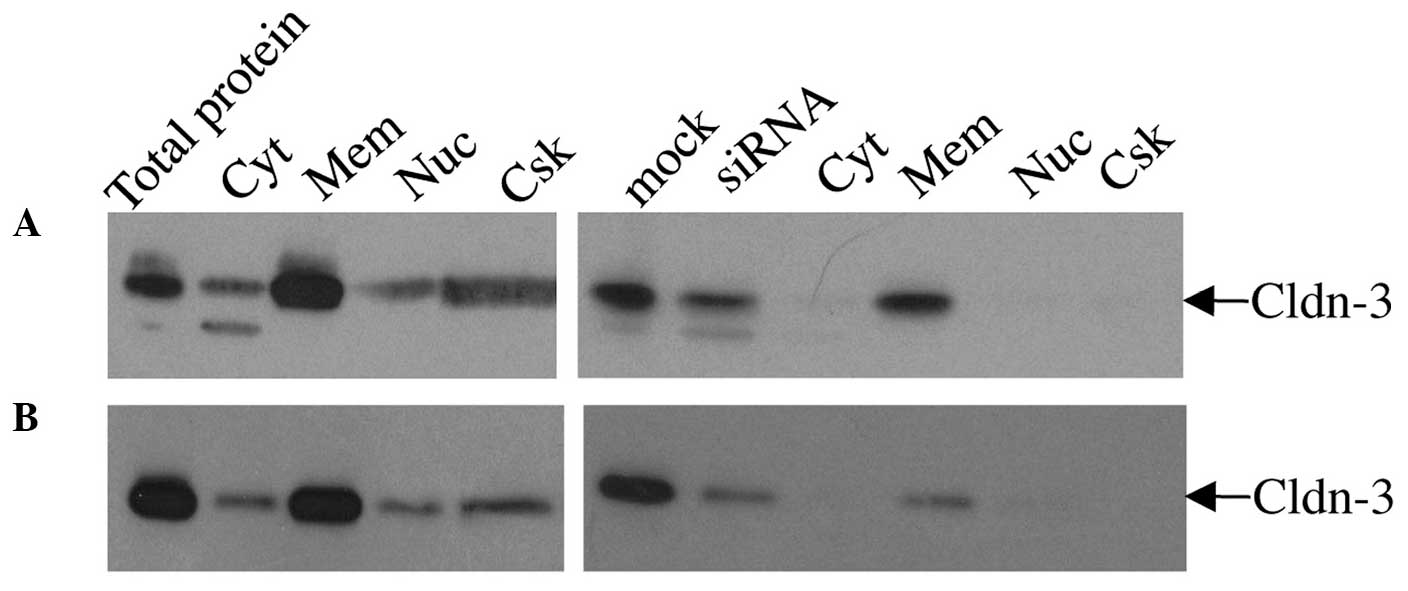
For the differential detergent cell fractionation
analysis, mock- and claudin-3 siRNA-transfected MCF-7 (Fig. 5A) and MDA-MB-415 (Fig. 5B) cells were incubated sequentially in
four extraction buffers to yield cytosolic, membranous, nuclear,
and cytoskeletal fractions. Immunoblot analysis of proteins from
each fraction of the mock-transfected cells indicated that the
majority of claudin-3 was localized in the membrane (Fig. 5). Lower levels of claudin-3 were also
observed in the cytosolic, nuclear and cytoskeletal fractions.
siRNA-mediated suppression of claudin-3 resulted in a marked
reduction in the overall level of claudin-3 protein, but did not
considerably affect its pattern of subcellular distribution
relative to the mock-transfected cells; however a more noticeable
decrease in signal was observed in the cytoskeletal fraction. In
addition to the 23 kDa claudin-3 protein, a lower molecular weight
band was detected in the mock- and siRNA-transfected whole cell
extracts and cytosolic fractions of the MCF-7 and, to a lesser
extent, MDA-MB-415 cells, which may correspond to a processed form
of the claudin-3 protein that is found in the cytosol (Fig. 5).
Effect of claudin-3 expression level
on cell motility
As normal breast epithelial cells are
characteristically non-motile, the Cytoselect wound-healing assay
was utilized to determine whether or not endogenous overexpression
of claudin-3 protein is involved in promoting cellular motility of
MCF-7 breast epithelial cells (Fig.
6). Whilst mock-transfected MCF-7 cells had fully migrated into
the wound field by day 3 (Fig. 6A), a
considerable lag was observed in the motility of the
siRNA-transfected cells (Fig. 6B),
associated with the sustained suppression of claudin-3 protein
expression (Fig. 6C).
Discussion
Although much is known about the role of claudins in
the regulation of paracellular transport across TJs, the extent of
the functions of these proteins has yet to be fully elucidated. In
particular, the increasing evidence associating deregulation of
claudins with tumorigenesis suggests that these proteins are
important in multiple cellular processes, including motility and
invasion (8–10), in addition to their established
functions.
The present study compared the level of claudin-3
protein in a panel of different normal tissues to ascertain the
variation in the basal levels of this protein. While the majority
of the tissues expressed undetectable to low levels of claudin-3,
normal breast tissue exhibited the highest, most readily detectable
level of expression. The assessment of claudin-3 expression in
three breast cancer cell lines revealed that, compared with normal
breast epithelial cells (HMEC), there was a considerable
overexpression of claudin-3 in two of the cell lines, MCF-7 and
MDA-MB-415. By contrast, the third breast cancer cell line,
MDA-MB-157, expressed a level of claudin-3 that was approximately
equivalent to that of the HMEC cells. Notably, overexpression of
claudin-4 protein was observed in the MCF-7 and MDA-MB-415 cell
lines, in addition to a less striking overexpression of claudin-1
protein in all three breast cancer cell lines. The finding of
abnormally elevated expression of claudin-3 and −4 proteins in
breast cancer cell lines is consistent with multiple studies that
have demonstrated overexpression of claudins in a variety of tumor
types. Specifically, elevated levels of claudin-3 and −4 have been
reported in ovarian (8) and
endometrial (11,12) cancers, while claudin-4 overexpression
has been reported in breast cancer (4). As previously stated, the MCF-7 and
MDA-MB-415 cell lines are metastatic breast adenocarcinomas derived
from pleural effusions. It is possible that the overexpression of
claudin-3 is involved in TJ disruption in these two breast tumors,
thereby facilitating metastasis. Consistent with this, Agarwal
et al (8) previously reported
that exogenous overexpression of claudins-3 and −4 in ovarian
cancer cell lines is associated with an increase in cell
invasiveness and survival.
The present study also assessed the subcellular
localization of claudin-3 protein in the MCF-7 and MDA-MB-415
breast cancer cell lines by immunofluorescence in order to
determine whether or not abnormally elevated levels of the protein
resulted in its delocalization. As predicted, both MCF-7 and
MDA-MB-415 cells revealed intense membrane staining at cell
junctions, consistent with the integral role of claudin-3 in TJs.
Notably, intracellular fluorescence was also observed in both cell
lines; this was, at least in part, attributable to the presence of
claudin-3, as siRNA-mediated suppression of claudin-3 in MCF-7
cells also resulted in a decrease in intracellular fluorescence
compared with mock-transfected cells. Consistent with the
immunofluorescence data, differential detergent cell fractionation
revealed low levels of claudin-3 in the cytoskeletal, nuclear and
cytosolic fractions of mock- and siRNA-transfected MCF-7 and
MDA-MB-415 cells, in addition to the predicted high level of
claudin-3 in the membrane fractions. Notably, siRNA-mediated
suppression of claudin-3 in MCF-7 and MDA-MB-415 cells resulted in
an overall decrease in the level of claudin-3 protein in every cell
fraction, however, the reduction appeared more prominent in the
cytoskeletal fraction. The finding of intracellular claudin-3 in
breast cancer cells expressing both elevated and suppressed levels
of the protein supports the notion of additional roles for claudin
proteins in cell signaling in tumor cells.
This abnormally elevated expression, coupled with
the delocalization of claudin-3, suggests a possible function for
TJ protein deregulation in local invasiveness of breast cancer
cells. Therefore, the current study investigated the effect of
claudin-3 overexpression on the motility of MCF-7 cells using a
wound-healing assay. siRNA-mediated suppression of claudin-3
protein expression resulted in a decrease in the motility of MCF-7
cells. These data indicate that the deregulated expression of
claudin-3 contributes to the invasive potential of breast cancer
cells. Agarwal et al (8)
previously reported enhanced cellular motility in ovarian
epithelial cell lines engineered to stably overexpress claudin-3
and claudin-4 proteins. Furthermore, siRNA-mediated suppression of
endogenously expressed claudin-3 and −4 in ovarian cancer cell
lines resulted in a decrease in cellular motility (8). Similarly, Oku et al (9), showed a correlation between endogenous
overexpression of claudin-1 protein and increased cellular invasion
in oral squamous carcinoma. siRNA-mediated suppression of claudin-1
in these same cell lines also resulted in a decrease in
invasiveness. Overexpression of claudin-1 has also been
demonstrated in primary and metastatic colon tumors and cell lines
(10). In the same study by Dhawan
et al (10), suppression of
claudin-1 expression in the metastatic colon cancer cell line,
SW620, resulted in decreased motility, invasiveness and
anchorage-independence (as assayed by soft agar). Consistent with
the above studies, exogenous expression of claudin-1 in malignant
melanoma cell lines results in delocalization of claudin-1 to the
cytosol with increased cellular migration and elevated MMP-2
activity (13). By contrast,
delocalization of exogenously expressed claudin-1 to the nucleus of
the same malignant melanoma cell lines (often demonstrated by
benign nevi) does not result in increased migration (13). These data suggest that the subcellular
localization, in addition to the level of expression of claudins,
may be important in the progression of tumors. Whereas the above
reports suggest a role for overexpression of claudins in enhancing
cellular motility, Michl et al (14) found that exogenous overexpression of
claudin-4 inhibited both the invasiveness and
anchorage-independence of the pancreatic cell line, SUIT-2. Thus,
it would appear that changes in claudin expression that deviate
from the expression levels normally exhibited by normal cells of
the same tissue type (including upregulation and downregulation of
TJ proteins) contribute to the regulation of cellular motility and
invasive potential.
Both the MCF-7 and MDA-MB-415 breast cancer cell
lines used in the current study were derived from metastases in the
lung, and thus it is possible that the overexpression and
delocalization of claudin-3 in these cells contributed to the
progression of the original tumors. The use of siRNA to suppress
claudin-3 expression may thus prove a beneficial gene therapeutic
approach for cancers that exhibit deregulated expression and
delocalization of claudins. If claudin-3 is involved in cell
signaling, the preferential reduction of claudin-3 in the
cytoskeletal fraction (observed in siRNA-transfected cells) may
mediate the mechanism by which transduction of growth-stimulatory
signals is attenuated in cancer cells.
Acknowledgements
This work was supported by the National Science
Foundation Major Research Instrumentation Grant (grant no.
0922258), Joe and Jessie Crump Fund at JP Morgan Bank, the ACS
Andrew W. Mellon Integrated Scholarly Grant and Howard Hughes
Medical Institute through the Undergraduate Science Education
Program (grant no. 52007558), and the Southwestern University
Faculty-Student Collaborative Projects fund.
The authors would also like to thank Ms. Bronwyn
Tyler for her technical assistance and Mr. Aidan Todd for his
assistance in the wound-healing assays.
References
|
1
|
American Cancer Society, . Breast Cancer
Facts & Figures. http://www.cancer.org/research/cancerfactsstatistics/breast-cancer-facts-figuresAccessed.
February 24–2014
|
|
2
|
Turksen K and Troy TC: Barriers built on
claudins. J Cell Sci. 117:2435–2447. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kominsky SL, Vali M, Korz D, Gabig TG,
Weitzman SA, Argani P and Sukumar S: Clostridium perfringens
enterotoxin elicits rapid and specific cytolysis of breast
carcinoma cells mediated through tight junction proteins claudin 3
and 4. Am J Pathol. 164:1627–1633. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lanigan F, McKiernan E, Brennan DJ,
Hegarty S, Millikan RC, McBryan J, et al: Increased claudin-4
expression is associated with poor prognosis and high tumour grade
in breast cancer. Int J Cancer. 124:2088–2097. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Tokés AM, Kulka J, Paku S, Szik A, Páska
C, Novák PK, Szilák L, Kiss A, Bögi K and Schaff Z: Claudin-1, −3
and −4 proteins and mRNA expression in benign and malignant breast
lesions: a research study. Breast Cancer Res. 7:R296–R305. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kominsky SL, Argani P, Korz D, Evron E,
Raman V, Garrett E, Rein A, Sauter G, Kallioniemi OP and Sukumar S:
Loss of the tight junction protein claudin-7 correlates with
histological grade in both ductal and carcinoma in situ and
invasive ductal carcinoma of the breast. Oncogene. 22:2021–2033.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Tokés AM, Kulka J, Paku S, Máthé M, Páska
C, Lódi C..Kiss A and Schaff Z: The expression of five different
claudins in invasive breast carcinomas: comparison of pT1pN1 and
pT1pN0 tumors. Pathol Res Pract. 201:537–544. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Agarwal R, D'Souza T and Morin PJ:
Claudin-3 and claudin-4 expression in ovarian epithelial cells
enhances invasion and is associated with increased matrix
metalloproteinase-2 activity. Cancer Res. 65:7378–7385. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Oku N, Sasabe E, Ueta E, Yamamoto T and
Osaki T: Tight junction protein claudin-1 enhances the invasive
activity of oral squamous cell carcinoma cells by promoting
cleavage of laminin-5 gamma2 chain via matrix metalloproteinase
(MMP)-2 and membrane-type MMP-1. Cancer Res. 66:5251–5257. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Dhawan P, Singh AB, Deane NG, No Y, Shiou
SR, Schmidt C, Neff J, Washington MK and Beauchamp RD: Claudin-1
regulates cellular transformation and metastatic behavior in colon
cancer. J Clin Invest. 115:1765–1776. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Pan XY, Wang B, Che YC, Weng ZP, Dai HY
and Peng W: Expression of claudin-3 and claudin-4 in normal,
hyperplastic, and malignant endometrial tissue. Int J Gynecol
Cancer. 17:233–241. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Konecny GE, Agarwal R, Keeney GA,
Winterhoff B, Jones MB, Mariani A, et al: Claudin-3 and claudin-4
expression in serous papillary, clear-cell, and endometroid
endometrial cancer. Gynecol Oncol. 109:263–269. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
French AD, Fiori JL, Camilli TC, Leotlela
PD, O'Connell MP, Frank BP, Subaran S, Indig FE, Taub DD and
Weeraratna AT: PKC and PKA phosphorylation affect the subcellular
localization of claudin-1 in melanoma cells. Int J Med Sci.
6:93–101. 2009.PubMed/NCBI
|
|
14
|
Michl P, Barth C, Buchholz M, Lerch MM,
Rolke M, Holzmann KH, et al: Claudin-4 expression decreases
invasiveness and metastatic potential of pancreatic cancer. Cancer
Res. 63:6265–6271. 2003.PubMed/NCBI
|