Introduction
Prostate cancer (PCa) is the most common malignancy
affecting men worldwide, and the second leading cause of
cancer-associated mortalities in western countries (1). The complexity of PCa is due to it
diverse metastasis profile, heterogeneous degree of aggressiveness
and variable response to conventional therapies (2). Although PCa may be controlled
temporarily by hormone deprivation, it eventually becomes
refractory to hormonal therapy, and no effective treatment has been
developed thus far for this type of hormone-insensitive cancer
(3). Therefore, the development of
novel drugs for the treatment of PCa, which offer higher
specificity and potentially improved prognosis and clinical
outcomes for patients with PCa, is required (4).
MicroRNAs (miRNAs) are a class of single-stranded
small non-coding RNAs, of 17–27 nucleotides in length, which
negatively regulate the expression of target genes by binding to
the 3′-untranslated regions (UTRs) of their messenger (m)RNAs, thus
inhibiting the process of protein translation or promoting the
degradation of these mRNAs (5). By
regulating the expression of genes associated with cancer, miRNAs
may act as oncogenes or tumour suppressors (6,7). Recent
studies have reported the dysregulation of miRNAs in human tumours,
which suggests that miRNAs participate in the pathogenesis of
cancer, including disease onset, progression and metastasis
(8,9).
miR-135b has been previously associated with human
colorectal (10) and breast cancer
(11), and its expression appears to
be downregulated in PCa cells (12).
However, functional analysis of miR-135b in PCa has not been
conducted thus far. In the present study, in silico analysis of
3′-UTRs identified signal transducer and activator of transcription
6 (STAT6) as a putative target of miR-135b. Activated STAT6 induces
the expression of various genes involved in cell differentiation,
proliferation, metastasis and resistance to apoptosis (13–15). It
has been previously observed that STAT6 is overexpressed and
activated in numerous malignancies, including PCa (16), colon cancer (17), lymphoma (18–20) and
leukemia (21,22). In addition, STAT6 was identified as a
robust marker gene for human PCa in previous DNA microarray studies
(23).
Therefore, the aim of the present study was to
investigate the association between miR-135b and its potential
target STAT6 in PCa cells.
Materials and methods
Cell culture and tissue
collection
PCa cell lines DU145 and PC3 were purchased from the
Shanghai Cell Bank, Chinese Academy of Sciences (Shanghai, China),
and maintained in Dulbecco's modified Eagle's medium (DMEM) (Gibco,
Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with
10% fetal bovine serum (FBS; GE Healthcare Life Sciences, Logan,
UT, USA), 100 U/ml penicillin and 100 µg/ml streptomycin (KeyGen
Biotech. Co. Ltd., Nanjing, China), in an atmosphere of 5% CO2 and
37°C. PCa and nonmalignant prostate tissues were collected from
patients who underwent radical prostatectomy at the Department of
Urology of The First Affiliated Hospital of Huzhou Teachers College
(Huzhou, China). The use of clinical specimens in the present study
was approved by the medical ethics committee of The First
Affiliated Hospital of Huzhou Teachers College. Detailed
information of each tissue donor is provided in Table I.
 | Table I.Clinical characteristics of the 32
patients with prostate cancer who participated in the present
study. |
Table I.
Clinical characteristics of the 32
patients with prostate cancer who participated in the present
study.
|
Characteristics | No. of
patients |
|---|
| Age (n=32) |
|
| Median
(range) | 70 (56.76) |
| T stage (n=32) |
|
| T1 | 2 |
| T2 | 17 |
| T3 | 12 |
| T4 | 1 |
| N stage |
|
| N0 | 27 |
| N1 | 5 |
| M stage |
|
| M0 | 0 |
| M1 | 32 |
| Total PSA at
diagnosis (n=29) |
|
| Median
(range) | 18.00
(6.66.63.74) |
| Free PSA at
diagnosis (n=26) |
|
| Median
(range) | 2.01
(0.65.21.00) |
| Gleason score
(n=32) |
|
|
<7 | 10 |
| =7 | 12 |
|
>7 | 10 |
Cell transfection
Homo sapiens (Hsa)-miRNA-135b and small
interfering (si)RNAs S-1 and S-2 were chemically synthesized by
Gene Pharma Co., Ltd. (Shanghai, China). The sequence of
Hsa-miRNA-135b mimics were as follows: Sense,
5′-UAUGGCUUUUCAUUCCUAUGUGA-3′, and anti-sense,
5′-ACAUAGGAAUGAAAGCCAUAUU-3′. The sequence of the negative controls
were as follows: Sense, 5′-UUCUCCGAACGUGUCACGUTT-3′, and
anti-sense, 5′-ACGUGACACGUUCGGAGAATT-3′. The sequences of
siRNA-STAT6 S-1 were, 5′-AAGCAGGAAGAACUCAAGUUUTT-3′ (sense) and
5′-AAACUUGAGUUCUUCCUGCUUTT-3′ (anti-sense), while for siRNA-STAT6
S-2 the sequences were 5′-AAACGAGAGUGUUAUAACUGUTT-3′ (sense) and
5′-ACAGUUAUAACACUCUCGUUUTT-3′ (anti-sense). Cells at 50–60%
confluence were transfected with the corresponding oligonucleotide
using Lipofectamine 2000 (Invitrogen, Thermo Fisher Scientific,
Inc.), according to the manufacturer's protocol.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted using TRIzol (Invitrogen,
Thermo Fisher Scientific, Inc.). For amplification of miR-135b,
RT-qPCR was performed with TaqMan MicroRNA Assays (Applied
Biosystems, Thermo Fisher Scientific, Inc.), using U6 as internal
control. The reaction conditions were as follows: 40 cycles of 95°C
for 10 min, 95°C for 15 sec and 60°C for 1 min. Relative expression
levels were calculated by the 2-∆∆Cq method.
Immunofluorescence analysis
Transfected DU145 cells were seeded on coverslips,
stimulated with 50 ng/ml human interleukin (IL)-4 (GE Healthcare
Life Sciences) for 30 min, fixed with 4% paraformaldehyde (KeyGen
Biotech. Co. Ltd.), blocked with 3% bovine serum albumin (BSA; GE
Healthcare Life Sciences) for 1 h, and incubated with primary
antibodies overnight at 4°C. Fluorescein isothiocyanate-labeled
secondary antibody was then added for 2 h at 37°C, and following
staining with DAPI (KeyGen Biotech. Co. Ltd.) for 5 min, the cells
were subjected to confocal laser scanning (Nikon Corporation,
Tokyo, Japan).
Migration and invasion assays
At 24 h post-transfection, PC3 and DU145 cells were
added to the upper chamber of a 24-well plate with 8-µm pores (BD
Biosciences, San Jose, CA, USA), which was coated with Matrigel (BD
Biosciences) for invasion assays, and 0.7 ml of 20% FBS-DMEM was
then added to the lower chamber. Following incubation for 48 h,
non-migrated or non-invaded cells were removed from the upper well
by cotton swabs, while the migrated or invaded cells were fixed
with 95% methanol, stained with 0.1% crystal violet (KeyGen
Biotech. Co. Ltd.), and photographed in five independent
fields/well.
Luciferase reporter assay
For the luciferase reporter assay, cells were
cotransfected with wild-type or mutant STAT6 3′-UTR reporter
plasmid and miR-135b, using Lipofectamine 2000. For normalization,
1 ng pRLSV40 Renilla reniformis luciferase construct (Promega
Corporation, Madison, WI, USA) was used. Luciferase assays were
performed at 48 h post-transfection, using Dual-Luciferase Reporter
Assay System (Promega Corporation).
Western blot analysis
Total protein was collected with Total Protein
Extraction Kit (KeyGen Biotech. Co. Ltd., Nanjing, China). Proteins
(30 mg) were separated on a 10% sodium dodecyl
sulfate-polyacrylamide gel, and transferred electrophoretically
onto polyvinylidene difluoride membranes (BD Biosciences). The
membranes were blocked in 5% skimmed milk for 1 h, and then
incubated for 2 h with rabbit anti-STAT6 (1:1,000; Abcam,
Cambridge, MA, USA), rabbit anti-phosphorylated (p)-STAT6 (1:1,000;
Cell Signaling Technology, Inc., Danvers, MA, USA) or rabbit
anti-GAPDH (1:1,000; Bioworld, Technology, Inc., St. Louis Park,
MN, USA), which was used as loading control. Following incubation
with goat anti-rabbit secondary antibody (Bioworld, Technology,
Inc.), the signals derived from the antibody-antigen binding
reaction were detected by enhanced chemiluminescence (BD
Biosciences).
Statistical analysis
Data are reported as the mean ± standard error of
the mean. All experiments were conducted in triplicate. The
correlation between downregulation of miR-135b and levels of total
or free PSA in the PCa samples was analysed using the Spearman rank
correlation test. All other data were compared by two-sided
t-test. Statistical analysis was performed with SPSS
software version 17.0 (SPSS, Inc., Chicago, IL, USA). P<0.05 was
considered to indicate a statistically significant difference.
Results
miR-135b is significantly
downregulated in PCa tissues
The expression of miR-135b was analyzed in 32
primary PCa samples and 14 nonmalignant samples by RT-qPCR. The
results revealed that the expression of miR-135b was significantly
downregulated in PCa tissues, compared with noncancerous tissues
(Fig. 1A). The results also indicated
that the expression levels of miR-135b were possibly correlated
with the pathological T stages of PCa (Fig. 1A). Furthermore, the results
demonstrated significant inverse correlations between the
expression levels of miR-135b and the levels of total and free PSA
(Fig. 1B and C, respectively). No
statistical significant difference was observed for the association
between the expression levels of miR-135b and the Gleason scores
(Fig. 1D).
miR-135b inhibited PCa cell migration
and invasion
The role of miR-135b on cell migration and invasion,
two key determinants of malignant progression and metastasis, was
assessed in human PCa cells. A significant decrease in cell
migration was observed in miR-135b-mimics-transfected PC3 and DU145
cells, compared with negative control (NC)-transfected cells
(Fig. 2A). The effect of miR-135b on
cell invasion across the extracellular matrix was evaluated, and it
was observed that the ectopic expression of miR-135b also reduced
the invasion ability of PCa cells (Fig.
2B). These results suggested a mechanism by which the
overexpression of miR-135b may contribute to the inhibition of
tumour progression and metastasis in human PCa.
STAT6 was targeted by miR-135b
To explore the mechanism by which miR-135b regulates
cell metastasis, a miRNA target search was performed with three
target prediction programs, including TargetScan (http://www.targetscan.org/), miRanda (http://www.microrna.org/microrna/home.do) and PicTar
(http://pictar.mdc-berlin.de/), and
highly conserved putative binding sites for miR-135b were
identified in the 3′-UTR of STAT6 (Fig.
3A). When miR-135b-mimics were cotransfected with the reporter
plasmids, the relative luciferase activity of the reporter plasmid
containing wild-type STAT6 3′-UTR was markedly reduced, while the
luciferase activity of the reporter plasmid containing mutant STAT6
3′-UTR was unaltered (Fig. 3D).
Furthermore, western blotting analysis was conducted to determine
whether the expression of STAT6 was regulated by miR-135b. As
presented in Fig. 3B, the protein
expression levels of STAT6 and p-STAT6 were markedly downregulated
following transfection with miR-135b mimics. These results
indicated that miR-135b may downregulate the expression of STAT6
through the miR-135-binding sequences located at the 3′-UTR of the
STAT6 gene.
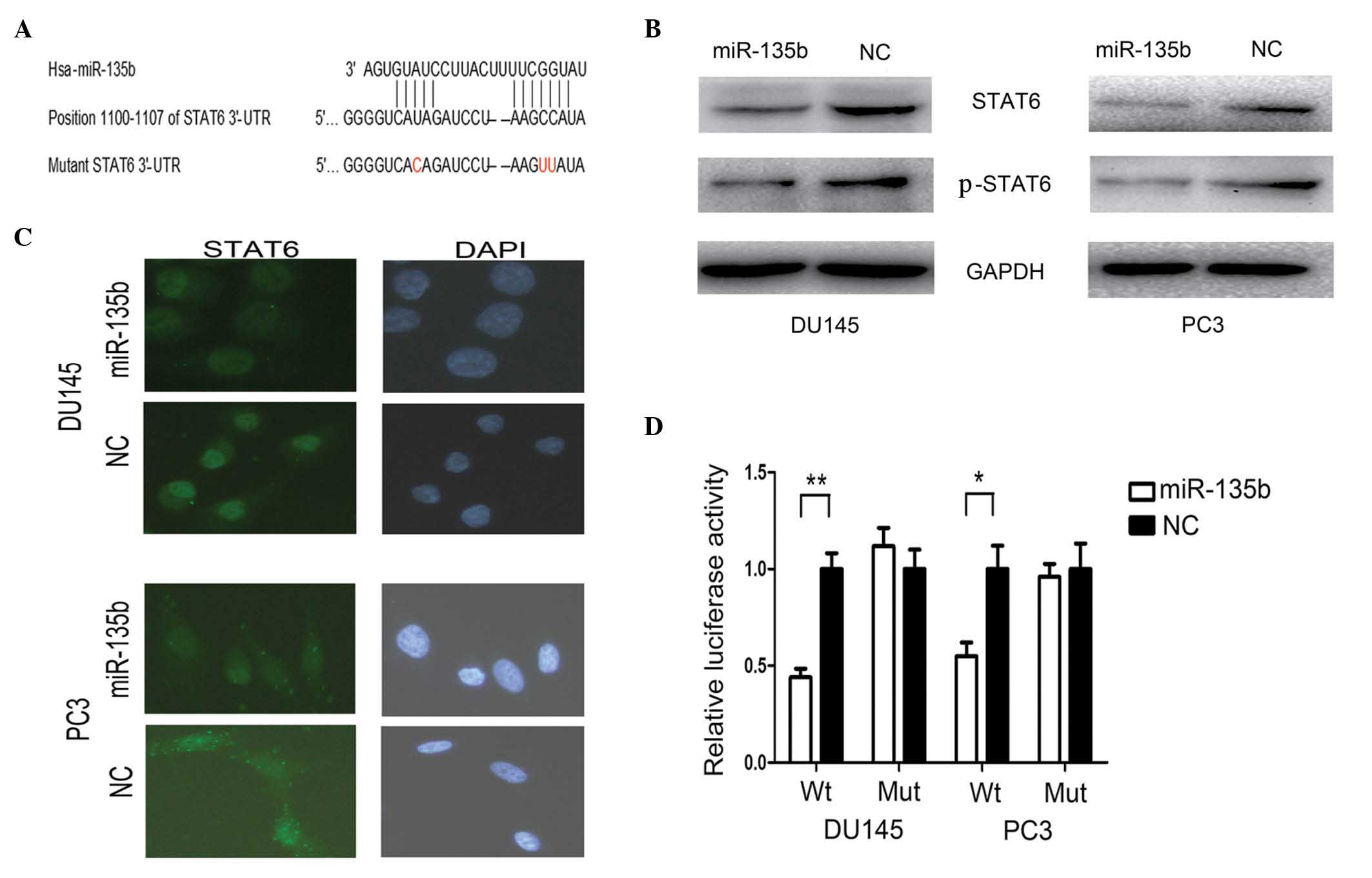 | Figure 3.STAT6 is a direct target gene of
miR-135b. (A) Schematic description of the 3′-untranslated region
of STAT6 with two putative binding sites for miR-135b. (B) The
protein levels of STAT6 and phosphorylated-STAT6 were reduced in
the human PCa cell lines DU145 and PC3 following transfection with
miR-135b. (C) Upregulation of miR-135b in PCa cells led to reduced
expression of STAT6 in the cytoplasm and nucleus of these cells.
(D) Luciferase reporter assay was performed in DU145 and PC3 cells
and indicated that miR-135b targets STAT6 through the
miR-135-binding sequences located at the 3′-UTR of the STAT6 gene.
Data are presented as the mean ± standard error of the mean.
*P<0.05 and **P<0.01 by t-test. STAT6, signal transducer and
activator of transcription 6; miR, microRNA; p, phosphorylated;
PCa, prostate cancer; Hsa, Homo sapiens; UTR, untranslated region;
NC, negative control; GAPDH, glyceraldehyde 3-phosphate
dehydrogenase; DAPI, 4′,6-diamidino-2-phenylindole; Wt, wild-type;
Mut, mutant. |
miR-135b reduced the IL-4-induced
nuclear translocation of STAT6
The effect of miR-135b on the cellular localization
of STAT6 was examined by immunofluorescence analysis. Under basal
conditions, the immunofluorescent signal corresponding to STAT6 was
located predominantly in the cytoplasm of PC3 cells (Fig. 3C). The levels of STAT6 were reduced in
the cytoplasm of PC3 cells following transfection with miR-135b
mimics (Fig. 3C). Treatment of DU145
cells with IL-4 (50 ng/ml) for 30 min resulted in marked
translocation of STAT6 to the nucleus (Fig. 3C). Furthermore, the IL-4-induced
nuclear translocation of STAT6 in DU145 cells was reduced following
transfection with miR-135b mimics (Fig.
3C).
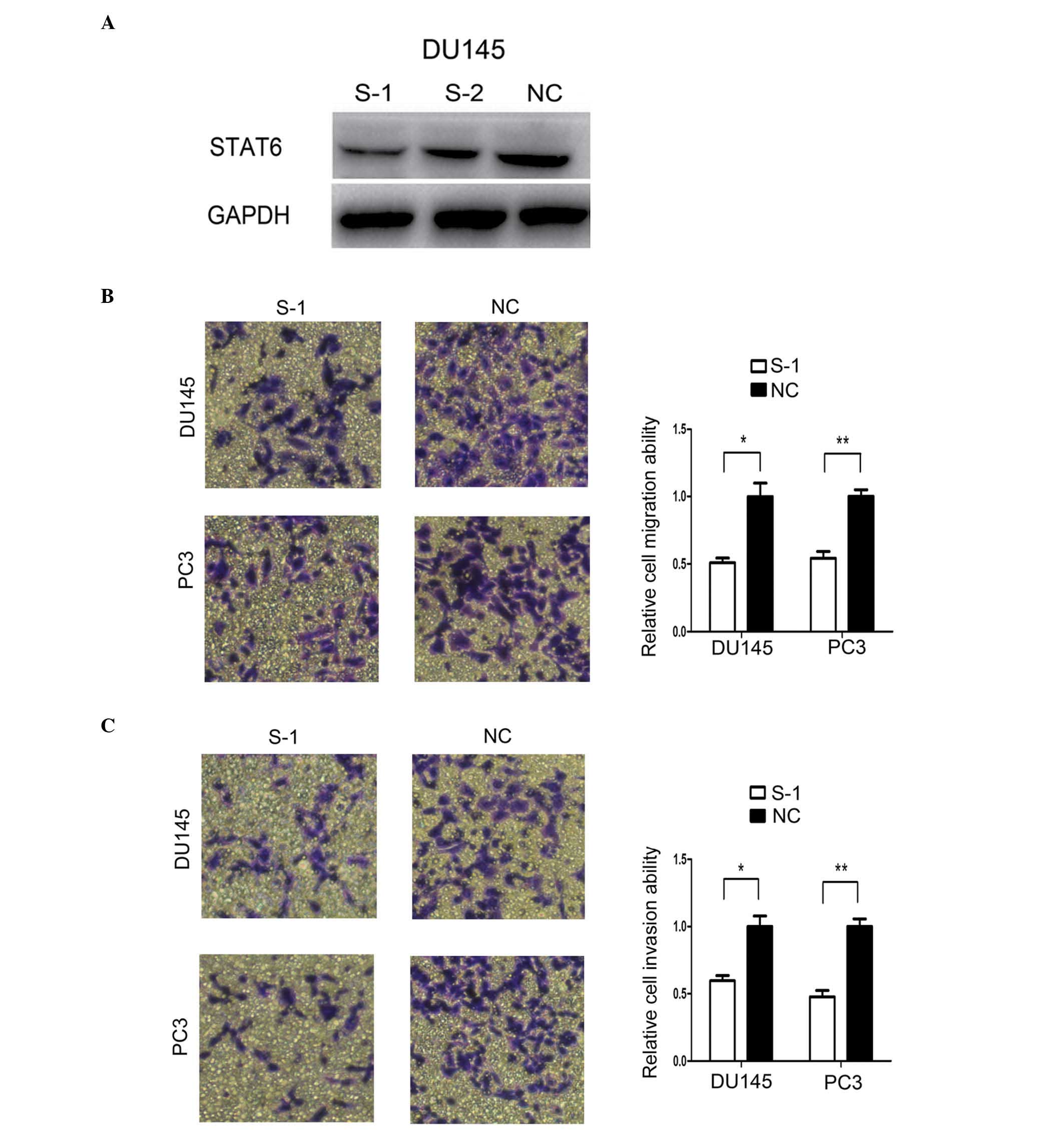
Knockdown of STAT6 mimicked miR-135b
inhibition
To investigate if miR-135b mediates its
metastasis-suppressive effects primarily through knocking down
STAT6, PC3 and DU145 cells were transfected with two siRNAs-STAT6
(S-1 and S-2). The results of western blot analysis indicated that
S-1 and S-2 knocked down the expression of STAT6 at the
protein level (Fig. 4A). In addition,
the effect of siRNA-STAT6 S-1 on the migration and invasion
abilities of PCa cells was analysed in subsequent experiments. As
expected, compared with NC-transfected cells,
STAT6-knockdown DU145 and PC3 cells exhibited reduced
migration and invasion abilities (Fig. 4B
and C), similarly to the phenotype displayed upon miR-135b
restoration.
Discussion
Previous studies have reported that miR-135b is
overexpressed in embryonic stem cells and colorectal cancer
(24,25), and a previous microarray profiling of
human head and neck squamous cell carcinoma samples demonstrated
that miR-135b was one of the most significantly upregulated miRNAs
in this type of cancer (26). The
expression of miR-135b in PCa is controversial, since miR-135b was
previously observed to be upregulated in certain PCa tissues
(27) but downregulated in certain
PCa cell lines (28). However, no
functional evidence for a role of miR-135b as an oncogenic or
tumour-suppressive miRNA has been documented thus far. In the
present study, miR-135b appeared to be markedly downregulated in
PCa tissues, compared with normal tissues. Previous clinical
observations have suggested that miR-135b may be downregulated in
oral squamous cell carcinoma, particularly in the advanced stage
(29), which is consistent with the
results of the present study. Furthermore, the expression levels of
miR-135b were observed to be inversely correlated with the levels
of total and free PSA. Previous studies have suggested that
miR-135b may be a potential diagnostic and prognostic marker for
colorectal cancer (30–32). These results indicate that, as a
tumour-suppressor gene, miR-135b may be used as a marker for early
diagnosis of PCa and for detecting tumour progression. Due to the
limited number of samples in the present study, it is not possible
to confirm whether the expression of miR-135b is associated with
the Gleason scores. Therefore, further studies with a larger number
of clinical samples are required.
Since the majority of cancer-associated mortalities
are caused by dissemination of the disease rather than the primary
tumour, metastasis has become the most prominent problem in the
clinical treatment of cancer (33).
There is increasing evidence suggesting that miRNAs participate in
tumour metastasis (34), since miRNAs
may influence multiple steps of the metastatic cascade, including
cell migration, invasion and intravasation. A previous study has
revealed that dysregulation of miR-221 and miR-222 was associated
with cancer progression, poor prognosis and development of
metastases in patients with PCa (35). It has been previously demonstrated
that miR-146a targets Rho-associated coiled-coil containing protein
kinase 1 (ROCK1), and that high levels of ROCK1 promote cell
proliferation, invasion and metastasis in PCa cells (36). The results of the gain-of-function
experiments conducted in the present study indicated that miR-135b
was positively correlated with the invasive and migratory abilities
of PCa cells in vitro, since upregulation of miR-135b in the
human PCa cell lines DU145 and PC3, which normally exhibit high
metastatic ability and low expression levels of miR-135b, reduced
the metastatic ability of these cells.
The STAT family of transcription factors consists of
seven members (STAT 1–4, 5a, 5b and 6), which can be activated by a
number of cytokines, hormones and growth factors (37). Upon activation, STATs dissociate from
their receptor, form dimmers, translocate to the nucleus and bind
to cognate DNA response elements, thus activating the transcription
of their target genes (38,39). STAT6 is activated through tyrosine
phosphorylation by the cytokines IL-4 and −13 (22). It has been previously reported that
the activity of STAT6 in PCa tissues and cell lines is
significantly higher than in normal tissues and cell lines
(40). In the present study,
STAT6 was confirmed to be a target of miR-135b in PCa cells
by luciferase reporter assay, and overexpression of miR-135b in
these cells was observed to reduce the expression levels of STAT6
by western blot assay. Previous immunofluorescence studies have
reported that STAT6 is located predominantly in the cytoplasm of
corneal fibroblasts, and IL-4 markedly induced the translocation of
STAT6 to the nucleus of these cells (41). These findings are consistent with the
results of the present study, whereby the expression levels of
STAT6 in the cytoplasm of the PCa cell lines tested, and the
IL-4-induced levels of STAT6 in the nucleus of these cells, were
observed to be reduced upon transfection with miR-135b mimics.
These results indicated that the upregulation of miR-135b inhibited
the expression of STAT6 and reduced the IL-4-induced nuclear
translocation of STAT6 in PCa cells. Previous studies have reported
that mice lacking STAT6 exhibited higher tumour immunity to primary
and metastatic mammary carcinoma, compared with normal mice
(42,43). In the present study, knocking down
STAT6 significantly reduced the metastatic potential of PCa cells
in vitro. This is consistent with the aforementioned
findings regarding the correlation between the upregulation of
miR-135b in PCa cells and the reduced aggressive phenotype
displayed by these cell lines. Taken together, the data of the
present study indicate that miR-135b modulates the metastatic
ability of PCa cells by regulating the expression of STAT6.
In conclusion, the findings of the present study
suggest that miR-135b functions as a tumour suppressor, affecting
the metastatic ability of PCa cells by targeting STAT6, since STAT6
knockdown resulted in reduced cell metastasis. In addition,
miR-135b was able to reduce the IL-4-induced nuclear translocation
of STAT6 in these cells. The findings of the present study suggest
that miR-135b functions as a tumour suppressor, reducing the
metastatic ability of PCa cells by targeting STAT6. Furthermore,
the expression of miR-135b was observed to be associated with the
pathological T stages and levels of total and free PSA in patients
with PCa. The present study indicates that miR-135b may offer an
attractive novel target for diagnostic and therapeutic intervention
in PCa.
References
|
1
|
Pflug BR, Pecher SM, Brink AW, Nelson JB
and Foster BA: Increased fatty acid synthase expression and
activity during progression of prostate cancer in the TRAMP model.
Prostate. 57:245–254. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Klein EA, Cooperberg MR, Magi-Galluzzi C,
Simko JP, Falzarano SM, Maddala T, Chan JM, Li J, Cowan JE, Tsiatis
AC, Cherbavaz DS, et al: A 17-gene assay to predict prostate cancer
aggressiveness in the context of Gleason grade heterogeneity, tumor
multifocality, and biopsy undersampling. Eur Urol. 66:550–560.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Feldman BJ and Feldman D: The development
of androgen-independent prostate cancer. Nat Rev Cancer. 1:34–45.
2001. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Best CJ, Gillespie JW, Yi Y, Chandramouli
GV, Perlmutter MA, Gathright Y, Erickson HS, Georgevich L, Tangrea
MA, Duray PH, et al: Molecular alterations in primary prostate
cancer after androgen ablation therapy. Clin Cancer Res.
11:6823–6834. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Valencia-Sanchez MA, Liu J, Hannon GJ and
Parker R: Control of translation and mRNA degradation by miRNAs and
siRNAs. Genes Dev. 20:515–524. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Esquela-Kerscher A and Slack FJ:
Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer.
6:259–269. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ventura A and Jacks T: MicroRNAs and
cancer: Short RNAs go a long way. Cell. 136:586–591. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Iorio MV and Croce CM: MicroRNAs in
cancer: Small molecules with a huge impact. J Clin Oncol.
27:5848–5856. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Baranwal S and Alahari SK: miRNA control
of tumour cell invasion and metastasis. Int J Cancer.
126:1283–1290. 2010.PubMed/NCBI
|
|
10
|
Bandrés E, Cubedo E, Agirre X, Malumbres
R, Zárate R, Ramirez N, Abajo A, Navarro A, Moreno I, Monzó M and
García-Foncillas J: Identification by Real-time PCR of 13 mature
microRNAs differentially expressed in colorectal cancer and
non-tumoural tissues. Mol Cancer. 5:292006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lowery AJ, Miller N, Devaney A, McNeill
RE, Davoren PA, Lemetre C, Benes V, Schmidt S, Blake J, Ball G and
Kerin MJ: MicroRNA signatures predict oestrogen receptor,
progesterone receptor and HER2/neu receptor status in breast
cancer. Breast Cancer Res. 11:R272009. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bhattacharya A, Ziebarth JD and Cui Y:
Systematic analysis of microRNA targeting impacted by small
insertions and deletions in human genome. PLoS One. 7:e461762012.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hebenstreit D, Wirnsberger G, Horejs-Hoeck
J and Duschl A: Signaling mechanisms, interaction partners and
target genes of STAT6. Cytokine Growth Factor Rev. 17:173–188.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Rothstein TL: Inducible resistance to
Fas-mediated apoptosis in B cells. Cell Res. 10:245–266. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ansel KM, Djuretic I, Tanasa B and Rao A:
Regulation of Th2 differentiation and Il4 locus accessibility. Annu
Rev Immunol. 24:607–656. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ni Z, Lou W, Lee SO, Dhir R, DeMiguel F,
Grandis JR and Gao AC: Selective activation of members of the
signal transducers and activators of transcription family in
prostate carcinoma. J Urol. 167:1859–1862. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li BH, Yang XZ, Li PD, Yuan Q, Liu XH,
Yuan J and Zhang WJ: IL-4/Stat6 activities correlate with apoptosis
and metastasis in colon cancer cells. Biochem Biophys Res Commun.
369:554–560. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Benekli M, Baer MR, Baumann H and Wetzler
M: Signal transducer and activator of transcription proteins in
leukemias. Blood. 101:2940–2954. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Skinnider BF, Elia AJ, Gascoyne RD,
Patterson B, Trumper L, Kapp U and Mak TW: Signal transducer and
activator of transcription 6 is frequently activated in Hodgkin and
Reed-Sternberg cells of Hodgkin lymphoma. Blood. 99:618–626. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Guiter C, Dusanter-Fourt I, Copie-Bergman
C, Boulland ML, Le Gouvello S, Gaulard P, Leroy K and Castellano F:
Constitutive STAT6 activation in primary mediastinal large B-cell
lymphoma. Blood. 104:543–549. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Melzner I, Bucur AJ, Brüderlein S, Dorsch
K, Hasel C, Barth TF, Leithäuser F and Möller P: Biallelic mutation
of SOCS-1 impairs JAK2 degradation and sustains phospho-JAK2 action
in the MedB-1 mediastinal lymphoma line. Blood. 105:2535–2542.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bruns HA and Kaplan MH: The role of
constitutively active Stat6 in leukemia and lymphoma. Crit Rev
Oncol Hematol. 57:245–253. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Xu L, Tan AC, Naiman DQ, Geman D and
Winslow RL: Robust prostate cancer marker genes emerge from direct
integration of inter-study microarray data. Bioinformatics.
21:3905–3911. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lin CH, Jackson AL, Guo J, Linsley PS and
Eisenman RN: Myc-regulated microRNAs attenuate embryonic stem cell
differentiation. EMBO J. 28:3157–3170. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Nagel R, le Sage C, Diosdado B, van der
Waal M, Oude Vrielink JA, Bolijn A, Meijer GA and Agami R:
Regulation of the adenomatous polyposis coli gene by the miR-135
family in colorectal cancer. Cancer Res. 68:5795–5802. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Liu CJ, Tsai MM, Hung PS, Kao SY, Liu TY,
Wu KJ, Chiou SH, Lin SC and Chang KW: miR-31 ablates expression of
the HIF regulatory factor FIH to activate the HIF pathway in head
and neck carcinoma. Cancer Res. 70:1635–1644. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Tong AW, Fulgham P, Jay C, Chen P, Khalil
I, Liu S, Senzer N, Eklund AC, Han J and Nemunaitis J: MicroRNA
profile analysis of human prostate cancers. Cancer Gene Ther.
16:206–216. 2009.PubMed/NCBI
|
|
28
|
Östling P, Leivonen SK, Aakula A, Kohonen
P, Mäkelä R, Hagman Z, Edsjö A, Kangaspeska S, Edgren H, Nicorici
D, et al: Systematic analysis of microRNAs targeting the androgen
receptor in prostate cancer cells. Cancer Res. 71:1956–1967. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Scapoli L, Palmieri A, Lo Muzio L,
Pezzetti F, Rubini C, Girardi A, Farinella F, Mazzotta M and
Carinci F: MicroRNA expression profiling of oral carcinoma
identifies new markers of tumour progression. Int J Immunopathol
Pharmacol. 23:1229–1234. 2010.PubMed/NCBI
|
|
30
|
Kanaan Z, Rai SN, Eichenberger MR, Roberts
H, Keskey B, Pan J and Galandiuk S: Plasma miR-21: A potential
diagnostic marker of colorectal cancer. Ann Surg. 256:544–551.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Gaedcke J, Grade M, Camps J, Søkilde R,
Kaczkowski B, Schetter AJ, Difilippantonio MJ, Harris CC, Ghadimi
BM, Møller S, et al: The rectal cancer microRNAome-microRNA
expression in rectal cancer and matched normal mucosa. Clin Cancer
Res. 18:4919–4930. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Xu XM, Qian JC, Deng ZL, Cai Z, Tang T,
Wang P, Zhang KH and Cai JP: Expression of miR-21, miR-31, miR-96
and miR-135b is correlated with the clinical parameters of
colorectal cancer. Oncol Lett. 4:339–345. 2012.PubMed/NCBI
|
|
33
|
Ma L and Weinberg RA: Micromanagers of
malignancy: Role of microRNAs inregulating metastasis. Trends
Genet. 24:448–456. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Khew-Goodall Y and Goodall GJ:
Myc-modulated miR-9 makes more metastases. Nat Cell Biol.
12:209–211. 2010.PubMed/NCBI
|
|
35
|
Pang Y, Young CY and Yuan H: MicroRNAs and
prostate cancer. Acta Biochim Biophys Sin (Shanghai). 42:363–369.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Lin SL, Chiang A, Chang D and Ying SY:
Loss of mir-146a function in hormone-refractory prostate cancer.
RNA. 14:417–424. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Calò V, Migliavacca M, Bazan V, Macaluso
M, Buscemi M, Gebbia N and Russo A: STAT proteins: From normal
control of cellular events to tumorigenesis. J Cell Physiol.
197:157–168. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Schindler C and Darnell JE Jr:
Transcriptional responses to polypeptide ligands: The JAK-STAT
pathway. Annu Rev Biochem. 64:621–651. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Darnell JE Jr: STATs and gene regulation.
Science. 277:1630–1635. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Das S, Roth CP, Wasson LM and Vishwanatha
JK: Signal transducer and activator of transcription-6 (STAT6) is a
constitutively expressed survival factor in human prostate cancer.
Prostate. 67:1550–1564. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Fukuda K, Fujitsu Y, Kumagai N and Nishida
T: Characterization of the interleukin-4 receptor complex in human
corneal fibroblasts. Invest Ophthalmol Vis Sci. 43:183–188.
2002.PubMed/NCBI
|
|
42
|
Ostrand-Rosenberg S, Grusby MJ and
Clements VK: Cutting edge: STAT6-deficient mice have enhanced
tumour immunity to primary and metastatic mammary carcinoma. J
Immunol. 165:6015–6019. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Ostrand-Rosenberg S, Sinha P, Clements V,
Dissanayake SI, Miller S, Davis C and Danna E: Signal transducer
and activator of transcription 6 (Stat6) and CD1: Inhibitors of
immunosurveillance against primary tumours and metastatic disease.
Cancer Immunol Immunother. 53:86–91. 2004. View Article : Google Scholar : PubMed/NCBI
|