Introduction
Breast cancer accounts for ~30% of all cancers in
women and is the most invasive form of cancer in women worldwide.
Following lung cancer, breast cancer has become the second leading
cause of cancer-associated mortality in women in developed nations
(1). Thus, it is urgent to explore
and define the mechanisms of breast cancer progression for the
development of effective therapeutic and preventative
methodologies.
The activation of Wnt/β-catenin signaling pathway
has been demonstrated to play important roles in breast
carcinogenesis and to be associated with a poorer prognosis in
breast cancer patients (2–5). Upon activation of this signaling
pathway, β-catenin accumulates and enters the nucleus where it
binds transcription factors of the transcription factor/lymphoid
enhancer-binding factor family and activates the transcription of
target genes associating with breast cancer progression (1). It is widely accepted that genetic
mutation is not the major contributing factor for β-catenin
activation in breast cancer, thus the aberrant expression of
signaling pathway components has become an area of intense focus
for detecting the association between β-catenin activation and
breast tumorigenesis (1,6). For example, numerous Wnt proteins,
including Wnt2, Wnt7b, and Wnt10b, were identified to be
upregulated in human breast carcinomas (6). It has also been reported that the
receptors of WNT, such as frizzled (FZD) and low-density
lipoprotein receptor-related protein 5/6 (LRP5/6), were upregulated
and results in aberrant activation of the β-catenin signaling
pathway in breast cancer (1).
Dishevelled-associated antagonist of β-catenin
homolog (DACT) 2 is a member of the DACT protein family that
interacts with dishevelled (Dvl), a key initiating factor of the
Wnt signaling pathway (7). DACT2
interacts with Dvl and promotes the degradation of Dvl; thus, DACT2
is an inhibitor of the WNT/β-catenin signaling pathway (8,9). However,
to the best of our knowledge, there have not yet been studies
investigating the dysregulation of DACT2 and its effect on the
signaling pathway in breast tumorigenesis. To explore the function
and modes of expressional regulation of DACT2 in breast cancer, the
present study assessed the status of the DACT2 gene in human breast
cancer and assessed its function in breast carcinogenesis.
Materials and methods
Cell and tissue samples
Breast cancer MCF-7, MDA-MB-231 and MDA-MB-468 cell
lines and the lentiviral vector packaging 293T/17 cell line were
bought from the Cell Center of Institute of Basic Medical Science,
Chinese Academy of Medical Science. Cells were grown in Dulbecco's
modified Eagle's medium (DMEM; Gibco; Thermo Fisher Scientific,
Inc., Waltham, MA, USA) containing 10% (v/v) fetal bovine serum
(FBS; Gibco; Thermo Fisher Scientific, Inc.), 100 U/ml penicillin
and 100 µg/ml streptomycin at 37°C and 5% CO2.
In total, 20 patients with breast cancer were
diagnosed at the Third Affiliated Hospital of the Harbin Medical
University between August 2009 and November 2010, their breast
cancer tissues and the paired adjacent normal breast tissues were
collected subsequent to surgery and immediately stored in liquid
nitrogen. The tissue slides of breast cancers and adjacent normal
tissues were obtained from the Pathology Department of the
hospital. The present study was approved by the Ethics Committee of
Harbin Medical University. Written informed consent was obtained
from all patients.
DNA extraction and
methylation-specific polymerase chain reaction (PCR) (MSP)
Genomic DNA extraction from cell lines, breast
cancer tissues and bisulfite modification was performed as
previously described (10). Briefly,
cells and tissues were digested with lysis buffer containing
proteinase K (Beyotime Institute of Biotechnology, Haimen, China),
and then genomic DNA was precipitated using isopropanol and
dissolved in nuclease-free water. For the modification of genomic
DNA, DNA was denatured with 0.2 M NaOH, and then modified with 10
mM hydroquinone (Sigma-Aldrich; Merck Millipore, Darmstadt,
Germany) and 3 M sodium bisulfite (Sigma-Aldrich; Merck Millipore).
Subsequent to purification, the modified DNA was treated with 0.3 M
NaOH followed by ethanol precipitation. DNA was finally resuspended
in nuclease-free water. The DNA polymerase, DNA ladder and Ethidium
bromide were obtained from (Beijing Transgen Biotech Co., Ltd.,
Beijing, China). MSP was performed as previously described
(10). MSP primers were designed
around transcription start sites and the sequences of primers are
listed in Table I. PCR products were
analyzed with 2% agarose gels, and the lengths of methylated and
unmethylated PCR products were identified to be 152 and 161 bp,
respectively. Experiments were repeated in triplicate.
 | Table I.Primers used in the present
study. |
Table I.
Primers used in the present
study.
| Primer name | Primer sequence
(5′-3′) |
|---|
| DACT2 qPCR |
|
|
Forward |
CGGTCGGTTGATGAGACTACT |
|
Reverse |
CAGGGCTCTGTCAAGATCACC |
| CyclinD1 qPCR |
|
|
Forward |
GCTGCGAAGTGGAAACCATC |
|
Reverse |
CCTCCTTCTGCACACATTTGAA |
| MMP7 qPCR |
|
|
Forward |
GAGTGAGCTACAGTGGGAACA |
|
Reverse |
CTATGACGCGGGAGTTTAACAT |
| GAPDH qPCR |
|
|
Forward |
ATGGGGAAGGTGAAGGTCG |
|
Reverse |
GGGGTCATTGATGGCAACAATA |
| DACT2 ORF |
|
|
Forward |
CGGGATCCGCCGCTCGTGGGGTTCGGGA |
|
Reverse |
CGACGCGTACCATGGTCATGACCTTCA |
| β-catenin reporter
(wide) |
|
|
Forward |
CGCGTAACTGACAGATCAAAGGGGGTAAGATCAAAGGGGGTAGTCAACTC |
|
Reverse |
TCGAGAGTTGACTACCCCCTTTGATCTTACCCCCTTTGATCTGTCAGTTA |
| β-catenin reporter
(mutant) |
|
|
Forward |
CGCGTAACTGACAGATCCCCTTTTTTAAGATCCCCTTTTTTAGTCAACTC |
|
Reverse |
TCGAGAGTTGACTAAAAAAGGGGATCTTAAAAAAGGGGATCTGTCAGTTA |
| MSP primers |
|
|
Methylated forward |
GCGCGTGTAGATTTCGTTTTTCGC |
|
Methylated reverse |
AACCCCACGAACGACGCCG |
|
Unmethylated forward |
TTGGGGTGTGTGTAGATTTTGTTTTTTGT |
|
Unmethylated reverse |
CCCAAACCCCACAAACAACACCA |
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from the harvested cell and
tissues using TRIzol reagent (Invitrogen; Thermo Fisher Scientific,
Inc.), according to the manufacturer's protocol. Subsequent to
quantification, the RNA was reversed to cDNA using M-MLV reverse
transcript kit (Invitrogen; Thermo Fisher Scientific, Inc.), and
the random primer was used as RT primer to synthesis cDNA. In
brief, 5X RT buffer (4 µl), primer (0.1 µg), transcriptase (1 µl),
10 mM RT dNTPs (1 µl), RNA sample (1 µg) and DEPC-treated water (to
make up 20 µl) mixture was incubated at 25°C for 5 min, 42°C for 30
min and 85°C for 5 min. All the reagents were obtained from
Invitrogen; Thermo Fisher Scientific, Inc. RT-qPCR was performed
using a Bio-Rad qPCR System (Bio-Rad Laboratories, Inc., Hercules,
CA, USA) using SYBR Premix Ex Taq kit (Takara Biotechnology Co.,
Ltd., Dalian, China), according to the manufacturer's protocol.
Gene relative expression is analyzed by the 2−∆∆Cq
method (11). GAPDH was used as the
endogenous controls for mRNA analysis. The primers used for qPCR
are summarized in Table I. The
experiment was repeated three times.
Western blot analysis
The cells were collected and treated with
radioimmunoprecipitation assay lysis buffer (Beyotime Institute of
Biotechnology). Then, the cell lysate was subjected to sodium
dodecyl sulfate-polyacrylamide gel electrophoresis and transferred
to a polyvinylidene fluoride membrane. Primary antibodies against
the following proteins were used: mouse anti-human β-catenin (Santa
Cruz Biotechnology, Inc., Santa Cruz, CA, USA, 1:500, sc-59737);
rabbit anti-human DACT2 (Abcam, Cambridge, MA, USA, 1:500,
ab79042); and mouse-anti-human β-actin (Santa Cruz Biotechnology,
Inc. 1:500, sc-130065). HRP-conjugated secondary antibodies were
purchased from Santa Cruz Biotechnology Inc. (bovine
anti-mouse/rabbit, 1:3000, sc-2371/2370). Signal was detected using
an enhanced chemiluminescence kit (EMD Millipore, Billerica, MA,
USA), according to the manufacturer's protocol.
Lentiviral vector construction and
packing
The coding sequence of DACT2 was amplified from cDNA
of MCF7 cells by PCR as described previously (12), using the primers listed in Table I. This was then sequenced and digested
with BamHI and MluI, followed by cloning into the
pC-1 plasmid lined with the same enzyme as previously described
(12). Lentiviral vectors were then
prepared using the lentiviral vector packing kit (System
Biosciences, Mountain View, CA, USA), according to the
manufacturer's protocol. MDA-MB-468 cells (1×105
cells/well) were plated into 24-well plate, 10 µ of
1×108 IU/ml lentiviral vectors were added into 24-well
plate and incubated for 12 h respectively. Then MDA-MB-468 cells
infected with lentiviral vectors were transferred to a 25
cm2 flask and grown for at least 72 h prior to be sorted
using a fluorescence-activated cell sorting Aria II flow cytometer
(BD Biosciences, Franklin Lakes, NJ, USA).
Luciferase assay
A β-catenin activity reporter plasmid was prepared
by inserting the synthesized β-catenin-recognizing DNA sequence
(Table I) into a pGL-3 basic vector
(Promega Corporation, Madison, WI, USA). MCF7 cells were plated
into 24-well plates to reach 50–70% confluency the following day.
The cells were co-transfected with 0.4 µg pGL3-basic-based
construct, 0.1 µg pRL-TK plasmid and 0.5 µg DACT2 overexpression
plasmid or the control vector to evaluate the effect of DACT2
overexpression on β-catenin activity, or with 10 µl of 20 µM DACT2
small interfering RNA (siRNA) or negative control siRNA to evaluate
the effect of DACT2 knockdown on β-catenin activity, using
Lipofectamine 2000 transfection reagent (Invitrogen; Thermo Fisher
Scientific, Inc.). Cells were then lysed and β-catenin activity was
assessed using the dual-luciferase reporter assay system (Promega
Corporation).
Transwell assay
Cell invasion was measured using the Biocoatmatrigel
Invasion Chamber kit (BD Biosciences, Franklin Lakes, NJ, USA). The
Matrigel-coated plates were rehydrated for 2 h, and then
2.5×104 cells were suspended in 500 ml DMEM medium and
placed on the insert. Subsequently, 750 ml complete DMEM medium was
added to the 24-well chamber. Cells were then incubated in 5%
CO2 at 37°C for 36 h. Subsequently, non-invading cells
on the upper surface of the membrane were scraped and invading
cells were fixed with formaldehyde and then stained with crystal
violet for counting.
Cell growth curve assay
For the cell proliferation assay, 2×105
cells were plated in a 6-well plate and cultured in complete medium
at 37°C and 5% CO2. The cells were counted using cell
counting chamber at time intervals and the cell growth curve was
drawn according to the cell numbers at different time points.
Statistical analysis
Data were presented as mean ± standard deviation and
subjected to one-way analysis of variance. Student's t test was
used to compare the differences of gene level and invasive ability
between two groups. One-way analysis of variance was used to test
the effect of DACT2 on the proliferation of breast cancer cells.
Multiple comparison between the groups was performed using the
Student-Newman-Keuls method. SPSS software was used for statistical
analysis (version 10.0; SPSS, Inc., Chicago, IL, USA). P<0.05
was considered to indicate a statistically significant
difference.
Results
DACT2 frequently decreases in human
breast cancer
To learn the expression status of DACT2 in breast
cancer, the present study first compared the mRNA expressional
level of DACT2 in breast cancer tissues and their paired adjacent
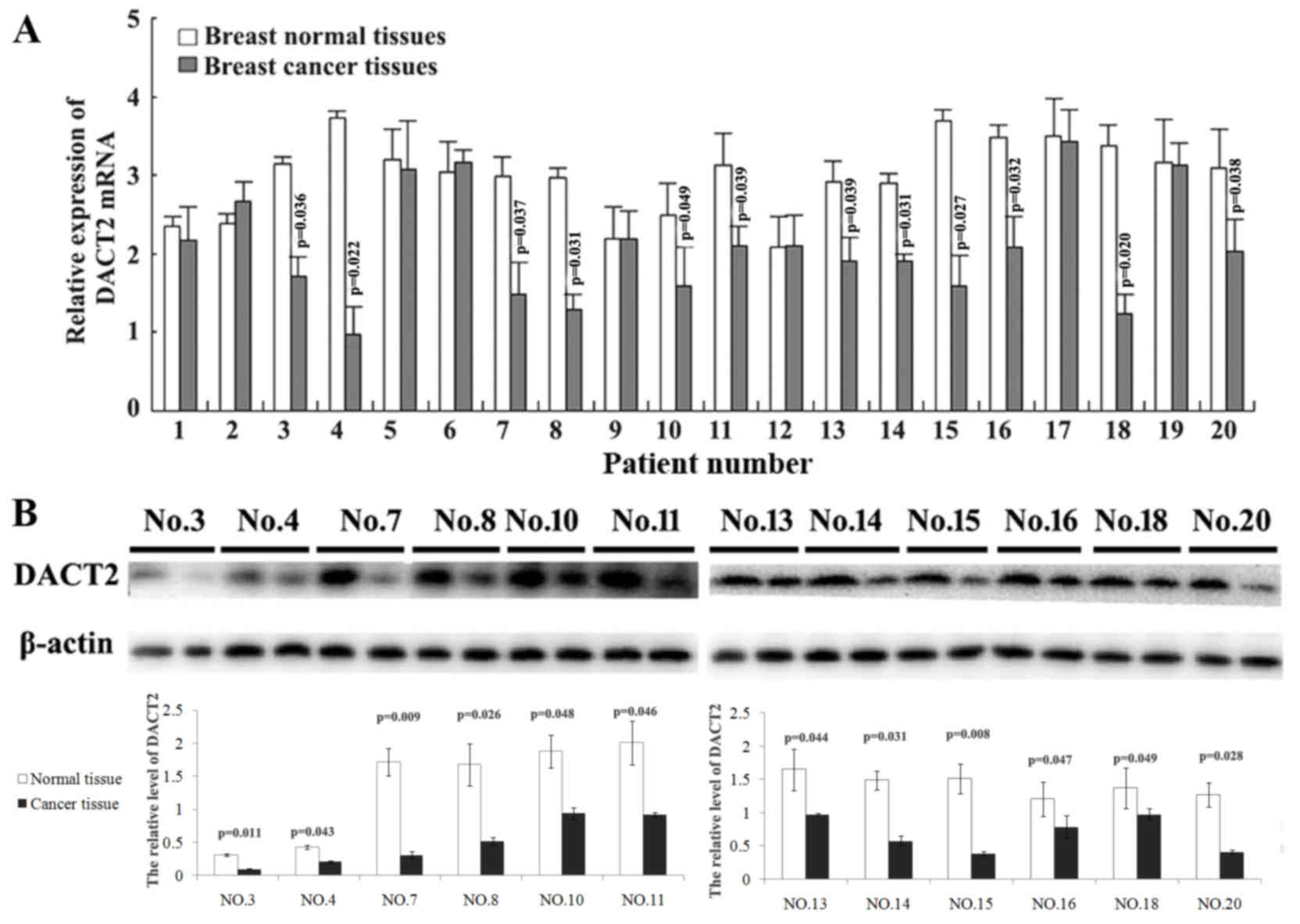
normal breast tissues using qPCR. It was identified that DACT2 mRNA
was decreased in 12/20 detected breast cancer tissues (Fig. 1A). Furthermore, the expressional
alters of DACT2 protein in these 12 breast cancer tissues was
validated using western blot analysis. The present study
demonstrated that DACT2 protein appeared to be decreased in these
breast cancer tissues with DACT2 mRNA decreasing compared with
their paired adjacent normal breast tissues (Fig. 1B). These results demonstrated that the
decrease of DACT2 occurred in 60% of observed human breast cancer
cases.
Hypermethylation of DACT2 promoter
majorly contributes to the loss of DACT2 in human breast
cancer
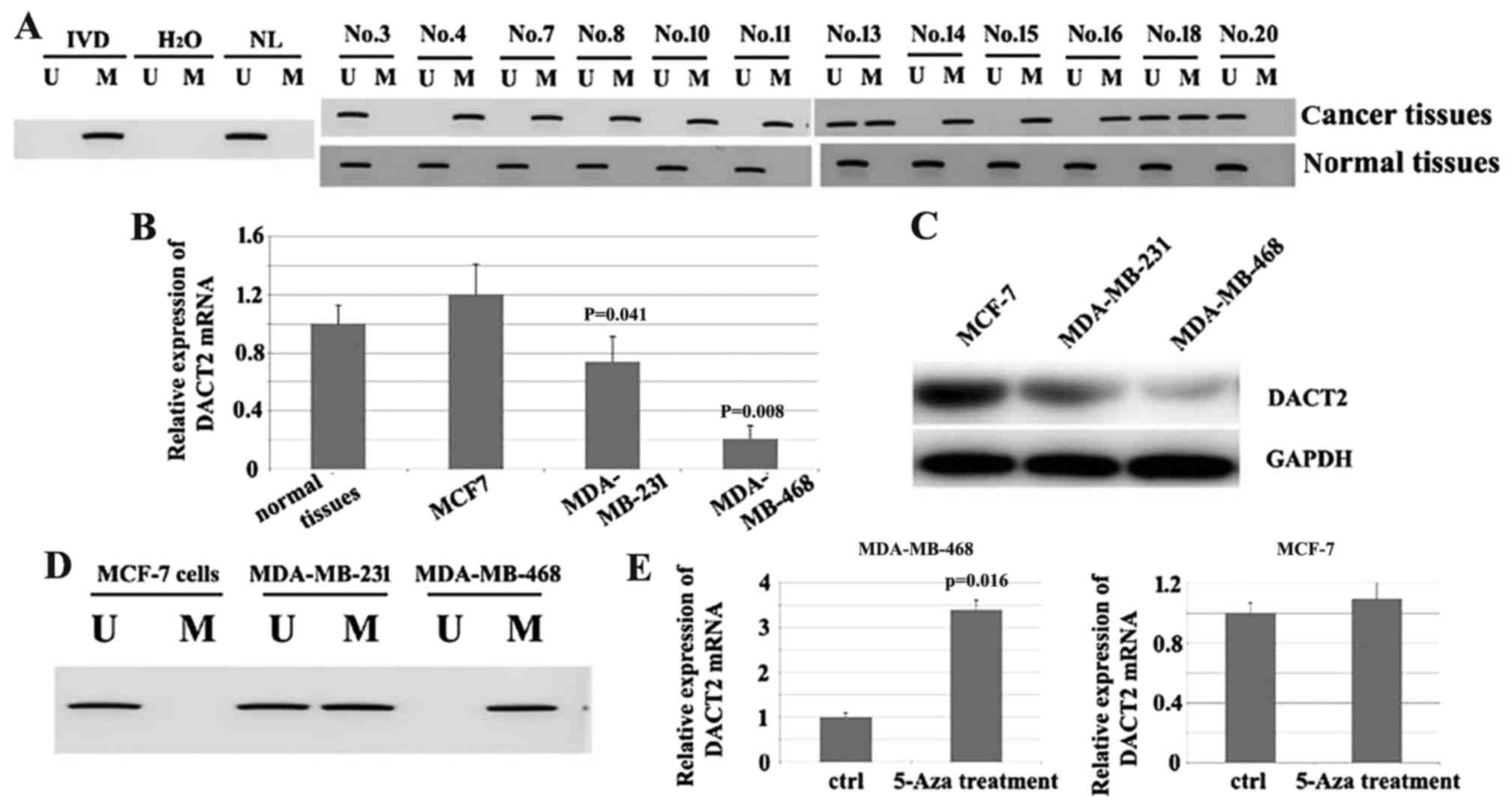
Since the decrease of DACT2 in breast cancer tissues
occurred at mRNA and protein level, it was suspected that the
decrease in DACT2 may result from the transcriptional inhibition.
To test this hypothesis, MSP was used to assess the methylation
status of DACT2 in these 12 cases of breast cancer tissues with
DACT2 decrease (Fig. 2A). The results
demonstrated that the promoter region of DACT2 was methylated in
10/12 tested breast cancer tissues, which indicated that promoter
methylation of DACT2 may contribute to the loss of DACT2 expression
in breast cancer patients. The expression of DACT2 was then
screened in the 3 well-established breast cancer MCF-7, MDA-MB-231
and MDA-MB-468 cell lines; it was identified that DACT2 is
significantly decreased in MDA-MB-468 cells at mRNA and protein
levels compared with normal tissues. (Fig. 2B and C). Additionally, MSP was
performed to identify if the loss of DACT2 in MDA-MB-468 resulted
from the methylation of the gene promoter; the results demonstrated
that the promoter region of DACT2 was methylated (Fig. 2D). To additionally confirm the
association between the methylation of DACT2 gene promoter and the
loss of DACT2 expression in the cell line 5-azacytidene (5-Aza) a
DNA methylation transferase inhibitor that can induce the
re-expression of methylated genes through de-methylation, was used
to treat MDA-MB-468. The results demonstrated that DACT2 expression
was induced in this cell line. In comparison, DACT2 expression was
not significantly affected in the MCF-7 cell line (Fig. 2E). Overall, these results indicate
that the loss or reduction of DACT2 in breast cancer may result
from the methylation of the DACT2 promoter region.
DACT2 inhibits the proliferation and
invasion of breast cancer cells
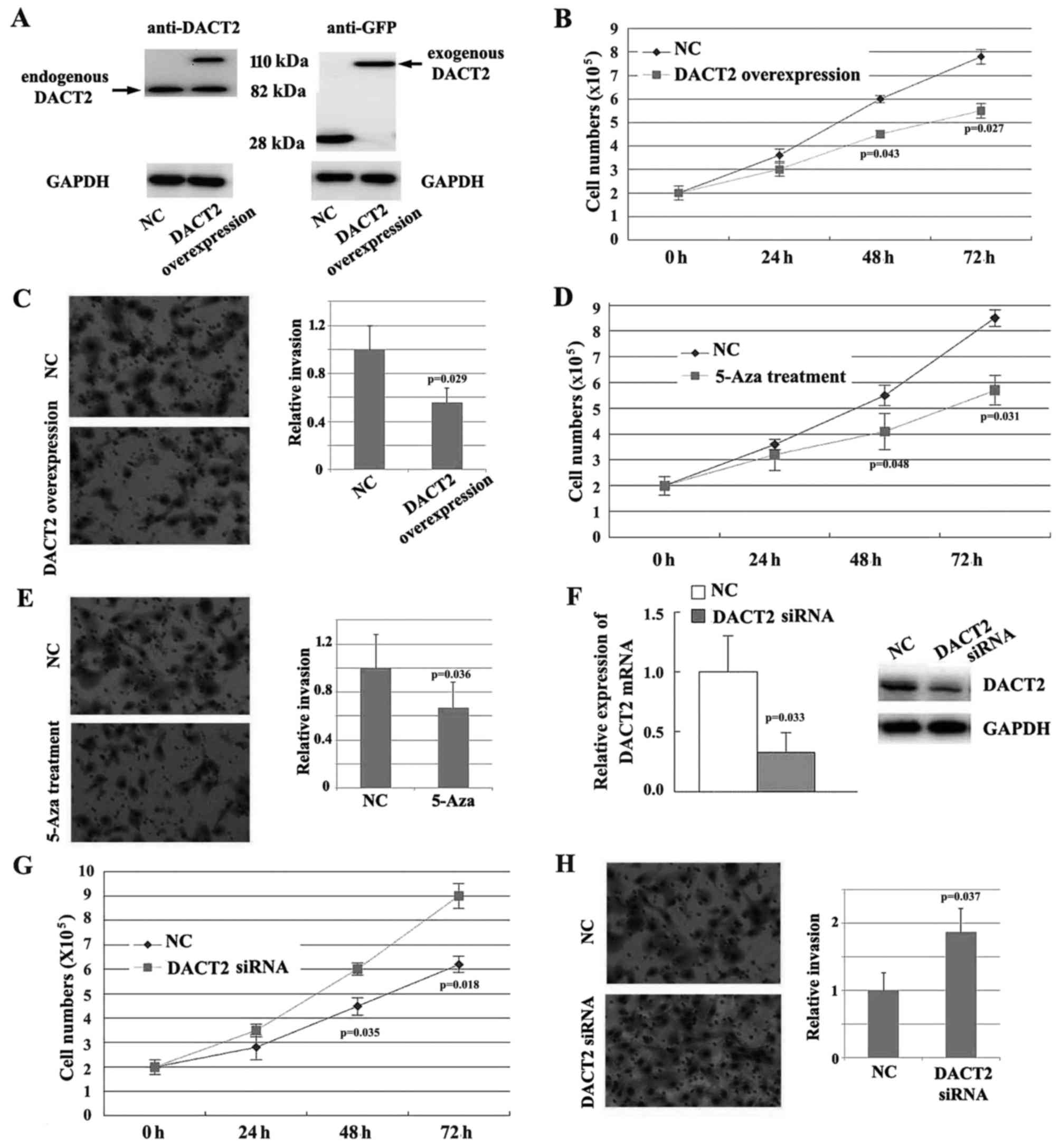
To learn the potential effect of DACT2 loss on the
breast cancer progression, the present study evaluated the role of
DACT2 in the proliferation and migration of breast cancer cells
in vitro. Firstly, DACT2 was overexpressed in MDA-MB-468
cells using a lentiviral vector (Fig.
3A), and it was demonstrated that enforced expression of DACT2
in MDA-MB-468 cells inhibits the proliferation (Fig. 3B) and invasion (Fig. 3C) of the cells. Similarly, treatment
with 5-Aza also inhibits the proliferation and invasion of the
cells (Fig. 3D and E). The expression
of DACT2 was knocked down in MDA-MB-231 cells using siRNA (Fig. 3F), and it was identified that the
repression of DACT2 promotes the proliferation and invasion of
MDA-MB-231 cells in vitro (Fig. 3G
and H). Overall, these results suggest that DACT2 acts as a
tumor suppressor and inhibits the progression of breast cancer.
DACT2 represses the expression of
β-catenin target genes in breast cancer cells
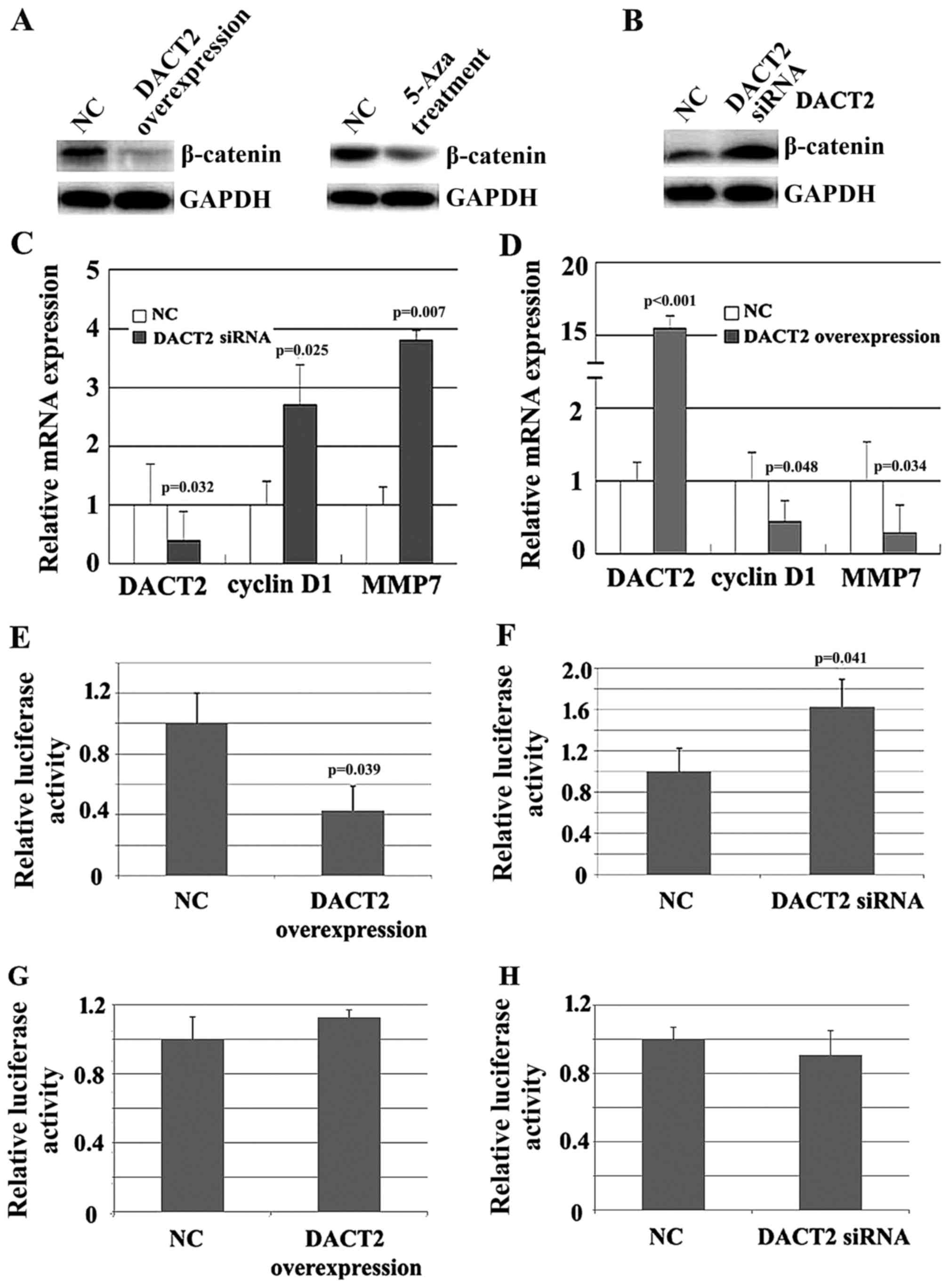
Since DACT2 promotes β-catenin degradation, the
change in β-catenin expression due to alteration of DACT2
expression was evaluated in vitro. As expected, either the
overexpression of DACT2 or 5-Aza treatment resulted in the decrease
of β-catenin in MDA-MB-468 cells (Fig.
4A). By contrast, knockdown of DACT2 resulted in the elevated
expression of β-catenin in MDA-MB-231 cells (Fig. 4B). Furthermore, the impact of DACT2
alteration on the expression of β-catenin target genes associated
with cell proliferation and invasion was detected. qPCR results
demonstrated that the knockdown of DACT2 in MDA-MB-231 cells
elevated the mRNA level of cyclin D1 and matrix metalloproteinase 7
(Fig. 4C), while the overexpression
of DACT2 attenuated the mRNA level of the two genes (Fig. 4D). Finally, β-catenin activity was
detected using a luciferase reporter assay. As shown in Fig. 4E and F, knockdown of DACT2 elevated
β-catenin activity, while overexpression of DACT2 attenuated
β-catenin activity. Mutation of β-catenin DNA binding sites
abolished the effect of DACT2 on β-catenin transcription activity
(Fig. 4G and H). The present results
suggested that DACT2 is a β-catenin signaling pathway inhibitor in
breast cancer cells.
Discussion
In the absence of Wnt ligands, β-catenin is degraded
in a proteasomal manner due to phosphorylation by a cytoplasmic
complex consisting of Axin, adenomatous polyposis coli, casein
kinase 1 and glycogen synthase kinase 3, and the signaling is
suppressed (13). The binding of Wnt
to its receptors FZD and LRP5/6 leads to the activation of Dvl,
which in turn inhibits the degradation complex and promotes the
accumulation of β-catenin, and thus the signaling is activated
(14). Wnt/β-catenin signaling
activation has been well established in breast carcinogenesis
(2–5).
Firstly, it has been identified that numerous Wnt proteins, such as
Wnt2, Wnt7b, and Wnt10b, are upregulated in human breast carcinomas
(6), and Wnt1 transgenic mice have
been shown to exhibit mammary gland hyperplasia and an increase in
adenocarcinomas (15). Secondly, it
was demonstrated that receptors of Wnt are also upregulated in
breast cancer. For example, FZD7 was identified to be overexpressed
in breast cancer, and downregulation of FZD7 inactivates
Wnt/β-catenin signaling and suppresses tumor formation (16). Overexpression of co-repressors LRP5
and LRP6 has been associated with the occurrence of breast cancer
(17,18), and animal models demonstrated that the
deletion of LRP5 or LRP6 delays mouse mammary tumor
virus-Wnt1-induced tumor formation (17,19).
Finally, certain inhibitors of this signaling pathway have been
revealed to be downregulated in breast carcinoma. For example, the
secreted proteins WNT inhibitory factor 1 (WIF1) and secreted
frizzled-related protein (sFRP) bind Wnt proteins and thus inhibit
their interaction with the FZD receptor. Additionally, it has been
revealed that WIF1 and sFRP1-5 are silenced in several types of
cancers, including breast cancer (20), whilst overexpression of sFRP markedly
represses the development of breast cancer (21,22).
DACT family proteins were initially identified in
Xenopus (8), and were
identified to suppress Wnt/β-catenin signaling activity through
interacting with or degrading Dvl, the initial activating factor of
the signaling pathway (7,9). The human DACT protein family has 3
members, and the encoding genes of DACT1, 2 and 3 are located on
human chromosome 14q22.3, 6q27 and 19q13.32, respectively (7,23).
Previously, studies have suggested that human DACT is frequently
silenced in human cancers. For example, deletion of chromosome 6q,
where the DACT2 gene is located, is one of the most frequent
chromosomal aberrations in human tumors (24,25). In
addition to deletion, epigenetic modifications of DACT genes were
also reported. For instance, DACT1 and DACT2 were reported to be
methylated in hepatocellular carcinoma, oral squamous, gastric
cancer, nasopharyngeal carcinoma, thyroid cancer, colon cancer and
lung cancer (10,26–31). In
addition, DACT3 was identified to have histone modifications in
colorectal cancer (25,26,32).
However, the function and expressional regulation of DACT in human
breast cancer is largely unknown. In the present study, the
expression of DACT2 was significantly decreased due to promoter
methylation in breast cancer.
To assess the expression status of DACT2 in breast
cancer cell lines and tissues, RT-qPCR and the western blot
analysis was used. The results show that DACT2 was frequently
silenced in breast cancer tissues and cell lines. MSP analysis of
these DACT2-silent breast cancer tissues and cell line demonstrated
promoter hypermethylation of the DACT2 gene. Furthermore, the DNMT
inhibitor 5-AZA induced the re-expression of DACT2 in DACT2-silent
breast cancer cells. These results indicated that the loss of DACT2
in breast cancer cells largely results from the methylation of the
DACT2 promoter region.
DACT2 was previously identified to bind Dvl and
promote Dvl degradation in a lysosome-dependent manner, thus
stabilizing the β-catenin degradation complex and decreasing
soluble β-catenin (9). Additionally,
it was also identified that DACT2 could inhibit β-catenin activity
by directly and firmly binding β-catenin in the cytoplasm (32). Consistently, the present study
demonstrated that the knockdown of endogenous DACT2 increases and
overexpression of DACT2 inhibits β-catenin target gene expression
and β-catenin/TCF reporter luciferase activity in the cell lines
studied. Furthermore, the current study evaluated the potential
roles of DACT2 in breast cancer progression. The effect of DACT2 on
breast cancer cell proliferation was evaluated by a cell growth
curve assay, and the effect on invasion of cancer cells was
evaluated by Transwell assay. The present results demonstrated that
DACT2 overexpression inhibits breast cancer cell proliferation and
invasion, while the knockdown of DACT2 promotes the proliferation
and invasion of breast cancer cells. The current data has
demonstrated that DACT2 acts as a tumor suppressor in breast
cancer.
In summary, the present study demonstrated that
DACT2 was frequently silent in breast cancer, and the methylation
of the DACT2 gene promoter largely contributes to the silencing of
the gene in human breast cancer. It was also identified that DACT2
acts as a tumor suppressor in breast cancer by inhibiting
Wnt/β-catenin activation and repressing cancer cell proliferation
and invasion. The present study indicates that the loss of DACT2
may contribute to breast cancer progression and provides a
promising therapeutic target for the treatment of breast
cancer.
References
|
1
|
King TD, Suto MJ and Li Y: The
Wnt/β-catenin signaling pathway: A potential therapeutic target in
the treatment of triple negative breast cancer. J Cell Biochem.
113:13–18. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Turashvili G, Bouchal J, Burkadze G and
Kolar Z: Wnt signaling pathway in mammary gland development and
carcinogenesis. Pathobiology. 73:213–223. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Geyer FC, Lacroix-Triki M, Savage K,
Arnedos M, Lambros MB, MacKay A, Natrajan R and Reis-Filho JS:
β-Catenin pathway activation in breast cancer is associated with
triple-negative phenotype but not with CTNNB1 mutation. Mod Pathol.
24:209–231. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Khramtsov AI, Khramtsova GF, Tretiakova M,
Huo D, Olopade OI and Goss KH: Wnt/beta-catenin pathway activation
is enriched in basal-like breast cancers and predicts poor outcome.
Am J Pathol. 176:2911–2920. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lin SY, Xia W, Wang JC, Kwong KY, Spohn B,
Wen Y, Pestell RG and Hung MC: Beta-catenin, a novel prognostic
marker for breast cancer: Its roles in cyclin D1 expression and
cancer progression. Proc Natl Acad Sci USA. 97:pp. 4262–4266. 2000;
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Howe LR and Brown AM: Wnt signaling and
breast cancer. Cancer Biol Ther. 3:36–41. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Fisher DA, Kivimäe S, Hoshino J, Suriben
R, Martin PM, Baxter N and Cheyette BN: Three Dact gene family
members are expressed during embryonic development and in the adult
brains of mice. Dev Dyn. 235:2620–2630. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Cheyette BN, Waxman JS, Miller JR,
Takemaru K, Sheldahl LC, Khlebtsova N, Fox EP, Earnest T and Moon
RT: Dapper, a Dishevelled-associated antagonist of beta-catenin and
JNK signaling, is required for notochord formation. Dev Cell.
2:449–461. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Teran E, Branscomb AD and Seeling JM: Dpr
acts as a molecular switch, inhibiting Wnt signaling when
unphosphorylated, but promoting Wnt signaling when phosphorylated
by casein kinase Idelta/epsilon. PLoS One. 4:e55222009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yu Y, Yan W, Liu X, Jia Y, Cao B, Yu Y, Lv
Y, Brock MV, Herman JG, Licchesi J, et al: DACT2 is frequently
methylated in human gastric cancer and methylation of DACT2
activated Wnt signaling. Am J Cancer Res. 4:710–724.
2014.PubMed/NCBI
|
|
11
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Li X, Zhang J, Gao L, McClellan S, Finan
MA, Butler TW, Owen LB, Piazza GA and Xi Y: MiR-181 mediates cell
differentiation by interrupting the Lin28 and let-7 feedback
circuit. Cell Death Differ. 19:378–386. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ha NC, Tonozuka T, Stamos JL, Choi HJ and
Weis WI: Mechanism of phosphorylation-dependent binding of APC to
beta-catenin and its role in beta-catenin degradation. Mol Cell.
15:511–521. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Du W, Liu X, Fan G, Zhao X, Sun Y, Wang T,
Zhao R, Wang G, Zhao C, Zhu Y, et al: From cell membrane to the
nucleus: An emerging role of E-cadherin in gene transcriptional
regulation. J Cell Mol Med. 18:1712–1719. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tsukamoto AS, Grosschedl R, Guzman RC,
Parslow T and Varmus HE: Expression of the int-1 gene in transgenic
mice is associated with mammary gland hyperplasia and
adenocarcinomas in male and female mice. Cell. 55:619–625. 1988.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yang L, Wu X, Wang Y, Zhang K, Wu J, Yuan
YC, Deng X, Chen L, Kim CC, Lau S, et al: FZD7 has a critical role
in cell proliferation in triple negative breast cancer. Oncogene.
30:4437–4446. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lindvall C, Zylstra CR, Evans N, West RA,
Dykema K, Furge KA and Williams BO: The Wnt co-receptor Lrp6 is
required for normal mouse mammary gland development. PLoS One.
4:e58132009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Badders NM, Goel S, Clark RJ, Klos KS, Kim
S, Bafico A, Lindvall C, Williams BO and Alexander CM: The Wnt
receptor, Lrp5, is expressed by mouse mammary stem cells and is
required to maintain the basal lineage. PLoS One. 4:e65942009.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lindvall C, Evans NC, Zylstra CR, Li Y,
Alexander CM and Williams BO: The Wnt signaling receptor Lrp5 is
required for mammary ductal stem cell activity and Wnt1-induced
tumorigenesis. J Biol Chem. 281:35081–35087. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Willemsen RH, de Kort SW, van der Kaay DC
and Hokken-Koelega AC: Independent effects of prematurity on
metabolic and cardiovascular risk factors in short
small-for-gestational-age children. J Clin Endocrinol Metab.
93:452–458. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Suzuki H, Toyota M, Carraway H, Gabrielson
E, Ohmura T, Fujikane T, Nishikawa N, Sogabe Y, Nojima M, Sonoda T,
et al: Frequent epigenetic inactivation of Wnt antagonist genes in
breast cancer. Br J Cancer. 98:1147–1156. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Matsuda Y, Schlange T, Oakeley EJ, Boulay
A and Hynes NE: WNT signaling enhances breast cancer cell motility
and blockade of the WNT pathway by sFRP1 suppresses MDA-MB-231
xenograft growth. Breast Cancer Res. 11:R322009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Katoh M and Katoh M: Identification and
characterization of human DAPPER1 and DAPPER2 genes in silico. Int
J Oncol. 22:907–913. 2003.PubMed/NCBI
|
|
24
|
Girard L, Zöchbauer-Müller S, Virmani AK,
Gazdar AF and Minna JD: Genome-wide allelotyping of lung cancer
identifies new regions of allelic loss, differences between small
cell lung cancer and non-small cell lung cancer, and loci
clustering. Cancer Res. 60:4894–4906. 2000.PubMed/NCBI
|
|
25
|
Steinemann D, Gesk S, Zhang Y, Harder L,
Pilarsky C, Hinzmann B, Martin-Subero JI, Calasanz MJ, Mungall A,
Rosenthal A, et al: Identification of candidate tumor-suppressor
genes in 6q27 by combined deletion mapping and electronic
expression profiling in lymphoid neoplasms. Genes, Chromosomes
Cancer. 37:421–426. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Jia Y, Yang Y, Brock MV, Zhan Q, Herman JG
and Guo M: Epigenetic regulation of DACT2, a key component of the
Wnt signalling pathway in human lung cancer. J Pathol. 230:194–204.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhang X, Yang Y, Liu X, Herman JG, Brock
MV, Licchesi JD, Yue W, Pei X and Guo M: Epigenetic regulation of
the Wnt signaling inhibitor DACT2 in human hepatocellular
carcinoma. Epigenetics. 8:373–382. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang S, Dong Y, Zhang Y, Wang X, Xu L,
Yang S, Li X, Dong H, Xu L, Su L, et al: DACT2 is a functional
tumor suppressor through inhibiting Wnt/β-catenin pathway and
associated with poor survival in colon cancer. Oncogene.
34:2575–2585. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhao Z, Herman JG, Brock MV, Sheng J,
Zhang M, Liu B and Guo M: Methylation of DACT2 promotes papillary
thyroid cancer metastasis by activating Wnt signaling. PLoS One.
9:e1123362014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Li L, Zhang Y, Fan Y, Sun K, Su X, Du Z,
Tsao SW, Loh TK, Sun H, Chan AT, et al: Characterization of the
nasopharyngeal carcinoma methylome identifies aberrant disruption
of key signaling pathways and methylated tumor suppressor genes.
Epigenomics. 7:155–173. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Schussel JL, Kalinke LP, Sassi LM, de
Oliveira BV, Pedruzzi PA, Olandoski M, Alvares LE, Garlet GP and
Trevilatto PC: Expression and epigenetic regulation of DACT1 and
DACT2 in oral squamous cell carcinoma. Cancer Biomark. 15:11–17.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kivimäe S, Yang XY and Cheyette BN: All
Dact (Dapper/Frodo) scaffold proteins dimerize and exhibit
conserved interactions with Vangl, Dvl, and serine/threonine
kinases. BMC Biochem. 12:332011. View Article : Google Scholar : PubMed/NCBI
|